Significance
More than 500 million people are persistently infected with hepatitis B virus (HBV) and/or hepatitis C virus (HCV) and are at a risk of developing chronic hepatitis, cirrhosis, and liver cancer. The absence of robust cell culture systems for both viral infections limits the understanding of the virus lifecycle and pathogenesis required for the development of vaccine and antivirals. We have established a novel human hepatoma cell line termed “HLCZ01” that supports the entire lifecycle of both HBV and HCV produced both in cell culture and clinically. This cell line provides a powerful tool for addressing the virus lifecycle and the development of antivirals and vaccines.
Keywords: cell culture model, primary human hepatocytes, cccDNA, interferon, ISGs
Abstract
The absence of a robust cell culture system for hepatitis B virus (HBV) and hepatitis C virus (HCV) infection has limited the analysis of the virus lifecycle and drug discovery. We have established a hepatoma cell line, HLCZ01, the first cell line, to the authors’ knowledge, supporting the entire lifecycle of both HBV and HCV. HBV surface antigen (HBsAg)-positive particles can be observed in the supernatant and the lumen of the endoplasmic reticulum of the cells via electron microscopy. Interestingly, HBV and HCV clinical isolates propagate in HLCZ01 cells. Both viruses replicate in the cells without evidence of overt interference. HBV and HCV entry are blocked by antibodies against HBsAg and human CD81, respectively, and the replication of HBV and HCV is inhibited by antivirals. HLCZ01 cells mount an innate immune response to virus infection. The cell line provides a powerful tool for exploring the mechanisms of virus entry and replication and the interaction between host and virus, facilitating the development of novel antiviral agents and vaccines.
More than 500 million people worldwide are persistently infected with hepatitis B virus (HBV) and/or hepatitis B virus (HCV) and are at risk of developing chronic liver diseases (1). There is no vaccine against HCV, and many patients who are persistently infected by HBV or HCV do not respond to currently available therapies (2, 3). Improved understanding of the biology and pathogenesis of these infections is required for the development of vaccine and antiviral drugs (4). The inability to grow HBV and HCV efficiently in cell culture has presented a major obstacle to understanding the virus lifecycle and pathogenesis and to developing improved therapeutics.
HBV is a member of the hepadnavirus families, and its genome is a relaxed circular, partially double-stranded DNA molecule. The negative strand has an invariable length of ∼3.2 kb, and the positive strand is 50–100% of this length. Several key issues about the biology of HBV remain to be explored, including the identification of the cellular receptors, the role of the X gene, and the mechanisms by which the viral minichromosome is formed. Covalently closed circular DNA (cccDNA) is responsible for the establishment of viral infection and persistence. Understanding the mechanisms underlying cccDNA formation and regulation is critical for understanding the HBV pathogenesis and finding a cure for hepatitis B. HepG2.2.15 cells derived from the hepatoma cell line HepG2 transfected with the full genome of HBV have been used to study HBV replication (5). Primary human hepatocytes (PHH) are susceptible to HBV infection (6, 7), but the use of this model is hampered by the limited availability and unpredictable variability of human liver. Several human hepatoma cell lines support HBV replication after HBV DNA transfection, and overexpression of sodium-taurocholate cotransporting polypeptide (NTCP) in HepG2 and Huh7 cells can render these cells able to support HBV produced in cell culture at low efficiency (5, 8, 9). HepaRG is a hepatoma cell line that is susceptible to HBV infection (10), but the susceptibility of HepaRG cells to HBV is strictly dependent on the differentiation state induced by DMSO, causing variable toxic side effects and irreproducible results for HBV replication levels. Moreover, poor viral replication, low viral yields, the absence of reinfection, and lack of cccDNA amplification in HepaRG cells make the study of HBV lifecycle and pathogenesis difficult (10). For reasons that still are unknown, HBV clinical isolates do not propagate in cell culture.
HCV is a positive-strand RNA virus of the Flaviviridae family. Its genome encodes a large polyprotein that is processed to produce viral structural proteins including core, E1, and E2 proteins as well as nonstructural proteins consisting of p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B. Genotype 2a, 2b, and 1a isolates were used to develop infectious cell culture systems for HCV study (11–15). However, other HCV genotypes cannot propagate in vitro. The permissive human hepatoma cell Huh7.5 may not mount an innate immune response to HCV infection, so it is difficult to explore the interaction between the virus and host cells. Moreover, clinical isolates of HCV do not propagate in cell culture. Although primary culture systems have been established for HCV (16, 17), these do not seem to extend the range of isolates that can be studied. We now report the establishment of a novel hepatoma cell line, HLCZ01, supporting the entire life cycles of HBV and HCV. Interestingly, the cells can be infected by the sera from patients with different HBV and HCV strains.
Results
Establishment of a Novel Hepatoma Cell Line, HLCZ01.
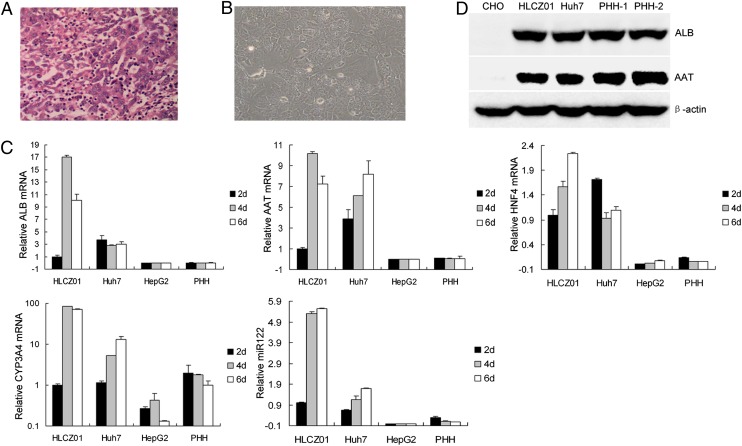
To understand better the virus lifecycle and the interaction between host and virus, we attempted to replicate HBV and HCV in hepatoma cells, which have histological features characteristic of hepatocellular carcinoma (Fig. 1A). Tumor tissue was harvested and cultured in DMEM/F12 medium, and several clones survived for long-time culture. We One of these clones, HLCZ01 (Fig. 1B), was derived from the grade 2 differentiated hepatocellular carcinoma of an HCV-infected male patient. To obtain hepatoma cell lines, we injected clones into immunodeficient mice and cultured tumor tissues from the mice in DMEM/F12 medium. HLCZ01 cells express liver-specific genes, such as human albumin (ALB), α1-antitrypsin (AAT), hepatocyte nuclear factor 4 (HNF4), cytochrome P450 3A4 (CYP3A4), and microRNA122 (miR-122) (Fig. 1C), and the liver-specific proteins human ALB and AAT (Fig. 1D). The presence of HCV RNA is not detectable by RT-PCR after the establishment of HLCZ01 cells in culture.
Fig. 1.
