Significance
Ancient DNA sequences from chickens provide an opportunity to study their human-mediated dispersal across the Pacific due to the significant genetic diversity and range of archaeological material available. We analyze ancient and modern material and reveal that previous studies have been impacted by contamination with modern chicken DNA and, that as a result, there is no evidence for Polynesian dispersal of chickens to pre-Columbian South America. We identify genetic markers of authentic ancient Polynesian chickens and use them to model early chicken dispersals across the Pacific. We find connections between chickens in the Micronesian and Bismarck Islands, but no evidence these were involved in dispersals further east. We also find clues about the origins of Polynesian chickens in the Philippines.
Keywords: Lapita, Pacific colonization, phylogeography, archaeology, migration
Abstract
The human colonization of Remote Oceania remains one of the great feats of exploration in history, proceeding east from Asia across the vast expanse of the Pacific Ocean. Human commensal and domesticated species were widely transported as part of this diaspora, possibly as far as South America. We sequenced mitochondrial control region DNA from 122 modern and 22 ancient chicken specimens from Polynesia and Island Southeast Asia and used these together with Bayesian modeling methods to examine the human dispersal of chickens across this area. We show that specific techniques are essential to remove contaminating modern DNA from experiments, which appear to have impacted previous studies of Pacific chickens. In contrast to previous reports, we find that all ancient specimens and a high proportion of the modern chickens possess a group of unique, closely related haplotypes found only in the Pacific. This group of haplotypes appears to represent the authentic founding mitochondrial DNA chicken lineages transported across the Pacific, and allows the early dispersal of chickens across Micronesia and Polynesia to be modeled. Importantly, chickens carrying this genetic signature persist on several Pacific islands at high frequencies, suggesting that the original Polynesian chicken lineages may still survive. No early South American chicken samples have been detected with the diagnostic Polynesian mtDNA haplotypes, arguing against reports that chickens provide evidence of Polynesian contact with pre-European South America. Two modern specimens from the Philippines carry haplotypes similar to the ancient Pacific samples, providing clues about a potential homeland for the Polynesian chicken.
The colonization of the remote Pacific was one of the last great human migrations, but despite the recent nature of the events, the timing and routes remain an area of considerable debate. The first colonization of Western Polynesia occurred around 3,250–3,100 calendar years before present (cal B.P.) as part of the eastward migration of Lapita pottery-bearing peoples (1). This migration occurred only a few hundred years after the emergence of this distinctive pottery tradition in the Bismarck Archipelago around 3,470–3,250 cal B.P., although its antecedents can be traced to Island Southeast Asia (ISEA) (2–5). Following the initial movement into Western Polynesia, a prolonged 1,800-y hiatus, or “pause,” is apparent before further colonization (6), potentially relating to the need to develop sailing technology essential for crossing the vast ocean barrier to the east (between Samoa and the Society Islands, 2,400 km; Fig. 1). The huge navigational achievement of colonizing the remote East Polynesian triangle (an oceanic region roughly the size of North America) then occurred rapidly (<300 y) (6). Although the overall chronology of the eastern Pacific island colonization has recently been further resolved, the precise details of this intensive migratory episode remain unclear (6).
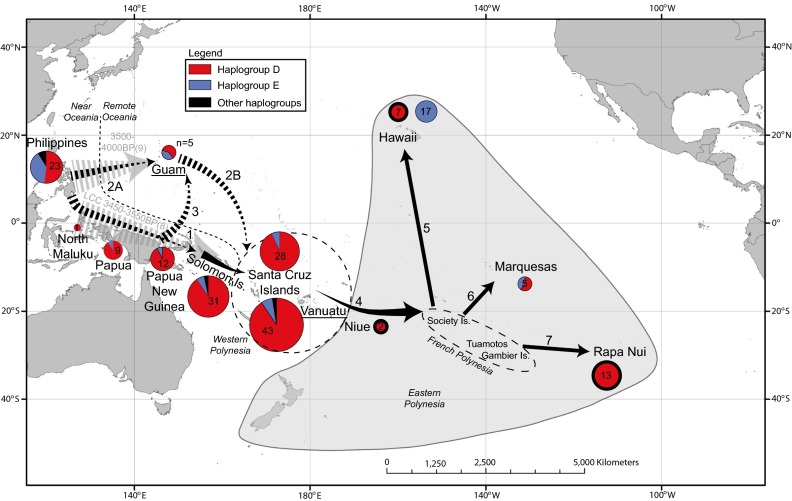
Fig. 1.
Map showing samples and localities mentioned in this study. Samples from Vanuatu and Guam previously published in Dancause et al. (26) are underlined, with haplogroup frequencies of the chicken specimens indicated by pie charts (thick outlines indicate ancient samples). Colors refer to haplotype/haplogroup: D haplogroup in red, E haplogroup in blue, and all other haplotypes are in black. Gray arrows represent movements inferred from archaeological data (49), whereas black arrows represent routes tested in BayeSSC analysis (dashed arrows indicate movements tested in different scenarios, whereas solid arrows are constant across the different scenarios). 1, introduction of chickens within Near Oceania; 2, alternate hypothesis proposed by (38); 3, introduction of chickens from New Guinea into Micronesia; 4–7, spread of chickens from Western Polynesia into, and within, Eastern Polynesia. Dashed line indicates demarcation between Near and Remote Oceania, Western Polynesia is defined by a dashed circle, and Eastern Polynesia is indicated by a gray shaded triangle.
