Significance
Neurexins are presynaptic cell-adhesion molecules that are essential for synapse formation and synaptic transmission. Extensive alternative splicing of neurexin transcripts may generate thousands of isoforms, but it is unclear how many distinct neurexins are physiologically produced. We used unbiased long-read sequencing of full-length neurexin mRNAs to systematically assess the alternative splicing of neurexins in prefrontal cortex. We identified a novel, abundantly used alternatively spliced exon of neurexins, and found that the different events of alternative splicing of neurexins appear to be independent of each other. Our data suggest that thousands of neurexin isoforms are physiologically generated, consistent with the notion that neurexins represent transsynaptic protein-interaction scaffolds that mediate diverse functions and are regulated by alternative splicing at multiple independent sites.
Keywords: schizophrenia, neuroligin, cerebellin, LRRTM, autism
Abstract
Neurexins are evolutionarily conserved presynaptic cell-adhesion molecules that are essential for normal synapse formation and synaptic transmission. Indirect evidence has indicated that extensive alternative splicing of neurexin mRNAs may produce hundreds if not thousands of neurexin isoforms, but no direct evidence for such diversity has been available. Here we use unbiased long-read sequencing of full-length neurexin (Nrxn)1α, Nrxn1β, Nrxn2β, Nrxn3α, and Nrxn3β mRNAs to systematically assess how many sites of alternative splicing are used in neurexins with a significant frequency, and whether alternative splicing events at these sites are independent of each other. In sequencing more than 25,000 full-length mRNAs, we identified a novel, abundantly used alternatively spliced exon of Nrxn1α and Nrxn3α (referred to as alternatively spliced sequence 6) that encodes a 9-residue insertion in the flexible hinge region between the fifth LNS (laminin-α, neurexin, sex hormone-binding globulin) domain and the third EGF-like sequence. In addition, we observed several larger-scale events of alternative splicing that deleted multiple domains and were much less frequent than the canonical six sites of alternative splicing in neurexins. All of the six canonical events of alternative splicing appear to be independent of each other, suggesting that neurexins may exhibit an even larger isoform diversity than previously envisioned and comprise thousands of variants. Our data are consistent with the notion that α-neurexins represent extracellular protein-interaction scaffolds in which different LNS and EGF domains mediate distinct interactions that affect diverse functions and are independently regulated by independent events of alternative splicing.
Neurons form highly specific and complex patterns of synaptic connections that underlie all brain function (1–5). Such specific connections require trillions of chemically differentiated synapses whose identity may be shaped by interactions of specific pre- and postsynaptic signaling molecules, especially cell-adhesion molecules. The genome size is insufficient to encode such diversity, but a mixture of combinatorial expression patterns that pair different synaptic cell-adhesion molecules with each other and of distinct alternative splicing patterns that amplify the number of cell-adhesion molecules into a large number of isoforms may generate the number of transsynaptic interactions needed to account for the enormous diversity of synaptic connections. In Drosophila melanogaster, alternative splicing of the mRNAs encoding the Down syndrome cell-adhesion molecule can generate nearly 20,000 protein isoforms whose structures specify axon bundling but not synapse formation (6). In mammals, alternative splicing of neurexin and some protocadherin mRNAs can also produce thousands of isoforms, which may at least in the case of neurexins be involved in synapse formation (7–10).
Neurexins are type I membrane proteins that were discovered as presynaptic receptors for α-latrotoxin (8, 9). Six principal neurexins (Nrxn1α–3α and Nrxn1β–3β) are synthesized from three genes (Nrxn1–Nrxn3), each of which expresses larger α- and shorter β-isoforms from independent promoters (11–13). All neurexin mRNAs are extensively alternatively spliced (8, 9, 14).
α- and β-neurexins contain different extracellular sequences but identical transmembrane regions and short cytoplasmic tails. Specifically, the extracellular sequences of α-neurexins are composed of six LNS (laminin-α, neurexin, sex hormone-binding globulin) domains with three interspersed EGF-like repeats followed by an O-linked sugar attachment sequence and a conserved cysteine loop sequence (8, 15). In contrast, the extracellular sequences of β-neurexins comprise a short β-neurexin–specific sequence, and then splice into the sixth LNS domain of α-neurexins, from which point on β-neurexins are identical to α-neurexins (11). All α-neurexins are subject to alternative splicing at five canonical sites [SS#1–SS#5 (14)], of which SS#4 and SS#5 are also found in β-neurexins. The alternatively spliced sequences for SS#1–SS#4 are highly homologous among the three neurexins, whereas those of SS#5 differ among neurexins. Here the variable sequence in Nrxn1 encompasses only 3 residues, whereas in Nrxn2 it is composed of 194 residues, and in Nrxn3 a variety of alternatively spliced sequences are observed that include inserts of 247 residues and sequences with in-frame stop codons, effectively producing secreted Nrxn3 isoforms (9, 14). Alternative splicing of neurexins is differentially regulated in different brain regions (10, 14, 16), exhibits a diurnal cycle (17), and is modulated by development, neurotrophins, and neuronal activity (18–21).
Neurexins are known to bind to a large number of ligands, of which neuroligins, leucine-rich repeat transmembrane proteins, and cerebellins are best-characterized (22–28). All of these ligands bind to both α- and β-neurexins because they interact with the sixth LNS domain; strikingly, these interactions are differentially regulated by alternative splicing of neurexins at SS#4 in the sixth LNS domain (24, 26–31). In addition to these ligands, neurexins were shown to bind to neurexophilin, dystroglycan, and CIRL/latrophilin (32–35).
