Significance
The process of DNA segregation is of central importance for all organisms. Although the basic mechanism of eukaryotic mitosis is relatively well established, the most common mechanism used for bacterial DNA segregation has been unclear. ParA ATPases form dynamic patterns on the bacterial nucleoid to spatially organize plasmids, chromosomes and other large cellular cargo, but the force-generating mechanism has been a source of controversy and debate. A dominant view proposes that ParA-mediated transport and cargo positioning occurs via a filament-based mechanism that resembles eukaryotic mitosis. Here, we present direct evidence against such models. Our cell-free reconstitution supports a non–filament-based mode of transport that may be as widely found in nature as filament-based mechanisms.
Keywords: bacterial chromosome segregation, ParA ATPase, spatial organization, protein gradients
Abstract
The faithful segregation of duplicated genetic material into daughter cells is critical to all organisms. In many bacteria, the segregation of chromosomes involves transport of “centromere-like” loci over the main body of the chromosome, the nucleoid, mediated by a two-protein partition system: a nonspecific DNA-binding ATPase, ParA, and an ATPase stimulator, ParB, which binds to the centromere-like loci. These systems have previously been proposed to function through a filament-based mechanism, analogous to actin- or microtubule-based movement. Here, we reconstituted the F-plasmid partition system using a DNA-carpeted flow cell as an artificial nucleoid surface and magnetic beads coated with plasmid partition complexes as surface-confined cargo. This minimal system recapitulated directed cargo motion driven by a surface ATPase gradient that propagated with the cargo. The dynamics are consistent with a diffusion-ratchet model, whereby the cargo dynamically establishes, and interacts with, a concentration gradient of the ATPase. A chemophoresis force ensues as the cargo perpetually chases the ATPase gradient, allowing the cargo to essentially “surf” the nucleoid on a continuously traveling wave of the ATPase. Demonstration of this non–filament-based motility mechanism in a biological context establishes a distinct class of motor system used for the transport and positioning of large cellular cargo.
Bacterial chromosomes and plasmids use active segregation, or partition (Par), mechanisms to ensure faithful inheritance. The ParA ATPase family forms dynamic gradients on the nucleoid while spatially organizing chromosomes and plasmids, as well as other large cargoes (1). ParA-type ATPases are ubiquitous in the microbial world, but the mechanism by which ParA patterning produces the driving force for cargo movement over the nucleoid remains controversial (2). The stability of plasmid (SOP) system of F plasmid is a canonical ParA-mediated cargo transport system. The ParA ATPase of F plasmid, SopA, is stimulated by SopB that assembles into a partition complex on the centromere-like locus, sopC, on the plasmid cargo (3, 4). SopA binds DNA nonspecifically in the presence of ATP and colocalizes with the nucleoid in vivo (5, 6). The partition complex locally removes SopA, forming a SopA depletion zone on the nucleoid in the vicinity of the plasmid (7), but how this patterning on the nucleoid results in cargo transport is a subject of considerable debate.
In vivo cytology of ParA-mediated transport is consistent with, and provides a foundation for, two competing models (2). The filament-pulling model is reminiscent of eukaryotic mitosis whereby partition complexes are mobilized by helical (8) or linear (9) contractile filaments composed of the ParA ATPase. The diffusion-ratchet model, however, proposes that ParA dimers, or small oligomers, independently bind the nucleoid and form concentration gradients in the vicinity of the partition complex, which generates the driving force for cargo movement (6, 10, 11). Here, we extend this model further by showing that a pulling force on the plasmid cargo can be mechanochemically coupled to the ParA concentration gradient on the nucleoid. This force can be modeled as a chemophoresis force (12): ParA provides a chemical potential gradient that is sensed by a macroscopic element, the partition complex, containing a large number of ParB molecules that bind weakly to ParA. The cumulative effect of the individual ParA–ParB interactions directs cargo motion toward regions of increased binding, that is, the partition complex moves up the gradient toward higher ParA concentrations.
We previously reconstituted the F- and P1-plasmid partition systems using purified components inside a DNA-carpeted flow cell (6, 11), which acted as an artificial nucleoid surface. The cell-free dynamics recapitulated the local removal of SopA (or P1 ParA) on the DNA carpet around a partition complex. However, the complexes were only transiently tethered to the DNA carpet and the SopA gradients surrounding a partition complex did not generate robust directed motion (6). Instead, the plasmid diffused away from the carpet once all tether points, i.e., SopB–SopA interactions, were released. We hypothesized our flow cell lacked the surface confinement needed to maintain contact between the plasmid and the DNA carpet, as the narrow gap between the nucleoid and the inner membrane in vivo would confine the plasmid to the nucleoid surface (13).