Human commensal and early domesticated species were widely, but not ubiquitously, dispersed as people colonized the Pacific. As a result, they provide an opportunity to study colonization events and subsequent movements for islands and regions where they were successfully introduced, especially through the use of biomolecular techniques, including ancient DNA. In the Asia–Pacific region, the complex histories of Pacific island colonizations have been investigated using the biological elements associated with these cultures, such as bottle gourds (7, 8), sweet potatoes (9), pigs (10, 11), dogs (12), Pacific rats (13), and chickens (14–17). However, studies of commensals and domesticates in the Pacific to date have provided limited resolution of dispersal routes, due to low amounts of genetic diversity in many groups and overwriting of genetic signals by subsequent introductions, especially for cotransported species like rats (10, 13, 18).
Ancient and modern DNA from chickens provide an opportunity to study human-mediated dispersal across the Pacific due to the extent of genetic and phenotypic diversity and the range of archaeological material available. Although recent studies of domestic chicken breeds have highlighted how the domestication process and subsequent breeding have resulted in a 70% loss of nuclear genetic diversity (19), substantial phylogeographic structure remains within the mitochondrial (mtDNA) sequences of chickens worldwide (20–23). Furthermore, an extensive reference dataset of >3,000 mtDNA control region (CR) sequences and >60 whole mtDNA genomes is available from across the natural range of wild and semiwild birds, as well as domestic breeds of chicken, permitting the reconstruction of phylogeographic patterns of domestic chickens and associated human cultures. Despite these intensive surveys, a resolved worldwide chicken mtDNA phylogeny has not been developed, and this is an essential prerequisite to interpreting short ancient DNA sequences. The current phylogenetic framework for chickens is based on Liu et al. (24), who identified nine highly divergent haplogroups (designated A–I) using mtDNA CR sequences, with an additional four recently described on the basis of whole mtDNA genomes (W–Z) (25). However, there is little information about the support for these topologies, and only neighbor-joining trees have been reported to date.
Phylogeographic studies have identified that one particular mtDNA lineage (CR haplogroup D) is largely limited to the Asia–Pacific region (24), whereas many of the other haplogroups are ubiquitous worldwide, potentially as a result of historical dispersal with European colonialists (e.g., haplogroups A, B, and E), and are therefore generally phylogeographically uninformative. Previous studies of modern and ancient chickens have identified both haplogroup D and E in the Pacific (14–17, 26), making interpretation of colonization history difficult due to potentially contrasting origins and dispersal histories (24). Indeed, the presence of haplogroup E in the Pacific has been used to infer a link between Polynesia and pre-Columbian South America, although both the phylogenetic signal and radiocarbon dating of the samples have been questioned (27–29). This issue has recently taken on more significance as other studies of ancient genetic diversity in South America emphasize the importance of evidence for pre-European Polynesian contact (17, 30).
In this study, we first quantify the support for previously defined chicken mtDNA CR haplogroups using recently published whole mitochondrial genomes (WMGs) (25). We then use the resulting robust evolutionary framework to analyze the spatial and temporal patterns of mtDNA CR haplotypes in ancient and modern Pacific chickens to examine their origins in ISEA (31), the dispersal of chickens into Near Oceania and Western Polynesia, potential connections between the New Guinea region and Micronesia, and the claimed introduction of Polynesian chickens to South America (14).
Results
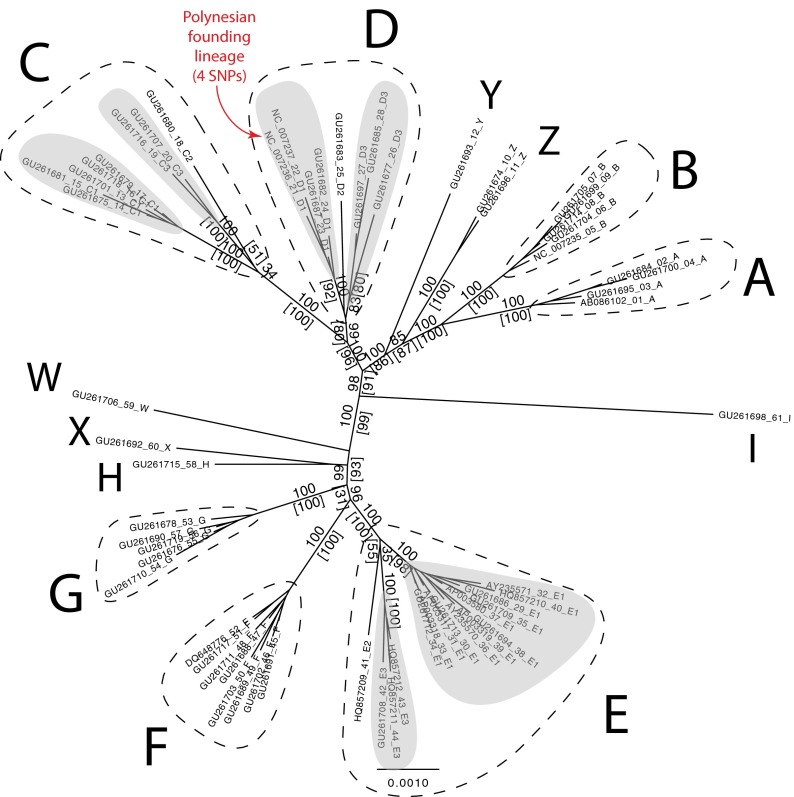
The 61 WMG dataset (25) contained 363 single-nucleotide polymorphisms (SNPs), of which 154 were potentially phylogenetically informative, with 62 (17%) located in the rapidly evolving CR (32). Bayesian and maximum likelihood inference analyses of the WMG dataset supported the haplogroup framework defined by Liu et al. (24) and Miao et al. (25) and, importantly, produced robust support for haplogroups A–G and Z (i.e., haplogroups where multiple individuals were sequenced), as shown in Fig. 2. Robust support values were also obtained for phylogenetic trees based on the WMG data without the CR sequences (SI Appendix, Fig. S1), but were less robust when based only on the highly variable CR sequences alone, likely due to issues with substitution rate heterogeneity (32) (SI Appendix, Fig. S2). However, the short (201 bp), hypervariable region of the CR used in previous studies contains >12× the average diversity per base compared with the rest of the WMG, and has the advantage of being available for a worldwide dataset of >1,000 chicken sequences. The comparative phylogenetic dataset constructed from these sequences identified 274 unique haplotypes, which we termed H001–H274 (SI Appendix, Dataset S6).