Despite extensive studies, the full extent of neurexin alternative splicing remains unclear. Full-length cDNA sequencing and PCR analysis of a small fraction of neurexin isoforms have suggested that thousands of neurexin isoforms may be created by combinatorial additions of various alternatively spliced sequences, but only a small fraction of these isoforms were actually identified in sequenced full-length cDNAs (14). The relatively large size of α-neurexin transcripts (∼4–5 kb) has made it difficult to obtain information about their full-length sequence, and hence about the use of alternative splice sites within single transcripts. Recent technological developments have enabled full-length sequencing of long DNA molecules (36, 37). In the present study, we applied long-read sequencing technology to map the cartography of neurexins (Fig. 1A). Read lengths of up to 30 kb enabled us to identify all of the splice combinations within a single transcript. We analyzed the alternative splicing landscape of Nrxn1α and Nrxn3α as well as of Nrxn1β, Nrxn2β, and Nrxn3β. Our data provide definitive evidence that neurexins are present in mammalian brain in an unprecedented diversity of alternatively spliced isoforms numbering in the thousands, and reveal the existence of a previously unknown sixth canonical site of alternative splicing of α-neurexins. Thus, our results support the notion that neurexins function as recognition molecules that contribute to the specification of synapses in the brain.
Fig. 1.
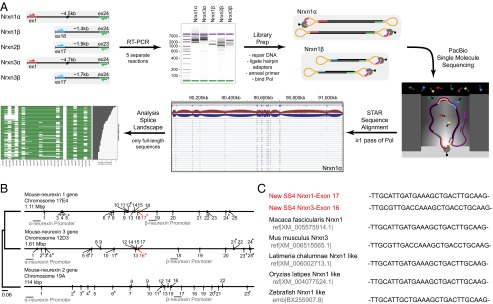
Single-molecule long-read sequencing of full-length neurexin mRNA transcripts. (A) Schema of the workflow of Pacific Biosciences sequencing of full-length neurexin transcripts for mapping neurexin diversity. Full-length transcripts of Nrxn1α, 1β, 2β, 3α, and 3β were separately reverse-transcribed and PCR-amplified (RT-PCR) from total RNA isolated from adult murine prefrontal cortex. PCR was performed with primer pairs specific to the first and last coding exons of each gene, with α- and β-specific the forward primers and identical reverse primers for the α- and β-versions of a given neurexin. The cDNA size distribution for each neurexin was examined by gel electrophoresis before preparation of Pacific Biosciences sequencing libraries by blunt-end ligation of hairpin adapters, annealing of the sequencing primer, and binding of biotinylated DNA polymerase. Subsequently, full-length transcripts were sequenced by single-molecule long-read sequencing (36), yielding an average read length of 3.6–5.8 kb. Reads containing at least two adapters (≥1 passes through circular DNA by the polymerase) were processed (quality filter, adapter removal) and then aligned to the mouse genome (mm10) using the STAR aligner (38) and used for further analysis. (B) Phylogenetic tree and structures of mouse neurexin genes. The diagrams depict the positions of exons and introns. Exons are numbered; asterisks mark exons subject to canonical alternative splicing. The positions of α- and β-neurexin promoters are indicated, and the position and size of the genes are shown above each gene diagram (see also Fig. S1). (C) Neurexin mRNAs from diverse species contain the newly identified SS#6. Sequences of murine Nrxn1α and Nrxn3α SS#6 that were identified in this study are colored in red.
Results
Sequencing Full-Length Neurexin mRNAs from Prefrontal Cortex RNA.
Three neurexin genes are dispersed in the mammalian genome but exhibit similar exon/intron structures (12, 13). The Nrxn1 and Nrxn3 genes are extraordinarily large with sizes over 1.1 Mbp, whereas the Nrxn2 gene measures only 0.1 Mbp (Fig. 1B). Comparison of protein sequence-based phylogenetic trees of neurexins indicates Nrxn1 and Nrxn3 proteins are evolutionarily more closely related to each other than to the Nrxn2 protein, suggesting that two separate duplication events generated the three neurexin genes (Fig. 1B).
In this study, we focused on the collective repertoire of full-length neurexin isoforms because only a small fraction of neurexins’ splice diversity has previously been experimentally identified. Earlier calculations of neurexin splice diversity were based on the assumption that all splice events are independent and splicing can occur only at five canonical positions in the gene and suggested the generation of more than 3,000 distinct neurexins (14), but no direct evidence supports this supposition. To understand the true diversity of neurexin mRNAs, we determined the full-length sequences of thousands of neurexin mRNAs (Fig. 1A).
All principal neurexin mRNAs (encoding Nrxn1α, Nrxn1β, Nrxn2β, Nrxn3α, and Nrxn3β) except for that of Nrxn2α were amplified by primers specific to the first and last exons, and separate sequencing libraries were prepared by ligating hairpin adapters to both ends of each DNA molecule such that the DNA polymerase can traverse each DNA molecule multiple times during rolling circle replication (Fig. 1A). Nrxn2α amplification was unsuccessful, possibly because of its lower abundance, sequence content, and/or larger size. After filtering out all reads that missed the exons targeted by the amplification primers (damaged or misprimed PCR products), we obtained a total of 23,943 full-length mRNA reads (2,574 Nrxn1α, 1,653 Nrxn1β, 10,283 Nrxn2β, 934 Nrxn3α, and 8,499 Nrxn3β cDNAs).
Identification of a New Conserved Alternatively Spliced α-Neurexin Exon.
We aligned filtered reads to the mouse genome (mm10) using the STAR sequence aligner (38). By comparing the full-length transcript sequences with the characterized neurexin gene structures (12), we observed a previously unidentified, alternatively spliced exon in the Nrxn1α and Nrxn3α genes [alternatively spliced sequence 6 (SS#6)]. The 9-residue amino acid sequence encoded by this alternatively spliced exon maps to the hinge region between EGF-C and the fifth LNS domains of Nrxn1α and Nrxn3α (Fig. 1B) (39, 40). Although this new alternatively spliced exon was not characterized previously, it is present in neurexin mRNAs in the public databases and is evolutionarily conserved in zebrafish and macaque neurexin mRNAs (Fig. 1C). However, this exon is missing in the Nrxn2 gene. Phylogenetic analysis (Fig. 1B) suggests that neurexins are the result of relatively recent gene duplication events, which first separated Nrxn2 from the two other neurexins and then diverged Nrxn1 and Nrxn3 from each other, accounting for the absence of the exon encoding SS#6 from Nrxn2α.
Splice Landscape of Nrxn1α.
