Significance
A fundamental question in developmental biology is how patterns are established in space and time. In plants, key differences in root system architecture are attributed to the spatial distribution pattern of lateral roots (LRs), yet how the pattern of LRs is established is only beginning to be understood. We demonstrate that the establishment of sites competent to form LRs roots requires carotenoid biosynthesis. Furthermore, our results implicate an uncharacterized carotenoid-derived molecule that functions non–cell-autonomously, specifically in LR formation. The results of this study reveal novel aspects of carotenoid biology and expand the roles of carotenoid-derived molecules into root developmental patterning.
Keywords: root development, patterning, secondary metabolite synthesis
Abstract
In plants, continuous formation of lateral roots (LRs) facilitates efficient exploration of the soil environment. Roots can maximize developmental capacity in variable environmental conditions through establishment of sites competent to form LRs. This LR prepattern is established by a periodic oscillation in gene expression near the root tip. The spatial distribution of competent (prebranch) sites results from the interplay between this periodic process and primary root growth; yet, much about this oscillatory process and the formation of prebranch sites remains unknown. We find that disruption of carotenoid biosynthesis results in seedlings with very few LRs. Carotenoids are further required for the output of the LR clock because inhibition of carotenoid synthesis also results in fewer sites competent to form LRs. Genetic analyses and a carotenoid cleavage inhibitor indicate that an apocarotenoid, distinct from abscisic acid or strigolactone, is specifically required for LR formation. Expression of a key carotenoid biosynthesis gene occurs in a spatially specific pattern along the root’s axis, suggesting spatial regulation of carotenoid synthesis. These results indicate that developmental prepatterning of LRs requires an uncharacterized carotenoid-derived molecule. We propose that this molecule functions non–cell-autonomously in establishment of the LR prepattern.
Anchorage and uptake of water and soluble nutrients are essential functions of plant root systems and key to plant productivity and survival. The capacity of a root system to carry out these functions can be maximized by iterative root branching. Root branches are formed de novo during primary root growth, which allows for the elaboration of a complex root system that effectively enables the plant to navigate and exploit the resources of diverse, locally variable subterranean environments. Understanding the developmental mechanisms underlying the pattern of root branches has broad significance in both basic and applied research.
As with other dicotyledonous plants, formation of a complex root system in the model plant Arabidopsis thaliana (Arabidopsis) occurs through iterative production of branches, or lateral roots (LRs), from the primary root. The simplified root system of Arabidopsis has yielded considerable insight into the cellular events and molecular regulators required for LR formation (recently reviewed in refs. 1–4). In Arabidopsis, LRs arise from an internal cell layer, the pericycle, which surrounds the cells of the vascular cylinder (i.e., cambium, xylem, phloem) (5) (Fig. S1 A and B). Pericycle cells adjacent to the xylem pole are unique among cells of this layer because they have a distinct morphology and gene expression profile, as well as the ability to give rise to LRs (6–8). The development of a lateral root primordium (LRP) occurs in seven sequential stages defined by cellular morphology (8). Initiation of an LRP begins when specific xylem pole pericycle cells, called LR founder cells, undergo asymmetrical divisions forming a series of small cells, termed a stage I primordium (7–9). Further cell divisions (stages II–VI) result in development of a dome-shaped LRP. At stage VII, the LRP closely resembles the primary root tip but remains confined within the primary root. The primordium is considered to be an LR only after it has emerged from the primary root and cell division is activated at its apex (8). Developmental progression of individual LRPs is sensitive to a vast range of developmental and environmental cues (reviewed in ref. 10), which, ultimately, allows for plasticity in root system architecture in variable subterranean environments.
In the root’s radial axis, LRs form strictly from a subset of xylem pole pericycle cells; however, for many years, it remained unclear how subsets of cells were specified along the longitudinal axis, that is, how the spacing between LRPs was determined. Because developmental progression and emergence of individual primordia are variable along the length of the primary root, assaying LR number is not necessarily a reliable measure of the total number of sites competent to form an LR or a root’s branching capacity (11, 12). Recent evidence shows that time is an important component in establishing the number of sites competent to form LRs (12, 13). In a region of the root tip termed the oscillation zone (OZ) (Fig. S1A), there is a highly dynamic pattern of gene expression that oscillates within a 6-h period (12). Each peak in expression results in formation of a prebranch site, which is competent to develop an LRP subsequently. During a fixed period, the number of prebranch sites formed in roots grown under various conditions was nearly identical, despite changes in primary root length (12). These results suggested that an LR prepattern is established by an endogenous clock-like mechanism, termed the LR clock (12). Because this periodic process is concurrent with primary root growth, the spatial distribution of prebranch sites, and eventually LRs, is not fixed, yet the total number of competent sites appears stable with time.
