Significance
The human microbiota represents the trillions of bacteria that live on the skin, in the oral, nasal, and aural cavities, and throughout the gastrointestinal tract. The species that live in the gastrointestinal tract, the gut microbiota, closely interact with host cells and have a profound impact on health. To develop tools to effectively monitor the gut microbiota and ultimately help in disease diagnosis, we have engineered Escherichia coli to sense and record environmental stimuli, and demonstrated that E. coli with such memory systems can survive and function in the mammalian gut. This work demonstrates that E. coli can be engineered into living diagnostics capable of nondestructively probing the mammalian gut.
Keywords: synthetic biology, genetic switch
Abstract
The mammalian gut is a dynamic community of symbiotic microbes that interact with the host to impact health, disease, and metabolism. We constructed engineered bacteria that survive in the mammalian gut and sense, remember, and report on their experiences. Based on previous genetic memory systems, we constructed a two-part system with a “trigger element” in which the lambda Cro gene is transcribed from a tetracycline-inducible promoter, and a “memory element” derived from the cI/Cro region of phage lambda. The memory element has an extremely stable cI state and a Cro state that is stable for many cell divisions. When Escherichia coli bearing the memory system are administered to mice treated with anhydrotetracycline, the recovered bacteria all have switched to the Cro state, whereas those administered to untreated mice remain in the cI state. The trigger and memory elements were transferred from E. coli K12 to a newly isolated murine E. coli strain; the stability and switching properties of the memory element were essentially identical in vitro and during passage through mice, but the engineered murine E. coli was more stably established in the mouse gut. This work lays a foundation for the use of synthetic genetic circuits as monitoring systems in complex, ill-defined environments, and may lead to the development of living diagnostics and therapeutics.
The gut microbiota interacts closely with the host, impacting health, disease, and metabolism. Changes in its behavior can lead to liver disease, inflammatory/autoimmune disease, transfer of antibiotic resistance, obesity and diabetes, inflammatory bowel disease, pathogenic infections, and cancer (1–3). Our ability to nondestructively interrogate the gut is limited. Recent synthetic biology efforts have yielded a number of genetic circuits that can sense and remember environmental stimuli (4–11). These synthetic circuits stand ready for practical applications and have begun to be used as such.
One class of artificial gene-based memory systems uses bistable transcriptional switches to record transient environmental signals transmitted either directly through one of the transcription factors in the switch or indirectly through a distinct trigger element (12–16). Such memory devices may be used as sophisticated reporters to record a biological event in an organism and interrogate the system at a later time. For example, O’Gorman et al. (17) developed a system with a trigger element expressing a flippase (FLP) recombinase and a memory element in which recombination between a pair of FLP sites led to excision of a gene-disrupting element and expression of β-galactosidase gene (lacZ). Developmental biologists subsequently used such trigger-memory systems to mark cells at early times in development, followed by later examination and inference about the fate of cells that had activated a certain promoter at an earlier time (18, 19). More recently, Endy and colleagues characterized reversible DNA rearrangement systems based on inversion rather than deletion; such systems could be used for more complex calculators (20).
Purely transcription-based bistable memory systems have also been constructed. Gardner et al. (14) generated a “toggle switch” in which the lac repressor (lacI-) and tetR-encoded repressors inhibit the synthesis of the other protein, such that the system exists in two stable states that can be interchanged by environmental exposure to either isopropyl-β-d-thiogalactopyranoside or tetracycline. Ajo-Franklin et al. (21) developed a more general system in which a formally identified trigger element was separated from a bistable transcriptional memory element in yeast; in this way, a wide variety of input signals can be recorded using a single memory element with diverse trigger promoters. Burrill et al. (4) used this type of memory system to characterize gene-expression profiles in cells that responded differentially to a uniform exposure to DNA damaging agents. Thus, memory devices can be used in laboratory applications under controlled conditions.
Microbes carrying memory elements have potential for broad use as nondestructive environmental sensing systems. To realize this potential, such memory systems will need to be able to function in real-world environments beyond the controlled conditions of a laboratory. Thus, a memory device must be stable in either of two states for long periods of time, even in the presence of basal expression from a trigger element. DNA rearrangement systems may undergo an uninduced change of state resulting from leaky expression of a trigger element if the chances of such rearrangement increase linearly with expression levels. This change is a particular risk in microbes where basal trigger expression levels may be noisy, as opposed to developing animals where the highly cooperative nature of repressed chromatin makes it easy to achieve tight OFF states. Finally, the memory system should have no deleterious fitness effects, so the cost of gene expression should be very low.
Here, we show that Escherichia coli engineered with a synthetic memory system based on the phage lambda cI/Cro genetic switch can sense and record antibiotic exposure during passage through the mouse gut. This work lays the foundation for the use of synthetic genetic circuits in living diagnostics.
