Protein transport is a fundamental activity for all living cells and an exciting area for scientific exploration (1, 2). In bacteria, the process depends on the concerted action of at least three membrane-embedded components: the ubiquitous SecYEG complex that forms the polypeptide-conducting membrane pore, the essential YidC insertase that works independently or in cooperation with SecYEG to insert hydrophobic protein segments into the lipid bilayer, and the auxiliary SecDF–yajC complex that associates with both SecYEG and YidC to accelerate the overall process. Depending on specific requirements of the substrate to be transported, SecYEG also associates with the ribosome or the cytosolic ATPase SecA, which mediate the cotranslational and posttranslational mode of translocation, respectively. Over the last decade, our understanding of the bacterial protein transport process has greatly advanced with the determination of structures for almost all of the individual components (3–5). However, our understanding of the interconnection between these components is still limited because the interactions are weak or transient, and certainly difficult to analyze because they occur in the membrane environment. In PNAS, Schulze et al. (6) report the isolation of a supercomplex that contains all seven subunits: the SecYEG–DFyajC–YidC holo-enzyme aka holo-translocon (HTL). This large membrane assembly of ∼250 kDa encompasses 34 transmembrane segments with three large loops exposed on the trans-side of the membrane. The isolation of the HTL is a breakthrough in the field, opening new avenues for investigation; it is also an elegant method that resolves major challenges in membrane biochemistry: membrane complex production and purification.
Schulze et al. (6) used a simple two-step approach to capture the holo-complex. Prior work showed that SecYEG, SecDF–yajC, and YidC interact with each other (7, 8), and each subcomplex could be purified individually, but their coreconstitution from separated components was hardly achievable. Schulze et al. recognized that if they first overproduced all of the subunits together in the bacterial envelope they might increase the chance of capturing the holo-complex. A daunting task, but by using progressive DNA recombinant technologies and plasmids with multiple expression promoters, the seven subunits were eventually cloned on two polycistronic operons (Fig. 1). Hypothetically, this concomitant overproduction strategy allows membrane proteins to insert efficiently, at the same time, in the same region of the cell, and with the right stoichiometry. The latter point may be crucial for the formation of the holo-translocon because SecYEG forms homo-dimers when overexpressed in the membrane (9). This oligomeric behavior appears functionally relevant, but it can also hinder or mask protein surfaces that are otherwise interacting with SecDF–yajC and YidC.
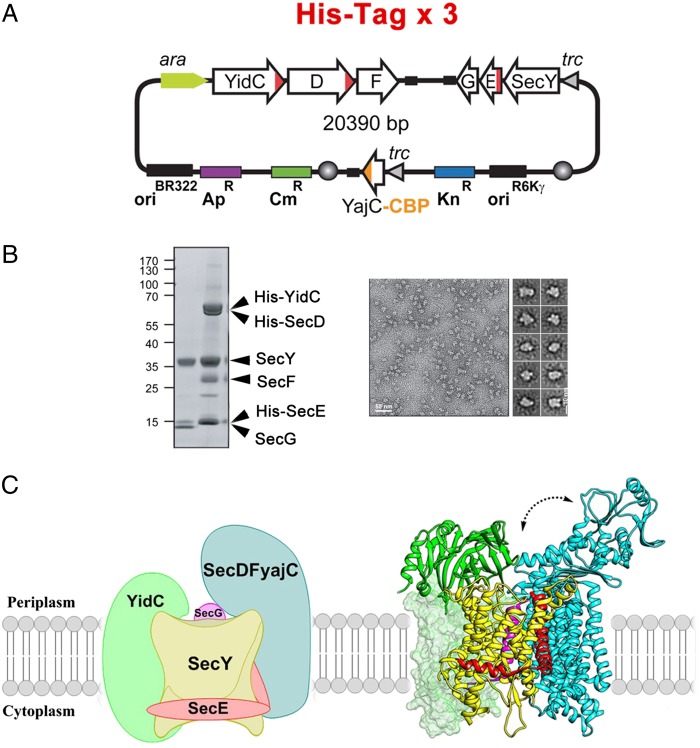
Fig. 1.