Here, we reconstituted and visualized directed cargo motion by the F-plasmid partition system with purified components using total internal reflection fluorescence microscopy (TIRFM) in a DNA-carpeted flow cell (6, 11). To mimic surface confinement on the nucleoid, we artificially confined magnetic beads, coated with sopC DNA, to the carpet. Our results demonstrate that physical confinement of the partition complex to the nucleoid surface is a key requirement for ParA-mediated transport by a diffusion-ratchet mechanism, where a chemophoresis force generates cargo motion.
Results
Sop Proteins Direct sopC-Bead Movement.
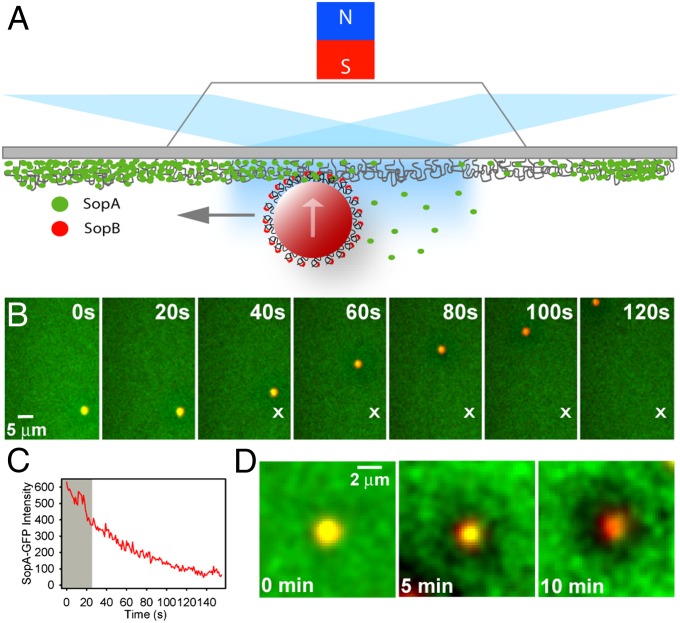
As in our previous reconstitutions of plasmid partition (6, 11), purified components of the partition system were assembled inside a DNA-carpeted flow cell, the surface of which acted as a nucleoid mimic. To confine partition complexes to this surface, magnetic beads coated with DNA encoding sopC were used as cargo (Fig. S1). An external magnetic field perpendicular to the slide surface was applied to confine the beads to the DNA carpet surface. SopB was preincubated with sopC-beads, and SopA-GFP with ATP. The two solutions were mixed and infused into the flow cell where the dynamics of the magnetic beads and the Sop proteins on the DNA carpet were visualized using prism-type TIRFM (Fig. 1A).
Fig. 1.
SopA- and SopB-mediated transport of a sopC-coated bead on a DNA carpet. (A) Schematic of the magnetic bead system in a DNA-carpeted flow cell visualized by TIRFM. The upward arrow represents magnetic force on the bead, and the gray arrow indicates Sop-mediated motion of the bead. (B) Time-lapse sequence of a directed sopC-bead (red) in the presence of SopA-GFP (green), SopB, and ATP. Time 0 is when flow was stopped. “X” marks the initial location of the traveling bead. (C) SopA disassembly is a prerequisite for bead movement. SopA-GFP intensity on the bead in B was plotted against time. The average SopA-GFP intensity on the carpet was subtracted. The bead was immobile (gray shading) until a threshold level of SopA was released, at which point the bead moved in a directed manner. (D) Sop protein dynamics in the vicinity of a directed bead. Time-lapse sequence of SopB-stimulated release of SopA-GFP (green) from a bead (red) and the surrounding DNA carpet over time. Time 0 is the point at which the bead tethered to the carpet.
sopC-beads initially formed tethers to the SopA-coated DNA carpet through protein-mediated cross-bridges (Fig. 1B). Bead tethering required both Sop proteins and ATP (Movie S1) as was the case when using a plasmid substrate (6). However, in striking contrast to plasmid cargo, which detached from the DNA carpet following SopA release, the sopC-beads began traveling across the DNA carpet after SopA disassembled in the vicinity of the bead (Fig. 1 B and C, and Movie S2). The traveling beads produced robust SopA depletion zones on the surrounding DNA carpet (Fig. 1D).