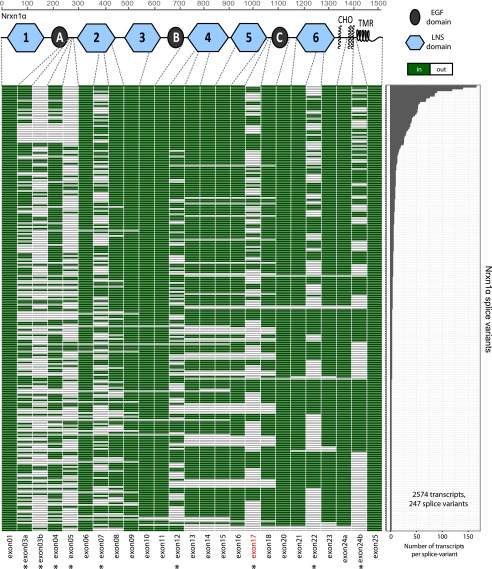
We used the sequencing reads to generate a transcript map of Nrxn1α. This map reveals that 247 unique splice isoforms (rows) are observed in 2,574 full-length transcripts, with 96 unique splice isoforms accounting for 90% of all detected transcripts (Fig. 2). In addition to events at the six canonical Nrxn1α sites of alternative splicing, we observed alternative splicing at noncanonical sites that include all Nrxn1α exons. As an example, exons 13, 14, and 15 were spliced out in 5% of the Nrxn1α mRNAs (Fig. 2; see also Fig. 6). The majority of novel alternative splicing events consisted of modular excisions of multiple exons: Whereas in some cases two canonical sites were spliced out together with interjacent exons, in other cases novel splice donors were used.
Fig. 2.
Splice landscape of Nrxn1α. The transcript map visualizes the 247 unique alternatively spliced isoforms (rows) observed for the 2,574 full-length Nrxn1α mRNAs sequenced. Exons (columns) are colored in green if present and in white if absent, and are numbered at the bottom (asterisks, exons with canonical alternative splicing; for an explanation of the numbering, see Fig. S1). The domain structure of Nrxn1α is shown at the top (light blue hexagons, LNS domains; black ovals, EGF-like domains; CHO, O-linked sugar modifications; TMR, transmembrane region) and is connected to the exons that encode the respective domains by dotted gray lines. The abundance of each splice isoform is shown in the bar graph (Right).
Fig. 6.
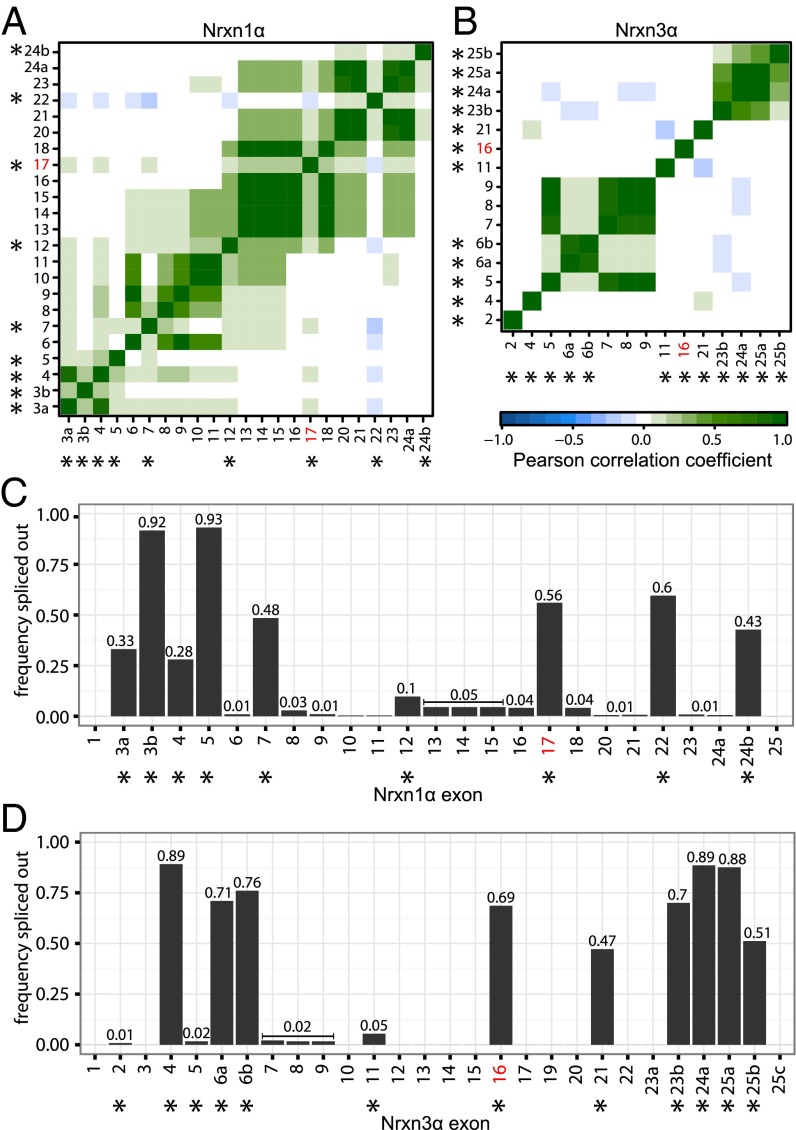
Independent and coordinated splicing of canonical and novel splice sites for Nrxn1α and Nrxn3α. (A and B) Correlograms showing the pairwise correlation in splice behavior between alternatively spliced exons of (A) Nrxn1α and (B) Nrxn3α. The color bar denotes the Pearson correlation coefficient from −1 (blue, anticorrelated splicing) through 0 (no correlation in splicing) to 1 (green, positively correlated splicing). Newly observed alternatively spliced exons tend to be spliced out in a coordinated way, whereas canonical events of alternative splicing appear to occur independent of each other. (C and D) Bar graphs visualizing the frequency of each exon to be spliced out for (C) Nrxn1α and (D) Nrxn3α. Canonical alternatively spliced exons (12) are marked by asterisks. Exons that are observed to be alternatively spliced in a coordinated way (A and B) show similar frequencies.