Results
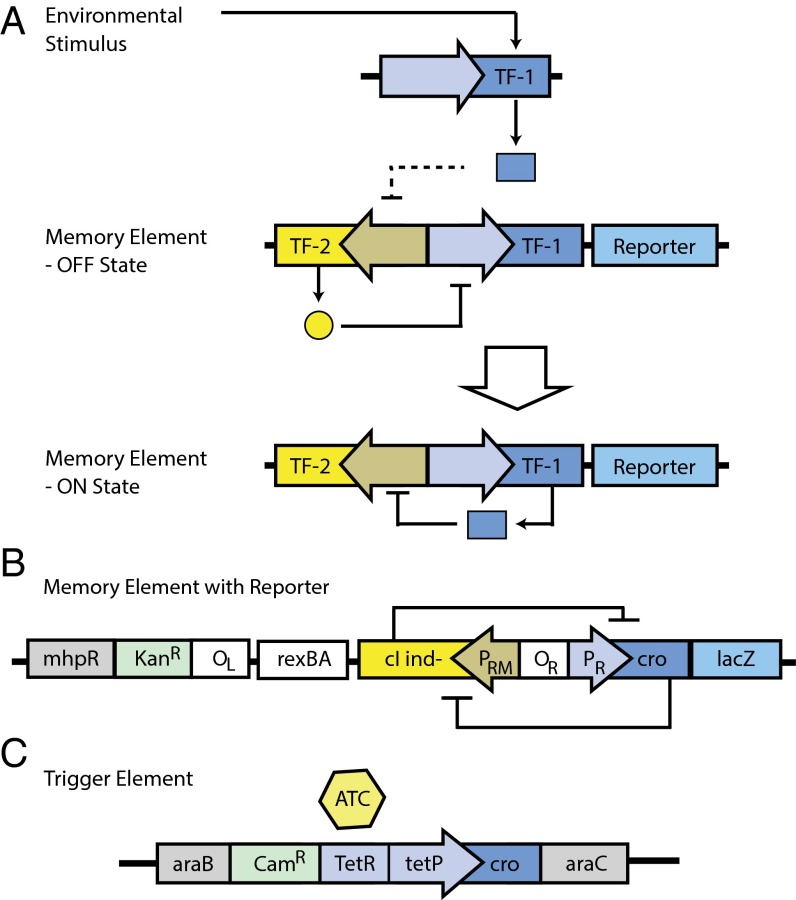
To engineer a bacterium to record an environmental signal in the mammalian gut, we set the following design specifications: (i) the initial “nonmemory” state should be highly stable, only failing as a result of mutation of the system; (ii) the “memory” state should also be highly stable; (iii) the engineered elements should be integrated into the chromosome rather than on plasmids to minimize the chance of loss; and (iv) the engineered elements should not impose a large fitness burden on the host (Fig. 1A).
Fig. 1.
Schematic of the memory circuit. (A) An abstraction of the genetic circuit used in this study combining the elements of a trigger/memory system and a toggle switch (4–10, 12–14, 17–23) (TF-1 and TF-2 symbolize generic antagonistic transcription factors). (B) The lambda cI/Cro-based transcriptional memory element used in this work. (C) The tetP-Cro trigger element used in this study. Construction of both elements is detailed in Methods.
We used the well-characterized cI/cro genetic switch from bacteriophage lambda (22–24) to construct a memory element for the circuit (Fig. 1B). Natural selection has already tuned the repressed cI state to be so stable that in an induction-deficient cIind- lysogen; the repressor state only fails because of spontaneous mutation of cI and not to fluctuations in cI protein levels (25). The presence of a lambda prophage causes little burden on the bacterial host as only 100–200 cI monomers per cell are present in a lysogen (26, 27). The well-characterized wild-type tetA promoter (tetP) was placed upstream of cro to generate a trigger element (Fig. 1C). The Tn10 tetracycline repressor is particularly sensitive to anhydrotetracycline (ATC), such that a low dose of 100 ng/mL ATC will cause full derepression of the promoter without inhibiting growth of tetracycline-sensitive E. coli (28).
To identify a well-behaved memory element, we constructed more than 10 candidate memory elements with the general structure as shown Fig. 1B, based on the elements of cI/cro regulation (22, 29). We inserted a DNA fragment from phage lambda from the left operator (OL), including the rexAB genes, cI, and cro, upstream of lacZ; these replaced lacI. The constructs should thus reproduce exactly the elements of cI expression, including the interaction between the OL-OR operator sites (29) and the natural downstream genes and terminators of the cI transcript, which may influence mRNA stability (Table 1 and Figs. S1–S4).
Table 1.
Strains used in this study
| Relevant Characteristics |
|||||
| Strain | Host | Trigger | Memory | rpsL | Source |
| PAS129 | MG1655 | araB::CAMR-tetP->cro | mphR::KanR-OL-rexBA -cI857-OR-cro-tR1::lacZ | This Study | |
| PAS130 | MG1655 | araB::CAMR-tetP->cro | mphR::KanR-OL-rexBA-cIind--OR-cro-tR1::lacZ | This Study | |
| PAS131 | MG1655 | araB::CAMR-tetP->cro | mphR::KanR-OL-rexBA-cI857-OR-cro::lacZ | This Study | |
| PAS132 | MG1655 | araB::CAMR-tetP->cro | mphR::KanR-OL-rexBA-cIind--OR-cro::lacZ | Lys42Arg | This Study |
| PAS133 | NGF-1 | araB::CAMR-tetP->cro | mphR::KanR-OL-rexBA-cIind--OR-cro::lacZ | Lys42Arg | This Study |
| TB10 | MG1655 | 32 | |||
E. coli MG1655 was also engineered to contain a trigger element driving Cro expression. tetP was placed upstream of cro, and a trigger element consisting of CAMR-tetR-tetP-cro was integrated between the divergent araC and araB promoters (Fig. S5) to minimize transcriptional effects from outside the element. When the concentration of cI falls below about 10% of its steady-state value in a lysogen (30), lambda switches from the lysogenic to lytic state, which leads to derepression of the PR promoter and the expression of Cro. When Cro levels reach about 100 molecules per cell, the activity of the PRM promoter decreases (30, 31). In the presence of Cro-mediated PRM repression, about four cell divisions are required for cI to be sufficiently diluted to allow switching from the cI state to the cro state (12, 23, 29). Thus, we expected that if tetP is induced via ATC for four consecutive cell divisions the memory element will switch from the cI state to the cro state.
