Significance
Excited states of proteins play functional roles, but their low population and conformational flexibility pose a challenge for characterization by most spectroscopic techniques. Here, this challenge is met by combining high hydrostatic pressure, which reversibly populates excited states, and site-directed spin labeling with double electron–electron resonance (DEER) spectroscopy, which resolves distinct conformational substates of proteins by measuring distances between spin-labeled pairs. We present a method for trapping high-pressure equilibria of proteins by rapid freezing under pressure, followed by depressurization and acquisition of DEER data at atmospheric pressure. The methodology is applied to myoglobin, revealing unique information on the length scale of helical fluctuations in the pressure-populated as compared with the pH-populated molten globule states of the apo-protein.
Keywords: EPR, dipolar spectroscopy, compressibility
Abstract
The dominance of a single native state for most proteins under ambient conditions belies the functional importance of higher-energy conformational states (excited states), which often are too sparsely populated to allow spectroscopic investigation. Application of high hydrostatic pressure increases the population of excited states for study, but structural characterization is not trivial because of the multiplicity of states in the ensemble and rapid (microsecond to millisecond) exchange between them. Site-directed spin labeling in combination with double electron–electron resonance (DEER) provides long-range (20–80 Å) distance distributions with angstrom-level resolution and thus is ideally suited to resolve conformational heterogeneity in an excited state populated under high pressure. DEER currently is performed at cryogenic temperatures. Therefore, a method was developed for rapidly freezing spin-labeled proteins under pressure to kinetically trap the high-pressure conformational ensemble for subsequent DEER data collection at atmospheric pressure. The methodology was evaluated using seven doubly-labeled mutants of myoglobin designed to monitor selected interhelical distances. For holomyoglobin, the distance distributions are narrow and relatively insensitive to pressure. In apomyoglobin, on the other hand, the distributions reveal a striking conformational heterogeneity involving specific helices in the pressure range of 0–3 kbar, where a molten globule state is formed. The data directly reveal the amplitude of helical fluctuations, information unique to the DEER method that complements previous rate determinations. Comparison of the distance distributions for pressure- and pH-populated molten globules shows them to be remarkably similar despite a lower helical content in the latter.
Although a well-defined native state is dominant for most proteins under physiological conditions, excited states involving large (multiangstrom) conformational changes relative to the native state have been shown to play critical roles in biological function (1–3). Even for low-lying excited states with relative energies on the order of a few kilocalories (1 kcal = 4.184 kJ) per mole, the equilibrium population is too small to be characterized by most spectroscopic methods, but such states can be reversibly populated by hydrostatic pressure (4–9). It is generally agreed that the mechanism for population of excited states by pressure is hydration of cavities in the excited state relative to the ground state (10–12), but other mechanisms are possible (13). The pressures required to populate low-lying excited states are typically a few kilobars, corresponding to perturbation energies of a few kilocalories per mole. At these low energies, pressure should not strongly remodel the energy landscape itself but simply shift the relative population of preexisting conformers (14). Thus, pressure should be a powerful perturbation technique for studying functional excited states at equilibrium (15).
With a funnel-shaped model for the energy landscape (16, 17), an excited state populated by pressure will have a greater configurational entropy than the native state and is expected to consist of an ensemble of conformations in exchange on a time scale that in principle can span from nanoseconds to milliseconds and beyond (3, 18). Structural characterization of the excited state thus is complicated by conformational heterogeneity, and how the ensemble is viewed depends on the intrinsic time scale of the experimental method used relative to the protein dynamics. High-pressure crystallography cannot reveal conformational equilibria directly because of lattice interactions that select a particular conformation, but it has been used to monitor compressibility in the crystal environment (15). Solution NMR has played a central role in illustrating the utility of high pressure in the study of the excited-state conformational ensemble, both in elucidating the physical basis for pressure effects and revealing the structure and dynamics of excited states in small, soluble proteins (11, 19). However, because of the time scale of the NMR experiment, conformational exchange on the microsecond time scale can limit information to average structures, whereas relaxation effects caused by exchange on the millisecond time scale can cause extreme broadening and loss of peaks in the commonly used 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum (20). Moreover, the application of these methods to the interesting case of membrane proteins with a high molecular mass in a native environment is challenging.
Conformational exchange is slow on the time scale of site-directed spin-labeling (SDSL) EPR, and continuous wave (CW) EPR spectra can provide a view of conformational equilibria frozen in time (21, 22), but the spectral resolution is insufficient to resolve all states present, and there is no information on the scale of structural differences between various conformations. However, double electron–electron resonance (DEER) spectroscopy (23, 24) in combination with SDSL can identify the individual states with high resolution and provide a length scale to compare structural differences. DEER spectroscopy determines long-range distance distributions (20–80 Å) between pairs of spin-labeled side chains selectively introduced in proteins. With a judiciously selected set of spin-labeled pairs, structural models can be checked and conformational changes detected with angstrom-level resolution (25). In the DEER experiment, the sample is flash-frozen, and data are acquired at cryogenic temperatures (50–80 K) (24, 26, 27). If different conformations are present in the sample, DEER will resolve each distinct distance and its probability distribution, thereby revealing the presence of heterogeneity, the magnitude of structural differences between members of the ensemble, and the inherent spatial order of each from the distribution width. Thus, SDSL-DEER is an ideal and unique spectroscopic technique for exploring the excited-state ensemble of proteins populated at high pressure. In order to use DEER with high pressure, an approach is introduced herein whereby the high-pressure equilibrium is kinetically trapped by flash-freezing samples under pressure followed by depressurization and subsequent data acquisition at 80 K.
In the present work, the above strategy is applied to the well-studied protein myoglobin (Mb) both to validate the methodology and to provide information on the nature of pressure-populated species, particularly regarding structural heterogeneity and the amplitude of structural rearrangements relative to the native state. Apomyoglobin (apoMb) has an excited state (the molten globule, MG) that can be populated at atmospheric pressure at pH 4.1, enabling structure determination by solution NMR (28). The pH 4.1 MG appears to be a folding intermediate of apoMb (29) and a true equilibrium state of the system, although it is present at very low concentrations at neutral pH (i.e., an excited state) (30). The NMR data show that the MG retains much of the helical structure of the native state of apoMb populated at pH 6 but with fluctuations to nonhelical states in particular regions (31). However, the NMR methods provided no information about the amplitude of the fluctuations, i.e., whether the nonhelical states involve large-scale unfolding or localized distortions.
If pressure does not alter the energy landscape substantially, the same MG state also should be populated at pH 6 with the application of high pressure. Indeed, solution NMR showed that pressurizing apoMb to 2 kbar at pH 6 produced a state accompanied by a loss of most cross-peak intensities in the 1H-15N HSQC spectrum, presumably because of the onset of global conformational exchange on the millisecond time scale. The state produced at 2 kbar was assumed to be an MG, but no information on the secondary or tertiary structure could be obtained because of the loss of cross peak intensity (20). Recently, high-pressure circular dichroism (CD) and SDSL together revealed that the MG state populated at 2 kbar, pH 6, has in fact the same helical content as the native state despite a tertiary fold that fluctuates on the millisecond time scale (13). The higher helical content in pressure-populated MG (2 kbar, pH 6) than in the pH-populated MG (0 bar, pH 4.1) (13) and the loss of peak intensities in the HSQC observed in the former but not in the latter (20, 31) raise questions about the similarity of the pH- and pressure-populated species.
In the present study, distance distributions were determined for pairs of spin labels in Mb selected to monitor motions of the helical segments as a function of pressure. In holomyoglobin (holoMb) at pH 6, there is little pressure dependence. For apoMb at pH 6, the interspin distance distributions are relatively narrow (∼5–10 Å) at atmospheric pressure, but broad distributions (>25 Å) implying large-scale collective motions of particular helices are seen at 2 kbar. Remarkably, the distinctive distance distributions of the pressure-populated MG are very similar to those in the pH-populated MG, despite the differences between them noted above. We conclude that the global topology of the folds is similar in the MG states, although there are clear differences in the local dynamics that likely are related to the compressibility of the excited state.
Results
Trapping High-Pressure Equilibria for DEER Spectroscopy.