Confirmation of a Novel Event of Modular Alternative Splicing.
To confirm novel splicing events, we focused on a modular event that excises Nrxn1α exons 12–18 (12–17 in ref. 12). Using specific primers, we amplified Nrxn1α exons 11–20 (11–19 in ref. 12) from prefrontal cortex total RNA and observed two distinct bands: a higher band of ∼1,500 bp matching the size of the mRNA including all exons, and a lower band of ∼300 bp matching the size of the mRNA lacking exons 12–18 (Fig. 3A). Sanger sequencing of the lower band confirmed that exons 12–18 are absent (Fig. 3B).
Fig. 3.
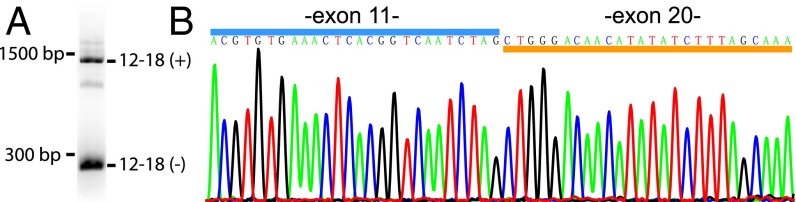
Validation of newly identified alternatively spliced isoform of Nrxn1α lacking exons 12–18 by RT-PCR. (A) Agarose gel electrophoresis analysis of the PCR product obtained with primers specific to the end of exon 11 and beginning of exon 20 of Nrxn1α. Two major splice variants are identified: a long DNA fragment (∼1,500 bp) corresponding to transcripts containing exons 12–18 as well as a short DNA fragment (∼300 bp) corresponding to transcripts lacking exons 12–18. Note that exon 19 encodes the N terminus of Nrxn1β and is always missing from Nrxn1α mRNAs (12). (B) Sanger sequencing of the short PCR product as obtained in A.
Splice Landscapes of Nrxn3α, 1β, 2β, and 3β.
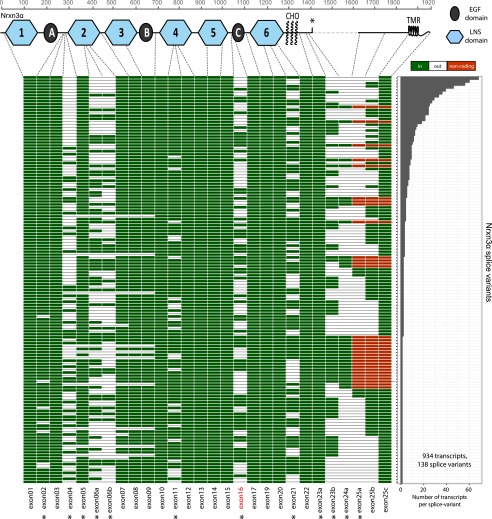
Using full-length reads of Nrxn3α, Nrxn1β, Nrxn2β, and Nrxn3β, we generated transcript maps of these neurexins to visualize their isoform diversity (Figs. 4 and 5). For Nrxn3α, we detected 934 full-length mRNA sequences that encode 138 different unique splice variants, with 67 unique splice isoforms accounting for 90% of all detected transcripts. Interestingly, we did not observe alternative splicing of the previously reported alternatively spliced exons 3 and 24b/c (23b/c in ref. 12), but we did detect alternative splicing of exons 5, 7, 8, 9, and 16, which have previously been reported to be invariant (Fig. 4). In contrast to Nrxn1α, Nrxn3α novel splicing events are limited to the second and third LNS and EGF-B domains.
Fig. 4.
Splice landscape of Nrxn3α. The transcript map visualizes the 138 unique alternatively spliced isoforms (rows) observed for the 934 full-length Nrxn3α mRNAs sequenced. Exons (columns) are colored in green for coding and red for exons with in-frame stop codons if present and in white if absent, and are numbered at the bottom (asterisks, exons with canonical alternative splicing; for an explanation of the numbering, see Fig. S1). The domain structure of Nrxn1α is shown at the top and is connected to the exons that encode the respective domains by dotted gray lines. In the domain structure, the asterisk denotes a stop codon that is present in some of the alternatively spliced exons and could produce secreted versions of neurexin-3. The abundance of each splice isoform is shown in the bar graph (Right).
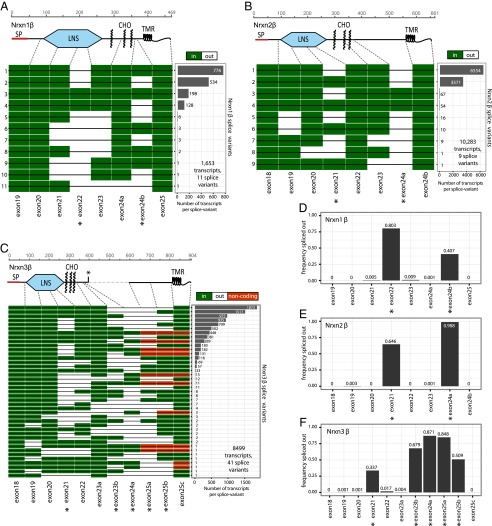
Fig. 5.
Splice landscape of β-neurexins. (A–C) Transcript maps visualizing unique splice isoforms (rows) observed for Nrxn1β (A; 1,653 transcripts, 11 splice variants), Nrxn2β (B; 10,283 transcripts, 9 splice variants), and Nrxn3β (C; 8,499 transcripts, 41 splice variants). Exons (columns) are colored in green if present and in white if absent, and are numbered at the bottom (asterisks, exons with canonical alternative splicing; for an explanation of the numbering, see Fig. S1). The domain structures of the respective β-neurexins are shown above the transcript maps and are connected to the exons that encode the respective domains by dotted gray lines. SP denotes the signal peptide; the asterisk in the domain structure of Nrxn3β indicates a stop codon encoded by one of the alternatively spliced exons. The abundance of each splice isoform is shown in the bar graph (Right). (D–F) Bar graphs visualizing the relative frequency of exon skipping for the indicated exons in the Nrxn1β (D), Nrxn2β (E), and Nrxn3β mRNAs (F). Canonical alternatively spliced exons are marked by asterisks.