The candidate memory elements were chromosomally inserted by “recombineering” into strain TB10, which automatically sets the element into the cI state (32). Upon removal of the prophage remnant in TB10 by P1vir transduction, several of these candidate memory elements showed frequent spontaneous switching to the cro state and were not characterized further. Four candidates (memory elements 11–14) (Figs. S1–S4) varying in the cI gene and the junction between cro and lacZ were characterized in detail (Fig. S6). Memory element 14, with a cIind- allele and the lacZ ATG, immediately follows the cro stop codon (TAA ATG) and was used in subsequent experiments.
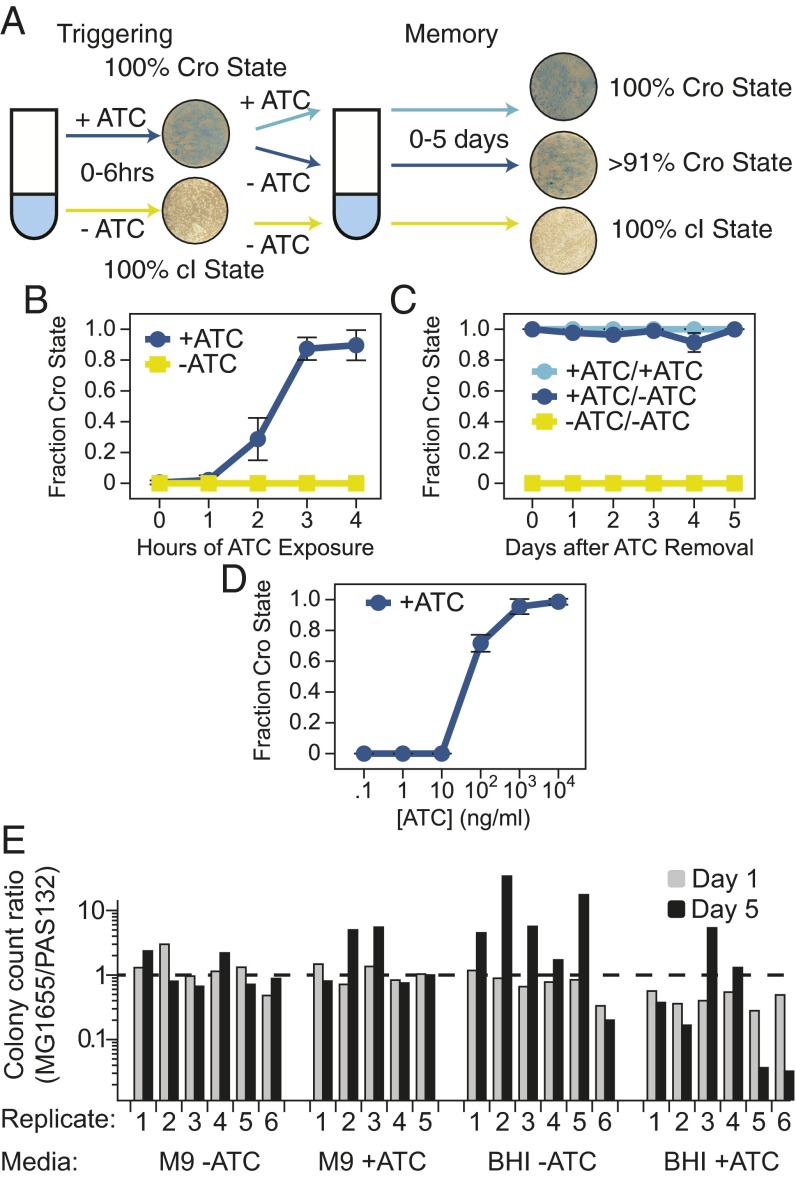
The genetic circuit consisting of the cI/cro memory element 14 and the tetP-cro trigger in strain PAS132 was capable of sensing and recording exposure to antibiotics (Fig. 2A). When these E. coli were exposed to ATC, they stably switched from the cI state to the Cro state after less than 4 h in cells grown in M9 glucose medium (Fig. 2B). This switching time is consistent with the memory element design, in which Cro expression from the trigger element represses further cI expression, and cI concentrations are diluted over about four cell divisions. After ATC removal, the memory element remained in the Cro state for at least 5 d of subculturing in M9 medium, representing about 150 cell divisions (Fig. 2C). The tetracycline repressor used here is particularly sensitive to ATC, such that 100 ng/mL ATC will derepress the promoter without inhibiting growth of tetracycline-sensitive E. coli (28) (Fig. 2 B and D). Previous reports noted that the Cro state of lambda N−O− prophages was unstable and spontaneously reverted to the cI state (12, 13). Those constructs did not express lambda N, causing the Cro transcripts to be terminated at tR1. Because PAS132 does not contain the natural post-cro termination sequences and the Cro transcript continues directly into lacZ, the half-life of the engineered transcript may correspond more closely to that of the longer N-antiterminated Cro transcript, which may be more stable and lead to higher Cro expression.
Fig. 2.
Engineered bacteria sense and remember ATC exposure in vitro. (A) Schematic of experimental design, as described in Methods. (B) In vitro triggering of PAS132. (C) In vitro memory of ATC exposure. (D) PAS132 was grown in liquid culture in the presence of ATC at the indicated concentration for 4 h, then plated on M9 glucose X-gal plates to determine the minimum dose of ATC required to switch the cells from the cI state to the Cro state. For all panels, means ± SD of three or more independent samples are shown. (E) Competitive growth assay to compare fitness of parental and engineered E. coli strains (Results, Methods). Mixed cultures of MG1655 rpsL and PAS132 were titered before (gray bars) and after (black bars) subculturing with a 1015-fold net serial dilution.