Fig. 2.
Phylogenetic tree based on WMG data from Miao et al. (25). Bayesian posterior probability and maximum likelihood bootstrap (in parentheses) support values are shown on branches. All haplogroups with more than one individual have robust support and concur with the designations of Liu et al. (24).
Of the 37 Polynesian archaeological chicken bones analyzed to study the temporal and spatial patterns within Polynesia, 22 (59%) yielded positive and repeatable PCR amplification and DNA sequencing results for a 330 bp region (which included the hypervariable 201 bp; Niue, n = 2/8; Hawai’i, n = 7/11; Rapa Nui, n = 13/18; SI Appendix, Table S1). All of the 22 positive ancient samples produced mtDNA CR sequences belonging to haplogroup D. Two samples that could not be reliably reproduced (from Niue and Rapa Nui) each generated a single PCR product with different non-D haplotypes (from haplogroup A and E, respectively; SI Appendix, Table S1 and Dataset S1). However, when DNase (double-strand–specific Shrimp DNase) pretreatment was used to remove potential contaminating DNA from reagents (33), these sequences were no longer detected (SI Appendix). Two of the 124 modern feather samples examined could not be successfully amplified (one from the Marquesas and another from Hawai’i). The large majority of the resulting 122 modern sequences belonged to haplogroup D (n = 90/122, 74%; SI Appendix, Figs. S3–S7), with haplogroup E sequences present at a lower frequency (n = 27/122, 22%). The remaining five samples fell within haplogroups A, B, and I (n = 1, 3, and 1, respectively, each <2.5%).
Previous studies of Pacific chickens have reported elevated levels of haplogroup E among ancient specimens (up to 48%) (14–16), in direct contrast to our results. However, the contamination of laboratory consumables with DNA from modern domestic species, including chickens, is a well-known problem in ancient DNA research (34), and this would also likely generate haplogroup E sequences, due to the ubiquity of the latter worldwide (SI Appendix, Dataset S2) (24). To examine this potential explanation, we reexamined key samples from a previous study that linked ancient Polynesian chickens to South American archaeological specimens (14). Four of the six bone samples from Rapa Nui used in the previous study were available for reexamination, but only three gave replicable results (SI Appendix, Dataset S1). However, these included the individual bone reported to have generated the critical single haplogroup E sequence (H268 of our unique haploptypes) used to link Rapa Nui and South America (sample PAQANA011; SI Appendix, Fig. S8) (14). In direct contrast to the previous results, our reanalysis of an independent sample of PAQANA011 using Shrimp DNase PCR pretreatment yielded a haplogroup D sequence (haplotype H239; SI Appendix, Dataset S6) identical to those of the other two Rapa Nui specimens we reexamined. This result was subsequently confirmed through independent replication of a subsample of the same specimen at Durham University (SI Appendix, Dataset S3).
Our results further revealed that the PAQANA011 specimen contained low amounts of DNA, with elevated levels of DNA template damage (SI Appendix, Dataset S4), and strongly suggests the previously reported haplogroup E sequence was the result of contamination with modern chicken DNA. A further 10 samples excavated from the same site on Rapa Nui (Anakena) were also examined, and all yielded replicable haplogroup D sequences (haplotype H239; SI Appendix, Dataset S1). Together with the haplogroup D results of the previous study (14), this means that all 15 different bones examined at the Anakena site have yielded H239 sequences.
To investigate the conflict between the results obtained here and those previously reported from ancient Pacific specimens (14–16), we calculated the probability of detecting the reported proportions of D and E haplogroups given the different datasets. If haplogroup E was authentically present within ancient Pacific chickens at the levels previously reported (48%) (14–16), then the probability that all 22 of our ancient samples would belong to haplogroup D is negligible (P value = 1.3 × 10−7). In contrast, our results suggest that if haplogroup E was present at all in ancient Pacific chickens, it must have been in less than 13% (at the 95% probability level; SI Appendix, Fig. S9). It is possible that if haplogroup E was present in very low frequencies among ancient Pacific chickens (e.g., <10%), we did not detect it within the 22 ancient samples we examined simply due to stochastic sampling effects (P value = 0.098). However, if E was actually present at only 10% in the ancient Pacific chickens, then it is also highly unlikely that haplogroup E sequences would have been detected in 15/31 (48%) of the specimens in previous studies (P value = 6.9 × 10−9).