Local signaling and long-distance signaling are fundamental mechanisms by which plants coordinate developmental processes across cell types and among organs. Many plant-signaling molecules are synthesized from secondary metabolic pathways, including the carotenoid biosynthesis pathway. Carotenoids play critical roles in organisms from all biological kingdoms; yet, synthesis of carotenoids occurs largely in plants and photosynthetic bacteria, where these pigments have essential functions in light harvesting during photosynthesis and in photoprotection (14, 15). Carotenoids also act as precursors in the synthesis of a range of small molecules (apocarotenoids) with diverse functions across the plant and animal kingdoms (16–18). For example, carotenoid pigments and volatile apocarotenoids, such as α- and β-ionone, are key aroma and flavor elements in attracting the agents of pollination and seed dispersal (18–20). Carotenoids serve as precursors for abscisic acid (ABA) and strigolactones, phytohormones that function in plant development as well as in response to the environment (21–25), and have also been implicated in the production of other regulatory signaling molecules (26, 27). Despite diverse roles in plant growth and development, functional analyses of carotenoids have largely been focused on aboveground organs.
In a chance observation, we discovered that Arabidopsis seedlings treated with carotenoid biosynthesis inhibitors produced very few LRs, suggesting that carotenoids have important functions in LR formation. Here, we show that carotenoids are required for prebranch site formation, indicating they are necessary for establishing the pattern of LRs along the root’s longitudinal axis. Additionally, we provide genetic and pharmacological evidence that an uncharacterized β-carotenoid–derived apocarotenoid functions in this process. Expression data suggest carotenoid biosynthesis is preferentially excluded from the region of the root tip encompassing the OZ and appears to peak in more differentiated regions. We suggest that this apocarotenoid provides a non–cell-autonomous cue necessary for the establishment or maintenance of the sites competent to form an LRP.
Results
LR Capacity Assay as a Measure of Competence for Root Branching.
The current paradigm for assessing changes in LR development in various genotypes and/or growth conditions is to determine LR density (the number of LRs and LRPs observed per unit length of the primary root) (28). Although this method provides insight into the development of an LRP based on cellular morphology, it cannot provide information regarding prebranch sites, because they are defined by expression of the pDR5:Luciferase (pDR5:LUC) reporter gene and thought to occur before morphological changes (12). Additionally, because it has been shown that prebranch site formation is periodic and that a consistent number of prebranch sites are formed even in conditions that decrease primary root length by nearly 50% (12), root length is a less critical parameter than time when considering establishment of competence to form LRs. Thus, we view LR density as an inadequate means to capture a root’s “capacity” (the total number of competent sites) for LR formation.
We observed that seedlings treated with two chemical inhibitors of the carotenoid biosynthesis pathway, norflurazon (NF; Chem Service) or 2-(4-chlorophenylthio)-triethylamine hydrochloride (CPTA) (Fig. 1A), had reduced primary root length and very few, if any, emerged LRs (Fig. 1 B and C and Fig. S2A). However, changes in LR number may occur as a result of changes in either the developmental progression of an LRP or the number of prebranch sites. Because we are primarily interested in the establishment of competence to form LRs over a given time period, as opposed to the developmental progression of LRPs, we sought an assay that would allow facile assessment of a root’s capacity for LR formation in different genotypes and growth conditions.