(A) Gene-expression system used for production of the HTL, as described in Schulze et al. (6). (B) SDS/PAGE and negative-stain electron microscopy analysis of the purified HTL, as shown in Schulze et al. (6). (C) Model of structural organization of the HTL. YidC (PDB ID code 3BLC) and SecDF (PDB ID code 3AQP) are docked on the structure of SecYEG (PDB ID code 1RH5). The structure of the transmembrane portion of YidC is unknown. The arrow illustrates the proposed rotation of the SecD periplasmic domain. Figure kindly provided by Mike Carlson.
Second, to capture the holo-complex from the crude membrane extract, Schulze et al. (6) appended a His6-tag on not just one but three different subunits from each subcomplex, namely SecE, SecD, and YidC (Fig. 1). His-tagging is a widespread and simple strategy to purify recombinant proteins via immobilized-metal affinity chromatography, and it is generally assumed that only one subunit must be tagged and attached to the resin for a specific purification. In reality, proteins that interact weakly in the lipid bilayer are prone to dissociation, especially when their purification involves extraction and delipidation with detergents. Evidently, the presence of multiple His6-tags at the surface of the HTL allowed the subunits to remain tightly associated on the chromatography matrix, preserving the holo-complex integrity during washes with detergents.
With the HTL complex at hand, Schulze et al. (6) could count one copy of SecYEG (∼75 kDa), one copy of SecDF–yajC (∼115 kDa), and one copy of YidC (∼60 kDa) per complex. The authors also determined its spatial organization using short-arm cross-linkers that can tether neighboring proteins together. The results showed that SecD is contacting SecE and SecG near the N-terminal half of SecY (TM1-5), whereas YidC is localized closer to the SecY’s lateral gate, which is the exit route of transmembrane segments into the lipid bilayer (10). Higher-resolution maps are needed, but these preliminary findings are significant as they rationalize earlier biochemical and genetic observations. For example, SecYEG forms a homo-dimer in the membrane through its SecEG interface, and accordingly SecDF–yajC overproduction can inhibit disulfide bond formation between two SecG molecules (11). Mutations in the transmembrane residues of YidC that are thought to weaken association with SecY lead to an increased dependence on SecDF–yajC for viability (12). These results can now be explained if SecDF–yajC is bridging YidC to SecYEG in the holo-translocon. Although the YidC membrane domain structure is not known, its large periplasmic domain (∼30 kDa) forms a β-fold sandwich that interacts with the periplasmic domain of SecF (13).
Schulze et al. (6) also show that the holo-translocon stimulates SecA ATPase as much as SecYEG, but translocation is less efficient and more dependent on the proton motive force (PMF). Membrane protein insertion is seemly more efficient as the ribosome binds with an apparent higher affinity. As pointed out by the authors, the mechanistic basis governing these effects is not immediately clear and will require additional analysis. Nevertheless, these experiments demonstrate conclusively that posttranslational export and cotranslational membrane insertion occur
The work in Schulze et al., combined with that from other laboratories, reveals once more the astonishing versatility and plasticity of the Sec translocon.
through the same unique complex. Significantly, the fact that YidC and SecDF–yajC associate with the same SecYEG unit strongly suggests that the two domains work in a concerted manner, either during the biogenesis of membrane proteins containing large extracellular domains or perhaps also during protein secretion. The recent crystal structure of Thermus thermophilus SecDF suggests that the large P1 domain of SecD captures the polypeptide chain as it emerges from the SecYEG channel (5). The rotation of the P1 domain is driven by the membrane PMF, and this rotation is thought to promote forward translocation. It is therefore interesting that membranes depleted for SecDF–yajC are unable to generate protein translocation intermediates that are normally formed when short polypeptide hydrophobic stretches enter the translocon (14, 15). Because YidC interacts with hydrophobic transmembrane segments passing through the lateral gate (16), YidC may also interact with these short hydrophobic stretches, thereby holding the polypeptide chain in the channel until SecDF promotes PMF-driven forward movement.