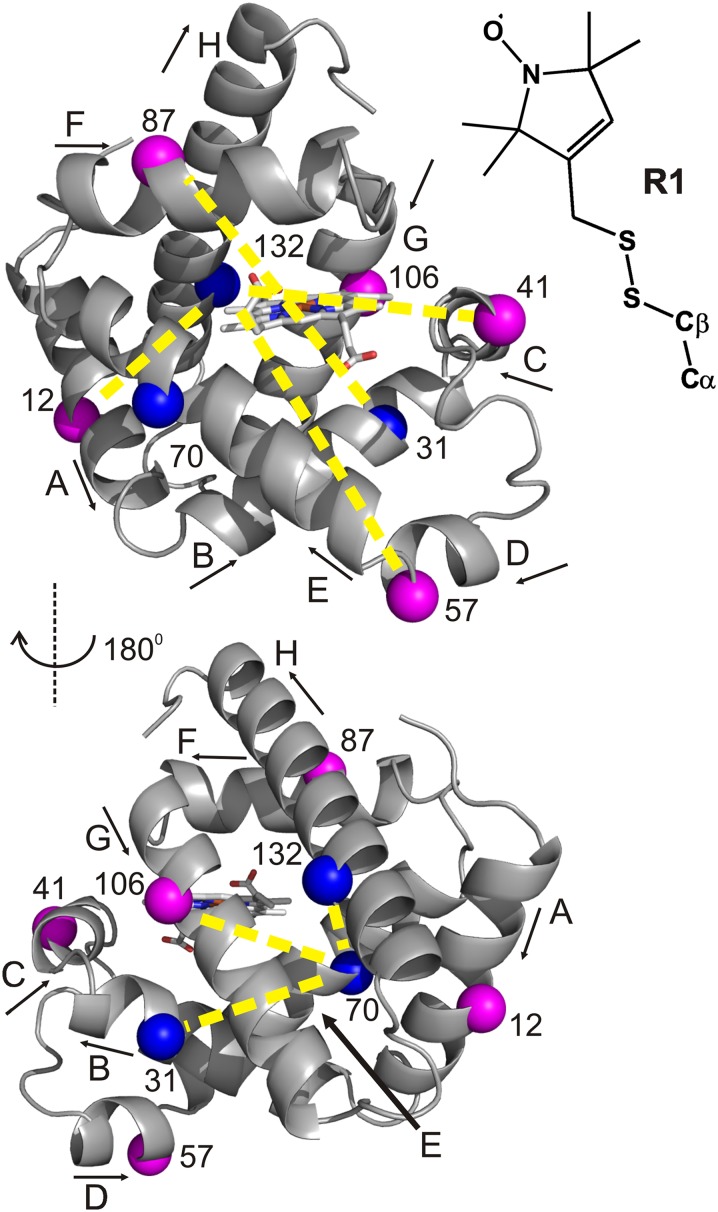
For DEER distance mapping of high-pressure states of Mb, seven doubly-labeled mutants were prepared containing the nitroxide side chain R1 (Fig. 1). Solvent-exposed sites for R1 were selected in each helix to monitor distance distributions relative to a selected reference site. The reference sites are in helices whose positions in the structure apparently are independent of pressure; as shown in Fig. 1, the selected sites are 31, 70, and 132 in helices B, E, and H, respectively.
Fig. 1.
Ribbon model of holoMb with spin-labeled sites used in DEER measurements (PDB ID code: 2MBW) (43). The spheres indicate the Cα of sites where the nitroxide spin label R1 (Inset) was introduced pairwise to measure the corresponding internitroxide distances (yellow dashed lines). Blue spheres indicate reference sites (see Results). The heme group is shown in stick representation. Arrows next to helix labels indicate the N-to-C terminus direction of each helix.
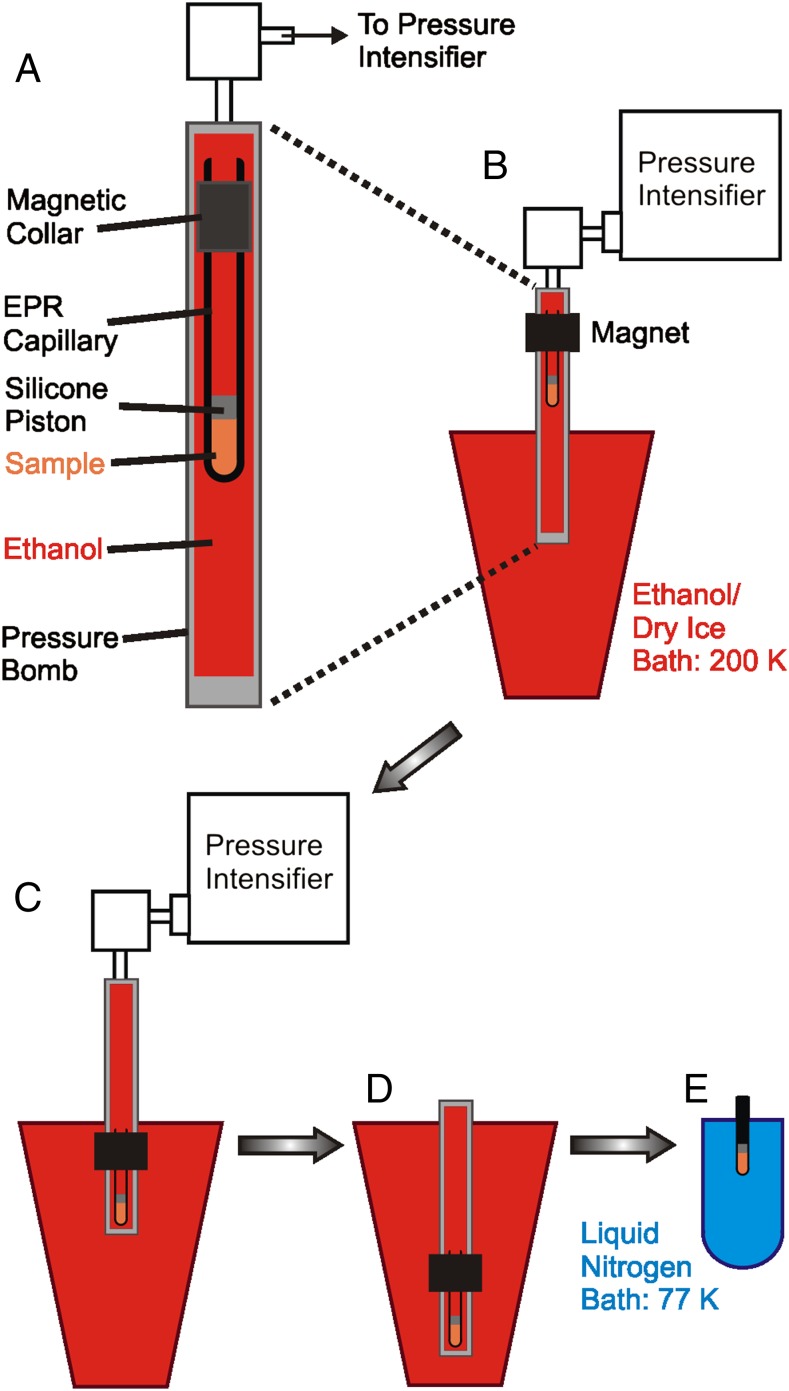
Standard DEER methodology requires flash-freezing of a sample, typically in liquid nitrogen, and data acquisition at 50–80 K (25, 26, 32, 33). To use DEER to monitor the structure of high-pressure states, a procedure is introduced here in which the pressurized sample is rapidly frozen in a dry ice/ethanol bath to 200 K. The frozen sample is then depressurized and cooled further to 77 K in liquid nitrogen for subsequent data acquisition at atmospheric pressure. This methodology is shown schematically in Fig. 2, and the details are provided in Methods. The freezing rate to ∼223 K is more rapid in dry ice/ethanol than in liquid nitrogen (Fig. S1), because liquid nitrogen boils when the borosilicate sample tube initially is submerged in the bath, resulting in the formation of an insulating gas sheath around the tube.
Fig. 2.
Methodology for freezing under pressure. (A) Detail of sample in the pressure bomb. A borosilicate capillary is modified by the addition of a magnetic collar near the top. A silicone piston separates the sample (orange) from the ethanol pressurization fluid (red) that fills the stainless steel pressure bomb. (B) The bomb is connected to the pressure intensifier with the lower portion immersed in dry ice/ethanol at 200 K. The sample is held at the top of the bomb using the magnet where the temperature is controlled at 298 K during pressurization. (C) Under pressure, the sample is moved quickly to the bottom of the bomb, triggering rapid cooling to 200 K. (D) The bomb is depressurized, disconnected from the pressure intensifier, and submerged in the dry ice/ethanol bath. (E) The sample capillary is transferred to liquid nitrogen (blue) for cooling to 77 K in preparation for DEER data acquisition at 80 K.
The evidence reported to date indicates that the effects of flash-freezing in liquid nitrogen on R1 rotameric equilibria are minimal (32) and that conformational equilibria are effectively trapped (25, 34–41). To compare the efficacy of rapid freezing in dry ice/ethanol with that in liquid nitrogen, we directly compared DEER distance distributions at atmospheric pressure for each doubly-labeled mutant of apoMb and holoMb using the two freezing methods (Fig. S2). The distance distributions are nearly identical, indicating the equivalence of the methods with respect to both protein conformational equilibria and rotameric equilibria of the R1 side chain.