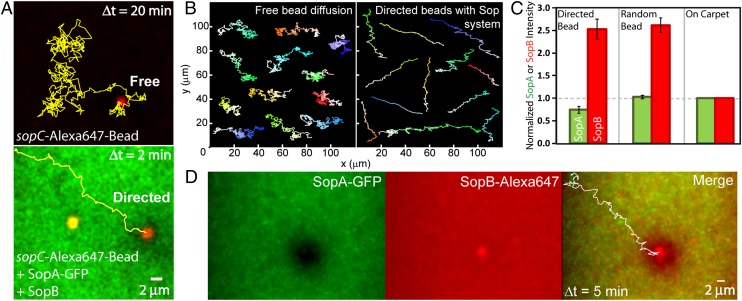
Not all beads displayed directed motion. The artificial confinement provided by the magnet produced a spectrum of bead behaviors ranging from directed transport to diffusive motion (Movie S3) similar to that observed with free beads—in the absence of either of the Sop proteins or ATP (Fig. 2 A and B, and Fig. S2). To determine whether the amount of protein associated with the “directed” and “diffusive” bead populations influenced their behavior, SopA-GFP and SopB-Alexa 647 (mixed 1:9 with unlabeled SopB) were covisualized with unlabeled sopC-beads (Movie S4). SopB content on both bead populations was similar, ∼2.5-fold higher than on the DNA carpet (Fig. 2C). However, SopA content on the directed beads was 25 ± 5% less than on diffusive beads. Also, the directed beads established robust SopA depletion zones on the surrounding carpet that propagated with the beads as they moved (Fig. 2D and Movie S4). The data suggest that diffusive beads do not maintain persistent interactions with the carpet because their high SopA content sequesters SopB from interaction with carpet-bound SopA. Consistent with this hypothesis, diffusive beads showed intermittent carpet contact, similar to free beads in the absence of Sop proteins, as measured by intensity fluctuations of beads “bouncing” in and out of the TIRF illumination (Fig. S2 D and E). The directed beads, however, have SopB available for persistent interaction with SopA molecules on the carpet. Fluorescence recovery after photobleaching analysis showed SopB exchange was faster on the nonspecific DNA carpet than on the sopC-coated bead (Fig. S3). SopB on the bead also showed a significant immobile fraction (23 ± 2%), which can be attributed to specific binding to sopC. The data show that SopB, concentrated at sopC sites, both generates and follows a SopA gradient on the carpet, resulting in directed bead motion.
Fig. 2.
SopB-induced patterning of SopA enables directed transport of sopC cargo. (A) Freeze-frame of a bead (red) in the absence (Upper) or presence (Lower) of SopA-GFP (green), SopB, and ATP. Bead trajectories are yellow lines. (B) Montage of free- and directed-bead trajectories in the absence (Left) or presence (Right) of the Sop system. Light to dark shading of the trajectories shows the progression from beginning to end. (C) Directed beads are depleted of SopA, but not SopB. The maximum intensity of SopA-GFP (green) and SopB-Alexa 647 (red) associated with directed or diffusive beads was normalized to the average intensity for each protein on the carpet. Error bars represent the SD. (D) One frame of the SopA (green) and SopB (red) channels for a directed bead. The bead trajectory is shown in white in the merge.
Directed Movement Requires Surface Confinement.
The magnet was required to maintain the beads on the carpet once SopA was depleted (Fig. S4 and Movie S5), indicating that without surface confinement of the cargo, SopA–SopB interactions alone are insufficient to maintain the persistent association between the plasmid and the nucleoid required for directed movement. Increasing the confinement force on the beads decreased the diffusive bead population. We conclude that persistent interactions between plasmid-bound SopB and nucleoid-bound SopA, facilitated by surface confinement of the cargo, are critical for nucleoid patterning of SopA and the resulting directed plasmid transport in vivo.
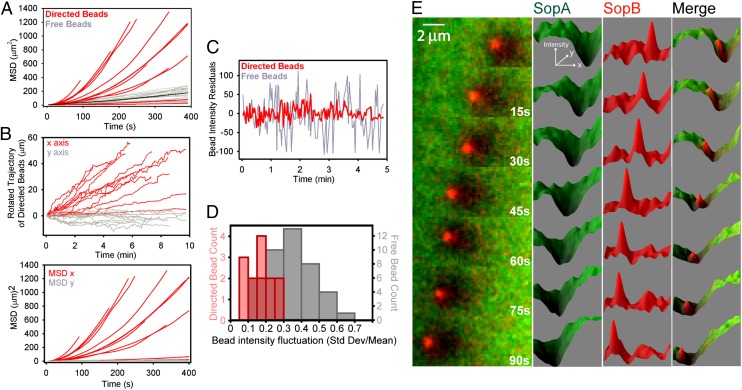
Beads with a distinct SopA depletion zone displayed directed and highly persistent motion with an average speed of 0.10 ± 0.02 μm/s (Fig. 3 A and B, and Fig. S2). The diffusion constant of the directed beads (0.03 ± 0.01 μm2/s) was 3.3-fold lower than that of free beads (0.10 ± 0.03 μm2/s). Vertical motion, or bead “bouncing,” was also suppressed compared with that of free beads (Fig. 3 C and D). These data suggest that a SopA-SopB–mediated interaction with the surface reduces bead diffusion and generates directed motion.