The trigger and memory elements were not significantly deleterious to growth of E. coli in diverse conditions, as inferred from competitive growth experiments in mixed cultures with the parental strain of E. coli (Fig. 2E) and growth curves (Fig. S7). Multiple independent mixed cultures with an initial ratio of about 1:1 E. coli MG1655strR and PAS132 were subcultured with and without ATC for about 50 cell divisions, and titered on indicator plates to distinguish the two strains. The change in ratios of parent cells to engineered cells varied from culture to culture but did not show a consistent overgrowth of parental cells (Fig. 2E). This observation suggests that in some cultures a spontaneous mutation enhancing growth under the conditions tested was arising in one strain or the other, and outgrowing the culture (33), rather than a fitness effect because of our engineered elements dominating the dynamics of the mixed cultures. These observations suggest that effects of the engineered elements on host fitness should not have a significant impact on detection of state switching.
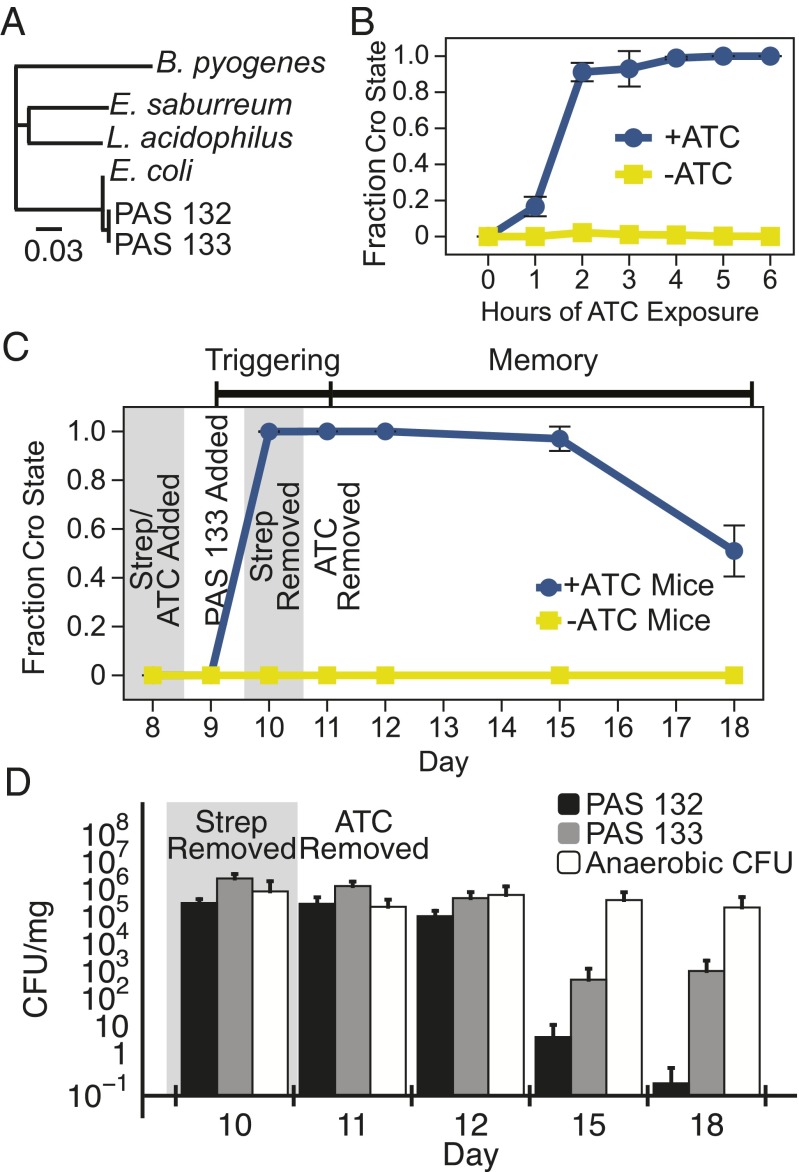
PAS132 detected ATC in the mammalian gut (Fig. 3A). To isolate E. coli containing genetic circuits after passage through the mouse gut, PAS132 contains a mutation in rpsL (Fig. S8) conferring resistance to >300 μg/mL streptomycin (34). Female Balb/C mice were given streptomycin (0.5 mg/mL in drinking water) to allow colonization by PAS132 (34); some mice also received ATC (0.1 mg/mL) in the water. About 107 bacteria were administered by oral gavage. Fecal samples were collected and titered on MacConkey lactose plates with streptomycin to select for PAS132, and on brain-heart infusion (BHI) plates (anaerobic) to titer culturable microbes. All of PAS132 isolated from mice that were given ATC stably switched from the cI state to the Cro state within 1 d of exposure (<1/104 Lac−) (Fig. 3B). The culturable endogenous gut flora began recolonizing the gut as soon as the streptomycin treatment ended, and the titer of the engineered bacteria decreased slowly thereafter (Fig. 3 C and D).
Fig. 3.