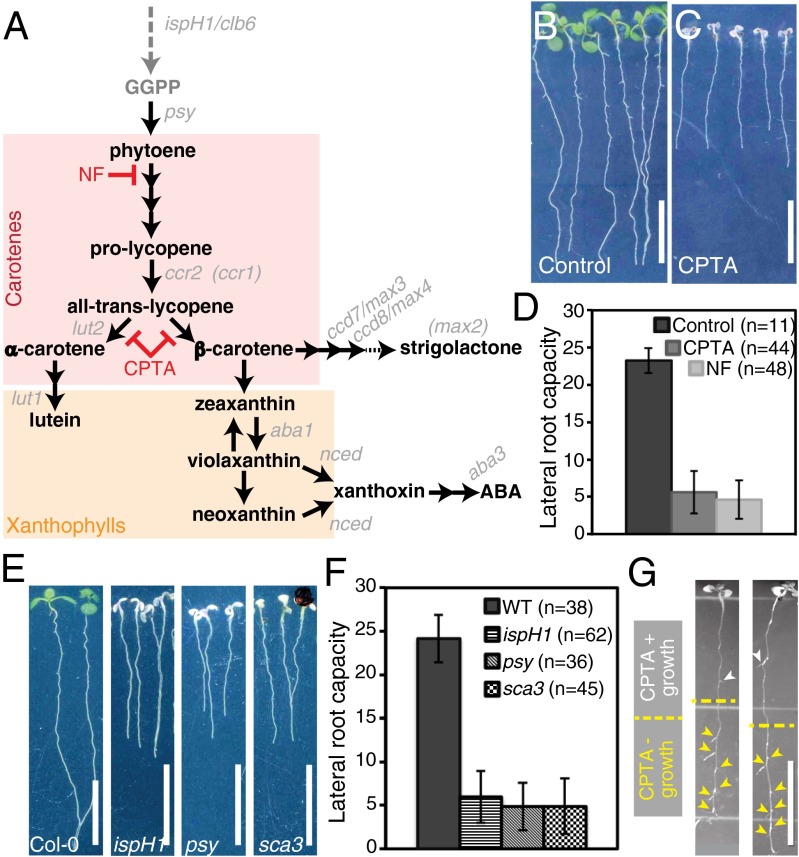
Fig. 1.
Carotenoid-deficient seedlings produce few LRs. (A) Simplified version of the carotenoid biosynthesis pathway. Carotenoid- and apocarotenoid-specific biosynthesis steps are depicted in black, and upstream steps are shown in dark gray. Carotenoid biosynthesis mutants used in this study are indicated (gray italics), and those with an indirect impact are shown in parentheses. The dotted arrow to strigolactone represents unknown final steps. Columbia-0 (Col-0) seedlings were grown under control (B) and CPTA treatment (C) conditions. (D) Quantification of LR capacity in control and NF- or CPTA-treated seedlings. (E) Col-0 and ispH1/clb6, psy, and sca3 albino seedlings. (F) Quantification of LR capacity in Col-0 and albino seedlings. The difference between control and treated or control and mutant in D and F, respectively, is statistically significant (Student t test, P < 1 × 10−6). (G) CPTA-treated seedlings transferred to control medium (yellow dotted line indicates root tip position at transfer). Arrowheads indicate emerged LRs: those formed in the presence of CPTA (white) and those formed after transfer (yellow). Error bars represent SD. (Scale bars: 1 cm.)
Prebranch site number is determined by the number of sites with pDR5:LUC activity along the primary root at 8 d after stratification (das) (12). To compare the number of prebranch sites with the number of LRs and LRPs at 8 das, seedlings were assayed for prebranch sites and then fixed and cleared to count LRs and LRPs. Upon clearing, fewer LR/LRP sites than prebranch sites were observed in the same roots, and the difference between these numbers on a root-by-root basis was variable (Table S1). This variability in the number of LR/LRP sites in cleared roots compared with the prebranch site number is likely to be due to natural variability in the developmental progression of competent sites to LRs between individual roots. Additionally, it is possible that early-stage LRPs are missed in cleared roots due to root orientation on the slide or tissue damage incurred during processing. These results indicate that although LR/LRP number can be interpreted as a measure of how many prebranch sites have progressed to form LRPs or LRs, it has limited reliability in accounting for a root’s total capacity to form LRs.
Although assessing prebranch site number on the basis of pDR5:LUC activity is straightforward, luciferase imaging systems of sufficient resolution are expensive and not readily available, and crossing this reporter to many genotypes with putative phenotypes in LR formation can be onerous. Therefore, we developed an assay that is simple and provides a more accurate measure of LR capacity. In brief, the root tip is excised just above the OZ, and the seedlings are then grown for several more days (Materials and Methods). We find root tip excision promotes the developmental progression of nearly all prebranch sites toward LR emergence. In the LR capacity assay, the number of prebranch sites is not significantly different from the number of LRs and late-stage LRPs, similar to previously published results (12), and the difference between these numbers on a root-by-root basis is less variable (Table S1). These results indicate that our LR capacity assay serves as an accurate reflection of the total number of prebranch sites and branching capacity of a given root and that it can be used to rapidly and accurately assess whether changes in LR prepatterning have occurred in various genotypes and/or growth conditions.
Inhibition of the Carotenoid Biosynthesis Pathway Reduces LR Capacity.