Fig. 3.
Quantitative characteristics of Sop-mediated directed motion. (A) Sop system induces highly directional bead movement. Mean square displacement (MSD) of directed and free beads was plotted against time. The black line is the average MSD for free beads. (B) Directed bead trajectories were rotated to achieve maximal projection along the x axis (Upper), and the displacements are plotted against time. Directed velocities were obtained from slopes of linear fits of the x motion as a function of time. The y motion as a function of time is essentially diffusive. MSD of rotated x and y motion plotted against time (Lower). (C) Directed beads maintain a persistent interaction with the carpet. Fluorescence intensity fluctuations of a free and directed bead as a function of time were fit to a linear equation and the residuals, which are indicative of the z position of the bead, are plotted. (D) Histogram of bead intensity fluctuations for free- and directed-bead populations. (E) A time-lapse image series of the fluorescence profile of SopA (green) and SopB (red) on and around a bead traveling from right to left.
A Chemophoresis Force for Directed Movement.
The Sop-mediated force driving directed movement was estimated from two independent measurements. The repulsive force between magnetic beads (SI Text) allowed directed beads to “bulldoze” diffusive beads out of their path (Movie S6). However, when a directed bead approached an immobile bead, it stalled or was deflected (Movie S7). Calculations of the repulsive magnetic force between beads at the distance of closest approach (9 ± 1 μm) suggest a deflection force of Sop-mediated transport of 5 ± 2 fN (Fig. S5B), similar to estimates of the driving force based on the viscous drag of ∼12 ± 4 fN calculated for directed beads (SI Text). These force estimates are compatible with estimates of the force generated by SopB interaction with the SopA gradient based on a chemophoresis model (12) where cargo-bound SopB senses the local SopA chemical potential gradient and is attracted toward higher SopA concentrations (SI Text and Fig. S6).
The magnetic force used to confine sopC-beads to the surface is not the major driving force for directed bead movement. The estimated magnetic force on the beads, ∼40 fN (Fig. S5A), is perpendicular to the surface. We estimate the magnetic axis alignment is typically within 1°. Even with 2° misalignment, the lateral magnetic force would be less than 1.5 fN. Furthermore, if the external force were the major contributor to bead motion, diffusive beads within the flow cell would have exhibited the same average directed motion, which was not the case. However, a slight lateral force due to magnet misalignment might have contributed to the extremely long persistence length of motion observed despite the local diffusive motion of the beads.
Discussion
It has been 60 years since Alan Turing first proposed the reaction-diffusion concept to explain how multiple diffusing species can interact and react to self-organize into patterns (14). Several groups have speculated that this concept might underlie ParA-directed plasmid/chromosome movement (15, 16). However, a more recently prevailing view favors the filament-pulling model of ParA action. We have argued that the in vivo cytological observations, upon which the contractile filament models have been based, are fully consistent with a diffusion-ratchet mechanism (6, 10, 11) that builds on the classical reaction-diffusion concept. Here, we have recapitulated the directed movement of a ParA system and shown that its features are well described by a diffusion-ratchet mechanism.