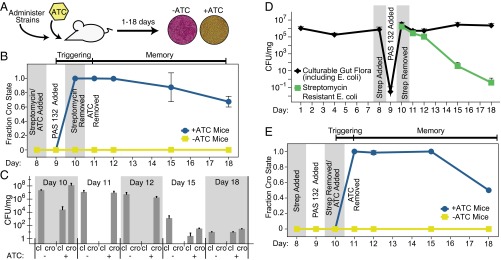
Engineered bacteria record, remember and report ATC exposure from the mammalian gut. (A) Schematic of experimental design, as described in Methods. (B) Mice were given ATC (0.1 mg/mL) and streptomycin (0.5 mg/mL) in drinking water on day 8. PAS132 cells were administered to the mice via oral gavage on day 9. Streptomycin and ATC were removed from the cage on days 10 and 11, respectively (means ± SD from four mice). (C) Total PAS132 cells in the cI and Cro states. Bars represent the average colony counts from four +ATC mice, and four −ATC mice. (D) The CFU of PAS132 and culturable bacteria (including PAS132) throughout the experiment (means ± SD from eight mice). (E) PAS132 cells were administered on day 9. ATC (0.1 mg/mL) was added to the drinking water on day 10 after streptomycin was removed. ATC was removed from the drinking water on day 11. All PAS132 cells triggered into the cro state within 1 d of ATC exposure, and remembered ATC exposure for more than 7 d (means ± SD from four mice).
PAS132 in ATC-treated mice remained in the Cro state for more than a week after termination of ATC treatment. After 24 h of ATC exposure, at which point 100% of the PAS132 in the mouse gut had switched to the Cro state (Fig. 3B), ATC was removed from the drinking water. More than 50% of the surviving PAS132 maintained a stable Cro memory state after more than a week in the mouse gut without further exposure to ATC (Fig. 3B).
In separate in vivo experiments, mice given ATC after PAS132 had colonized the mouse gut and streptomycin was removed (Fig. 3E). Again, PAS132 switched from the cI state to the Cro state within 24 h and remained in the Cro state for several days (Fig. 3E). This result indicates that PAS132 cells that have already colonized the gut are able to record subsequent changes to their environment. Mouse weight was not affected by antibiotic treatment or administration of PAS132 (Fig. S9), indicating that these bacteria are not grossly deleterious to their host. After ATC removal, there was not sufficient ATC in the filtered fecal samples to activate the memory circuit in cultured PAS132. Tetracycline (Tc) is undetectable in the serum, kidneys, and liver of female mice after less than 8 h of administration (35). Thus, the ATC was likely cleared from the mouse when we evaluated our engineered bacteria for memory at later times.
The genetic memory circuit functioned essentially identically in an uncharacterized coliform bacterium from the mouse gut. From an untreated mouse we isolated a bacterium that fermented lactose on MacConkey Lactose plates, and confirmed that its 16S ribosomal RNA matched that of E. coli (Fig. 4A and Fig. S10). P1vir transduction was used to insert the memory circuit, trigger, and streptomycin resistance mutation into this isolate from natural gut flora, termed NGF-1. The engineered NGF-1 strain (PAS133) behaved similarly to the engineered K12 strain PAS132 in vitro, registering ATC exposure within 4 h (Fig. 4B).
Fig. 4.
Memory behavior of an endogenous murine E. coli strain engineered to contain the memory circuit. (A) The 16S ribosomal subunits of PAS132 and PAS133 were sequenced and compared against known gut microbes (46). (B) PAS133 was grown in M9 glucose + casamino acids liquid medium +ATC (100 ng/mL), or −ATC for 0–6 h. PAS133 was unable to grow in M9 glucose media without casamino acids. Aliquots were titered hourly on MacConkey Lactose plates to evaluate switching from the cI state to the cro state in response to ATC (means ± SD of three independent samples). (C) PAS133 was administered by oral gavage to mice exposed to antibiotics, and gut flora were characterized following the same protocol as in Fig. 3. On day 11 the ATC was removed from the cage (means ± SD from four mice). (D) Comparison of survival of PAS133 and PAS132, engineered E. coli K12 in the mouse gut. Shown are PAS132, PAS133, and anaerobic gut flora CFU per milligram of fecal sample on corresponding days (means ± SD from eight mice).
PAS133 sensed and remembered exposure to ATC in the mouse gut. Female Balb/C mice were given PAS133 and treated as described above. Cells were collected and analyzed as above for lacZ expression on lactose indicator streptomycin plates. PAS133 detected ATC exposure within 1 d, and remembered exposure of mice to ATC for more than 7 d after ATC withdrawal (Fig. 4C). Moreover, PAS133 stably colonized the mouse gut longer than PAS132, the K12-derived strain. Although we administered roughly equal amounts of PAS132 and PAS133, after 1 d in the mouse we recovered 10-fold more PAS133 per milligram of fecal sample (Fig. 4D). Between 5 and 8 d postinoculation, the PAS133 population stabilized to around 1,000 colony-forming units per milligram, which was comparable to the coliform titers in most of the pretreated mice obtained from Charles River Laboratories over the course of these experiments. In contrast, PAS132, the K12 based-strain, was almost completely outcompeted by the natural gut flora after 8 d.
Discussion
Microbe-based recording systems have the potential to be used as diagnostics in health care, environmental monitoring, and other applications. In this work, we sought to construct a bacterium that could sense and record exposure to a chemical cue, ATC, in the mammalian gut. Based on the strengths and limitations of previously constructed genetic memory systems, we built a system in E. coli strains based on the bistable lambda cI/Cro switch (4–10, 12–14, 17–23, 31). This circuit was designed to begin in the cI state and switch to the Cro state upon induction of a “trigger” element in which a tetracycline-responsive promoter directs transcription of a cro gene. We demonstrated that the cI and Cro states were stable both in bacterial cultures and when the bacteria were passaged through the mouse gut. Switching from the cI to the Cro state occurred efficiently upon exposure to ATC in E. coli in laboratory culture or in the mouse gut. In addition, the entire circuit was transferred from E. coli K12 to an uncharacterized murine E. coli strain; in this context the system behavior was virtually identical, but the engineered murine strain was more stably established in the mouse gut compared with the engineered K12 strain.