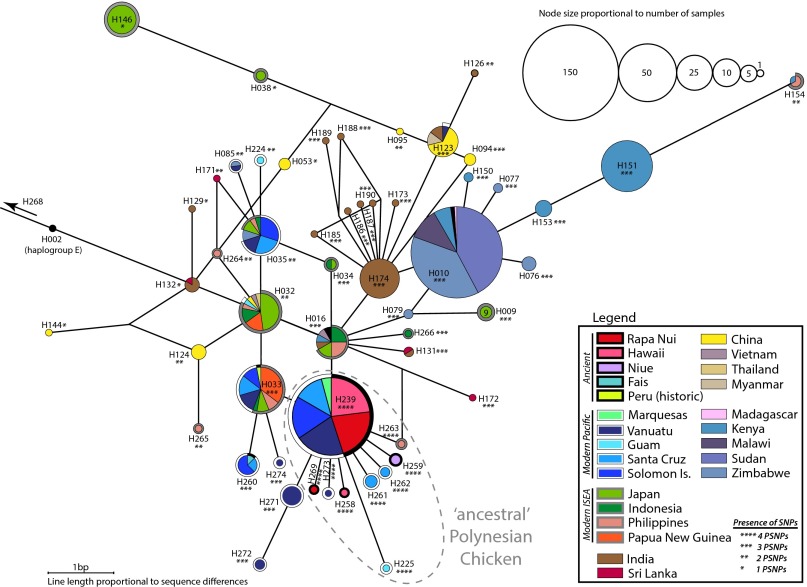
A median-joining network of the haplogroup D chicken sequences revealed that all of the ancient Pacific sequences generated in this study (n = 22) and those from previous studies (n = 16) (14, 16) together comprise only five different haplotypes (Fig. 3), none of which have been found outside the Pacific region. Four of these five are from Polynesia and cluster together, possessing a diagnostic motif of four SNPs (A → G at base 281, C → T at base 296, T → C at base 306, A → G at base 342 compared with NC_007235; SI Appendix, Dataset S5). The four diagnostic SNPs were also detected in four additional haplotypes within the diversity of sequences from modern chickens sampled across the western Pacific and the Philippines, but only from Vanuatu, Santa Cruz, Philippines, and Guam (Fig. 3) (26). Indeed one of the previously published WMGs, from the Philippines (NC_007236; 25), contains all four of these diagnostic SNPs (SI Appendix, Figs. S1 and S2). Fig. 3 shows that the most common ancient haplotype, H239, forms the central node from which the other three ancient Polynesian D haplotypes radiate, consistent with a recent rapid expansion. The central haplotype was also the most common sequence in modern Pacific chicken populations, being present on almost all Pacific islands sampled.
Fig. 3.
Unrooted haplogroup D network generated using 201 bp of mitochondrial CR analyzed in this study, including 144 sequences generated in this study and 1,226 sequences from GenBank (14, 21–24, 26, 27, 43–46). Labels on nodes are unique haplotype numbers from this study (SI Appendix, Dataset S6). Colors reflect sampling location, with outline around pie charts representing ancient samples (black), modern Pacific (white), and modern ISEA (gray).
South America.
Given that at least some of the previously reported ancient Pacific chicken data appear to be due to contamination, and the fact that all of the authenticated or reliable ancient Pacific chicken sequences are restricted to the unique Pacific group of haplogroup D sequences, we performed approximate Bayesian coalescent simulations to evaluate the evidence for the pre-Columbian introduction of chickens to South America. The coalescent simulations provided no evidence to support prehistoric dispersal of chickens from Polynesia to South America either when the datasets included (i) ancient sequences only from haplogroup D or (ii) all sequences reported from ancient specimens (Haplogroups B, E, and D) (14–16) (SI Appendix, Fig. S11 and Tables S2–S5). The analyses reveal that the more likely route and explanation for South American chicken diversity appears to be via Europe and early historical introductions, or as modern DNA contamination of experiments (SI Appendix, Fig. S11). A single D haplotype sequence (H033) has been reported from post-European contact Peru (16), but this sequence is common within ISEA populations, and could have been associated with early-colonial Spanish trade. Importantly, it has not been found among the ancient Pacific chicken sequences.
Micronesia and Western Polynesia.
To investigate early human-mediated dispersal patterns within the Asia–Pacific region, we examined modern chickens from islands across ISEA, Micronesia, and Western Polynesia, because few specimens of ancient chickens were available from this area [however, see Fais D haplotype sample from Storey et al. (16)]. Although the ISEA sequences are scattered across the haplogroup D network, the majority of haplotypes from modern Pacific chickens are genetically clustered together (H032–35, H085, H224–225, H260, H262, H271–274; Fig. 3). Within Micronesia, haplogroup D has been reported from modern chickens in Guam (n = 3/5; H032, H224, and H225; 26), although interestingly, these particular D haplotypes are not shared with any other Pacific island group. In fact, two of these haplotypes have only ever been found in Guam (H224 and H225), whereas the third Guam haplotype is shared with the Philippines, Japan, Indonesia, and Papua New Guinea (H032). The modern haplogroup D chickens in Guam do not appear to be significantly genetically differentiated from those in the Philippines, Japan, and Indonesia (SI Appendix, Table S6).