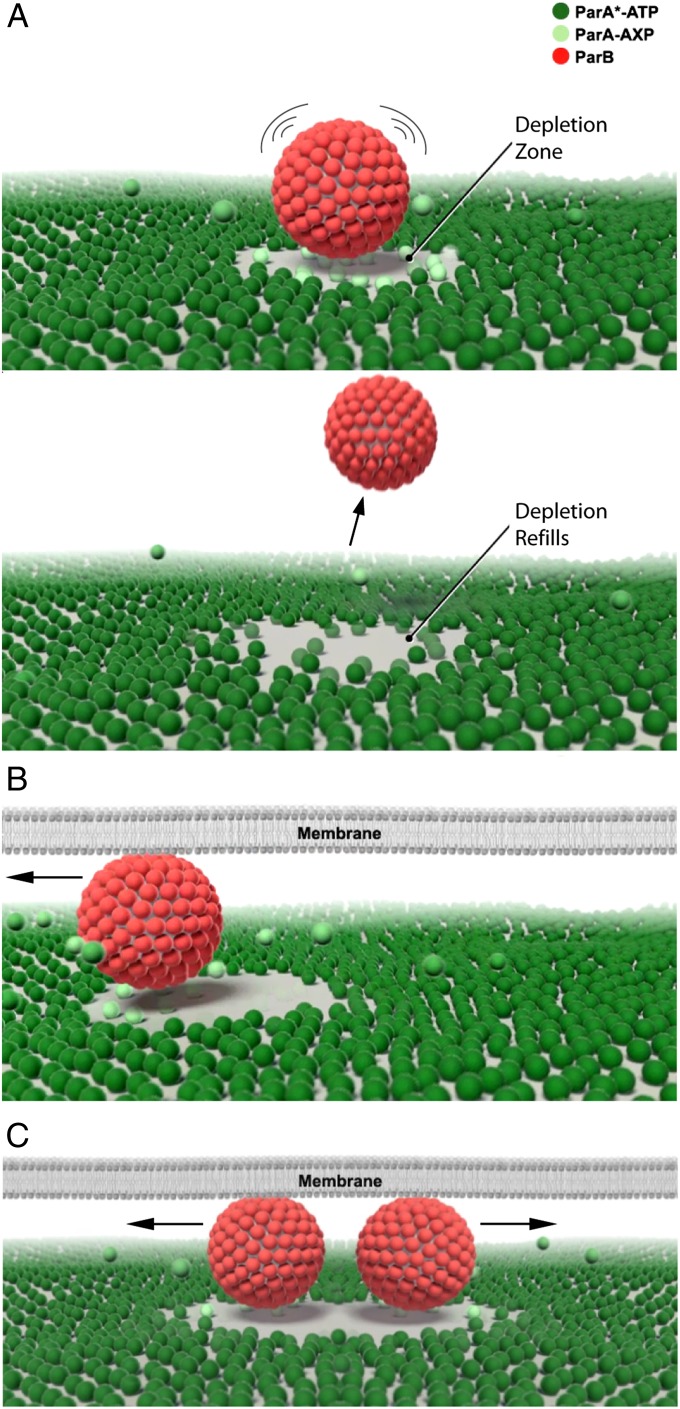
The protein distribution on and around a bead undergoing directed movement (Fig. 3E and Movie S8) remarkably mirrors the nucleoid patterning and cargo transport activities observed in vivo for several ParA-type systems (7, 8, 17, 18). SopA uniformly binds the DNA carpet away from SopB-bound cargo (sopC-bead). We have previously shown that, under these conditions, SopA binding density on the DNA carpet is less than 1% of saturation (6), which is comparable to the in vivo estimate of 0.2% binding density per bacterial chromosome (19). SopB stimulates the release of SopA on and around the cargo, forming a local SopA gradient on the carpet by a reaction-diffusion process. At the center of the SopA-depleted zone, the cargo randomly diffuses (Fig. 4A and Movie S9). When surface-confined cargo drifts to one side of the depletion zone, forward motion is promoted by the lowered free energy state resulting from an increased number of SopB–SopA interactions at the edge of the depletion zone, whereas reverse motion is suppressed by decreased SopB–SopA interaction in its wake (Fig. 4B and Movie S9). The SopA gradient, which mechanochemically drives motility via a chemophoresis force, is maintained by a delay in nucleoid rebinding by the SopA molecules released upon interaction with the partition complex. This delay prevents SopA from immediately rebinding the nucleoid in the vicinity of the cargo that stimulated its release. We recently identified one of the biochemical delays in the ATPase cycle of ParA from P1 plasmid (10). This diffusion-ratchet mechanism allows the cargo to essentially “surf” the nucleoid on a continually redistributing wave of the ATPase. Also, bidirectional cargo segregation is built into the above mechanism (Fig. 4C and Movie S9). After replication, the SopB-bound daughter plasmids would split as they chase the nucleoid-bound ATPase gradient on opposite sides of the existing depletion zone. Similarly, when multiple cargoes share the same nucleoid, their respective ATPase depletion zones would distribute the cargoes equidistantly.
Fig. 4.
A diffusion-ratchet model for ParA-mediated transport and partition. (A) The DNA-binding form of ParA (ParA*-ATP) binds the nucleoid and the ParB stimulator protein loads onto the cargo. ParB transitions ParA into a state (ParA-AXP) that weakens its interaction with the nucleoid. ParA release around the cargo creates a ParA depletion zone on the nucleoid. The cargo undergoes Brownian motion as ParA anchor points are removed. Without surface confinement, the cargo diffuses away once all ParA anchor points are released and the depletion zone refills. ParA-AXP is recycled through ATP hydrolysis, nucleotide exchange, and conformational changes back to ParA*-ATP. Time delays in the recycling process allow diffusion of ParA in solution away from the point of nucleoid release, resulting in slow refilling of the ParA depletion zone. (B) In a bacterial cell, the narrow cytosolic gap between the membrane and nucleoid confines large cargoes to the nucleoid surface and maintains a ParA depletion zone in the vicinity of the cargo. Cargo movement is directed by the large number of ParA contacts at its movement front and reversal is suppressed by the ParA depletion zone in the wake of movement. (C) Replicated cargoes, or cargoes in close proximity, would bidirectionally segregate as they chase ParA gradients in opposite directions.