Establishment of a novel hepatoma cell line HLCZ01. (A) H&E-stained section of moderately differentiated hepatocellular carcinoma from a surgical resection specimen. (B) Morphology of the HLCZ01 cell line derived from hepatocellular carcinoma. (C) Detection of the markers of human hepatocytes by real-time PCR. Total cellular RNA was isolated from HLCZ01, Huh7, and HepG2 cells and from PHH. Human AAT, ALB, HNF4, and CYP3A4 mRNA were detected by real-time-PCR and normalized with GAPDH; miR-122 was determined by real-time PCR and normalized to U6. (D) HLCZ01 cells express the liver-specific proteins AAT and ALB. Protein was isolated from HLCZ01, Huh7, and CHO cells and from PHH. AAT and ALB were detected by Western blot.
Infection of HLCZ01 Cells by HBV.
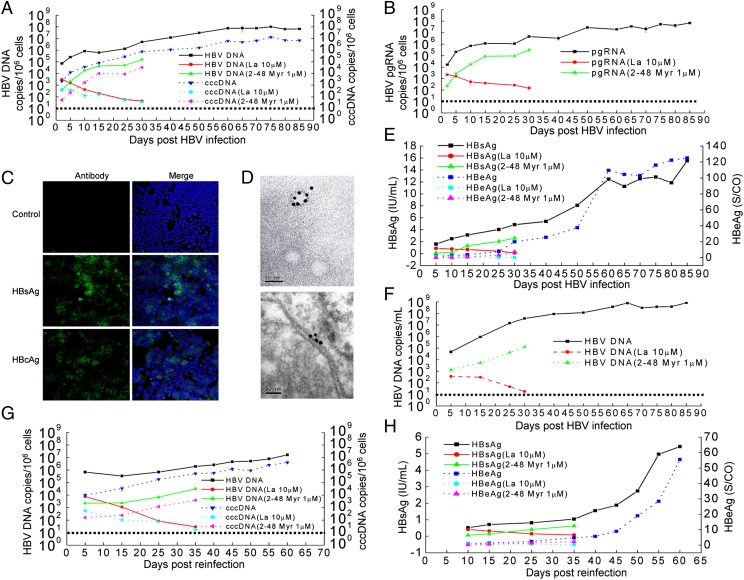
To determine whether the various hepatoma cell lines we established are permissive for HBV infection, we inoculated those cell lines with the filtered supernatant of HepG2.2.15 cells producing infectious HBV. Intracellular viral DNA and cccDNA could be detected only in HLCZ01 cells (Fig. 2A and Fig. S1A). When we attempted to infect Huh7 cells under identical conditions, no signal corresponding to viral DNA could be observed (Fig. S1 A and B). Similarly, we confirmed the presence of the pregenomic RNA of the virus in HLCZ01 cells inoculated with HBV (Fig. 2B and Fig. S1C), whereas viral RNA could not be detected in Huh7 cells under similar conditions (Fig. S1 C and D). For comparison with other HBV-permissive cells, we inoculated PHH with the supernatant of HepG2.2.15 cells. HBV DNA could be identified in PHH inoculated with the supernatant of HepG2.2.15 cells (Fig. S1F). HBV surface antigen (HBsAg) and HBV core antigen (HBcAg)-positive HLCZ01 cells could be observed clearly (Fig. 2C). Around 3.2% of HLCZ01 cells were infected at 5 d postinfection (dpi), and HBV-infected cells increased up to 30% at 65 dpi (Fig. S2). HBsAg-positive particles were observed in the supernatant and in the lumen of the endoplasmic reticulum of HBV-infected HLCZ01 cells under electron microscopy (Fig. 2D and Fig. S1G).
Fig. 2.
Infection of HLCZ01 cells by HBV. (A) Kinetics of intracellular viral DNA and cccDNA in HLCZ01 cells inoculated with the supernatant of HepG2.2.15 cells. The supernatant from HepG2.2.15 cells was mixed with the HBV large-surface, protein-derived peptide (amino acids 2–48). Then HLCZ01 cells were inoculated with the mixture at an MOI of 20 Geq per cell. Alternately, HLCZ01 cells were inoculated with the supernatant of HepG2.2.15 cells at an MOI of 20 Geq per cell and were cultured in the presence of 10 μM lamivudine. Intracellular HBV DNA or cccDNA measured by real-time PCR is shown as the number of HBV DNA or cccDNA copies per 106 cells. (B) The kinetics of viral pregenomic RNA within HLCZ01 cells inoculated with the supernatant from HepG2.2.15 cells. HLCZ01 cells were treated as described in A. The viral pregenomic RNA level determined by real-time PCR is shown as the number of HBV pregenomic RNA copies per 106 cells. (C) Immunofluorescence of HBsAg or HBcAg in HBV-infected HLCZ01 cells. HLCZ01 cells were inoculated with the supernatant of HepG2.2.15 cells. Cells were harvested for immunostaining at 40 dpi using mouse monoclonal anti-HBsAg or anti-HBcAg antibody. DAPI was used for nuclei counterstaining. Identical settings were maintained for image capture. (D) Immunoelectron micrographs of HBsAg-positive particles in the supernatant and the lumen of the endoplasmic reticulum of HBV-infected HLCZ01 cells. HLCZ01 cells were treated as described in C. The supernatants (Upper) or cells (Lower) were immunostained for observation under electron microscopy. Dense, dark particles are HBsAg-positive particles. (Scale bar, 50 nm.) (E) ELISA for HBV-specific proteins. HLCZ01 cells were treated as described in A. HBsAg and HBeAg in the supernatant of HBV-infected HLCZ01 cells were detected by ELISA. (F) Kinetics of HBV DNA in the supernatant of HLCZ01 inoculated with the supernatant of HepG2.2.15 cells. HLCZ01 cells were treated as described in A. The viral DNA in the supernatant as determined by real-time PCR is shown as the number of HBV copies per milliliter of supernatant. (G) Detection of viral DNA and cccDNA in HLCZ01 cells inoculated with the supernatant of HBV-infected HLCZ01 cells. HLCZ01 cells were inoculated with the supernatant of HBV-infected HLCZ01 cells in the presence of the HBV large-surface, protein-derived peptide or lamivudine. The viral DNA or cccDNA replication was measured by real-time PCR. (H) ELISA for HBV-specific proteins. HLCZ01 cells were treated as described in G. HBsAg and HBeAg in the supernatant were detected by ELISA. Horizontal dashed lines indicate the low limit of quantification (LLOQ) of the assay.
To test whether HLCZ01 cells support the entire HBV life cycle, we examined the production of viral particles in the supernatant by analyzing both the secretion of HBsAg or HBeAg and the production of virions. HBsAg and HBeAg increased and reached a plateau at 60 dpi (Fig. 2E). HBV DNA could be identified in the supernatant of HBV-infected HLCZ01 cells and reached at plateau at 65 dpi (Fig. 2F and Fig. S1E). To explore whether the virus particles from HBV-infected HLCZ01 cells could be transferred to naive HLCZ01 cells, we harvested the supernatant of HBV-infected HLCZ01 cells and inoculated naive HLCZ01 cells. Viral DNA and cccDNA could be detected in HLCZ01 cells inoculated with the supernatant of HBV-infected HLCZ01 cells (Fig. 2G). The titers of HBV preparations applied to and released from HLCZ01 cells inoculated with the supernatant of HepG2.2.15 cells are shown in Fig. S1H. HBsAg and HBeAg also could be identified in the supernatant of HLCZ01 cells inoculated with the supernatant of HBV-infected HLCZ01 cells (Fig. 2H). Taken together, these data indicate that HLCZ01 cells support the entire HBV lifecycle.