An investigation of the discordant haplogroup D lineages in Micronesia and Polynesia using coalescent simulations identified an early movement of chickens between New Guinea and Micronesia as the most likely of five models tested (SI Appendix, Fig. S12 and Tables S7 and S8). The simulations suggest that chickens were transported between Micronesia and islands in the Bismarck Sea off the coast of New Guinea and New Britain around 3,850 years ago (ya), without further onward transportation of chickens into Western and Eastern Polynesia (SI Appendix, Fig. S12). In contrast, the origins of the chickens currently found in Polynesia appear to be via the standard southern route from New Guinea to the Solomon Islands, the Santa Cruz Islands, Vanuatu, and further eastward (Fig. 1, arrows 1 and 4–7, and SI Appendix, Fig. S12).
Discussion
Our results indicate that a small cluster of mtDNA haplogroup D sequences, defined by a diagnostic combination of four CR SNPs (which we term the “ancestral Polynesian motif”), represent the founding lineages of chickens transported as prehistoric domesticates across the Pacific and ultimately ending up in Polynesia (i.e., “Polynesian chickens”). We suggest that the most common haplotype in ancient samples (H239) represents the core mtDNA lineage of Polynesian chickens, and that the one- or two-step derivatives in ancient Pacific island specimens (Fig. 3) represent in situ evolution following colonization. This hypothesis is supported by the geographic distribution of the ancient daughter lineages, which are unique to each Pacific island group, and the elevated frequency of lineages with the four diagnostic SNPs in the eastern Pacific (SI Appendix, Fig. S10). Although mtDNA is maternally inherited as a single genetic locus, limiting the ability to recover complex colonization histories, our data establish clear hypotheses that can be tested with genomic data from both modern and ancient chickens, and other groups such as humans, commensals, and other domesticates. It is important to note that in situations like the Pacific, phylogeographic signals in domestic species are likely to represent processes of initial human dispersal and later trade patterns.
Our findings contrast substantially with previous studies (14–16), which we suggest stems from our strict adherence to contamination reduction measures—for example, the use of Shrimp DNase. By removing a key source of potential contamination with domestic chicken DNA (PCR reagents), the use of Shrimp DNase has allowed us to recharacterize the crucial ancient Rapa Nui sample from a prior study (PAQANA011) as haplogroup D and not, as previously reported, haplogroup E. Consequently, we cast doubt on the authenticity of other haplogroup E sequences reported from ancient Pacific chicken specimens, where such procedures were not used. Perhaps more importantly, we suggest it will be very difficult to categorically rule out contamination as the source of haplogroup E sequences in ancient samples, due to the sporadic presence of domesticate DNA in laboratory consumables (34) and the likelihood that any such contamination would result in haplogroup E sequences. Importantly, sequencing longer stretches of such contaminating templates (17) does not provide any additional support for authenticity.
Our recharacterization of the Rapa Nui PAQANA011 specimen as haplogroup D has implications for the other E sequences reported by Storey et al. (14), including the putative ancient Chilean chicken sequence from El Arenal-1 used to propose a prehistoric link between Polynesia and South America. Coalescent simulations using “all ancient haplogroups” and the modern data found that a European–South America route was more likely than a direct link between haplogroup E chicken sequences in Polynesia and South America, due to the phylogeographic signals within the worldwide dataset showing more similarities between chickens from Europe and South America. Perhaps more generally, these findings highlight how haplogroup E sequences are uninformative in nature and lack phylogeographic signal worldwide. A clear understanding of the nature and extent of Polynesian contact with South America will require genomic analyses of both ancient and modern populations of humans, commensals, and domesticates.
The distribution of the nine D haplotypes currently known to share the ancestral motif provides a unique genetic signature that can be used to trace the human dispersal of chickens through ISEA and the Pacific islands. Our reconstruction of the chicken colonization history of Micronesia highlights how simulations with CR data can provide sufficient phylogeographic signal to generate new hypotheses regarding trade and migration scenarios. Although it has been proposed that many commensals and domesticates are late arrivals to the Micronesian islands compared with humans (35), we have reconstructed a link between chickens from islands in the Bismarck Sea and Micronesia that dates to ∼3,850 B.P. Such an early date is broadly consistent with archaeological evidence for human settlement of Saipan at 3,300–3,500 B.P. (36) and Palau at almost 4,000 B.P. (35), however few comparably early zoo-archaeological remains have been found in Micronesia to date (10, 13, 37). The inferred link between chickens from the Bismarcks and Micronesia without subsequent eastward movement does not support a two-wave model of Polynesian origins (14, 15, 38) where an earlier Lapita migration wave (2,800–3,500 ya) was mixed with a second, later wave moving through Micronesia to Western Polynesia (1,500–2,000 ya). Our simulations suggest that there was little interaction between chickens from Micronesia and the islands further eastward. One caveat concerning the power of the simulation analysis is the small number of Micronesian samples [one ancient Fais (16) and five modern Guam (26) specimens] and the expected historical and recent turnover of chicken populations in the region. Reassuringly, the ancient Fais haplotype H260 is present in modern chickens from the Santa Cruz (n = 2) and Solomon Islands (n = 5), apparently surviving any later introgression. Our reconstruction of the colonization history of Micronesian chickens demonstrates the potential power of coalescent simulations to test hypothesized migration and trade routes in archaeology and anthropology.
The only ISEA location where the ancestral SNP motif has been detected are Camiguin and Manila in the Philippines, and a link with this area is consistent with other lines of evidence about early Polynesian origins (3, 4, 31). The other Philippine chicken haplotypes are spread throughout the haplogroup D network (Fig. 3), reflecting relatively high genetic diversity (haplotype diversity = 0.89; SI Appendix, Table S9).