Our results strongly implicate the extreme spatial confinement within a bacterial cell as an underappreciated requirement for ParA-type transport, and likely for other forms of intracellular spatial organization (20). In our diffusion-ratchet model, the motive force is not generated by a self-supporting filament, as it is for the actin-like class of partition ATPases (21). Rather, it is based on a biased Brownian walk of the cargo, bound by a sufficiently large number of ATPase stimulator molecules (SopB/ParB), which individually interact weakly and transiently with the nucleoid-bound ATPase (SopA/ParA). The cumulative contact density between ATPase and stimulator is continually probed via Brownian dynamics of the cargo, which occurs at a much slower timescale than the individual contacts. We further propose that for the Brownian walk to be sufficiently directionally biased, the diffusion of the cargo must be intrinsically slow and/or slowed by the drag caused by contacts with the ATPase. Otherwise, the cargo could diffuse out of the ATPase depletion zone and lose its positional memory. Therefore, surface confinement, along with molecular crowding that increases viscosity and slows diffusion inside the cell, likely play pivotal roles in the transport mechanism. Despite the confinement inside a bacterial cell, small cargoes may not be able to fulfill these requirements. Thus, it is perhaps not a coincidence that all cargoes using ParA-type transport are massive bodies that, without active transport, are excluded from the nucleoid region of the cell.
The results show ParA-mediated systems can transport very large cargoes by a diffusion-ratchet mechanism in a defined cell-free reaction setup. The mechanism requires many ATPase stimulator molecules to be bound to the cargo. For low-copy plasmids, the stimulator not only loads specifically onto the plasmid centromeres but also spreads onto surrounding DNA regions, and clusters sister copies into groups by a mechanism that is still unclear (22). Limiting the spread of the stimulator on the plasmid compromises partition in vivo. Thus, the partition complex contains many more stimulator molecules than the number of specific binding sites on the plasmid. In our cell-free reaction, we have not accomplished high loading of ParB/SopB molecules around parS/sopC sites (refs. 6 and 11, and this study). Instead, the requisite number of stimulator molecules per unit cargo was achieved by clustering a large number of plasmid copies (6, 11), or in this study, by using beads coated with sopC DNA. Although the nature of a bead is indeed different from a low-copy plasmid, the finding that ParA-type systems transport a variety of DNA- and protein-based cargoes suggest that the transport mechanism is indifferent to cargo composition (2). A unifying feature of the cargoes is their size, which we propose accommodates the binding of a large number of ATPase stimulator molecules.
The magnetic force used here to confine the beads to the DNA-carpeted surface differs in many respects from plasmid confinement between the nucleoid and membrane in vivo. Nevertheless, we could tune the magnetic force to find conditions that supported robust directional motion of beads coated with partition complexes. Too strong a force impeded bead motion, whereas too weak a force left most beads freely diffusing near the surface. Thus, we believe the conditions used here qualitatively mimic the spatial confinement of the cargo in vivo.
Reaction-diffusion mechanisms have been proposed to underlie many biological patterns from surface patterning of animals and digit number determination in mammals (23, 24) to divisome site selection and faithful genomic inheritance in bacteria (2). Here, we demonstrate that biomolecular patterning can drive the transport of cellular cargo under confinement. In the diffusion-ratchet mechanism of cargo transport, the system components function as a dispersed ensemble of transient interactions, which is fundamentally distinct from classical motor systems that rely on stable molecular assemblies. It is not difficult to imagine similar principles for a special class of burnt-bridge motion systems. Take, for example, surface-bound ligand molecules that are destroyed by a cargo-bound enzyme. If a large number of enzyme molecules are bound to the cargo, and if the enzyme–ligand interaction kinetics is appropriate, a chemophoresis force could drive cargo motion on the surface while the ligand molecules are mowed down. Influenza viruses bind and subsequently destroy the surface receptors of a host cell to prevent viral aggregation and allow for efficient viral spread in the respiratory tract (25). Could the association of a viral particle with the surface of a host cell exemplify a similar mechanism? A diffusion-ratchet mechanism would certainly be more efficient than one that is purely diffusion based. We anticipate that the two major components of our diffusion-ratchet model, patterning by reaction-diffusion and force generation by chemophoresis, can explain directed motion and positioning for mesoscale spatial organization of many biological systems.
Materials and Methods
Proteins.
Protein expression, purification, and labeling were performed as described previously (6).
Flow Cell.
Flow cell assembly, pacification, and carpeting with sonicated salmon sperm DNA was done as described previously (11).
Biotinylated sopC Construction.