To investigate the role of carotenoids in LR formation, we examined LR capacity in seedlings treated with NF, which inhibits the desaturation of phytoene, the second step of carotenoid biosynthesis. We also treated seedlings with CPTA, which blocks the cyclization of lycopene, which is required for the synthesis of the cyclized carotenoids, α- and β-carotene (Fig. 1A and Fig. S1C). Seedlings grown in the presence of either of these inhibitors had decreased LR capacity (Fig. 1D). In the absence of photoprotective pigments, these seedlings also had smaller albino shoots (Fig. 1C). Thus, it was possible that reduced LR capacity in carotenoid-deficient seedlings occurred as a secondary consequence of either albinism or an unknown toxicity of the chemical treatment.
To determine if these chemical inhibitors were eliciting a toxic effect that reduced LR formation, we examined Arabidopsis mutants that phenocopied the albino shoot phenotype of the chemically treated plants. Mutations in genes encoding enzymes involved directly in isoprenoid and carotenoid biosynthesis, as well as genes involved in chloroplast biogenesis, were tested. Mutants in the biosynthesis genes 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase/CHLOROPLAST BIOGENESIS 6 (HDR/IspH/CLB6) (29, 30) and PHYTOENE SYNTHASE (PSY) (31) each showed a similar reduction in LR capacity as the inhibitor treatments (Fig. 1 E and F). Mutation of a nuclear-encoded, plastid-targeted RNA polymerase SCABRA 3 (SCA3) (32), which is required for chloroplast biogenesis and has an indirect impact on carotenoid content, also showed similarly reduced LR capacity (Fig. 1 E and F). These results indicate that chemical treatment or genetic lesions that directly or indirectly impair carotenoid biosynthesis result in a reduced LR capacity phenotype. Thus, the possibility that decreased LR capacity under chemical treatment is merely a toxic side effect of the chemical appears unlikely; however, if LR capacity were reduced as an indirect phenotypic consequence of albinism, it might be expected that roots of albino seedlings would exhibit additional phenotypes.
CPTA-treated roots were examined for additional phenotypes associated with general defects in root growth and development. The reduction in primary root length upon CPTA treatment is due to a decrease in root growth rate beginning at 3 das (Fig. S2 A and E). Additionally mature cell size is reduced, with cells of CPTA-treated roots being ∼30% shorter than controls (Fig. S2F). The size of the meristematic zone in CPTA-treated roots is comparable to WT at 5 das and begins to decrease in size by 7 das; however, the cellular morphology of the root meristem appeared normal at all time points examined (Fig. S2 B–D). Moreover, carotenoid-deficient roots show normal growth responses because they are both gravitropic and responsive to reorientation to the gravity vector (Fig. 1 C and E and Fig. S2 G and H). Also, if CPTA treatment resulted in substantial damage, the seedlings might not be expected to recover if CPTA were removed (33, 34). Seedlings germinated on CPTA-containing growth medium were transferred to standard growth medium at 5 das and assayed for recovery. Within 3 d of transfer, the newly formed leaves were green, with the first leaf pair and cotyledons remaining albino; however, more importantly, LR formation resumed, and the pattern of LR formation appeared nearly normal (Fig. 1G and Fig. S2I). These results show that exposure to carotenoid biosynthesis inhibitors has minimal lasting consequences on seedling growth and development, particularly on the formation of new organs. Additionally, we observed very few LRs in the portion of the root that formed in the presence of CPTA, suggesting that carotenoid biosynthesis is required for an early step in LR formation. The possibility that an indirect effect of albinism was responsible for reducing LR capacity was further discounted because a carotenoid cleavage inhibitor (see below) was identified that similarly reduced LR capacity while the shoot remained green. Taken together, our results reveal a more direct role for carotenoid biosynthesis in LR formation.
Carotenoid Biosynthesis Is Required for Prebranch Site Formation.
The LR clock functions at the earliest known step of LR formation with a periodic oscillation in gene expression as the key temporal feature, which leads to competence to initiate an LRP at a prebranch site (12). To determine if carotenoids are functioning at this early step in LR formation, we treated seedlings expressing the pDR5:LUC transcriptional reporter (12) with CPTA and examined the number of prebranch sites formed. CPTA-treated seedlings showed dramatically reduced numbers of prebranch sites (Fig. 2). Importantly, the average number of prebranch sites closely corresponded with the average LR capacity observed in carotenoid-deficient seedlings (compare Fig. 2E and Fig. 1D). Thus, even under carotenoid-deficient conditions, once a prebranch site is formed, it can go on to initiate an LRP, suggesting that carotenoid biosynthesis is necessary to establish the normal number of prebranch sites.