Despite extensive European settlement in the Pacific region over the last few centuries, many native chicken populations appear to contain relatively high frequencies of founding mitochondrial lineages—for example, the Marquesas, Solomon Islands, Vanuatu (26), and the Santa Cruz Islands—suggesting a high level of genetic continuity on these islands since prehistoric times. In addition to the two ancient haplotypes detected in modern samples, many other D haplotypes are also present in modern Pacific chicken populations, from the Santa Cruz Islands, Solomon Islands, and Vanuatu (26). Therefore, Polynesian chickens may be one of the few examples where ancestral genetic patterns can still be observed in a domesticated species. Chickens on remote Pacific islands may also contain Polynesian nuclear genomic lineages, and if so, would represent one of the few surviving examples of precolonial domestic chickens.
Conclusion
Although mtDNA lacks the power of genomic loci to reconstruct complex evolutionary histories, we show that an informative region of the chicken mitochondrial genome can be used to trace their human dispersal in the Pacific. The analysis of ancient and modern specimens reveals a unique Polynesian genetic signature, which can be traced back to ISEA, and promises to allow further resolution of migration and trading routes in the area. Importantly, we reveal that a previously reported connection between pre-European South America and Polynesian chickens most likely resulted from contamination with modern DNA, and that this issue is likely to confound ancient DNA studies involving haplogroup E chicken sequences. These observations reaffirm the potential of coalescent simulations of genetic data to evaluate new hypotheses regarding the dispersal of humans, commensals, and domesticates derived from archaeology. These hypotheses can be further grounded using genomic-scale studies in combination with direct dating and genetic investigation of new archaeological samples.
Materials and Methods
Samples.
Thirty-seven ancient chicken bones were collected for analysis, comprising eight from Niue, 11 from Hawai’i, and 18 from Rapa Nui excavated from deposits at Anakena by T.L.H. [including the six samples previously analyzed by Storey et al. (14); SI Appendix, Dataset S1]. Modern feather samples from ISEA and the Pacific (n = 124) were also examined to investigate current phylogeographic patterns (for location details, see Fig. 1 and SI Appendix, Figs. S3–S6, Table S1, and Dataset S6). The ancient samples were extracted, amplified (using primers in SI Appendix, Fig. S13), and sequenced at the Australian Centre for Ancient DNA (ACAD) in Adelaide, South Australia, according to a range of strict protocols (39), including numerous controls. Importantly, we included Shrimp DNase pretreatment in all PCR reactions, before adding template DNA, to remove any contaminating double-stranded DNA introduced via PCR reagents and plastic-ware (SI Appendix) (33). Independent external replication with direct sequencing of the PAQANA011 ancient sample was performed in a dedicated ancient DNA laboratory in the Archaeology Department at Durham University following strict laboratory procedures (39). The initial and independently replicated PCR fragments from bone sample PAQANA011 were also cloned and sequenced at the ACAD laboratories (SI Appendix, Dataset S4). Modern samples were extracted, with the highly variable 201 bp of the CR amplified and sequenced in a physically separate pre-PCR clean laboratory at the University of Adelaide and in the Archaeology Department at Durham University, following standard protocols (39).
WMG Analysis.
To determine the robustness of the current standard chicken phylogenetic framework for the analysis of the short ancient sequences, all 61 WMG sequences (25) were downloaded and aligned; PartitionFinder (40) was used to identify the number of preferred partitions and their substitution model; and phylogenetic trees were produced using both Bayesian (MrBayes v3.2; 41) and maximum likelihood estimation (RaxML v7.0.4; 42). See SI Appendix for more details.
CR Sequence Analysis.
In addition to the 144 CR sequences generated in this study, we downloaded 1,226 worldwide mtDNA CR chicken sequences from GenBank to establish the geographic distribution for each chicken haplogroup (14, 21–24, 26, 27, 43–46). To allow direct comparisons of the CR haplotypes, the 1,370 chicken sequences were aligned and trimmed to the highly variable 201 bp common to all of our 144 newly generated sequences (referred to as “201 bp CR dataset”). The 201 bp CR dataset was collapsed to unique haplotypes using Collapse v1.2, resulting in 274 unique haplotypes (H001–H274; SI Appendix, Dataset S6; referred to as “unique CR haplotype dataset”). ModelGenerator (47) was used to establish the best model to fit the unique CR haplotype dataset (GTR+I+G). The haplogroup of each of our 144 newly generated sequences was established by comparison with sequences of known haplogroup designation from Liu et al. (24) (SI Appendix, Dataset S6). As the majority of the new 144 CR sequences were identified as haplogroup D, a Median Joining Network (using Network v4.6; 48) was generated for just the D haplogroup (SI Appendix). All new sequences were uploaded to GenBank (KJ000585–KJ000642; SI Appendix, Dataset S6).
Statistical Analysis.
To examine the discrepancies between the composition and phylogeographic distribution of haplogroups reported by Storey et al. (14, 16) and those generated in this study, we tested the likelihood of detecting the reported proportions under different scenarios. A linear regression plot was also generated to visualize the correlation between occurrence of the four characteristic CR SNPs of the Polynesian chicken and longitude using the standard plotting function in R.
Bayesian Coalescent Simulations.