The glass transition temperature of the glycerol/water mixture used here is ∼150 K (42). Thus, it is possible that above this temperature, at 200 K, the conformational ensemble could relax after depressurization and before cooling to 77 K. Complete relaxation at 200 K during the time between depressurization and cooling to 77 K (∼3 min) is ruled out by the observation of distinct distance distributions for individual spin pairs held at different pressures before freezing to 200 K. However, to check for partial relaxation at 200 K, samples of 57R1/132R1 were pressurized to 2 kbar, frozen in dry ice/ethanol as described above, and depressurized. Then the samples were held at 200 K for 3, 10, or 120 min before further cooling to 77 K in liquid nitrogen for data acquisition. The resulting distance distributions are similar (Fig. S3A), indicating that conformational relaxation does not occur at 200 K on the experimental time scale used here. Reversibility of pressure-populated changes was verified using a 57R1/132R1 sample prepared at 2 kbar, as described above. After data acquisition, the sample was thawed and refrozen at atmospheric pressure, and DEER was performed. The distance distribution is nearly identical to that for 57R1/132R1 at 0 bar, before pressurization (Fig. S3B), confirming the reversibility of the observed pressure-populated conformational changes.
DEER Distributions for HoloMb Suggest Local Regions of Compressibility in an Otherwise Rigid Structure.
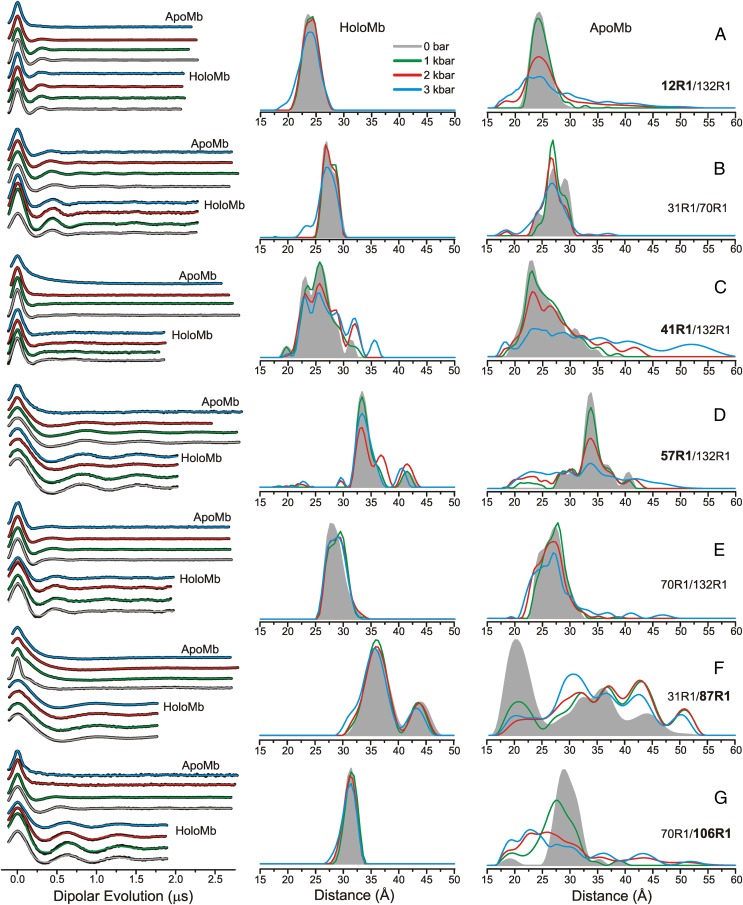
Fig. 3 shows the background-corrected dipolar evolution functions (Left) and the corresponding distance distributions (Center) for holoMb at pressures in the range of 0 bar to 3 kbar (gauge pressure; 0 bar gauge is atmospheric pressure). At atmospheric pressure, the distributions (shaded gray areas) are dominated by a single distance with distribution widths of ∼3–5 Å (FWHM), except for 41R1/132R1 and 31R1/87R1, which have broader bimodal distributions that likely arise from two conformations of the protein involving the helices C and F, respectively (Discussion). In all cases, the most probable distance corresponds closely to that for the R1 side chains modeled in the holoMb crystal structure [Protein Data Bank (PDB) ID code 2MBW] (43). As the pressure is increased, there is a slight broadening of the distance distributions, but the relative populations at different distances are unchanged for most pairs (Fig. 3). For 12R1/132R1, 31R1/70R1, 31R1/87R1, 70R1/106R1, and 70R1/132R1 the distance distributions are nearly superimposable up to 2 kbar, and only a slight degree of broadening in the distance widths is observed at 3 kbar (Fig. 3), reflecting the incompressibility of holoMb.
Fig. 3.
Variable-pressure DEER distance distributions for holoMb and apoMb at pH 6. (Left) Background-corrected dipolar evolutions are shown in black, with fits to the data (Methods) overlaid and color-coded by pressure. (Center and Right) The area-normalized distance distributions are shown for holoMb (Center) and apoMb (Right) for (A) 12R1/132R1, (B) 31R1/70R1, (C) 41R1/132R1, (D) 57R1/132R1, (E) 70R1/132R1, (F) 31R1/87R1, and (G) 70R1/106R1. The Inset indicates the color coding of the distance distributions and fits to the dipolar evolutions. Dipolar evolutions are vertically offset for clarity. Bold labels indicate sites in each mutant in which structural motion as a function of pressure is attributed.
Although the distance distributions for most pairs are nearly independent of pressure, the distributions for 57R1/132R1 and 41R1/132R1 exhibited a shift in populations toward longer distances at 2 kbar but without the appearance of additional populations; at 3 kbar, an additional population appears in 41R1/132R1 at ∼36 Å (Fig. 3 C and D).
ApoMb Structure Exhibits Strong and Nonuniform Pressure Dependence.
Removal of the heme to form apoMb produces structural changes relative to holoMb at atmospheric pressure (Fig. 3, Right) sensed by interspin distance distributions. In all cases except for 12R1/132R1, the distributions are broadened and modified in shape relative to holoMb. The increase in distribution width reflects heterogeneity in the population relative to holoMb, which in turn mirrors increased flexibility and structural fluctuations at ambient temperature. In 31R1/87R1 the effect is extreme, with a dominant population appearing at a distance that is 15 Å shorter and the full distribution extending over a range of 25 Å. This result is consistent with the dynamic disorder in helix F, where 87R1 resides, as deduced from both NMR and SDSL in apoMb (22, 30).
For the purposes of the present study, the effect of pressure at 2 kbar on apoMb is the most interesting, because 2 kbar is the pressure at which the putative MG state is populated, as inferred from extreme broadening in the HSQC NMR spectrum (20) and the appearance of multiple components in the EPR spectra of R1 (13). There are detectable changes in the distance distributions for all R1 pairs at 2 kbar, but the effect is small for 70R1/132R1, and the most probable distance for 31R1/70R1 is unchanged, although there is a shift in relative populations. Moreover, the distance distributions for 31R1/70R1 and 70R1/132R1 display a total width of ∼10 Å, substantially narrower than that of any other spin pair in apoMb reported here at high pressure. Based on these data, we assign residues 31R1, 70R1, and 132R1 as reference sites located in helical segments that apparently do not change position substantially as a function of pressure up to at least 3 kbar (Fig. 3, Right). These reference sites do not lack conformational flexibility, but they are spatially restricted on the angstrom-length scale. With this tentative assignment, and because the helical content of apoMb is unchanged up to 2.4 kbar (13), the interspin distance distributions can be discussed in terms of rigid body movement of helical segments relative to the reference sites.
In the context of the above model, helices B, E, and H (in which the reference sites reside) remain relatively localized at 2 kbar compared with atmospheric pressure; i.e., additional states with substantial populations and different structures do not appear. Helix A, monitored by 12R1 relative to the 132R1 reference, is not displaced at 2 kbar but has an increased spatial disorder indicated by the breadth of the distance distribution (Fig. 3A). Helix F, monitored by 87R1 relative to the 31R1 reference, shows a dramatic shift in population from the main 20-Å peak at atmospheric pressure to a broad distribution of states spread from ∼20–50 Å at 2 kbar, with only small additional changes at 3 kbar. Residue 41R1 relative to the 132R1 reference monitors the position of helix C. At 2 kbar, the most probable distance at 23 Å remains unchanged, and although additional distances appear at 36 and 42 Å, they have relatively small populations. Helices D and G, monitored by 57R1 and 106R1 relative to references 132R1 and 70R1, respectively (Fig. 3 D and G), show striking displacements to shorter distances with broad distributions at high pressure. Collectively, the data show that the high-pressure state of apoMb at pH 6 is spatially disordered, but not uniformly so, relative to its state at atmospheric pressure. Interestingly, the distance distributions at high pressure also contain the discrete populations present at atmospheric pressure, suggesting that the high-pressure ensemble includes a population corresponding to the native apoMb structure. A structural model of the high-pressure state consistent with these changes is presented in Discussion.