HBV Clinical Isolates Do Propagate in HLCZ01.
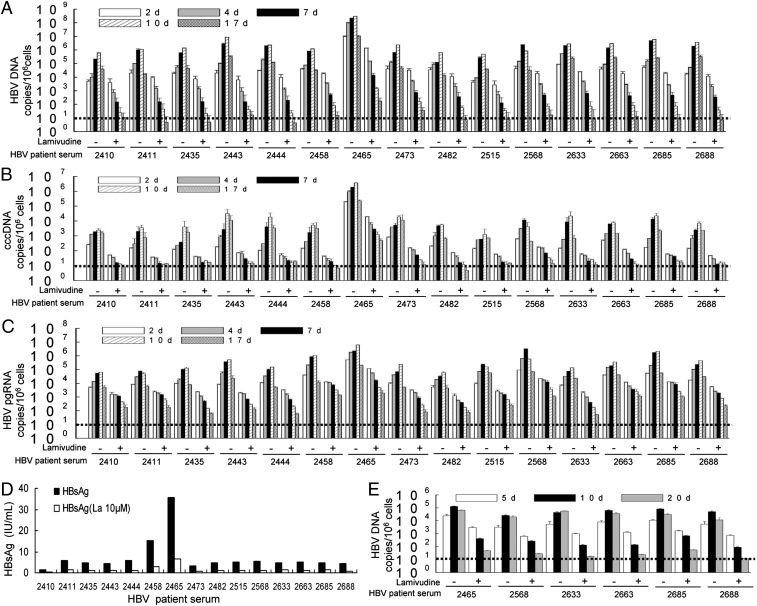
To determine whether clinical isolates of HBV can propagate in HLCZ01 cells, we inoculated HLCZ01 cells with the sera from HBV-infected donors. HBV DNA and cccDNA could be detected in HLCZ01 cells (Fig. 3 A and B and Fig. S3A). When we attempted to infect Huh7 cells under same conditions, no signal corresponding to viral DNA could be observed (Fig. S3B). When the infectious HBV sera were heat-inactivated, only HBV DNA and cccDNA at the background level were observed in HLCZ01 cells (Fig. S3C). The levels of HBV DNA and cccDNA in HLCZ01cells were dose-dependent when the cells were inoculated with a dilution series of sera from hepatitis B patients (Fig. S3 D and E). For comparison with other HBV-permissive cells, we inoculated PHH with the sera from HBV-infected donors. HBV DNA and cccDNA could be detected in PHH (Fig. S3 D and E). We also ascertained the presence of viral pregenomic RNA within HLCZ01 cells inoculated with the sera from HBV-infected donors (Fig. 3C and Fig. S3F). Similar to viral DNA, viral RNA was undetectable in Huh7 cells inoculated with HBV sera (Fig. S3F). Flow cytometry results showed that HBsAg-positive HLCZ01 cells increased from ∼3.5% at 7 dpi to 7% at 10 dpi (Fig. S3G), and when the cells were inoculated with the serum of patient #2465, which had high viral titer, HBsAg-positive HLCZ01 cells increased from 8.7% at 7 dpi to 10.9% at 10 dpi (Fig. S3G). HBsAg could be found in the supernatant of HLCZ01 cells inoculated with the sera from hepatitis B patients (Fig. 3D). Moreover, viral particles in the supernatant of HLCZ01 cells infected by HBV sera could be transferred to naive HLCZ01 cells (Fig. 3E). However, when we attempted to infect Huh7 cells under similar conditions, no signal corresponding to viral DNA could be observed (Fig. S3H). These findings clearly show that HBV clinical isolates can propagate in HLCZ01 cells.
Fig. 3.
HBV clinical isolates do propagate in HLCZ01. (A and B) HBV clinical isolates propagate in HLCZ01. HLCZ01 cells were inoculated with the sera from hepatitis B patients in the presence of 10 µM lamivudine. Total cellular DNA was isolated. Real-time PCR was performed for HBV DNA (A) and cccDNA (B). The viral DNA or cccDNA replication is shown as the number of HBV or cccDNA copies per 106 cells. (C) Viral pregenomic RNA is present in HLCZ01 cells inoculated with the sera from HBV-infected donors. HLCZ01 cells were treated as described in A. Intracellular HBV pregenomic RNA measured by real-time PCR is shown as the number of HBV pregenomic RNA copies per 106 cells. (D) HBsAg is detected in the supernatant of sera-infected HLCZ01 cells. HLCZ01 cells were treated as described in A. HBsAg in the supernatant of HLCZ01 cells at 17 d pi was detected by ELISA. (E) Viral particles in the supernatant of HBV sera-infected HLCZ01 cells can be transferred to naive HLCZ01 cells. HLCZ01 cells were treated as described in A. HLCZ01 cells were inoculated with the supernatant of HBV sera-infected HLCZ01 cells in the presence of lamivudine. Cellular DNA was isolated, and real-time PCR for HBV DNA was performed. The viral DNA replication is shown as the number of HBV copies per 106 cells. Horizontal dashed lines indicate the LLOQ of the assay.
Inhibition of HBV Infection in HLCZ01 Cells by Antiviral Drugs in Vitro and in Vivo.