Regarding the shorter β-neurexins, we detected 1,653 full-length mRNA sequences for Nrxn1β, 10,283 full-length sequences for Nrxn2β, and 8,499 full-length sequences for Nrxn3β, encoding 11, 9, and 41 splice variants in total, respectively. For β-neurexins, we were able to detect all previously identified alternative splicing events except for exons 24b and 24c (23b and 23c in ref. 12) of Nrxn3. We detected 146 splicing events of the previously unreported alternatively spliced exon 22 in Nrxn3β but not in the Nrxn3α isoforms.
Alternative Splicing Events of Nonadjacent Exon Clusters Are Independent of Each Other.
To examine whether alternative splicing at different splice sites is coordinated, we analyzed the pairwise correlation between events of alternative splicing at different sites in Nrxn1α and Nrxn3α. We detected no correlation and hence independent splicing of different splice sites (Fig. 6 A and B). However, correlations were observed within canonical splice sites, such as Nrxn1α exons 3a and 4 (SS#1), or in the case of modular excision events of exon blocks such as Nrxn3α exons 5–9 (Fig. 6 A and B). In these cases, correlations are the result of neighboring exons being part of the same splicing reaction.
Effect of the Novel Alternatively Spliced Exon on the Structure of Nrxn1α and Nrxn3α.
By using single-molecule long-read sequencing, we have identified not only a new alternatively spliced exon (SS#6 Fig. 7) but also observed splicing of exons that were previously not considered to be subject to alternative splicing. In most of these newly identified events of alternative splicing, multiple exons were excised, resulting in the deletion of repeated modules in the neurexin protein domain structure. The question arises whether these events of alternative splicing represent accidents of nature or whether they are physiologically relevant. If they were accidents, they should be random and disturb the protein domain structure of a neurexin.
Fig. 7.
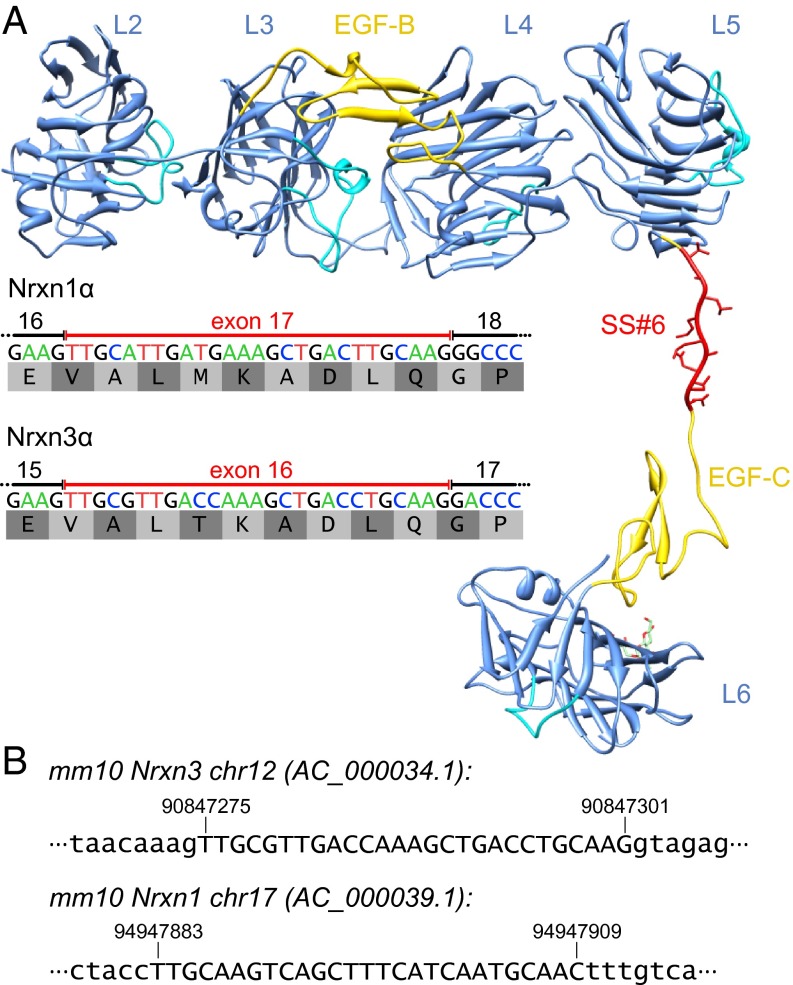
Sequence and structure of newly identified alternatively spliced exons for Nrxn1α and Nrxn3α. (A) mRNA sequence and translated 9-residue amino acid sequence of the newly identified alternatively spliced Nrxn1α exon 17 and Nrxn3α exon 16 (SS#6). A possible extended conformation of Nrxn1α exon 17 was modeled into the crystal structure of Nrxn1α [PDB ID 3QCW (39)] using Coot (42). SS#6 is located between the LNS5 and EGF-C domains, thereby extending the flexible hinge region between these structural features. The image of the Nrxn1α protein structure was created in Chimera (43). (B) Location of the newly identified exon of Nrxn1α and Nrxn3α in the mouse genome (mm10). Exonic nucleotides are shown in capital letters, whereas intronic nucleotides are shown in lowercase letters.
To visualize the effect of novel alternative splicing events on protein domain structure, we determined the location of novel splice sites within the domain structure (Fig. 8). As neurexin protein domains mostly consist of multiple exons, alternative splicing events mostly lead to the partial deletion of domains and produce nonfunctional proteins. However, some of the novel alternative splicing events completely remove large structural modules, thereby creating new neurexins that are likely to have new functional properties. Strikingly, we observed very few large-scale domain structure-changing events of alternative splicing in Nrxn3α, but several independent events in Nrxn1α with dramatic effects. For example, some rare but repeatedly observed events of large-scale alternative splicing of Nrxn1α completely remove all domains between the first and last LNS and EGF-like domains (Fig. 8), which are likely to form functional neurexin isoforms with novel protein structures. We estimated that 97.4% of Nrxn1α transcripts and 99.6% of Nrxn3α transcripts generate functional proteins, based on the assumption that a transcript can be translated into a functional protein if the correct reading frame is maintained and predicted protein modules are likely either present or absent.