Comparison of Pressure- and pH-Populated MG States of ApoMb.
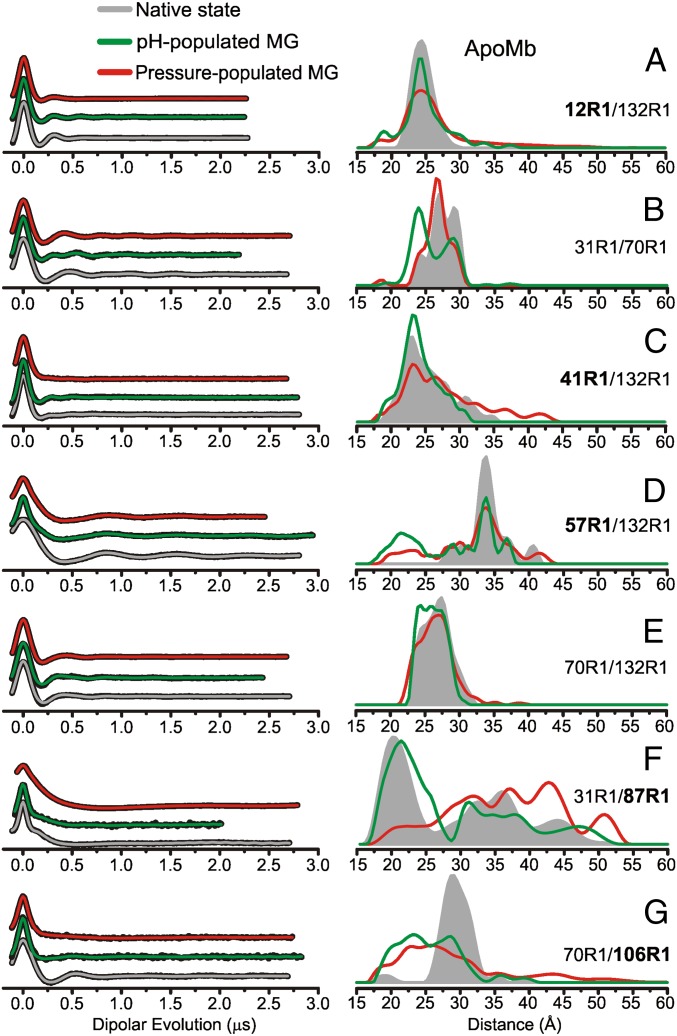
As reviewed in the introduction, the NMR 1H-15N HSQC spectrum was well resolved in the pH-populated MG, and analysis showed local unfolding in the C, D, E, and F helices as well as in the N-terminal half of helix B (31, 44). On the other hand, the pressure-populated MG state showed extreme broadening of cross peaks in the 1H-15N HSQC spectrum (20), and CD and SDSL showed that this state maintains the full helical content of the native state (13). Thus, the two states cannot be identical, and their structural relationship is revealed in the DEER distance distributions for the two states shown in Fig. 4. Despite the differences in secondary structure and dynamics exposed by the studies described above, the DEER data show a surprising similarity in the topology of the two MG states, except for the location of helix F in the structure. In the pH-populated MG state, the distance distribution resembles that of native apoMb, whereas the dominant population at 20 Å is largely depleted in the high-pressure state, being shifted to a broad distribution centered at greater distances (Fig. 4F). Despite their similarity, there are interesting differences in the distributions of particular pairs. For example, for 12R1/132R1, 41R1/132R1, and 70R1/106R1, the overall distribution widths are greater for the pressure-populated MG, reflecting greater spatial disorder. For 31R1/70R1, the same discrete distances are present in the two states, but high pressure favors the longer distances.
Fig. 4.
DEER distance distributions for the conformational ensemble of apoMb in the native state at 0 bar and pH 6, the pH-populated MG at 0 bar and pH 4.1, and the pressure-populated MG at 2 kbar and pH 6. (Left) Background-corrected dipolar evolutions of the indicated mutants are shown in black with fits to the data (Methods) overlaid and color coded by state. (Right) The area-normalized distance distributions from are shown for (A) 12R1/132R1, (B) 31R1/70R1, (C) 41R1/132R1, (D) 57R1/132R1, (E) 70R1/132R1, (F) 31R1/87R1, and (G) 70R1/106R1. The Inset indicates the color coding of the distance distributions and fits to the dipolar evolutions. Dipolar evolutions are vertically offset for clarity. Bold labels indicate sites in each mutant in which structural motion as a function of pressure is attributed.
Discussion
One aim of this study was to develop a methodology to kinetically trap high-pressure conformational ensembles of proteins by flash-freezing, so that DEER distance measurements may be used to determine structure and structural heterogeneity in the ensemble. If successful, DEER in this context offers the unique advantage of providing global distance constraints and analytical distance distributions that directly reveal structural heterogeneity and the amplitude of the corresponding fluctuations that are presumed to occur at ambient temperatures (25). The success of this approach relies on the ability to kinetically trap the high-pressure conformational equilibrium by flash-freezing to 200 K in the presence of glycerol as a cryoprotectant.
In evaluating the method, two questions are relevant. First, is conformational exchange slow enough at 200 K so that thermal and pressure equilibration do not occur following depressurization during the time required to cool the sample below the glass transition temperature (150 K)? Second, is the cooling time to 200 K sufficiently short compared with the characteristic relaxation time for conformational movement so that the trapped ensemble is representative of the one at ambient temperature?
To address the first question, we should consider the environment of the protein at 200 K. At the cooling rates achieved here (Fig. S1), the amount of ice crystal formation will be much less than that computed from the equilibrium-phase diagram of the two-component glycerol–water mixture at atmospheric pressure (45). If the system were at thermal equilibrium during cooling, ∼80% of the water would be ice crystal as predicted by the phase diagram, thereby concentrating the protein in a supercooled water–glycerol mixture by a factor of approximately three. Analysis of the exponential background in the DEER data gives a total spin concentration similar to that of the starting solution (∼300–400 μM) (46, 47), demonstrating that such concentration does not occur at either atmospheric pressure or high pressure. Thus, the bulk of the sample is assumed to be a supercooled solution whose viscosity at 200 K can be estimated as ≥3,000 cP at atmospheric pressure (42) and presumably is higher under pressure. In the context of Kramer’s theory (48), rate constants for conformational exchange scale as η−1⋅e−Ea/RT, where η is solvent viscosity and Ea is the activation energy (49). Thus, both the low thermal energy and the high viscosity conspire to slow conformational exchange at 200 K dramatically. Indeed, as delineated above, no relaxation of the distinct ensemble of apoMb frozen under high pressure occurs at 200 K after depressurization, even on a time scale of 120 min. Because the pressure effect is fully reversible (i.e., thawing a pressurized sample and refreezing at atmospheric pressure gives the native-like ensemble), this result shows that conformational movements are very slow at 200 K. Because the sample is cooled below the glass transition temperature within ∼3 min after depressurization, we tentatively conclude that the high-pressure conformational ensemble has been trapped effectively. On the other hand, small side-chain motions can occur above the glass transition temperature on the time scale of seconds, and it is likely that side-chain rotamers will equilibrate with respect to both temperature and pressure at 200 K (50). If so, the rotamer population of R1 in the DEER experiment is unchanged from that at atmospheric pressure. Under any circumstance, the pressure independence of CW spectra for R1 in holoMb to 2 kbar strongly suggests that rotameric equilibria of R1 are not pressure sensitive in the pressure range investigated (13).
In regard to the second question above, the cooling time to reach 200 K is, in general, long compared with the characteristic relaxation time for conformational motions at ambient temperature. However, the large temperature coefficient for the viscosity of glycerol–water mixtures (42) and, in some cases, large enthalpies of activation likely slow the exchange rates sufficiently during cooling to result in kinetic entrapment. This kinetic entrapment is a basic assumption in all published DEER experiments, and considerable empirical evidence is available to justify the assumption (25, 34–41). In addition, distance distributions between spin pairs in T4 lysozyme were found to be only weakly dependent on cooling rates (32). Typically, DEER samples are frozen in liquid nitrogen, which in fact takes longer to reach ∼223 K than the dry ice/ethanol used here (Fig. S1), and we rely on the same body of evidence found in conventional DEER to justify kinetic entrapment during the cooling phase. For relatively unstable MG states, one must consider the possibility of cold denaturation of the protein during the early stages of the freezing process. The few studies of the rate of cold denaturation that have been published found the process to be extremely slow, on the order of minutes to hours (51–53). Although no measurements have been made for the MG state of apoMb, we tentatively assume that the rate of cold denaturation, like that of conformational exchange, is sufficiently slow so that this process does not occur during the freezing to 200 K.