DNA rearrangement, such as deletion, is an alternative method for genetic information storage, with the obvious advantage that the rearranged state is stable. Developmental biologists previously used trigger elements based on expression of a FLP recombinase from a promoter of interest, followed by permanent excision of a DNA to mark the event in daughter cells for fate-mapping (18, 19). These studies were carried out in the mouse, in which the expression of developmental regulatory proteins is tightly controlled and thus false-positive deletions because of low-level expression of FLP may have been avoided. Mammalian genes are generally highly cooperative, in part a result of self-reinforcing states of chromatin that involve large numbers of adjacent nucleosomes. Eubacteria lack these mechanisms and it remains to be seen if the “transfer function” relating levels of FLP protein expression to DNA excision is well-behaved.
In the context of a microbial gene-based sensor, stability of the starting state is particularly important so that false-positives can be avoided, and low-level stochastic expression from a trigger element must not be enough to flip the switch. The natural lambda-epigenetic switch is extremely stable. Gimble and Sauer (25) noted that lambda-lysogens with a DNA damage-insensitive cIind- mutation only spontaneously induce via inactivating a mutation in cI. Thus, “epigenetic failure,” which might be the result of a rare stochastic failure to express enough cI protein or segregate it evenly to both daughter cells, was not observed. Natural selection may have acted on this system to be particularly tightly regulated, with a highly cooperative structure defining the repressed state, involving eight cI proteins bound to operators left (OL) and right (OR) with DNA looping (29).
The use of the lambda cI/Cro system as a memory element also had the advantages that it imposed a low burden on the host and was largely predesigned by nature. In the lysogenic state, the 256-AA lambda cI protein is only present in 100–200 copies per cell (23), whereas the 66-AA Cro protein is likely present in <1,000 copies per cell (31); thus, the burden of expressing these proteins was expected to be minimal. We found no detectable selective burden on hosts carrying the trigger-memory system in either epigenetic state (Fig. 2E).
The natural design of the cI/Cro element directly provides genetic circuit builders with a highly stable cI state. In our most successful memory elements, we reproduced the cI control elements as precisely as possible, even including the rexA and rexB genes because, although they have no functional role in the circuit, their presence may influence the stability of the cI transcript.
In contrast, the precise sequence requirements for a stable Cro state were not clear when we started. The Cro state in a single-copy anti-immune lysogen is unstable (12). In the natural Cro state, the phage is growing lytically and its DNA is replicating. The element with the most stable Cro state lacks the tR1 terminator at the end of the Cro gene; this Cro mRNA thus appears be more stable and may correspond to the N-antiterminated longer transcript. This system has a completely stable cI state and a Cro state that is stable over many cell divisions (Figs. 2–4). For environmental detection and recording, this behavior will often be adequate because the presence of state-switched bacteria will definitively indicate that a detection event has occurred.
When orally administered to mice and then recovered, E. coli cells carrying the memory circuit responded to ATC administered to the mice by switching the memory element into the Cro state. This result occurred efficiently in several experimental permutations: for example, ATC was administered for varying periods during or after establishment of the bacteria in the gut. The complete circuit worked in both E. coli K12 and in an uncharacterized E. coli strain isolated from a mouse during the course of the experiments. The cI state was completely stable, and Cro state was stable for many days in the context of the mouse gut. It is particularly noteworthy that the memory system behaved as desired in an uncharacterized murine bacterium, which colonized the mouse gut more stably than E. coli K12. These results indicate that artificial genetic circuits can be designed and characterized in well-understood but attenuated laboratory strains, and then transferred to a related isolate from the environment of interest.
The ability to engineer bacteria to report on environmental cues in variable, incompletely understood environments has implications for the development of living diagnostics and therapeutics. For example, bacteria native to the human gut or skin might be used to monitor for exposure to chemicals characteristic of specific disease states. The system described herein is sufficiently modular that the trigger and memory circuits could be reengineered to respond to chemical signatures of inflammation, cancer, parasites, or environmental toxins in the gut. In combination with additional genetic circuits, such as the recent search-and-destroy circuits (36–38), cells could be designed with the memory element described here to diagnose a specific pathogen, and emit a therapeutic. Together, these approaches may allow construction of a new class of engineered probiotic bacteria that serve as benign and transient diagnostics and therapeutics.
Methods
Strain Construction.
The chromosomally integrated memory and trigger elements were constructed by a combination of commercial synthesis (Genscript) and PCR amplification of component elements from source DNAs, assembly in vitro through overlap extension PCR (39), and introduction directly into E. coli TB10 (40) by recombineering without plasmid intermediates (32). A spontaneous high-level streptomycin-resistance mutation was isolated in MG1655 and confirmed to be rpsL (lys42arg) (41, 42). Memory and trigger elements and the rpsL mutation were moved between strains by P1vir transduction (43).
Memory element 14 consists of a kanamycin-resistance cassette transcribed away from cI and cro, and phage lambda sequences from 35561 to 38241, including the cIind- mutation (44). This DNA was inserted between bases 366802 and 365529 in the E. coli K12 MG1655 genome (45). The resulting construct contains a potential terminator downstream of the mhp gene upstream of lacI, but lacks sequences from the lacI promoter up to the start codon for lacZ. These genes are replaced by phage sequences, including PL, OL, rexB, rexA, cI, OR, and cro through the cro stop codon, such that lacZ is now transcribed from PR after cro. Memory elements 11–13 were constructed similarly (Figs. S1–S4).
The trigger element consists of a chloramphenicol-resistance cassette, a tetR-tetP segment from Tn10 that includes the divergent tetracycline promoters, and the cro gene transcribed from the tetA promoter. This segment was inserted into the MG1655 genome at base 70165, in a CAP binding site between araB and araC promoters to minimize aberrant read-through from external promoters (Fig. S5).