Fig. 8.
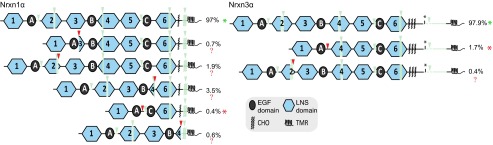
Protein domain structures of Nrxn1α and Nrxn3α splice isoforms. Protein domain structures corresponding to groups of splice isoforms are shown for Nrxn1α and Nrxn3α. Used splice sites are denoted by downward-pointing triangles colored in green for previously known canonical splice sites and red for novel splice sites. Protein parts that are spliced out only in a subset of splice variants of a given group are shown in semitransparent green. The percentage of total transcripts detected for each protein isoform is given. In the majority of detected transcripts only canonical splice sites are used, which are known to produce functional proteins (green asterisks). Most protein isoforms resulting from transcripts in which novel alternative splice sites are used appear to be nonfunctional (red question marks), because substantial parts of LNS domains are removed. However, a small subset of transcripts using novel alternative splice sites is suggested to produce functional, truncated proteins (red asterisks), because structural neurexin domains (LNS and EGF) are completely removed.
Discussion
We used a single-molecule long-read sequencing approach for profiling neurexin isoform diversity. Previous analyses of neurexins were based on sequencing of a limited number of cDNAs and on PCR-based methods (14). These methods either did not have enough depth to map the complete diversity of neurexin mRNAs (cDNA sequencing) or analyzed individual splice sites separately (PCR), and thus did not provide information about the combination of different splice sites within full-length transcripts. In contrast, the long-read sequencing approach overcomes these limitations by sequencing thousands of independent full-length mRNA transcripts. Our study provides more than 25,000 full-length neurexin mRNA reads, allowing us to analyze all splicing events within single mRNA molecules and to detect low-expressed isoforms.
We found that neurexins are likely even more polymorphic than previously thought. Full-length mRNA sequencing revealed an enormous diversity of α-neurexin isoforms in which no single splice variant dominates (Figs. 2 and 4). The situation is much simpler for β-neurexins in which diversity is limited, and nearly all variants can be explained by alternative splicing at the previously described SS#4 and SS#5 (14). Based on our sequencing data, how many different neurexins are there? For this calculation, we need to know whether all alternative splicing events are independent of each other. To test this, we calculated the Pearson correlation coefficient between all alternatively spliced sequences. We did not observe any correlation between canonical splice sites, including the newly identified alternatively spliced exon, suggesting that these sites of alternative splicing are used independent of each other. Cumulatively, our results suggest that the various events of alternative splicing can be used independent of each other in all possible combinations. Based on this conclusion, we calculated a total number of possible neurexin variants, taking into account only splice events that were observed more than twice. We observed in this manner a minimal diversity of 1,159 isoforms for Nrxn1α, 1,120 isoforms for Nrxn3α, and a total of 152 isoforms for all three β-neurexins. Thus, earlier estimates of 2,000–3,000 neurexin variants created by alternative splicing (8, 9, 12, 14) may have been an underestimate, because our present study arrived at the same numbers by analyzing only one brain region and one developmental stage (prefrontal cortex of adult mice) and because Nrxn2α mRNAs were not examined, suggesting that the true number of neurexin variants may be even higher.
An important observation of our present study is that we found a new evolutionarily conserved exon that is alternatively spliced in Nrxn1α and Nrxn3α, which we propose referring to as alternatively spliced sequence 6. Although we did not analyze Nrxn2α mRNAs in the current study, the exon encoding SS#6 is absent from the Nrxn2α gene, suggesting that Nrxn2α lacks SS#6. According to the phylogenetic tree of neurexins, the Nrxn2 gene separated from the common precursor gene to Nrxn1 and Nrxn3 early in evolution, whereas Nrxn1 and Nrxn3 diverged later (Fig. 1). In mammals, the Nrxn2 gene is strikingly smaller (10-fold) than the Nrxn1 and Nrxn3 genes, and the intronic region of Nrxn1 and Nrxn3 containing the exon encoding SS#6 is 14 and 60 times larger, respectively, than the corresponding intron in the Nrxn2 gene (327 bp for the mouse genome).
SS#6 encodes 9 residues that localize to the flexible hinge region between the fifth LNS domain (LNS5) and the third (C) EGF-like sequence of neurexins (Fig. 7A) (39, 40). This is the only region of the Nrxn1α structure in which there are no interdomain contacts between the LNS and EGF-like domains. The flexible hinge region has been proposed to explain existing biochemical data suggesting that the sixth LNS domain (LNS6) behaves differently depending on whether it is examined on its own (as it is present in β-neurexins) or together with other LNS domains [as it is present in α-neurexins (39, 40)]. The hinge region may contribute to the regulation of the interactions of the LNS5 and LNS6 domains with their binding partners, and the location of SS#6 suggests that this novel splice site plays a role in this regulation (Fig. 8).
In addition to SS#6, our results revealed other novel events of alternative splicing that could significantly increase the number of possible neurexin isoforms. These events consist of the excision of multiple exons, resulting in mRNAs that encode for Nrxn1α or Nrxn3α isoforms, in which multiple domains are deleted. Five such large excision events were repeatedly observed for Nrxn1α (Fig. 2), deleting exons 7 and 8, exons 2–9, exons 2–15, exons 13–19 (13–18 in ref. 12), and exons 13–25 (13–24 in ref. 12). A single such larger skipping event was repeatedly observed for Nrxn3α, deleting exons 7–9 (Fig. 4). All of these large-scale skipping events create in-frame junctions in the mRNAs, such that the resulting mRNAs encode proteins in which the sequences (and domains) encoded by the skipped exons are simply deleted (Fig. 8). These large-scale exon excision events are relatively rare but not negligible, accounting for more than 5% of the Nrxn1α mRNAs. Their physiological significance is unclear, but at least for the event-deleting exons 12–18 (12–17 in ref. 12) in the Nrxn1α mRNAs, we have confirmed by PCR and Sanger sequencing that this event is real and not an artifact of unknown provenance.