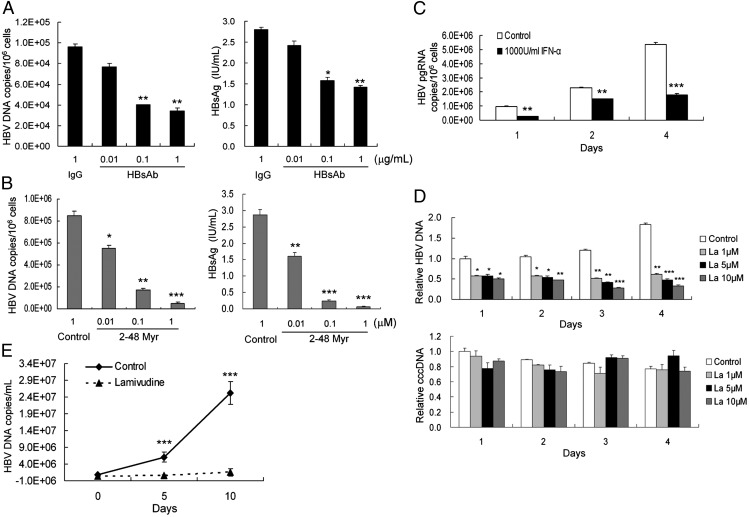
To verify further that infection of HLCZ01 cells by HBV follows the authentic entry pathway, we incubated HBV with various concentrations of the anti-HBsAg antibody before incubation with HLCZ01 cells. Anti-HBsAg antibody blocked viral infection (Fig. 4A). In addition, we examined the inhibition effect of myristoylated peptide representing the N-terminal 48 amino acids of the HBV large-surface protein required for HBV infectivity. As expected, HBV infection could be inhibited in HLCZ01 cells when the peptide was added during the infection process (Figs. 2 and 4B). One recent study showed that NTCP is a putative HBV receptor (9). The expression of NTCP protein in HLCZ01 and PHH is comparable (Fig. S4A). Silencing of NTCP inhibited HBV infection in HLCZ01 cells (Fig. S4 B and C). A previous study indicated that IFN-α inhibits HBV transcription and replication in vitro and in vivo (18), so we examined the ability of IFN-α to inhibit HBV replication in HBV-infected HLCZ01 cells. In our culture system IFN-α inhibits HBV replication by decreasing the transcription of viral pregenomic RNA (Fig. 4C). Lamivudine is widely used for hepatitis B treatment, so we tested the effect of lamivudine on HBV DNA replication in our cultured cells and in HBV-infected mice. Lamivudine markedly decreased the HBV DNA level in our cell culture system in a dose-dependent manner (Figs. 2A, 3A, and 4D). Moreover, lamivudine inhibited HBV infection in mice implanted with HBV-infected HLCZ01 cells (Fig. 4E). All these data indicate that HBV infection of HLCZ01 may be a powerful model studying for HBV and for the development of antivirals and therapeutic vaccines.
Fig. 4.
Inhibition of HBV infection in HLCZ01 cells by antiviral drugs in vitro and in vivo. (A and B) Blockage of HBV infection by anti-HBsAg antibody and the HBV large-surface, protein-derived peptide. Viruses were incubated with anti-HBsAg antibody (A) or with the HBV large-surface, protein-derived peptide (amino acids 2–48) bearing myristic acid on glycine 2 (B) before incubation with HLCZ01 cells. Intracellular HBV DNA at 10 dpi measured by real-time PCR is shown as the number of HBV copies per 106 cells (Left). HBsAg in the supernatant was detected by ELISA (Right). The results are the average of three independent experiments performed in triplicate. (C) Inhibition of HBV pregenomic RNA transcription in HBV-infected HLCZ01 cells by IFN-α. HBV-infected HLCZ01 cells were treated with IFN-α. Intracellular viral pregenomic RNA was measured by real-time PCR. The results are the average of three independent experiments performed in triplicate. (D) Inhibition of HBV infection in HLCZ01 cells by lamivudine. HLCZ01 cells were inoculated with the supernatant of HepG2.2.15 cells and cultured for 40 d. Then HBV-infected HLCZ01 cells were treated by lamivudine. Intracellular HBV DNA or cccDNA was measured by real-time PCR and normalized to GAPDH. The results are the average of three independent experiments performed in triplicate. (E) Lamivudine inhibits HBV infection in mice implanted with HBV-infected HLCZ01 cells. Sera were collected from NOD/SCID mice implanted with HBV-infected HLCZ01 cells, and viral DNA levels were analyzed by real-time PCR. *P < 0.05, **P < 0.01, ***P < 0.001 versus control.
Infection of HLCZ01 Cells by HCV.
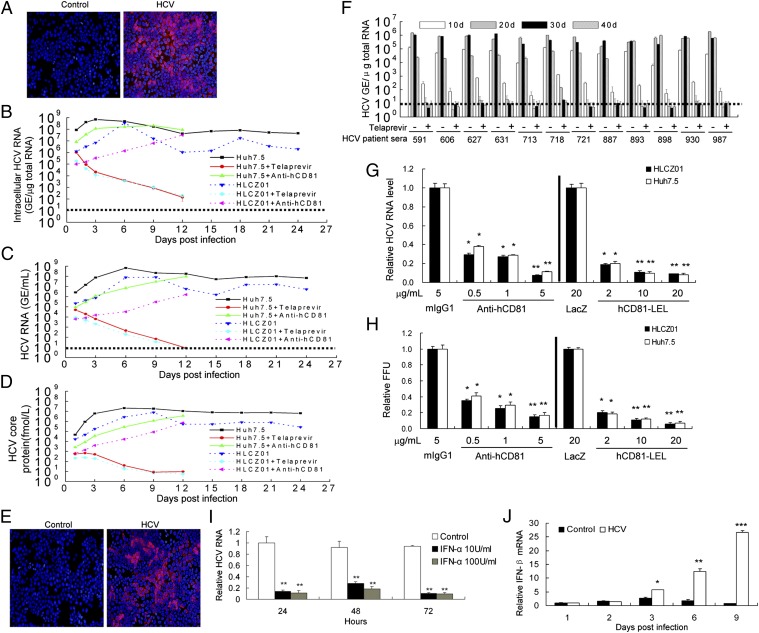
Infection of the host cell by HCV is initiated by the interactions between the viral envelop protein and several previously identified HCV entry receptors, including CD81, scavenger receptor class B type I (SR-BI), claudin-1 (CLDN1), and occludin (OCLN) (19–23). ApoE is critical for HCV assembly (24). The expression of these receptors in HLCZ01 and permissive Huh7.5 cells is comparable (Fig. S5). To understand better the interaction between HCV and host cells, we attempted to replicate HCV in HLCZ01 cells. We inoculated HLCZ01 cells with HCV in cell culture (HCVcc) and found that NS5A-positive HLCZ01 cells could be observed readily (Fig. 5A). Intracellular viral RNA was detectable in the cells at 1 dpi and increased rapidly (Fig. 5B). Viral replication was slightly less efficient in HLCZ01 cells than in Huh7.5 cells (Fig. 5B). Extracellular viral RNA and core protein levels in the supernatant of HLCZ01 and Huh7.5 cells were comparable (Fig. 5 C and D). To prove that HCV-infected HLCZ01 cells indeed release infectious viruses, we inoculated naive HLCZ01 cells with the supernatant collected from HCV-infected HLCZ01 cells and detected NS5A-positive signal and viral RNA in the infected naive cells (Fig. 5E and Fig. S6A). Moreover, HCV-infected HLCZ01 and Huh7.5 cells produced comparable infectivity titers (Fig. S6B).
Fig. 5.