Induction of Cro Expression with ATC (Triggering).
Overnight M9 glucose cultures of engineered bacteria in were diluted 100-fold into the same medium with or without ATC (100 ng/mL) and grown aerobically with shaking for up to 6 h. For each condition, at least three cultures were tested. At the indicated times, aliquots were diluted and plated on M9 glucose X-gal plates to evaluate the fraction of cells that switched from the cI state to the cro state in response to ATC.
Maintenance of the Cro State in the Absence of Inducer (Memory).
PAS132 that had been induced into the cro state by prolonged exposure to ATC were pelleted, washed, resuspended in M9 glucose media without ATC, and grown aerobically with shaking for up to 5 d, with 1,000-fold dilutions performed every 8 h. At the indicated times, aliquots were plated on M9 glucose X-gal plates to evaluate the fraction of cells that remained in the cro state. PAS132 continuously grown in the presence of ATC or never exposed to ATC were used as controls.
Competitive Growth Assays.
To compare the relative growth rates of engineered and nonengineered bacteria, mixed cultures were grown as follows. Six overnight cultures of MG1655rpsLstrR and six of PAS132rpsLstrR were grown in M9 glucose or BHI media, from which six mixed cultures were created in the same media with and without 100 ng/mL ATC. Cultures were titered on indicator plates immediately after mixing, and after 5 d of daily growth to saturation followed by 1,000-fold dilution.
Analysis of Engineered Bacteria in Mouse Fecal Samples.
Because E. coli administered orally to mice will generally not colonize the gut unless the endogenous bacteria are displaced, we followed the protocol of Foucault et al. (34), in which mice are given a low dose of streptomycin in drinking water to reduce the endogenous flora. Before administration of engineered bacteria, fecal samples were collected from acclimated female BALB/c mice (Charles River Laboratories) on days 1, 4, and 8, weighed, solubilized in 0.85% NaCl, and titered on anaerobic BHI plates and on aerobic MacConkey Lactose Streptomycin plates at 37 °C. On day 8 the mice were fasted overnight and given water with 5% sucrose, 0.5mg/mL streptomycin (34), with or without 0.1 mg/mL ATC. On day 9, 107 engineered bacteria were administered by oral gavage, and food was returned. On day 10 the streptomycin was removed from the water. On day 11 the ATC and sucrose were removed from the water. Throughout all experiments, mice were fed a grain-based chow without lactose. Each mouse experiment represents the cumulative data from eight mice; four without and four with ATC. For some experiments, fecal samples were tested for residual activity by spotting filter-sterilized solubilized fecal samples onto indicator plates spread with PAS132; no ATC activity was observed. The animal protocol was approved by the Harvard Medical Area Standing Committee on Animals, protocol 04966.
Supplementary Material
Acknowledgments
We thank Amanda Graveline, DVM for her assistance and training with the mouse experiments, especially with oral gavage. This work was supported by Defense Advanced Research Projects Agency Grant N66001-11-C-4203 and funds from the Wyss Institute for Biologically Inspired Engineering.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321321111/-/DCSupplemental.
References
- 1.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336(6086):1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Modi SR, Lee HH, Spina CS, Collins JJ. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499(7457):219–222. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrill DR, Inniss MC, Boyle PM, Silver PA. Synthetic memory circuits for tracking human cell fate. Genes Dev. 2012;26(13):1486–1497. doi: 10.1101/gad.189035.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon TS, Lou C, Tamsir A, Stanton BC, Voigt CA. Genetic programs constructed from layered logic gates in single cells. Nature. 2012;491(7423):249–253. doi: 10.1038/nature11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying genetic logic gates. Science. 2013;340(6132):599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- 7.Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. 2013;31(5):448–452. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- 8.Friedland AE, et al. Synthetic gene networks that count. Science. 2009;324(5931):1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi H, et al. Programmable cells: Interfacing natural and engineered gene networks. Proc Natl Acad Sci USA. 2004;101(22):8414–8419. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litcofsky KD, Afeyan RB, Krom RJ, Khalil AS, Collins JJ. Iterative plug-and-play methodology for constructing and modifying synthetic gene networks. Nat Methods. 2012;9(11):1077–1080. doi: 10.1038/nmeth.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia JR, Cha HJ, Rao G, Marten MR, Bentley WE. Microbial nar-GFP cell sensors reveal oxygen limitations in highly agitated and aerated laboratory-scale fermentors. Microb Cell Fact. 2009;8:6. doi: 10.1186/1475-2859-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neubauer Z, Calef E. Immunity phase-shift in defective lysogens: Non-mutational hereditary change of early regulation of lambda prophage. J Mol Biol. 1970;51(1):1–13. doi: 10.1016/0022-2836(70)90265-2. [DOI] [PubMed] [Google Scholar]
- 13.Toman Z, Dambly-Chaudière C, Tenenbaum L, Radman M. A system for detection of genetic and epigenetic alterations in Escherichia coli induced by DNA-damaging agents. J Mol Biol. 1985;186(1):97–105. doi: 10.1016/0022-2836(85)90260-8. [DOI] [PubMed] [Google Scholar]
- 14.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403(6767):339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 15.Wittrup KD, Bailey JE. Mathematical modeling of a single-cell enzyme assay. Biotechnol Bioeng. 1990;35(5):525–532. doi: 10.1002/bit.260350511. [DOI] [PubMed] [Google Scholar]