In addition to the cryoprotective function mentioned above, glycerol is a stabilizing osmolyte that favors folded states of proteins (54–57), and the effect of glycerol on the structure and conformational equilibria of Mb must be considered. A systematic SDSL-EPR study of the osmolyte effect on holoMb, apoMb, and the MG state of apoMb has been carried out for each of the labeled R1 sites used in the present experiments with sucrose as the osmolyte. In each case, addition of osmolyte had observable effects only where multicomponent EPR spectra signaled the existence of conformational equilibria. The effects were a simple shift in the equilibrium and were relatively small; for example, the largest effects were observed for equilibria involving order–disorder equilibria in helix F, where 30% (wt/wt) sucrose resulted in a stabilization of the ordered state by approximately −0.3 kcal/mol (58). Glycerol is a considerably weaker stabilizing osmolyte than sucrose (59–61), and in general the perturbation of 25% (vol/vol) glycerol on conformational equilibria should be even smaller and acceptable in exchange for the cryoprotective function.
Collectively, the data presented here provide a tentative empirical validation of the methodology for trapping high-pressure states of proteins for exploration with DEER spectroscopy. In addition to providing validation, the data add another dimension to the characterization of the pressure-populated MG state of apoMb, namely, the amplitude of structural changes, and allow a comparison with the extensively characterized pH-populated MG state. The results of the experiments, beginning with holoMb, and a comparison with expectations from other methods are discussed below, leading to a model for the ensemble of apoMb in the high-pressure state.
The Apparent Compressibility of HoloMb Is Nonuniform in the Structure.
Five of the seven mutants in holoMb reported here exhibit minimal changes in distance distributions as a function of pressure (Fig. 3), consistent with previous studies that showed holoMb to be a highly ordered and well-packed protein (22, 31, 43) with minimal pressure sensitivity at pH 6 and in the pressure range used here (13, 62). The only cases in which significant pressure-dependent changes in holoMb are seen are the mutants with R1 in the short helices C and D, which show a shift in population toward longer distances at high pressure. Crystallographic and SDSL studies found a relatively high degree of flexibility in these two helices (22, 63, 64), and the most attractive model consistent with the data is that the alternative packing modes of the structure around helices C and D, identified by the discrete populations in the DEER distance distributions, have different molar volumes. The population shift at high pressure reflects local protein compressibility. Interestingly, compression does not result in a decrease in the total width of the distributions, as one might expect for simple compaction, but rather drives a slight increase in the distribution widths. A possible mechanism may be the hydration of packing imperfections, which lowers the partial molar volume, but enables alternative packing arrangements of the local core (65).
DEER Reveals the Structural Diversity of the Pressure-Populated MG State.
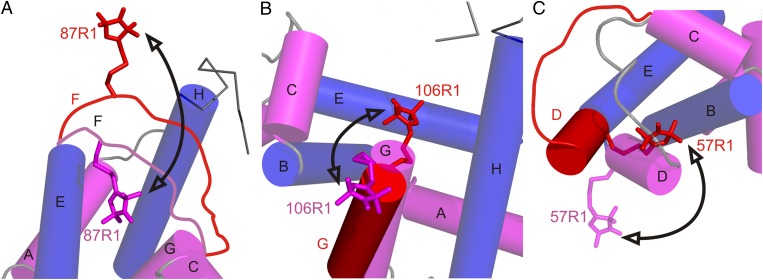
Unlike the relatively pressure-insensitive structure of holoMb, pressurizing apoMb to 2 kbar results in extremely broad distance distributions in particular mutants, covering >25 Å in five of the seven spin pairs studied here. The increased widths reflect the increase of conformational heterogeneity in the protein. The distance distributions include discrete states corresponding to native apoMb, indicating that the excited-state ensemble includes the native conformation. In all cases, the static distributions resolved with DEER are taken to reflect conformational fluctuations at ambient temperature. Because the high-pressure state has the full helical content of the native protein (13), it can be concluded that the fluctuations are comprised of nanometer-scale rigid-body motions of helices or helical segments (with the exception of helix F, which is locally unfolded in the native state). A tentative model for the structural origins of conformational fluctuations involving helices D, F, and G in the pressure-populated MG of apoMb is presented in Fig. 5 and discussed below. The model builds on earlier data showing that the helical content of the pressure-populated MG is the same as that of native apoMb. Because there are no secondary structural changes, the simple models of Fig. 5 assume that the distance changes monitored by a single site in a helix reflect the motion of the entire helix. In reality, independent segmental motions within individual helices are clearly possible, but the sparse dataset presented here does not permit this level of interpretation.
Fig. 5.
Models for the conformational changes of selected sequences in the transition to the pressure-populated MG of apoMb at 2 kbar, pH 6. Shown are the proposed changes relative to the native structure in particular regions. In each panel, the model of the native state is based on the crystal structure of holoMb (PDB ID code: 2MBW), except for the contiguous sequence of the F helix and N terminus of G, which is locally unfolded in the native state of apoMb and is drawn in as a disordered loop in A. This sequence is omitted from B for clarity (66). Helices are shown in cylinder representation; helices that contain reference sites are colored blue, and helices that contain sites that undergo large-amplitude motions in the pressure-populated MG are in magenta. Models for a dominant conformation in the pressure-populated MG based on DEER data at 2 kbar are superimposed and shown in red. Arrows indicate a trajectory of motion followed by each segment in the pressure-populated native-to-MG transition. (A) The sequence of helix F moves out of the heme pocket. The native-state position of the F helix is modeled to satisfy the most probable distance (20 Å) of 31R1/87R1 in apoMb at 0 bar. (B) Helix G fluctuates between the native state and a twist/tilt which inserts R1 into the heme pocket; alternatively, additional fraying of the N-terminal end of helix G at high pressure could result in the disordered sequence containing 106R1 rearranging in a number of distinct conformations that insert the nitroxide into the heme pocket. (C) Helix D fuses with the E helix in a motion that involves a concurrent rotation placing the 57R1 side chain near helix B.
In the native state of apoMb, NMR, and SDSL-DEER data showed that the sequence corresponding to helix F undergoes conformational exchange involving nonhelical states that probably occupy the otherwise empty heme pocket (22, 30). The extremely broad distribution of 87R1 (helix F) with respect to the reference site 31R1 in apoMb reflects a conformational ensemble that involves motions of ∼25 Å, consistent with conformational exchange events of this amplitude at ambient temperature. Moreover, the dominant 20-Å peak in the distance distribution requires a 15-Å displacement of 87R1 toward 31R1 from its position in holoMb (Fig. 3F), a motion that would place 87R1 in the heme cavity. At high pressure, the 20-Å peak diminishes sharply, whereas multiple populations at longer distances grow in a broad distribution, indicating that 87R1 moves out of the heme pocket at high pressure (Fig. 5A), forming a distribution more similar to that of holoMb.
At the N terminus of helix G, the distribution of 106R1 relative to 70R1 at atmospheric pressure is broader than that of holoMb (6 vs. 3 Å width at half-maximum; Fig. 3G), mirroring the disorder in the N-terminal segment of the G helix identified by NMR (30, 66, 67). High pressure produces a dramatic increase in overall breadth of the distribution, to more than 35 Å. The most dominant populations now are centered at shorter distances, revealing a displacement of 106R1 by ∼5 Å toward 70R1, requiring 106R1 to be located in the heme cavity. The model shown in Fig. 5B to account for this shift in the distribution is a twisting motion of the N-terminal segment of helix G. Because 106R1 is near the end of the helical portion of G at atmospheric pressure, it is possible that the sequence is disordered in the pressure-populated MG, so that many other specific configurations could place 106R1 in the cavity. In both the 31R1/87R1 and 70R1/106R1 distributions, there are populations corresponding to native apoMb at atmospheric pressure, suggesting that the F and G helical segments fluctuate between positions in and out of the heme cavity.