Another approach for assessing the potential biological significance of large-scale events of alternative splicing is to examine whether the resulting proteins are likely to have correctly folded domains. Indeed, some of these rare mRNAs encode potentially very interesting Nrxn1α protein variants that lack entire regions containing multiple domains. However, others encode protein variants with only partial deletions of domains, and their biological significance remains unclear (Fig. 8). It is possible that the proteins resulting from large-scale events of alternative splicing may be expressed at certain developmental stages or in specific cell subpopulations, and may regulate specific functions.
Neurexins are likely involved in multiple functions at the synapse that are mediated by different domains and are regulated by distinct events of alternative splicing. At present, only the functional importance of SS#4 in LNS6 has been characterized, and was found to have a massive effect on the transsynaptic regulation of synaptic strength in Nrxn3 (10). However, it seems likely that at least some of the other events of alternative splicing are also functionally significant and that at least α-neurexins form an extracellular protein-interaction scaffold, in which different protein interactions mediate distinct functions and are regulated by independent events of alternative splicing.
Materials and Methods
Library Preparation and Pacific Biosciences Sequencing.
Reverse transcription (RT) was performed for each neurexin isoform separately, and cDNAs were amplified by PCR as described in detail in SI Materials and Methods. SMRT bell sequencing libraries were prepared using Pacific Biosciences DNA Template Prep Kit 2.0 (001-540-835) according to the 2- or 5-kb template preparation and sequencing protocol (Pacific Biosciences) with a minimum of 1 µg and 500 ng of cDNA input into each library preparation for α-neurexins and β-neurexins, respectively. SMRT bell templates were bound to polymerases using DNA/Polymerase Binding Kit XL 1.0 (100-150-800) and v2 primers. Sequencing was carried out on the Pacific Biosciences real-time sequencer using C2 sequencing reagents with 90-min movies. Each α-neurexin was sequenced on six SMRT cells, yielding a total of 151,776 raw reads for Nrxn1α and 198,505 raw reads for Nrxn3α. β-Neurexins were sequenced on two or three SMRT cells, each yielding a total of 56,449 raw reads for Nrxn1β, 73,637 raw reads for Nrxn2β, and 92,537 raw reads for Nrxn3β.
Confirmation of a Newly Identified Splicing Event.
RT was performed using a primer specific to Nrxn1 exon 20 (19 in ref. 12) (5′-TCCAGGTAGTCACCCAGTCC-3′). PCR was followed using 2 µL RT product as template in 25 µL PrimeSTAR polymerase (Clontech) and a PCR primer pair specific to exons 11 and 20 (19 in ref. 12) (forward, 5′-TGAGAGAGAGGCAACGGTTT-3′; reverse, 5′-AATCTGTCCACCACCTTTGC-3′).
Processing, Alignment, and Analysis of Pacific Biosciences Sequencing Data.
Subread filtering was performed using Pacific Biosciences SMRT analysis software (v1.3.3). For all β-neurexins, circular consensus (CCS) reads were constructed from molecules that the DNA polymerase passed at least twice (reads containing four times the hairpin adapter sequence), whereas CCS reads for α-neurexins were constructed from molecules that the polymerase fully passed at least once. CCS reads were mapped to the mouse genome (mm10) and analyzed using STAR 2.2.0 (38) and R (41) as described in detail in SI Materials and Methods.
Transcript Maps, Bar Graphs, and Correlograms.
Transcript maps were generated in R (41) by first aggregating the binary splice matrix to obtain a matrix of all unique splice variants and then drawing a heatmap of all unique splice variants using the heatmap.2 function. Bar graphs were generated using ggplot2. Correlograms were created by calculating pairwise Pearson correlations between all alternatively spliced exons across all unique splice variants (between columns in the aggregated splice matrix) and plotting the resulting correlation matrix using the R function levelplot.
Modeling of the New Exon 17 into the Nrxn1α Structure.
The newly identified Nrxn1α exon 17 with the amino acid sequence VALMKADLQ was modeled in an arbitrary, extended conformation into the Nrxn1α crystal structure [Protein Data Bank (PDB) ID code 3QCW (39)] between E1088 and G1089 using Coot (42). To accommodate the additional 9 amino acids of the new exon, Nrxn1α C-terminal domains EGF-C and LNS6 (G1089–V1355) were dislocated from the N-terminal part of the protein by about 27 Å.
Effects of Modular Splicing Events on Protein Structure.
Aggregated binary splice matrices were created for the examined regions of Nrxn1α and Nrxn3α in R to identify all unique splice variants and their abundances. Splice variants were grouped based on similarity of the corresponding protein domain structure. All splice variants containing only previously detected canonical splice events were grouped together because they are known to produce functional neurexin proteins with only small changes in the overall protein domain structure. All remaining splice variants corresponded to proteins with more substantial changes in the protein domain structure that are suggested to be either nonfunctional (substantial parts of LNS domains spliced out) or functional (full LNS/EGF modules spliced out).
Supplementary Material
Acknowledgments
We thank Jody Puglisi for sharing equipment. This study was supported by Grants R37 MH052804 from the National Institute of Mental Health and R01 NS077906 from the National Institute of Neurological Disorders and Stroke (to T.C.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Sequence Read Archive (accession no. SRP039451).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403244111/-/DCSupplemental.