Given the importance of pre- and post-Columbian mtDNA sequences from Chile and Peru, respectively (14, 16), we tested whether coalescent simulations and approximate Bayesian computation of the 201 bp CR dataset could reconstruct a prehistoric link between the Pacific and South America (SI Appendix). To explore likely demographic histories for chickens in Micronesia and Polynesia, we also used BayeSSC to simulate alternate hypotheses of migration routes for comparison with the observed phylogeographic patterns within the Pacific.
Supplementary Material
Acknowledgments
We thank Jessica Metcalf, Peggy Macqueen, and other members of the Australian Centre for Ancient DNA for assistance; John Terrell for manuscript discussions; Richard Walter (Department of Anthropology, University of Otago) and Atholl Anderson (Australian National University) for providing access to the Niue samples; and Will Millard for collecting modern feather samples from New Guinea. G.L. and K.D. also thank Atholl Anderson, Hanneke Boon, James Wharram, Klaus Hymphendahl, Matt Fletcher, Ingo Isensee, and the Lapita Expedition for collecting feather samples from ISEA and Western Polynesia. This work was funded by the Australian Research Council and the University of Adelaide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KJ000585–KJ000642).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320412111/-/DCSupplemental.
References
- 1.Denham T, et al. Dating the appearance of Lapita pottery in the Bismarck Archipelago and its dispersal to Remote Oceania. Archaeology in Oceania. 2012;47(1):39–46. [Google Scholar]
- 2.Green RC. The Lapita Cultural Complex: Current evidence and proposed models. Indo-Pacific Prehistory Association Bulletin. 1991;11(1991):295–305. [Google Scholar]
- 3.Kirch P. The Lapita Peoples: Ancestors of the Oceanic World. Oxford: Blackwell; 1997. [Google Scholar]
- 4.Spriggs M. The Island Melanesians. Oxford: Blackwell; 1997. [Google Scholar]
- 5.Summerhayes G. Lapita Interaction. Canberra, Australia: Australian National Univ Press; 2000. [Google Scholar]
- 6.Wilmshurst JM, Hunt TL, Lipo CP, Anderson AJ. High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. Proc Natl Acad Sci USA. 2011;108(5):1815–1820. doi: 10.1073/pnas.1015876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke AC, Burtenshaw MK, McLenachan PA, Erickson DL, Penny D. SMBE Tri-National Young Investigators Proceedings of the SMBE Tri-National Young Investigators’ Workshop 2005. Reconstructing the origins and dispersal of the Polynesian bottle gourd (Lagenaria siceraria) Mol Biol Evol. 2006;23(5):893–900. doi: 10.1093/molbev/msj092. [DOI] [PubMed] [Google Scholar]
- 8.Whitaker T, Carter G. Oceanic drift of gourds—Experimental observations. Am J Bot. 1954;41(9):697–700. [Google Scholar]
- 9.Roullier C, Benoit L, McKey DB, Lebot V. Historical collections reveal patterns of diffusion of sweet potato in Oceania obscured by modern plant movements and recombination. Proc Natl Acad Sci USA. 2013;110(6):2205–2210. doi: 10.1073/pnas.1211049110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larson G, et al. Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proc Natl Acad Sci USA. 2007;104(12):4834–4839. doi: 10.1073/pnas.0607753104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larson G, et al. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc Natl Acad Sci USA. 2010;107(17):7686–7691. doi: 10.1073/pnas.0912264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savolainen P, Leitner T, Wilton AN, Matisoo-Smith E, Lundeberg J. A detailed picture of the origin of the Australian dingo, obtained from the study of mitochondrial DNA. Proc Natl Acad Sci USA. 2004;101(33):12387–12390. doi: 10.1073/pnas.0401814101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matisoo-Smith E, Robins JH. Origins and dispersals of Pacific peoples: Evidence from mtDNA phylogenies of the Pacific rat. Proc Natl Acad Sci USA. 2004;101(24):9167–9172. doi: 10.1073/pnas.0403120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storey AA, et al. Radiocarbon and DNA evidence for a pre-Columbian introduction of Polynesian chickens to Chile. Proc Natl Acad Sci USA. 2007;104(25):10335–10339. doi: 10.1073/pnas.0703993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storey AA, et al. Mitochondrial DNA from 3000-year old chickens at the Teouma site, Vanuatu. J Archaeol Sci. 2010;37(10):2459–2468. [Google Scholar]
- 16.Storey AA, et al. Investigating the global dispersal of chickens in prehistory using ancient mitochondrial DNA signatures. PLoS ONE. 2012;7(7):e39171. doi: 10.1371/journal.pone.0039171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storey AA, et al. Polynesian chickens in the New World: A detailed application of a commensal approach. Archaeology in Oceania. 2013;48(2):101–119. [Google Scholar]
- 18.Oskarsson MCR, et al. Mitochondrial DNA data indicate an introduction through Mainland Southeast Asia for Australian dingoes and Polynesian domestic dogs. Proc Biol Sci. 2012;279(1730):967–974. doi: 10.1098/rspb.2011.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muir WM, et al. Review of the initial validation and characterization of a 3K chicken SNP array. Worlds Poult Sci J. 2008;64(2):219–225. [Google Scholar]