The distance of 57R1 relative to 132R1 monitors the position of helix D. In native apoMb, the distance distribution is broader than that for holoMb because of the appearance of additional populations, reflecting the flexibility of this short helical segment in the native apoMb state. The striking increase in width of the distribution at 2 kbar identifies conformational disorder involving fluctuations of this helix with an amplitude of ∼20 Å (Fig. 3D). The shorter distances in the distribution suggest a large rotation of helix D to place the nitroxide of 57R1 closer to 132R1. In a previously reported model for the pH-populated MG state of apoMb, the D helix sequence fuses with the end of E to form a transient helical structure (30, 68); this structural rearrangement is consistent with the 22-Å distance in the high-pressure state and is tentatively proposed for one conformer of the pressure-populated MG ensemble.
Structural Similarity of the pH- and Pressure-Populated MG States of ApoMb.
Overall, the pH- and pressure-populated MG states are more similar than different as assessed by distance mapping. The major difference lies in the spatial distribution of the F helix segment, which is located outside the heme cavity in the high-pressure state but is dominated by an intracavity conformation similar to that in native apoMb in the pH-populated MG. However, the N terminus of the G helix still occupies part of that cavity in the pH-populated MG, in a location similar to that in the pressure-populated MG, indicating either that the F and G segments occupy unique sites within the cavity or that the two segments alternate between collapsed and native-like conformations out of phase with one another.
The differences between the pH- and pressure-populated MG states identified by NMR, CD, and SDSL may appear to be at odds with the obvious global similarity of the two MGs as viewed by DEER distance mapping. For example, NMR describes local unfolding in the noncore C, D, and E helices with dynamic disorder on the nanosecond time scale in the pH-populated MG, whereas CD shows the helical content of the high-pressure state to be the same as that of the native apoMb. Based on these data, one might expect the distance distributions in the pH-populated MG between sites in helices found to undergo unfolding by NMR to be broader than those in the pressure-populated state. This expectation is rooted in a model for partial unfolding of helices identified by NMR in which the entire helix collectively fluctuates between a folded and unfolded conformation. However, this model is incompatible with the DEER data, shown in Fig. 4, in which the distance distributions between residues in the C, D, and E helices and stable references are similar in the two MGs. An alternative interpretation consistent with the DEER results is that dynamic disorder revealed by NMR and SDSL is not necessarily correlated with large-amplitude motions. Partial unfolding of secondary structure elements as deduced by NMR 13Cα shifts may involve relatively small-amplitude local distortions that migrate along helical sequences, fluctuating on a nanosecond-to-microsecond time scale, that do not contribute to a large-amplitude motion that would be observed in the DEER distance distributions.
Structural Transitions Responsible for the Reduction of Volume at High Pressure.
Typically, pressure effects are attributed primarily to the elimination of internal cavities through hydration (12, 69), leading to local and eventually global unfolding (20). Based on the similar helical content of apoMb and the pressure-populated MG state, it was suggested that pressure populates alternative packing arrangements of the helical core rather than producing local unfolding to reduce internal cavity volume (13), and this notion is consistent with recent studies showing that pressure shifts helix–coil equilibria toward the helical conformation rather than an unfolded state (70, 71). The high-pressure DEER data add the amplitude of conformational rearrangements involved to this model. The large degree of heterogeneity reported by the DEER distance distributions, along with the long distances populated (40–50 Å in some cases), suggest that perhaps some helices have lost close packing entirely in the MG state because of the insertion of solvent between native hydrophobic contacts (72).
Concluding Remarks
Hydrostatic pressure is expected to access protein excited states of higher configurational entropy than the ground state. NMR and CW SDSL-EPR spectroscopy can identify sites undergoing conformational fluctuations in proteins under pressure and identify a time scale, but they do not provide a length scale for the motion or resolve the individual members in an ensemble. The results presented here establish the feasibility of kinetically trapping high-pressure conformational equilibria of spin-labeled proteins for analysis by conventional DEER instrumentation operated at atmospheric pressure. High-pressure EPR sample cells are not required, and pressures near 7 kbar can be explored with DEER using standard high-pressure equipment. The important feature of DEER is that the distance distributions directly reveal the structural heterogeneity of the excited states populated by pressure and provide a length scale for structural fluctuations. The initial results from application to the pressure-populated MG of apoMb serve both to validate the methodology and reveal fluctuations of helices or segments thereof with amplitudes as large as 25 Å. Although the dataset presented is sparse, and the model of Fig. 5 is highly schematic, systematic application of the method with judiciously selected spin pairs can increase the resolution. The method should be general and serve to map structure and structural heterogeneity in pressure-populated excited states in proteins of any degree of complexity, including membrane proteins.
Methods
Mb Preparation.
The plasmid pET17b (EMD Millipore) containing the WT sperm whale Mb gene was generously provided by Steven Boxer (Stanford University, Stanford, CA). Double-cysteine mutants were generated in the WT background using the QuikChange Method (Agilent) and were confirmed by DNA sequencing. Four (H12C/N132C, R31C/T70C, R31C/K87C, and A57C/N132C) of the seven mutants reported here have been published previously (22). The mutant plasmids were transformed into and expressed in Escherichia coli BL21(DE3) cells and then were purified from inclusion bodies, spin-labeled with (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl) methanethiosulfonate reagent, gel-filtered (apoMb), and heme-reconstituted (holoMb) as previously described (22). The samples were concentrated to ∼250 μM for apoMb or ∼360 μM for holoMb and were switched into a buffer containing 20 mM MES, pH 6.0, using Amicon ultra concentrators (10 kDa molecular weight cutoff) (Millipore) for variable-pressure DEER experiments. The pH-populated MG state was prepared by switching the apoMb into a buffer containing 10 mM sodium acetate, pH 4.1, using Amicon ultra concentrators. Protein concentration was measured via absorption spectroscopy at 280 nm (ε280 = 15,400 L⋅mol−1⋅cm−1) for apoMb and 409 nm (ε409 = 157,000 L⋅mol−1⋅cm−1) for holoMb. Finally, glycerol was added to a final concentration of 25% (vol/vol) just before DEER studies. The typical volume of DEER samples was 12–16 µL.
Pressurization and Freezing of DEER Samples.
Silicone pistons were made using a two-part encapsulant (Sylgard 184; Dow Corning), prepared according to the manufacturer’s instructions and allowed to cure inside clean quartz capillaries (1.4 mm i.d. × 1.7 mm o.d.) (VitroCom, Inc.). After curing, the capillaries were broken open to release the silicone, which then was cut into ∼3-mm-long pistons. DEER samples were loaded into a capillary tube of the same size as that used to prepare the silicone pistons. A silicone piston was loaded into the capillary and pushed into place just above the sample, minimizing air bubbles between the sample and the piston. Ethanol was used to wash the inside of the capillary above the piston, and then fresh ethanol was used to fill the capillary. A custom-built magnetic collar (416 stainless steel) was glued onto the outside of the capillary near the top end to allow external control of the sample position within the pressure bomb using a neodymium-iron-boron ring magnet on the outside of the bomb. The loaded capillary was sealed inside a pressure bomb (60-9H4-316; High Pressure Equipment Company) connected to a pressure intensifier (Barocycler HUB440; Pressure BioSciences Inc.). The pressurization fluid inside the bomb was ethanol; water was used in the rest of the high-pressure system.
The sample capillary was positioned at the top of the bomb using the magnet. The bottom portion of the bomb was submerged in a dry ice/ethanol bath for 2 min to cool the lower half of the bomb and pressurization fluid to 200 K, while the upper half of the bomb was maintained at room temperature using a heat gun (the temperature setting and placement of the heat gun were calibrated using a mock setup of the pressure bomb and cooling bath, with a thermocouple in place of the capillary). The bomb was pressurized, and the sample was allowed to equilibrate for 30 s before freezing. The sample was moved rapidly to the cooled portion of the pressure bomb using the magnet. After 30 s, the bomb was depressurized, disconnected from the pressure intensifier, and fully submerged in the dry ice/ethanol bath for 2 min.
The capillary was removed from the pressure bomb, and the portion of the capillary containing the sample was kept immersed in the dry ice/ethanol bath while the ethanol above the piston was removed with a syringe. The capillary was removed from the dry ice/ethanol bath and immediately was submerged in liquid nitrogen to cool the sample from 200 K to 77 K. Finally, the magnetic collar was cut, and the sample tube was loaded into the resonator for data acquisition.
Q-Band DEER.