References
- 1.Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci USA. 1963;50(4):703–710. doi: 10.1073/pnas.50.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Essen DC. Cartography and connectomes. Neuron. 2013;80(3):775–790. doi: 10.1016/j.neuron.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargmann CI, Marder E. From the connectome to brain function. Nat Methods. 2013;10(6):483–490. doi: 10.1038/nmeth.2451. [DOI] [PubMed] [Google Scholar]
- 4.Meinertzhagen IA, Lee CH. The genetic analysis of functional connectomics in Drosophila. Adv Genet. 2012;80:99–151. doi: 10.1016/B978-0-12-404742-6.00003-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleinfeld D, et al. Large-scale automated histology in the pursuit of connectomes. J Neurosci. 2011;31(45):16125–16138. doi: 10.1523/JNEUROSCI.4077-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hattori D, et al. Robust discrimination between self and non-self neurites requires thousands of Dscam1 isoforms. Nature. 2009;461(7264):644–648. doi: 10.1038/nature08431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97(6):779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 8.Ushkaryov YA, Petrenko AG, Geppert M, Südhof TC. Neurexins: Synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257(5066):50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- 9.Ushkaryov YA, Südhof TC. Neurexin III α: Extensive alternative splicing generates membrane-bound and soluble forms. Proc Natl Acad Sci USA. 1993;90(14):6410–6414. doi: 10.1073/pnas.90.14.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC. Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell. 2013;154(1):75–88. doi: 10.1016/j.cell.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ushkaryov YA, et al. Conserved domain structure of β-neurexins. Unusual cleaved signal sequences in receptor-like neuronal cell-surface proteins. J Biol Chem. 1994;269(16):11987–11992. [PubMed] [Google Scholar]
- 12.Tabuchi K, Südhof TC. Structure and evolution of neurexin genes: Insight into the mechanism of alternative splicing. Genomics. 2002;79(6):849–859. doi: 10.1006/geno.2002.6780. [DOI] [PubMed] [Google Scholar]
- 13.Rowen L, et al. Analysis of the human neurexin genes: Alternative splicing and the generation of protein diversity. Genomics. 2002;79(4):587–597. doi: 10.1006/geno.2002.6734. [DOI] [PubMed] [Google Scholar]
- 14.Ullrich B, Ushkaryov YA, Südhof TC. Cartography of neurexins: More than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14(3):497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 15.Gokce O, Südhof TC. Membrane-tethered monomeric neurexin LNS-domain triggers synapse formation. J Neurosci. 2013;33(36):14617–14628. doi: 10.1523/JNEUROSCI.1232-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrmann I, et al. The tissue-specific RNA binding protein T-STAR controls regional splicing patterns of neurexin pre-mRNAs in the brain. PLoS Genet. 2013;9(4):e1003474. doi: 10.1371/journal.pgen.1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapiro-Reznik M, Jilg A, Lerner H, Earnest DJ, Zisapel N. Diurnal rhythms in neurexins transcripts and inhibitory/excitatory synapse scaffold proteins in the biological clock. PLoS ONE. 2012;7(5):e37894. doi: 10.1371/journal.pone.0037894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Z, Sharpe CR, Simons JP, Górecki DC. The expression and alternative splicing of α-neurexins during Xenopus development. Int J Dev Biol. 2006;50(1):39–46. doi: 10.1387/ijdb.052068zz. [DOI] [PubMed] [Google Scholar]
- 19.Rozic-Kotliroff G, Zisapel N. Ca2+-dependent splicing of neurexin IIalpha. Biochem Biophys Res Commun. 2007;352(1):226–230. doi: 10.1016/j.bbrc.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Patzke H, Ernsberger U. Expression of neurexin Ialpha splice variants in sympathetic neurons: Selective changes during differentiation and in response to neurotrophins. Mol Cell Neurosci. 2000;15(6):561–572. doi: 10.1006/mcne.2000.0853. [DOI] [PubMed] [Google Scholar]
- 21.Iijima T, et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147(7):1601–1614. doi: 10.1016/j.cell.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ichtchenko K, et al. Neuroligin 1: A splice site-specific ligand for beta-neurexins. Cell. 1995;81(3):435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- 23.Ichtchenko K, Nguyen T, Südhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 1996;271(5):2676–2682. doi: 10.1074/jbc.271.5.2676. [DOI] [PubMed] [Google Scholar]
- 24.Ko J, Fuccillo MV, Malenka RC, Südhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64(6):791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Wit J, et al. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64(6):799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, Craig AM. LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J Neurosci. 2010;30(22):7495–7506. doi: 10.1523/JNEUROSCI.0470-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uemura T, et al. Trans-synaptic interaction of GluRdelta2 and neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141(6):1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda K, Yuzaki M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci. 2011;33(8):1447–1461. doi: 10.1111/j.1460-9568.2011.07638.x. [DOI] [PubMed] [Google Scholar]
- 29.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to α- and β-neurexins. Neuron. 2005;48(2):229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 30.Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51(2):171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Comoletti D, et al. Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for β-neurexins. Biochemistry. 2006;45(42):12816–12827. doi: 10.1021/bi0614131. [DOI] [PubMed] [Google Scholar]
- 32.Petrenko AG, et al. Structure and evolution of neurexophilin. J Neurosci. 1996;16(14):4360–4369. doi: 10.1523/JNEUROSCI.16-14-04360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Missler M, Hammer RE, Südhof TC. Neurexophilin binding to α-neurexins. A single LNS domain functions as an independently folding ligand-binding unit. J Biol Chem. 1998;273(52):34716–34723. doi: 10.1074/jbc.273.52.34716. [DOI] [PubMed] [Google Scholar]
- 34.Sugita S, et al. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 2001;154(2):435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boucard AA, Ko J, Südhof TC. High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J Biol Chem. 2012;287(12):9399–9413. doi: 10.1074/jbc.M111.318659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eid J, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 37.English AC, et al. Mind the gap: Upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE. 2012;7(11):e47768. doi: 10.1371/journal.pone.0047768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen F, Venugopal V, Murray B, Rudenko G. The structure of neurexin 1α reveals features promoting a role as synaptic organizer. Structure. 2011;19(6):779–789. doi: 10.1016/j.str.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller MT, et al. The crystal structure of the α-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function. Structure. 2011;19(6):767–778. doi: 10.1016/j.str.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. R Development Core Team (2009) R: A Language and Environment for Statistical Computing (R Found Stat Comput, Vienna)
- 42.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.