Infection of HLCZ01 cells by HCV. (A) Immunofluorescent staining of HLCZ01 cells inoculated with JFH1 virus. HLCZ01 cells were inoculated with JFH1 virus at an MOI of 0.1. Cells were fixed at 6 dpi for immunostaining using monoclonal anti-HCV NS5A antibody (red). DAPI (blue) was used to counterstain nuclei. (B) Kinetics of viral RNA determined by real-time PCR in HCV-infected HLCZ01 cells. HLCZ01 cells and Huh7.5 cells were exposed to 5 μg/mL anti-hCD81 antibody (5A6) before inoculation with JFH1 virus at an MOI of 0.1, or inoculated with JFH1 virus at an MOI of 0.1 in the presence of 5 μM telaprevir. Intracellular HCV RNA determined by real-time PCR is shown as the HCV RNA genomic equivalence (GE) per microgram of total cellular RNA. (C and D) Kinetics of extracellular viral RNA and core protein in the supernatant of HCV-infected HLCZ01 cells. HLCZ01 and Huh7.5 cells were treated as described in B. HCV RNA in the supernatant measured by real-time PCR is shown as the HCV RNA GE per milliliter of supernatant (C). Core protein in the supernatant of HCV-infected HLCZ01 cells was determined by ELISA (D). (E) Release of infectious viruses from HCV-infected HLCZ01 cells. HLCZ01 cells were inoculated with the supernatant from HCV-infected HLCZ01 cells. Cells were fixed for immunostaining at 3 dpi using anti-HCV NS5A antibody. (F) HCV clinical isolates propagate in HLCZ01 cells. HLCZ01 cells were inoculated with the different genotypes of sera from HCV-infected donors in the presence of 5 μM telaprevir. The intracellular viral RNA level was measured by real-time PCR. Horizontal dashed lines indicate the LLOQ of the assay. (G and H) Blockage of HCV infection in HLCZ01 cells by anti-human CD81 antibody or hCD81-LEL. HLCZ01 cells were exposed to increasing concentrations of anti-hCD81 antibody before inoculation with JFH1 virus, or viruses were incubated with different concentrations of hCD81-LEL protein before incubation with HLCZ01 cells. (G) Intracellular HCV RNA determined by real-time PCR at 5 d pi was normalized with GAPDH. The results are the average of three independent experiments performed in triplicate. (H) The supernatant was harvested and tittered by focus-forming unit assay on naive Huh7.5 cells. Data shown are mean ± SD of three independent experiments performed in triplicate. (I) Inhibition of HCV RNA replication in HLCZ01 cells by IFN-α. HCV-infected HLCZ01 cells were treated with IFN-α. Intracellular HCV RNA measured by real-time PCR was normalized with GAPDH. The results are the average of three independent experiments performed in triplicate. (J) Induction of type I IFN in HLCZ01 cells by HCV infection. HLCZ01 cells were infected by HCV. IFN-β mRNA measured by real-time PCR was normalized with GAPDH. The results are the average of three independent experiments performed in triplicate. *P < 0.05, **P < 0.01, ***P < 0.001 versus control.
To examine whether clinical isolates of HCV can propagate in HLCZ01 cells, we inoculated HLCZ01 with different genotypes of sera from patients with hepatitis C. HCV RNA and core protein could be observed in the cells (Fig. 5F and Fig. S7A). Recently, the protease NS3 inhibitor telaprevir has entered clinical use for treatment of HCV-infected individuals (25). IFN-α or telaprevir decreased viral RNA levels in HLCZ01 cells under identical conditions (Fig. S7 B and C). When the infectious HCV sera were inactivated by UV, viral RNA could be detected in HLCZ01 only at the background level (Fig. S7B). Viral particles in the supernatant of HLCZ01 cells infected by sera from HCV-infected donors could be transferred to naive HLCZ01 cells (Fig. S7D). However, IFN-α or telaprevir inhibited the passage of sera-derived HCV from sera-inoculated HLCZ01 cells to naive HLCZ01 cells (Fig. S7D). The quantification of infectious units in HLCZ01 cells infected with HCV sera is shown in Fig. S7E. Flow cytometry results further demonstrated that HLCZ01 cells can be infected by sera from hepatitis C patients (Fig. S7F). All these data suggest that HLCZ01 cells indeed can be infected by HCVcc and by the sera from hepatitis C patients.
It has been reported that human CD81 is an essential coreceptor for HCV entry (19). To provide evidence that the uptake of HCV into HLCZ01 cells follows the authentic entry pathway, we performed infection competition assays using mouse monoclonal anti-CD81 antibody or soluble CD81 large extracellular loop (hCD81-LEL). HCVcc infectivity could be blocked with either anti-CD81 antibody or hCD81-LEL (Fig. 5 G and H), indicating that the virus enters via the authentic HCV entry pathway.
The current standard of care for chronic hepatitis C involves IFN-α–based therapy. We found that IFN-α significantly inhibited HCV RNA replication in HLCZ01 cells (Fig. 5I and Fig. S7B), demonstrating that HLCZ01 cells infected by HCV may be useful for testing novel drugs.
To understand better the interaction between HCV and host cells, we infected HLCZ01 cells with HCVcc and tested the innate immune response in the cells. IFN-β and IFN-stimulated genes (ISGs) were induced in viral-infected cells (Fig. 5J and Fig. S8), indicating that HLCZ01 cells mount an innate immune response to HCV infection.
HBV/HCV Coinfection in HLCZ01 Cells.
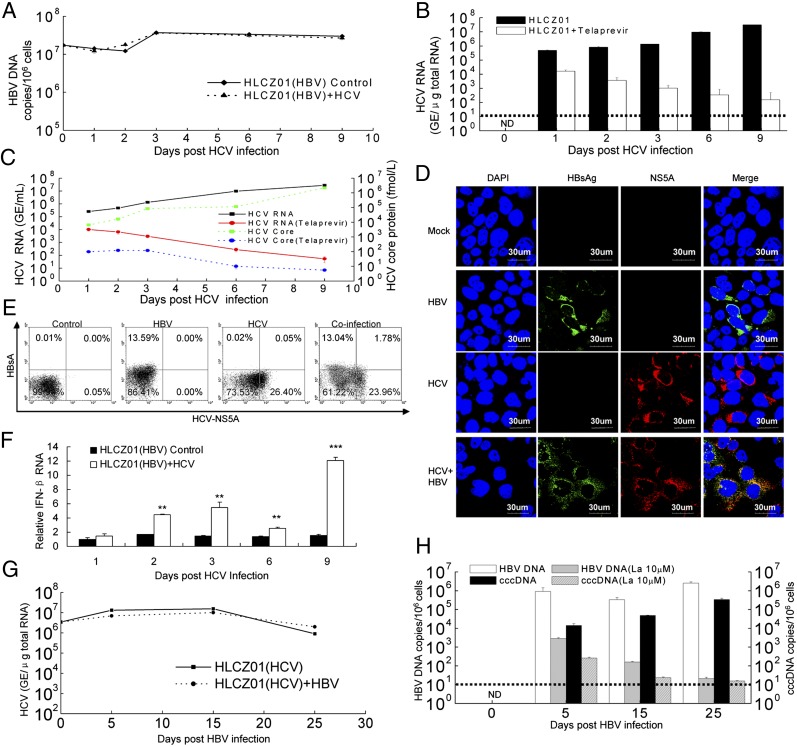
HBV/HCV coinfection is common, with an estimated 7–20 million individuals affected worldwide. Patients with HBV/HCV coinfection have an increased risk for cirrhosis, hepatocellular carcinoma, and death (26). The virological and molecular aspects of HBV/HCV coinfection are poorly understood. The lack of appropriate model systems has made the study of the interactions between HBV and HCV difficult. Our novel cell culture system allows us to investigate the interactions between HBV and HCV. HCV infection did not affect HBV replication in HLCZ01 cells (Fig. 6 A and B and Fig. S9A). HCV RNA and core protein in the supernatant of the cells could be verified (Fig. 6C). Interestingly, HBV and HCV infected the same cells (Fig. 6 D and E and Fig. S9A), providing new insights into the pathogenesis of HBV/HCV coinfection. The frequency of coinfected cells increased when HLCZ01 cells were infected with HBV and HCV at higher virus titers (Fig. S9A). Moreover, HBV had little effect on the innate host response of HLCZ01 cells to HCV infection (Fig. 6F). HBV infection did not affect HCV replication in HLCZ01 cells (Fig. 6G). HBV infection was confirmed in the cells (Fig. 6H). Finally, simultaneous infection of HLCZ01 cells by HBV and HCV did not affect either HBV (Fig. S9B) or HCV (Fig. S9C) replication. These data indicate that HBV and HCV replicate in the same cells without evidence of direct interference in vitro (27). Superinfection exclusion generally is restricted to homologous viruses (28), whereas nonrelated viruses are able to replicate normally. In agreement with the general concept, our results showed that there is no superinfection exclusion of HBV by HCV or of HCV by HBV.