- 20.Fumihito A, et al. Monophyletic origin and unique dispersal patterns of domestic fowls. Proc Natl Acad Sci USA. 1996;93(13):6792–6795. doi: 10.1073/pnas.93.13.6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanginakudru S, Metta M, Jakati RD, Nagaraju J. Genetic evidence from Indian red jungle fowl corroborates multiple domestication of modern day chicken. BMC Evol Biol. 2008;8:174. doi: 10.1186/1471-2148-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oka T, et al. Analysis of mtDNA sequences shows Japanese native chickens have multiple origins. Anim Genet. 2007;38(3):287–293. doi: 10.1111/j.1365-2052.2007.01604.x. [DOI] [PubMed] [Google Scholar]
- 23.Silva P, et al. Mitochondrial DNA-based analysis of genetic variation and relatedness among Sri Lankan indigenous chickens and the Ceylon junglefowl (Gallus lafayetti) Anim Genet. 2009;40(1):1–9. doi: 10.1111/j.1365-2052.2008.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu YP, et al. Multiple maternal origins of chickens: Out of the Asian jungles. Mol Phylogenet Evol. 2006;38(1):12–19. doi: 10.1016/j.ympev.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 25.Miao YW, et al. Chicken domestication: An updated perspective based on mitochondrial genomes. Heredity (Edinb) 2013;110(3):277–282. doi: 10.1038/hdy.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dancause KN, Vilar MG, Steffy R, Lum JK. Characterizing genetic diversity of contemporary pacific chickens using mitochondrial DNA analyses. PLoS ONE. 2011;6(2):e16843. doi: 10.1371/journal.pone.0016843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gongora J, et al. Indo-European and Asian origins for Chilean and Pacific chickens revealed by mtDNA. Proc Natl Acad Sci USA. 2008;105(30):10308–10313. doi: 10.1073/pnas.0801991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gongora J, et al. Reply to Storey et al.: More DNA and dating studies needed for ancient El Arenal-1 chickens. Proc Natl Acad Sci USA. 2008;105(48):E100. [Google Scholar]
- 29.Storey AA, et al. Pre-Columbian chickens, dates, isotopes, and mtDNA. Proc Natl Acad Sci USA. 2008;105(48):E99. doi: 10.1073/pnas.0807625105. author reply E100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonçalves VF, et al. Identification of Polynesian mtDNA haplogroups in remains of Botocudo Amerindians from Brazil. Proc Natl Acad Sci USA. 2013;110(16):6465–6469. doi: 10.1073/pnas.1217905110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellwood P. Prehistory of the Indo-Malaysian Archipelago. 3rd Ed. Canberra, Australia: Australian National Univ Press; 2007. p. 385. [Google Scholar]
- 32.Barker FK, Benesh MK, Vandergon AJ, Lanyon SM. Contrasting evolutionary dynamics and information content of the avian mitochondrial control region and ND2 gene. PLoS ONE. 2012;7(10):e46403. doi: 10.1371/journal.pone.0046403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Champlot S, et al. An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLoS ONE. 2010;5(9):e13042. doi: 10.1371/journal.pone.0013042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leonard J, et al. Animal DNA in PCR reagents plagues ancient DNA research. J Archaeol Sci. 2007;34(9):1361–1366. [Google Scholar]
- 35.Wickler S. Modelling colonisation and migration in Micronesia from a zooarchaeological perspective. In: Mondini M, Munoz S, Wickler S, editors. Colonisation, Migration, and Marginal Areas: A Zooarchaeological Approach. Oxford: Oxbow Books; 2004. pp. 28–40. [Google Scholar]
- 36.Carson M. Refining earliest settlement in Remote Oceania: Renewed archaeological investigation at Unai Bapot, Saipan. Journal of Island & Coastal Archaeology. 2008;3(1):115–139. [Google Scholar]
- 37.Anderson A. The rat and the octopus: Initial human colonization and the prehistoric introduction of domestic animals to Remote Oceania. Biol Invasions. 2009;11(7):1503–1519. [Google Scholar]
- 38.Addison DJ, Matisoo-Smith E. Rethinking Polynesians origins: A West-Polynesia Triple-I model. Archaeology in Oceania. 2010;45(1):1–12. [Google Scholar]
- 39.Cooper A, Poinar HN. Ancient DNA: Do it right or not at all. Science. 2000;289(5482):1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 40.Lanfear R, Calcott B, Ho SY, Guindon S. Partitionfinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29(6):1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 41.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 43.Adebambo A, et al. Lack of phylogeographic structure in Nigerian village chickens revealed by mitochondrial DNA D-loop sequence analysis. Int J Poult Sci. 2010;9(5):503–507. [Google Scholar]
- 44.Berthouly-Salazar C, et al. Vietnamese chickens: A gate towards Asian genetic diversity. BMC Genet. 2010;11:53. doi: 10.1186/1471-2156-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muchadeyi FC, et al. Mitochondrial DNA D-loop sequences suggest a Southeast Asian and Indian origin of Zimbabwean village chickens. Anim Genet. 2008;39(6):615–622. doi: 10.1111/j.1365-2052.2008.01785.x. [DOI] [PubMed] [Google Scholar]
- 46.Mwacharo JM, et al. Mitochondrial DNA reveals multiple introductions of domestic chicken in East Africa. Mol Phylogenet Evol. 2011;58(2):374–382. doi: 10.1016/j.ympev.2010.11.027. [DOI] [PubMed] [Google Scholar]
- 47.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6(29):29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 49.Specht J. Small islands in the big picture: The formative period of Lapita in the Bismarck Archipelago. In: Bedford S, Sand C, Connaughton SP, editors. Oceanic Explorations: Lapita and Western Pacific Settlement, Terra Australis. Vol 26. Canberra, Australia: Australian National Univ Press; 2007. pp. 51–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.