DEER measurements were performed on a Bruker ELEXSYS 580 spectrometer equipped with a SuperQFT Q-band bridge and a 10-W AmpQ Q-band amplifier. An ER 5107 D 2 Q-Band resonator (Bruker Biospin) was used for all data acquisition. The cryostat temperature was maintained at 80 K using liquid nitrogen and an Oxford ITC503S temperature controller (Oxford Instruments). The standard four-pulse DEER sequence [(π/2)υ,obs - τ1 - (π)υ,obs - T - (π)υ,pump - τ2 - (π)υ,obs - τ1 - echo, where υ, obs and υ, pump indicate the observed and the pump frequency, respectively] was used. The pulse durations of the (π/2)υ,obs, (π)υ,obs, and (π)υ,pump pulses were 16 ns, 32 ns, and 36 ns, respectively. The τ1 was selected to be 200 ns; τ2 varied from 2 to 3.2 μs depending on the sample with a constant step size of 16 ns. Interval T was set to 100 ns to collect a dead-time−free DEER trace. A two-step phase cycling (+x, −x) was carried out on the first (π/2) pulse. The microwave frequency for all measurements was in the range of 33.4–33.8 GHz, depending on coupling conditions. The pump pulse frequency always was adjusted to be consistent with the maximum peak of the Q-band absorption spectrum of the nitroxide labels, and the observe pulses always were applied at ∼50 MHz lower than the pump frequency. Typical signal acquisition time was 30–60 min depending on sample concentration. All DEER data were analyzed using the program “LongDistances,” written in LabVIEW and available at www.chemistry.ucla.edu/directory/hubbell-wayne-l.
Supplementary Material
Acknowledgments
We thank Matthias Elgeti for a careful reading of the manuscript and Christian Altenbach for assistance with analysis of the DEER data using his program “LongDistances.” This work was supported by National Institutes of Health Grants 2R01 EY05216, 2P30 EY00331, and T32 GM 008496 and by the Jules Stein Professorship Endowment.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403179111/-/DCSupplemental.
References
- 1.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol. 2009;5(11):808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 2.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450(7172):964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 4.Kitahara R, et al. High pressure NMR reveals active-site hinge motion of folate-bound Escherichia coli dihydrofolate reductase. Biochemistry. 2000;39(42):12789–12795. doi: 10.1021/bi0009993. [DOI] [PubMed] [Google Scholar]
- 5.Kitahara R, Yokoyama S, Akasaka K. NMR snapshots of a fluctuating protein structure: Ubiquitin at 30 bar-3 kbar. J Mol Biol. 2005;347(2):277–285. doi: 10.1016/j.jmb.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 6.Girard E, et al. Structure-function perturbation and dissociation of tetrameric urate oxidase by high hydrostatic pressure. Biophys J. 2010;98(10):2365–2373. doi: 10.1016/j.bpj.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dellarole M, Roumestand C, Royer C, Lecomte JT. Volumetric properties underlying ligand binding in a monomeric hemoglobin: A high-pressure NMR study. Biochim Biophys Acta. 2013;1834(9):1910–1922. doi: 10.1016/j.bbapap.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 8.Kapoor S, et al. Revealing conformational substates of lipidated N-Ras protein by pressure modulation. Proc Natl Acad Sci USA. 2012;109(2):460–465. doi: 10.1073/pnas.1110553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sineva EV, Davydov DR. Cytochrome P450 from Photobacterium profundum SS9, a piezophilic bacterium, exhibits a tightened control of water access to the active site. Biochemistry. 2010;49(50):10636–10646. doi: 10.1021/bi101466y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Royer CA. Revisiting volume changes in pressure-induced protein unfolding. Biochim Biophys Acta. 2002;1595(1-2):201–209. doi: 10.1016/s0167-4838(01)00344-2. [DOI] [PubMed] [Google Scholar]
- 11.Akasaka K. Probing conformational fluctuation of proteins by pressure perturbation. Chem Rev. 2006;106(5):1814–1835. doi: 10.1021/cr040440z. [DOI] [PubMed] [Google Scholar]
- 12.Roche J, et al. Cavities determine the pressure unfolding of proteins. Proc Natl Acad Sci USA. 2012;109(18):6945–6950. doi: 10.1073/pnas.1200915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lerch MT, Horwitz J, McCoy J, Hubbell WL. Circular dichroism and site-directed spin labeling reveal structural and dynamical features of high-pressure states of myoglobin. Proc Natl Acad Sci USA. 2013;110(49):E4714–E4722. doi: 10.1073/pnas.1320124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akasaka K, Kitahara R, Kamatari YO. Exploring the folding energy landscape with pressure. Arch Biochem Biophys. 2013;531(1-2):110–115. doi: 10.1016/j.abb.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 15.Fourme R, Girard E, Akasaka K. High-pressure macromolecular crystallography and NMR: Status, achievements and prospects. Curr Opin Struct Biol. 2012;22(5):636–642. doi: 10.1016/j.sbi.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: A kinetic approach to the sequence-structure relationship. Proc Natl Acad Sci USA. 1992;89(18):8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: The energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 18.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254(5038):1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 19.Kitahara R, Hata K, Li H, Williamson MP, Akasaka K. Pressure-induced chemical shifts as probes for conformational fluctuations in proteins. Prog Nucl Magn Reson Spectrosc. 2013;71:35–58. doi: 10.1016/j.pnmrs.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Kitahara R, Yamada H, Akasaka K, Wright PE. High pressure NMR reveals that apomyoglobin is an equilibrium mixture from the native to the unfolded. J Mol Biol. 2002;320(2):311–319. doi: 10.1016/S0022-2836(02)00449-7. [DOI] [PubMed] [Google Scholar]
- 21.Bridges MD, Hideg K, Hubbell WL. Resolving conformational and rotameric exchange in spin-labeled proteins using saturation recovery EPR. Appl Magn Reson. 2010;37(1-4):363–390. doi: 10.1007/s00723-009-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.López CJ, Oga S, Hubbell WL. Mapping molecular flexibility of proteins with site-directed spin labeling: A case study of myoglobin. Biochemistry. 2012;51(33):6568–6583. doi: 10.1021/bi3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milov AD, Ponomarev AB, Tsvetkov YD. Electron-electron double resonance in electron spin echo: Model biradical systems and the sensitized photolysis of decalin. Chem Phys Lett. 1984;110(1):67–72. [Google Scholar]
- 24.Pannier M, Veit S, Godt A, Jeschke G, Spiess HW. Dead-time free measurement of dipole-dipole interactions between electron spins. J Magn Reson. 2000;142(2):331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- 25.Mchaourab HS, Steed PR, Kazmier K. Toward the fourth dimension of membrane protein structure: Insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19(11):1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeschke G. DEER distance measurements on proteins. Annu Rev Phys Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, et al. Pulsed ESR dipolar spectroscopy for distance measurements in immobilized spin labeled proteins in liquid solution. J Am Chem Soc. 2012;134(24):9950–9952. doi: 10.1021/ja303791p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eliezer D, Jennings PA, Dyson HJ, Wright PE. Populating the equilibrium molten globule state of apomyoglobin under conditions suitable for structural characterization by NMR. FEBS Lett. 1997;417(1):92–96. doi: 10.1016/s0014-5793(97)01256-8. [DOI] [PubMed] [Google Scholar]
- 29.Jennings PA, Wright PE. Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science. 1993;262(5135):892–896. doi: 10.1126/science.8235610. [DOI] [PubMed] [Google Scholar]
- 30.Meinhold DW, Wright PE. Measurement of protein unfolding/refolding kinetics and structural characterization of hidden intermediates by NMR relaxation dispersion. Proc Natl Acad Sci USA. 2011;108(22):9078–9083. doi: 10.1073/pnas.1105682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eliezer D, Yao J, Dyson HJ, Wright PE. Structural and dynamic characterization of partially folded states of apomyoglobin and implications for protein folding. Nat Struct Biol. 1998;5(2):148–155. doi: 10.1038/nsb0298-148. [DOI] [PubMed] [Google Scholar]
- 32.Georgieva ER, et al. Effect of freezing conditions on distances and their distributions derived from Double Electron Electron Resonance (DEER): A study of doubly-spin-labeled T4 lysozyme. J Magn Reson. 2012;216:69–77. doi: 10.1016/j.jmr.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. de Vera IMS, Blackburn ME, Galiano L, Fanucci GE (2013) Pulsed EPR distance measurements in soluble proteins by site-directed spin labeling (SDSL). Curr Protoc Prot Sci 74:17.17.1–17.17.29. [DOI] [PMC free article] [PubMed]