SopB binds specifically to sopC and nonspecifically to DNA flanking the sopC site. Therefore, the pBR322::sopC template (6) was used to amplify a 3.36-kb DNA fragment containing a sopC site (12 × 43 bp tandem repeat = 516 bp) flanked by 1.7 and 1.1 kb of pBR322 sequence upstream and downstream of sopC, respectively. Primers (IDT) were designed such that the sopC-fragment was biotinylated (three biotin molecules in tandem) at the upstream end for conjugation to streptavidin-coated beads (DNA–Bead Coupling), and Alexa 647 labeled (one dye per DNA fragment) at the downstream end for fluorescence visualization when used (Table S1). The restriction enzyme PstI was used to digest fragments off the bead after conjugation to estimate the number of fragments/bead (see below).
DNA–Bead Coupling.
Fifty microliters of 10 mg/mL MyOne Steptavidin C1 Dynabeads (Invitrogen) were washed in wash buffer (10 mM Tris⋅HCl, pH 8.2, 1 M NaCl, 1 mM EDTA) according to manufacturer’s specifications, and resuspended in 1.3 mL of wash buffer plus 0.2% Tween20. Biotinylated sopC-DNA (10 pmol in 200 μL) was added to the 1.3 mL of beads (cf = 0.33 mg/mL) and incubated for 1 h with gentle rotation. The beads were then washed with wash buffer according to manufacturer’s guidelines, and resuspended in 50 μL of 30 mM Tris⋅HCl, pH 7, 100 mM KCl, 1 mM EDTA, and stored on ice. A 5-μL aliquot of sopC-beads was digested with PstI, the beads were pelleted, and sopC-DNA fragments in the supernatant were ethanol precipitated to quantify the mean number of fragments/bead by A260 on a Nanodrop spectrophotometer (1,000 ± 200 DNA fragments per bead; n = 8).
Sample Handling and Preparation.
All experiments were performed in Sop buffer: 50 mM Hepes, pH 7.5, 100 mM KCl, 10% (vol/vol) glycerol, 5 mM MgCl2, 2 mM DTT, 0.1 mg/mL α-casein, and 0.6 mg/mL ascorbic acid. Two millimolar phosphoenolpyruvate (Sigma) and 10 μg/mL pyruvate kinase (Sigma) were also added for ATP regeneration.
SopA-GFP (5 μM) was preincubated with 2 mM ATP, and 10 μM SopB (mixed 1:9 with SopB-Alexa 647, when specified) was preincubated with 1.6 μg/μL sopC-beads (Alexa 647-labeled, when specified) for 20 min at 23 °C. The two samples were then mixed and diluted to the final concentrations of 0.5 μM SopA-GFP, 1 μM SopB, 0.16 μg/μL sopC-beads, and 2 mM ATP. The sample was then immediately loaded into a syringe (Hamilton), and TFZL 1/16” × 0.02” tubing (UpChurch) was used to connect to a Micro-Metering Valve (UpChurch), which was connected to the flow cell inlet nanoport (UpChurch). The same valve type was also connected to the flow cell outlet nanoport. The sample was infused into the flow cell at 20 μL/min for 1 min, and flow was stopped for data acquisition. The Micro-Metering Valves were closed to ensure that all residual flow was stopped. Using a goniometer mount, a magnet [N 52, cylindrical magnet 1.5” (38.1 mm) in length and 1/4” (6.35 mm) in diameter; K&J Magnetics] was centered 12 mm above the objective lens of the inverted microscope. Movies were then acquired.
Imaging and Analysis.
The illumination, microscope, and camera settings were described previously (11). Movies were acquired using Metamorph 7 (Molecular Devices) and transferred to ImageJ (National Institutes of Health) for conversion to QuickTime file format (.mov). For bead trajectories, the background was subtracted using a sliding window length of 40 pixels with the Mosaic Plug-in (26) for ImageJ. The Octane plug-in (27) for ImageJ was then used to track bead trajectories and fluorescence intensity transients of the tracked bead. The brightness and contrast were set for each picture or movie individually for the best representation of the features of interest. Movie frame rates are indicated in the figure legends. Adobe Illustrator was used to convert movies into figures.
Supplementary Material
Acknowledgments
We are particularly thankful to Ethan Tyler for model animation and graphics, Hemai Parthasarathy for editorial assistance, and John Silver and Yeonee Seol for comments. This work was supported by the intramural research fund for National Institute of Diabetes and Digestive and Kidney Diseases (to K.M.) and National Heart, Lung, and Blood Institute (to K.C.N.), National Institutes of Health US Department of Health and Human Services, and the Nancy Nossal Fellowship (to A.G.V.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 4741.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401025111/-/DCSupplemental.