Fig. 6.
HBV/HCV coinfection in HLCZ01 cells. (A and B) HCV infection has no effect on HBV replication. HBV-infected HLCZ01 cells were infected with JFH1 viruses in the presence of 5 µM telaprevir. Intracellular HBV DNA (A) and HCV RNA (B) were measured by real-time PCR. (C) Detection of HCV RNA and core protein in the supernatant of HCV-infected HLCZ01 preinfected with HBV. HBV-infected HLCZ01 cells were treated as in A. HCV RNA and core protein in the supernatant were examined by real-time PCR and ELISA, respectively. (D and E) Infection of HLCZ01 cells by both viruses. HLCZ01 cells were infected with HBV for 10 d. Then HBV-infected cells were infected with HCV for another 6 d. (D) Cells were fixed for immunostaining using monoclonal anti-HCV NS5A (red) and anti-HBsAg (green) antibodies. DAPI (blue) was used to counterstain nuclei. Identical settings were maintained for image capture. Representative images are shown. (E) Frequency of HLCZ01 cells that are coinfected with both HBV and HCV as determined by flow cytometry. Cells were collected for flow cytometry using anti-HBsAg and anti-HCV NS5A antibodies. The data are from one experiment that is representative of three independent experiments. (F) HBV has little effect on the innate immune response of HLCZ01 cells to HCV infection. HBV-infected HLCZ01 cells were infected with JFH1 virus. IFN-β mRNA was examined by real-time PCR and normalized with GAPDH. The results are the average of three independent experiments performed in triplicate. (G and H) HBV infection does not affect HCV replication in HLCZ01 cells. HCV-infected HLCZ01 cells were infected with HBV in the presence of 10 µM lamivudine. Intracellular HCV RNA was verified by real-time PCR (G). HBV DNA and cccDNA in the cells were examined by real-time PCR (H). ND, not detectable. Horizontal dashed lines indicate the LLOQ of the assay. **P < 0.01, ***P < 0.001 versus control.
Discussion
We have established that HLCZ01 is a robust cell culture model of HBV infection by showing the kinetics of several markers of viral infection, including viral DNA replication, the formation and amplification of cccDNA, newly synthesized pregenomic viral RNA, the secretion of HBsAg and HBeAg, and the production and release of infectious viral particles from HBV-infected HLCZ01 cells. In addition, evidence that HBV infection is blocked by specific anti-HBsAg antibody or by pre-S1–blocking peptide strongly indicates that HBV infection of HLCZ01 cells follows the authentic entry pathway and that the process of viral adsorption and entry of HBV can be studied in this system. That the expression of NTCP protein is comparable in HLCZ01, HepG2, and Huh7 cells and in PHH but only HLCZ01 and PHH are susceptible to HBV infection suggests that other HBV receptors exist. Our data show that HBV infection in HLCZ01 cells results in the formation of foci of infected cells and that the percentage of HBV-infected cells increases, indicating that HBV may spread via cell-to-cell transmission and/or by attaching preferentially to the adjacent cells after secretion. Interestingly, HBV clinical isolates can propagate HLCZ01 cells, providing a very useful tool for the analysis of clinical isolates of HBV and for the development of antiviral drugs and vaccines. The HLCZ01 cell line provides a powerful tool for improving our understanding of the HBV life cycles, including the identification of the still unknown receptors and the mechanisms by which cccDNA is formed and amplified.
The HLCZ01 cell line also is susceptible to HCV infection, as shown by the kinetics of intracellular viral RNA replication, the expression of viral protein, and the production and release of infectious virus particles. HCVcc infectivity could be blocked with either anti-CD81 antibody or hCD81-LEL, indicating that virus enters via the authentic HCV entry pathway. The remarkable feature of HLCZ01 cells is their susceptibility to different genotypes of sera from hepatitis C patients, providing a useful tool for the analysis of clinical isolates of HCV and for the development of vaccines.
Our novel culture system allows us to investigate the interactions between HBV and HCV. Interestingly, the two viruses can infect the same cells without evidence for direct interference, providing new insights into the pathogenesis of HBV/HCV coinfection.
In summary, we have established a robust cell culture model of HBV and HCV infection in which infectious HBV and HCV can be produced and passaged to naive cells. Supporting the entire life cycles of both viruses, the HLCZ01 cell line provides a powerful tool for addressing aspects of the virus life cycles, including the identification of the yet unknown receptors of HBV, virus entry, formation and amplification of cccDNA, virus assembly, the analysis of clinical isolates of HBV and HCV, and HBV/HCV coinfection. It will enable us to explain functionally the role of viral surface proteins in the entry process and consequently will facilitate the development of antiviral drugs interfering with the early steps of viral infection. Evidence that type I IFN and ISGs are induced in HCV-infected HLCZ01 cells and that different genotypes of HBV and HCV clinical isolates can propagate in HLCZ01 cells suggests that this cell culture system may be useful for the analysis of host–virus interactions that should facilitate the discovery of antiviral drugs and vaccines.
Materials and Methods
Isolation and Establishment of a Novel Hepatoma Cell Line.
Cells were isolated from the tissue of a liver tumor from a male patient with chronic HCV infection. Experimental procedures were performed in accordance with the provisions of the Ethics Committee of Hunan Provincial Tumor Hospital (Changsha, China). The cells were isolated and cultured as described previously (29). Briefly, immediately after surgical resection, the tumor tissue was stored in PBS, and cells were dissociated within 1 h by two-step perfusions. Visible vessels were perfused first for 15 min with Liver Perfusion Medium (Invitrogen) to eliminate the blood cells. A second perfusion was performed with collagenase- and dispase-containing Liver Digest Medium (Invitrogen) until the tissue was digested. Then the liver tissue was cut into small pieces and shaken gently in Hepatocytes Wash Medium (Invitrogen). The cells and small pieces of liver tissue were cultured with DMEM/F12 medium supplemented with 10% (vol/vol) FBS (Invitrogen). The cell clones were selected and cultured in a six-well plate until cell growth filled the culture well. To obtain the hepatoma cell line, 5 × 106 cells were injected into NOD/SCID immunodeficient mice. Two months later, the tumor tissues were removed from the mice, cut into small pieces, and cultured in DMEM/F12 medium supplemented with 40 ng/mL of dexamethasone (Sigma) and 10 ng/mL of EGF (BD). Several clones were obtained, one of which we designated HLCZ0. All cells were cultured in collagen-coated tissue culture plates. HLCZ01 cells were passaged every 5 d (1/3 dilution) by trypsinization.