- 34.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105(21):7439–7444. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dawidowski D, Cafiso DS. Allosteric control of syntaxin 1a by Munc18-1: Characterization of the open and closed conformations of syntaxin. Biophys J. 2013;104(7):1585–1594. doi: 10.1016/j.bpj.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Vera IMS, et al. Elucidating a relationship between conformational sampling and drug resistance in HIV-1 protease. Biochemistry. 2013;52(19):3278–3288. doi: 10.1021/bi400109d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dockter C, et al. Rigid core and flexible terminus: Structure of solubilized light-harvesting chlorophyll a/b complex (LHCII) measured by EPR. J Biol Chem. 2012;287(4):2915–2925. doi: 10.1074/jbc.M111.307728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgieva ER, Borbat PP, Ginter C, Freed JH, Boudker O. Conformational ensemble of the sodium-coupled aspartate transporter. Nat Struct Mol Biol. 2013;20(2):215–221. doi: 10.1038/nsmb.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krishnamani V, Hegde BG, Langen R, Lanyi JK. Secondary and tertiary structure of bacteriorhodopsin in the SDS denatured state. Biochemistry. 2012;51(6):1051–1060. doi: 10.1021/bi201769z. [DOI] [PubMed] [Google Scholar]
- 40.Sattig T, Rickert C, Bamberg E, Steinhoff HJ, Bamann C. Light-induced movement of the transmembrane helix B in channelrhodopsin-2. Angew Chem Int Ed Engl. 2013;52(37):9705–9708. doi: 10.1002/anie.201301698. [DOI] [PubMed] [Google Scholar]
- 41.Van Eps N, et al. Interaction of a G protein with an activated receptor opens the interdomain interface in the alpha subunit. Proc Natl Acad Sci USA. 2011;108(23):9420–9424. doi: 10.1073/pnas.1105810108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez JAT, Longinotti MP, Corti HR. The viscosity of glycerol-water mixtures including the supercooled region. J Chem Eng Data. 2011;56(4):1397–1406. [Google Scholar]
- 43.Brucker EA, Olson JS, Phillips GN, Jr, Dou Y, Ikeda-Saito M. High resolution crystal structures of the deoxy, oxy, and aquomet forms of cobalt myoglobin. J Biol Chem. 1996;271(41):25419–25422. doi: 10.1074/jbc.271.41.25419. [DOI] [PubMed] [Google Scholar]
- 44.Uzawa T, et al. Hierarchical folding mechanism of apomyoglobin revealed by ultra-fast H/D exchange coupled with 2D NMR. Proc Natl Acad Sci USA. 2008;105(37):13859–13864. doi: 10.1073/pnas.0804033105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris GJ, Goodrich M, Acton E, Fonseca F. The high viscosity encountered during freezing in glycerol solutions: Effects on cryopreservation. Cryobiology. 2006;52(3):323–334. doi: 10.1016/j.cryobiol.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 46.Raitsimring AM, Salikhov KM, Umanskii BA, Tsvetkov YD. Instantaneous diffusion in electron-spin echo of paramagnetic centers stabilized in solid matrices. Fiz Tverd Tela (St. Petersburg) 1974;16(3):756–766. [Google Scholar]
- 47.Ruthstein S, et al. Distribution of guest molecules in Pluronic micelles studied by double electron electron spin resonance and small angle X-ray scattering. Phys Chem Chem Phys. 2009;11(1):148–160. doi: 10.1039/b812475b. [DOI] [PubMed] [Google Scholar]
- 48.Kramers HA. Brownian motion in a field of force and the diffusion model of chemical reactions. Physica. 1940;7(4):284–304. [Google Scholar]
- 49.Ramos CH, Weisbuch S, Jamin M. Diffusive motions control the folding and unfolding kinetics of the apomyoglobin pH 4 molten globule intermediate. Biochemistry. 2007;46(14):4379–4389. doi: 10.1021/bi602574x. [DOI] [PubMed] [Google Scholar]
- 50.Nienhaus GU, Muller JD, McMahon BH, Frauenfelder H. Exploring the conformational energy landscape of proteins. Phys D (Amsterdam) 1997;107(2-4):297–311. [Google Scholar]
- 51.Romero-Romero ML, Inglés-Prieto A, Ibarra-Molero B, Sanchez-Ruiz JM. Highly anomalous energetics of protein cold denaturation linked to folding-unfolding kinetics. PLoS ONE. 2011;6(7):e23050. doi: 10.1371/journal.pone.0023050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cho KC, Chan KK. Kinetics of cold-induced denaturation of metmyoglobin. Biochim Biophys Acta. 1984;786(1-2):103–108. doi: 10.1016/0167-4838(84)90159-6. [DOI] [PubMed] [Google Scholar]
- 53.Chen BL, Baase WA, Schellman JA. Low-temperature unfolding of a mutant of phage T4 lysozyme. 2. Kinetic investigations. Biochemistry. 1989;28(2):691–699. doi: 10.1021/bi00428a042. [DOI] [PubMed] [Google Scholar]
- 54.Gekko K, Timasheff SN. Mechanism of protein stabilization by glycerol: Preferential hydration in glycerol-water mixtures. Biochemistry. 1981;20(16):4667–4676. doi: 10.1021/bi00519a023. [DOI] [PubMed] [Google Scholar]
- 55.Saunders AJ, Davis-Searles PR, Allen DL, Pielak GJ, Erie DA. Osmolyte-induced changes in protein conformational equilibria. Biopolymers. 2000;53(4):293–307. doi: 10.1002/(SICI)1097-0282(20000405)53:4<293::AID-BIP2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 56.Flores Jiménez RH, Do Cao MA, Kim M, Cafiso DS. Osmolytes modulate conformational exchange in solvent-exposed regions of membrane proteins. Protein Sci. 2010;19(2):269–278. doi: 10.1002/pro.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee JC, Timasheff SN. The stabilization of proteins by sucrose. J Biol Chem. 1981;256(14):7193–7201. [PubMed] [Google Scholar]
- 58.López CJ, Fleissner MR, Guo ZF, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18(8):1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Auton M, Rösgen J, Sinev M, Holthauzen LMF, Bolen DW. Osmolyte effects on protein stability and solubility: A balancing act between backbone and side-chains. Biophys Chem. 2011;159(1):90–99. doi: 10.1016/j.bpc.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Street TO, Bolen DW, Rose GD. A molecular mechanism for osmolyte-induced protein stability. Proc Natl Acad Sci USA. 2006;103(38):13997–14002. doi: 10.1073/pnas.0606236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Silvers TR, Myers JK. Osmolyte effects on the self-association of concanavalin a: Testing theoretical models. Biochemistry. 2013;52(51):9367–9374. doi: 10.1021/bi401049s. [DOI] [PubMed] [Google Scholar]
- 62.Zipp A, Kauzmann W. Pressure denaturation of metmyoglobin. Biochemistry. 1973;12(21):4217–4228. doi: 10.1021/bi00745a028. [DOI] [PubMed] [Google Scholar]
- 63.Phillips GN., Jr Comparison of the dynamics of myoglobin in different crystal forms. Biophys J. 1990;57(2):381–383. doi: 10.1016/S0006-3495(90)82540-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frauenfelder H, et al. Thermal expansion of a protein. Biochemistry. 1987;26(1):254–261. doi: 10.1021/bi00375a035. [DOI] [PubMed] [Google Scholar]
- 65.Baldwin RL, Rose GD. Molten globules, entropy-driven conformational change and protein folding. Curr Opin Struct Biol. 2013;23(1):4–10. doi: 10.1016/j.sbi.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Eliezer D, Wright PE. Is apomyoglobin a molten globule? Structural characterization by NMR. J Mol Biol. 1996;263(4):531–538. doi: 10.1006/jmbi.1996.0596. [DOI] [PubMed] [Google Scholar]
- 67.Lecomte JT, Sukits SF, Bhattacharya S, Falzone CJ. Conformational properties of native sperm whale apomyoglobin in solution. Protein Sci. 1999;8(7):1484–1491. doi: 10.1110/ps.8.7.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao J, Chung J, Eliezer D, Wright PE, Dyson HJ. NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding. Biochemistry. 2001;40(12):3561–3571. doi: 10.1021/bi002776i. [DOI] [PubMed] [Google Scholar]
- 69.Kamatari YO, Smith LJ, Dobson CM, Akasaka K. Cavity hydration as a gateway to unfolding: An NMR study of hen lysozyme at high pressure and low temperature. Biophys Chem. 2011;156(1):24–30. doi: 10.1016/j.bpc.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 70.Neumaier S, Büttner M, Bachmann A, Kiefhaber T. Transition state and ground state properties of the helix-coil transition in peptides deduced from high-pressure studies. Proc Natl Acad Sci USA. 2013;110(52):20988–20993. doi: 10.1073/pnas.1317973110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Imamura H, Kato M. Effect of pressure on helix-coil transition of an alanine-based peptide: An FTIR study. Proteins. 2009;75(4):911–918. doi: 10.1002/prot.22302. [DOI] [PubMed] [Google Scholar]
- 72.Hummer G, Garde S, García AE, Paulaitis ME, Pratt LR. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc Natl Acad Sci USA. 1998;95(4):1552–1555. doi: 10.1073/pnas.95.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.