The work in Schulze et al. (6), combined with that from other laboratories, reveals once more the astonishing versatility and plasticity of the Sec translocon, with the core SecYEG interacting with itself, with two other auxiliary membrane components, three different cytosolic partners, and even specific membrane lipids (17). Additional work is needed to understand these dynamic assemblies, their actual lifetime in the cell membrane, and their relationship with the substrate characteristics (18). Meanwhile, given the homogeneity and stability of the HTL preparation (Fig. 1), it is expected that high-resolution electron-microscopy analysis will soon unravel the intimate details of its organization. The HTL will appear as a beautiful membrane machinery.
Footnotes
The author declares no conflict of interest.
See companion article on page 4844.
References
- 1.Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310(5753):1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
- 2.Park E, Rapoport TA. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu Rev Biophys. 2012;41:21–40. doi: 10.1146/annurev-biophys-050511-102312. [DOI] [PubMed] [Google Scholar]
- 3.Van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427(6969):36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 4.Samuelson JC, et al. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406(6796):637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 5.Tsukazaki T, et al. Structure and function of a membrane component SecDF that enhances protein export. Nature. 2011;474(7350):235–238. doi: 10.1038/nature09980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schulze RJ, et al. Membrane protein insertion and proton-motive-force-dependent secretion through the bacterial holo-translocon SecYEG–SecDF–YajC–YidC. Proc Natl Acad Sci USA. 2014;111:4844–4849. doi: 10.1073/pnas.1315901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16(10):2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nouwen N, Driessen AJ. SecDFyajC forms a heterotetrameric complex with YidC. Mol Microbiol. 2002;44(5):1397–1405. doi: 10.1046/j.1365-2958.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 9.Bessonneau P, Besson V, Collinson I, Duong F. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 2002;21(5):995–1003. doi: 10.1093/emboj/21.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachelaru I, et al. YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. J Biol Chem. 2013;288(23):16295–16307. doi: 10.1074/jbc.M112.446583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato Y, Nishiyama K, Tokuda H. Depletion of SecDF-YajC causes a decrease in the level of SecG: implication for their functional interaction. FEBS Lett. 2003;550(1-3):114–118. doi: 10.1016/s0014-5793(03)00847-0. [DOI] [PubMed] [Google Scholar]
- 12.Li Z, Boyd D, Reindl M, Goldberg MB. Identification of YidC residues that define interactions with the Sec apparatus. J Bacteriol. 2014;196(2):367–377. doi: 10.1128/JB.01095-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie K, Kiefer D, Nagler G, Dalbey RE, Kuhn A. Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry. 2006;45(44):13401–13408. doi: 10.1021/bi060826z. [DOI] [PubMed] [Google Scholar]
- 14.Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 1997;16(16):4871–4879. doi: 10.1093/emboj/16.16.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato K, Mori H, Yoshida M, Tagaya M, Mizushima S. Short hydrophobic segments in the mature domain of ProOmpA determine its stepwise movement during translocation across the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1997;272(9):5880–5886. doi: 10.1074/jbc.272.9.5880. [DOI] [PubMed] [Google Scholar]
- 16.Beck K, et al. YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2001;2(8):709–714. doi: 10.1093/embo-reports/kve154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moser M, Nagamori S, Huber M, Tokuda H, Nishiyama K. Glycolipozyme MPIase is essential for topology inversion of SecG during preprotein translocation. Proc Natl Acad Sci USA. 2013;110(24):9734–9739. doi: 10.1073/pnas.1303160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao C, et al. Stoichiometry of SecYEG in the active translocase of Escherichia coli varies with precursor species. Proc Natl Acad Sci USA. 2013;110(29):11815–11820. doi: 10.1073/pnas.1303289110. [DOI] [PMC free article] [PubMed] [Google Scholar]