Isolation of PHH.
All procedures were performed in accordance with the provision of the Ethical Commission of Hunan Provincial Tumor Hospital. PHH were obtained from healthy peritumoral liver resection specimens from non-HBV–, non-HCV–, and non-HIV–infected patients undergoing partial hepatectomy for primary hepatocellular carcinoma or secondary metastatic lesions caused by other cancers. PHH were isolated and cultured as described previously (30).
Serum Samples.
HBV and HCV sera were collected between 2009 and 2011 in Xiangya Hospital of Central South University, China, with the approval of the Ethics Committee of Xiangya Hospital. Information regarding these donors can be found in Tables S1 and S2.
HBV Infection of Cell Culture.
The supernatant of HepG2.2.15 cells derived from human hepatoma cell line HepG2 transfected with the full genome of HBV was collected and filtered. HLCZ01 cells were inoculated overnight with the filtered supernatant at a multiplicity of infection (MOI) of 20 genome equivalents (Geq) per cell. The cells were washed three times with PBS, were maintained in DMEM/F12 medium, and were harvested at the indicated times. HLCZ01 cells were inoculated with the different strains of sera from hepatitis B patients diluted at 1:20 and were cultured for the indicated time periods.
Quantitative Assay for HBV-Specific Proteins.
HBsAg and HBV e antigen (HBeAg) in the supernatant were quantified by a chemiluminescent microparticle immunoassay using the Architect i2000SR Analyzer according to the manufacturer’s instructions. Quantitative HBsAg levels are reported in infectious units (IU) per milliliter, with a reactive range of 0.05–250.00 IU/mL Samples with HBsAg titers beyond 250 IU/mL were diluted at 1:20 and 1:500 before further testing. In the Architect platform, the HBeAg assay is designed to be semiquantitative and is reported as sample per cutoff (S/CO). Samples with S/CO values greater than 1.0 were considered to be HBeAg positive.
Isolation and Analysis of Viral DNA.
DNA was isolated from the whole-cell lysates and supernatant. Real-time PCR for total HBV DNA and cccDNA was performed as described previously (31). Intracellular hepatic HBV DNA and cccDNA were measured using real-time PCR analysis. The primers used for PCR to detect HBV DNA were 5′-CACCTCTGCCTAATCATC-3′ (sense) and 5′-GGAAAGAAGTCAGAAGGCAA-3′ (antisense). The primers used for PCR to detect cccDNA were 5′-GTGCCTTCTCATCTGCCGG-3′ (sense) and 5′-GGAAAGAAGTCAGAAGGCAA-3′ (antisense). Plasmid-safe ATP-dependent DNase (Epicentre) was used to digest the single-strand region of HBV genome, allowing enrichment of cccDNA for subsequent real-time PCR detection.
Reverse Transcription and Analysis of HBV-Specific Transcripts.
Total RNA was extracted from the cells using TRIzol reagent (Invitrogen) as recommended by the manufacturer. RNA samples were treated with RNase-free DNase (Promega) for 1 h at 37 °C to remove genomic DNA. Quantitative measurement of HBV pregenomic RNA was performed using real-time PCR as described previously (31). The primers for the detection of pregenomic were 5′-CTCAATCTCGGGAATCTCAATGT-3′ (sense) and 5′-TGGATAAAACCTAGCAGGCATAAT-3′ (antisense).
HCV Infection of HLCZ01 Cells.
Huh7.5 cells were kindly provided by Charles Rice (Rockefeller University, New York). pJFH1 was a gift from Takaji Wakita (National Institute of Infectious Diseases, Tokyo) (13). HCVcc was generated as described previously (12). HLCZ01 cells were inoculated overnight with HCVcc at an MOI of 0.1, and the inoculum was removed. The cells were maintained and harvested at the indicated times. HLCZ01 cells were inoculated with the sera from hepatitis C patients diluted at 1:20 and were cultured for various time periods.
Detection of HCV RNA and IFN-β by Real-Time PCR.
Total cellular RNA was isolated using TRIzol. The primers targeting HCV, IFN-β, G1P3, 1-8U, and GAPDH have been reported, and real-time PCR was performed as described previously (30). The primers for amplification of ALB, AAT, HNF4, and CYP3A4 are given shown in SI Materials and Methods. The cDNA of miR-122 was synthesized from total RNA using the stem-loop reverse-transcription primer 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCAAACA-3′, and miR-122 was quantitated by real-time PCR using primers 5′-GGGTGGAGTGTGACAATGG-3′ and 5′-TGCGTGTCGTG GAGTC -3′. The internal control was U6. The cDNA of U6 was synthesized from total RNA using the stem-loop RT primer 5′-CGCTTCACGAATTTGCGTGTCAT-3′, and U6 was quantitated by real-time PCR using primers 5′-GCTTCGGCAGCACATAT ACAAAAT and 5′-CGCTTCACGAATTTGCGTGTCAT-3′. Fold variations were calculated after normalization to U6.
Southern Blot Analysis.
HBV DNA was isolated from whole-cell lysates. Cells in a 60-mm dish recovered after trypsinization and one washing were lysed overnight at 37 °C in 1 mL lysis buffer [50 mM Tris⋅HCl (pH 8.0), 10 mM EDTA, 1% SDS, 150 mM NaCl] supplemented with proteinase K (200 g/mL). The cccDNA was selectively extracted from cells in a 60-mm dish recovered by trypsinization. Cells were lysed for 1 h at 37 °C in 1 mL lysis buffer not supplemented with proteinase K, followed by the addition of 0.25 mL of 2.5 M KCl and incubation at 4 °C overnight. The lysate then was clarified by centrifugation at 12,000 × g for 30 min at 4 °C. In both cases, the lysate was extracted with phenol and phenol:chloroform, followed by ethanol precipitation. For cccDNA detection, the prepared DNA sample was treated with plasmid-safe, ATP-dependent DNase (Epicentre Technologies) following the manufacturer’s instructions. HBV viral particles in cell supernatants were concentrated by ultracentrifugation at 28,000 rpm in a SW28 rotor (Beckman Coulter) for 16 h at 4 °C. Fifteen milliliters of supernatant per sample were used for the concentration and extraction of HBV viral DNA. Nucleic acids were separated on 1% agarose gel and analyzed by Southern blot procedures with modifications (32). HBV-specific nucleic acids were detected with a digoxygenin (DIG)-labeled probe obtained by random priming (DIG-High primer DNA labeling and detection kit; Roche Diagnostics) on a 3.2-kb EcoRI fragment containing a complete linear HBV genome from HepG2.2.15 cells, according to the manufacturer’s instructions. Biodyne B Nylon transfer membranes (0.45 μm) were from PALL.