References
- 1.Lutkenhaus J. The ParA/MinD family puts things in their place. Trends Microbiol. 2012;20(9):411–418. doi: 10.1016/j.tim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vecchiarelli AG, Mizuuchi K, Funnell BE. Surfing biological surfaces: Exploiting the nucleoid for partition and transport in bacteria. Mol Microbiol. 2012;86(3):513–523. doi: 10.1111/mmi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ah-Seng Y, Lopez F, Pasta F, Lane D, Bouet JY. Dual role of DNA in regulating ATP hydrolysis by the SopA partition protein. J Biol Chem. 2009;284(44):30067–30075. doi: 10.1074/jbc.M109.044800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez A, Rech J, Gasc C, Bouet JY. Insight into centromere-binding properties of ParB proteins: A secondary binding motif is essential for bacterial genome maintenance. Nucleic Acids Res. 2013;41(5):3094–3103. doi: 10.1093/nar/gkt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castaing JP, Bouet JY, Lane D. F plasmid partition depends on interaction of SopA with non-specific DNA. Mol Microbiol. 2008;70(4):1000–1011. doi: 10.1111/j.1365-2958.2008.06465.x. [DOI] [PubMed] [Google Scholar]
- 6.Vecchiarelli AG, Hwang LC, Mizuuchi K. Cell-free study of F plasmid partition provides evidence for cargo transport by a diffusion-ratchet mechanism. Proc Natl Acad Sci USA. 2013;110(15):E1390–E1397. doi: 10.1073/pnas.1302745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hatano T, Yamaichi Y, Niki H. Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol Microbiol. 2007;64(5):1198–1213. doi: 10.1111/j.1365-2958.2007.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci USA. 2009;106(46):19369–19374. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ptacin JL, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12(8):791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vecchiarelli AG, et al. ATP control of dynamic P1 ParA-DNA interactions: A key role for the nucleoid in plasmid partition. Mol Microbiol. 2010;78(1):78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang LC, et al. ParA-mediated plasmid partition driven by protein pattern self-organization. EMBO J. 2013;32(9):1238–1249. doi: 10.1038/emboj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sugawara T, Kaneko K. Chemophoresis as a driving force for intracellular organization: Theory and application to plasmid partitioning. Biophys Soc Japan. 2011;7:77–88. doi: 10.2142/biophysics.7.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mika JT, Poolman B. Macromolecule diffusion and confinement in prokaryotic cells. Curr Opin Biotechnol. 2011;22(1):117–126. doi: 10.1016/j.copbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Turing AM. The chemical basis of morphogenesis. Philos Trans R Soc Lond Ser B. 1952;237(641):37–72. [Google Scholar]
- 15.Hunding A, Ebersbach G, Gerdes K. A mechanism for ParB-dependent waves of ParA, a protein related to DNA segregation during cell division in prokaryotes. J Mol Biol. 2003;329(1):35–43. doi: 10.1016/s0022-2836(03)00401-7. [DOI] [PubMed] [Google Scholar]
- 16.Adachi S, Hori K, Hiraga S. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J Mol Biol. 2006;356(4):850–863. doi: 10.1016/j.jmb.2005.11.088. [DOI] [PubMed] [Google Scholar]
- 17.Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327(5970):1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 18.Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29(18):3068–3081. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouet JY, Rech J, Egloff S, Biek DP, Lane D. Probing plasmid partition with centromere-based incompatibility. Mol Microbiol. 2005;55(2):511–525. doi: 10.1111/j.1365-2958.2004.04396.x. [DOI] [PubMed] [Google Scholar]
- 20.Pinot M, et al. Confinement induces actin flow in a meiotic cytoplasm. Proc Natl Acad Sci USA. 2012;109(29):11705–11710. doi: 10.1073/pnas.1121583109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315(5816):1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodionov O, Yarmolinsky M. Plasmid partitioning and the spreading of P1 partition protein ParB. Mol Microbiol. 2004;52(4):1215–1223. doi: 10.1111/j.1365-2958.2004.04055.x. [DOI] [PubMed] [Google Scholar]
- 23.Kondo S, Miura T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science. 2010;329(5999):1616–1620. doi: 10.1126/science.1179047. [DOI] [PubMed] [Google Scholar]
- 24.Sheth R, et al. Hox genes regulate digit patterning by controlling the wavelength of a Turing-type mechanism. Science. 2012;338(6113):1476–1480. doi: 10.1126/science.1226804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alymova IV, Taylor G, Portner A. Neuraminidase inhibitors as antiviral agents. Curr Drug Targets Infect Disord. 2005;5(4):401–409. doi: 10.2174/156800505774912884. [DOI] [PubMed] [Google Scholar]
- 26.Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151(2):182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Niu L, Yu J. Investigating intracellular dynamics of FtsZ cytoskeleton with photoactivation single-molecule tracking. Biophys J. 2008;95(4):2009–2016. doi: 10.1529/biophysj.108.128751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.