- 16.Wittrup KD, Bailey JE, Ratzkin B, Patel A. Propagation of an amplifiable recombinant plasmid in Saccharomyces cerevisiae: Flow cytometry studies and segregated modeling. Biotechnol Bioeng. 1990;35(6):565–577. doi: 10.1002/bit.260350604. [DOI] [PubMed] [Google Scholar]
- 17.O’Gorman SF, Fox DT, Wahl GM. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251(4999):1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 18.Dymecki SMT, Tomasiewicz H. Using Flp-recombinase to characterize expansion of Wnt1-expressing neural progenitors in the mouse. Dev Biol. 1998;201(1):57–65. doi: 10.1006/dbio.1998.8971. [DOI] [PubMed] [Google Scholar]
- 19.Zinyk DLM, Mercer EH, Harris E, Anderson DJ, Joyner AL. Fate mapping of the mouse midbrain-hindbrain constriction using a site-specific recombination system. Curr Biol. 1998;8(11):665–668. doi: 10.1016/s0960-9822(98)70255-6. [DOI] [PubMed] [Google Scholar]
- 20.Bonnet J, Subsoontorn P, Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc Natl Acad Sci USA. 2012;109(23):8884–8889. doi: 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ajo-Franklin CM, et al. Rational design of memory in eukaryotic cells. Genes Dev. 2007;21(18):2271–2276. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ptashne M, et al. How the lambda repressor and cro work. Cell. 1980;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- 23.Shea MA, Ackers GK. The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J Mol Biol. 1985;181(2):211–230. doi: 10.1016/0022-2836(85)90086-5. [DOI] [PubMed] [Google Scholar]
- 24.Arkin A, Ross J, McAdams HH. Stochastic kinetic analysis of developmental pathway bifurcation in phage lambda-infected Escherichia coli cells. Genetics. 1998;149(4):1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gimble FS, Sauer RT. Mutations in bacteriophage lambda repressor that prevent RecA-mediated cleavage. J Bacteriol. 1985;162(1):147–154. doi: 10.1128/jb.162.1.147-154.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reichardt L, Kaiser AD. Control of lambda repressor synthesis. Proc Natl Acad Sci USA. 1971;68(9):2185–2189. doi: 10.1073/pnas.68.9.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bentley WE, Mirjalili N, Andersen DC, Davis RH, Kompala DS. Plasmid-encoded protein: The principal factor in the “metabolic burden” associated with recombinant bacteria. Biotechnol Bioeng. 1990;35(7):668–681. doi: 10.1002/bit.260350704. [DOI] [PubMed] [Google Scholar]
- 28.Moyed HSB, Bertrand KP. Mutations in multicopy Tn10 tet plasmids that confer resistance to inhibitory effects of inducers of tet gene expression. J Bacteriol. 1983;155(2):557–564. doi: 10.1128/jb.155.2.557-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB. Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes Dev. 2001;15(22):3013–3022. doi: 10.1101/gad.937301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailone A, Levine A, Devoret R. Inactivation of prophage lambda repressor in vivo. J Mol Biol. 1979;131(3):553–572. doi: 10.1016/0022-2836(79)90007-x. [DOI] [PubMed] [Google Scholar]
- 31.Morelli MJ, Ten Wolde PR, Allen RJ. DNA looping provides stability and robustness to the bacteriophage lambda switch. Proc Natl Acad Sci USA. 2009;106(20):8101–8106. doi: 10.1073/pnas.0810399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomason L, et al. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol. 2007;Chapter 1:16. doi: 10.1002/0471142727.mb0116s78. [DOI] [PubMed] [Google Scholar]
- 33.Novick A, Szilard L. Experiments with the Chemostat on spontaneous mutations of bacteria. Proc Natl Acad Sci USA. 1950;36(12):708–719. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foucault ML, Thomas L, Goussard S, Branchini BR, Grillot-Courvalin C. In vivo bioluminescence imaging for the study of intestinal colonization by Escherichia coli in mice. Appl Environ Microbiol. 2010;76(1):264–274. doi: 10.1128/AEM.01686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Böcker R, Warnke L, Estler CJ. Blood and organ concentrations of tetracycline and doxycycline in female mice. Comparison to males. Arzneimittelforschung. 1984;34(4):446–448. [PubMed] [Google Scholar]
- 36.Gupta S, Bram EE, Weiss R. Genetically programmable pathogen sense and destroy. ACS Synth Biol. 2013;2(12):715–723. doi: 10.1021/sb4000417. [DOI] [PubMed] [Google Scholar]
- 37.Hwang IY, et al. ACS Synth Biol. 2013. Reprogramming microbes to be pathogen-seeking killers. [DOI] [PubMed] [Google Scholar]
- 38.Duan F, March JC. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc Natl Acad Sci USA. 2010;107(25):11260–11264. doi: 10.1073/pnas.1001294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timms AR, Steingrimsdottir H, Lehmann AR, Bridges BA. Mutant sequences in the rpsL gene of Escherichia coli B/r: Mechanistic implications for spontaneous and ultraviolet light mutagenesis. Mol Gen Genet. 1992;232(1):89–96. doi: 10.1007/BF00299141. [DOI] [PubMed] [Google Scholar]
- 42.Springer B, et al. Mechanisms of streptomycin resistance: Selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob Agents Chemother. 2001;45(10):2877–2884. doi: 10.1128/AAC.45.10.2877-2884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller JH. 1972. Experiments in Molecular Genetics (Cold Spring Harbor Lab Press, Cold Spring Harbor, N.Y.), pp xvi, 466 pp.
- 44.Sanger F, Coulson AR, Hong GF, Hill DF, Petersen GB. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982;162(4):729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 45.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277(5331):1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 46.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173(2):697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.