Northern Blot Analysis.
Total RNA was isolated by using the TRIzol reagent and treated with RNase-free DNase I. Thirty micrograms of total cellular RNA per sample denatured for 5 min at 100 °C was separated on 1.2% agarose gel and analyzed by Northern blot according to the procedures published previously (33) and using the DIG-labeled HBV probe described above.
Immunofluorescence of Viral Protein and Human Hepatocyte-Specific Markers.
Cells were seeded on glass coverslips and fixed with ice-cold acetone for 10 min. Cells were blocked with 1:50 goat serum for 30 min and then were incubated for 1 h with mouse monoclonal anti-NS5A(HL1126), a gift from Chen Liu (University of Florida, Gainesville, FL), mouse monoclonal anti-HBsAg (S26) or anti-HBcAg (10E11) antibody (Pierce), mouse monoclonal anti-CD81 antibody (5A6) (Santa Cruz Biotechnology), mouse monoclonal anti–claudin-1 (2H10D10) or anti-occludin (OC-3F10) (Invitrogen) antibody, or rabbit anti–SR-BI antibody (ab137829) (Abcam). Cells were washed three times with PBS and stained with fluorescence-labeled secondary antibodies (Invitrogen) for 45 min. Finally, the coverslips were washed with PBS, and the nuclei were counterstained with DAPI (Vector Laboratories, Inc.). Fluorescent images were obtained with a fluorescent microscope (Olympus). Titration of infectious HCV was reported previously (12).
Statistical Analyses.
The data were analyzed using a two-tailed Student t test and are presented as means ± SD.
Supplementary Material
Acknowledgments
We thank Charles M. Rice for the Huh7.5 cell line; Takaji Wakita for pJFH1; and Chen Liu for sharing research materials and helpful discussions. This work was supported by National Science and Technology Major Project of the Ministry of Science and Technology of China Grant 2009ZX10004-312 and National Natural Science Foundation of China Grant 81271885 (to H.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320071111/-/DCSupplemental.
References
- 1.El-Serag HB. EI-Serag HB Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–1273, e1. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tujios SR, Lee WM. New advances in chronic hepatitis B. Curr Opin Gastroenterol. 2012;28(3):193–197. doi: 10.1097/MOG.0b013e32835297ef. [DOI] [PubMed] [Google Scholar]
- 3.Rice CM. New insights into HCV replication: Potential antiviral targets. Top Antivir Med. 2011;19(3):117–120. [PMC free article] [PubMed] [Google Scholar]
- 4.Guidotti LG, Chisari FV. Immunobiology and pathogenesis of viral hepatitis. Annu Rev Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- 5.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84(4):1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ochiya T, et al. An in vitro system for infection with hepatitis B virus that uses primary human fetal hepatocytes. Proc Natl Acad Sci USA. 1989;86(6):1875–1879. doi: 10.1073/pnas.86.6.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galle PR, et al. In vitro experimental infection of primary human hepatocytes with hepatitis B virus. Gastroenterology. 1994;106(3):664–673. doi: 10.1016/0016-5085(94)90700-5. [DOI] [PubMed] [Google Scholar]
- 8.Sureau C, Romet-Lemonne JL, Mullins JI, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47(1):37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 9.Yan H, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hantz O, et al. Persistence of the hepatitis B virus covalently closed circular DNA in HepaRG human hepatocyte-like cells. J Gen Virol. 2009;90(Pt 1):127–135. doi: 10.1099/vir.0.004861-0. [DOI] [PubMed] [Google Scholar]
- 11.Lindenbach BD, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 12.Zhong J, et al. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102(26):9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li YP, et al. Robust full-length hepatitis C virus genotype 2a and 2b infectious cultures using mutations identified by a systematic approach applicable to patient strains. Proc Natl Acad Sci USA. 2012;109(18):E1101–E1110. doi: 10.1073/pnas.1203829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YP, et al. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proc Natl Acad Sci USA. 2012;109(48):19757–19762. doi: 10.1073/pnas.1218260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinmann E, Pietschmann T. Cell culture systems for hepatitis C virus. Curr Top Microbiol Immunol. 2013;369:17–48. doi: 10.1007/978-3-642-27340-7_2. [DOI] [PubMed] [Google Scholar]
- 17.Sheahan T, Jones CT, Ploss A. Advances and challenges in studying hepatitis C virus in its native environment. Expert Rev Gastroenterol Hepatol. 2010;4(5):541–550. doi: 10.1586/egh.10.53. [DOI] [PubMed] [Google Scholar]
- 18.Belloni L, et al. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J Clin Invest. 2012;122(2):529–537. doi: 10.1172/JCI58847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pileri P, et al. Binding of hepatitis C virus to CD81. Science. 1998;282(5390):938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 20.Scarselli E, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21(19):5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans MJ, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446(7137):801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 22.Ploss A, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457(7231):882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorner M, et al. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501(7466):237–241. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang KS, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J Virol. 2007;81(24):13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlütter J. Therapeutics: New drugs hit the target. Nature. 2011;474(7350):S5–S7. doi: 10.1038/474S5a. [DOI] [PubMed] [Google Scholar]
- 26.Potthoff A, Manns MP, Wedemeyer H. Treatment of HBV/HCV coinfection. Expert Opin Pharmacother. 2010;11(6):919–928. doi: 10.1517/14656561003637659. [DOI] [PubMed] [Google Scholar]
- 27.Bellecave P, et al. Hepatitis B and C virus coinfection: A novel model system reveals the absence of direct viral interference. Hepatology. 2009;50(1):46–55. doi: 10.1002/hep.22951. [DOI] [PubMed] [Google Scholar]
- 28.Lee YM, Tscherne DM, Yun SI, Frolov I, Rice CM. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J Virol. 2005;79(6):3231–3242. doi: 10.1128/JVI.79.6.3231-3242.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu H, et al. Primary human hepatocyte culture for HCV study. Methods Mol Biol. 2009;510:373–382. doi: 10.1007/978-1-59745-394-3_28. [DOI] [PubMed] [Google Scholar]
- 30.Yang D, et al. Innate host response in primary human hepatocytes with hepatitis C virus infection. PLoS ONE. 2011;6(11):e27552. doi: 10.1371/journal.pone.0027552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong DK, et al. Occult hepatitis B infection and HBV replicative activity in patients with cryptogenic cause of hepatocellular carcinoma. Hepatology. 2011;54(3):829–836. doi: 10.1002/hep.24551. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsh EF, Maniatis T. Molecular Cloning: A Laboratory Manual. NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 33.Zhu H, et al. Gene expression associated with interferon alfa antiviral activity in an HCV replicon cell line. Hepatology. 2003;37(5):1180–1188. doi: 10.1053/jhep.2003.50184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.