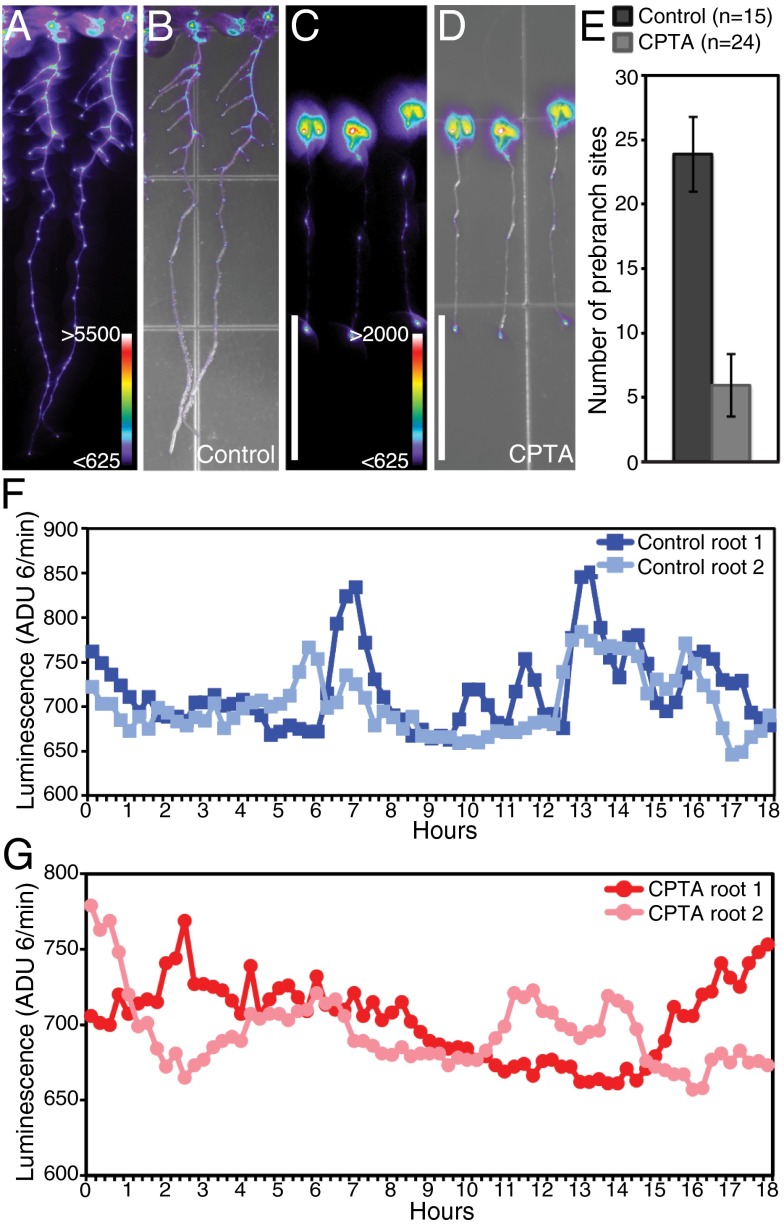
Fig. 2.
Carotenoid-deficient plants have few prebranch sites. pDR5:LUC expression in control (A and B) and CPTA-treated seedlings (C and D). Luciferase activity (A and C) and overlays of luciferase activity and bright-field images (B and D) are shown. Color bars represent the range of analog-digital units (ADU) for luciferase activity. (Scale bars: 1 cm.) (E) Quantification of prebranch site number in control and CPTA-treated seedlings; the difference is statistically significant (Student t test, P < 1 × 10−6). Error bars represent SD. pDR5:LUC expression in the OZ is shown over time in control (F) and CPTA-treated (G) seedlings.
To determine if carotenoids are important for the oscillatory process that establishes prebranch sites, we examined real-time pDR5:LUC expression in carotenoid-deficient seedlings. In CPTA-treated roots, the oscillation of pDR5:LUC was arrhythmic and appeared poorly organized compared with controls (Fig. 2 F and G and Movies S1 and S2). These results suggest that the LR clock is disrupted in carotenoid-deficient seedlings, rendering them unable to establish the LR prepattern.
Expression of Carotenoid-Related Genes Supports a Role for Carotenoids in LR Formation Independent of the LR Clock’s Oscillation in Gene Expression.
Carotenoid-related gene expression was not predicted to be part of the oscillatory mechanism of the LR clock because carotenoid-related gene ontology (GO) terms are not significantly enriched among the oscillating gene expression dataset (12). To examine more closely how carotenoid-related genes are expressed in the oscillating dataset, a list of “carotenoid metabolism” genes was assembled (Table S2). Of the 155 carotenoid metabolism genes, 13% can be described as oscillating, a similar proportion as found in the whole genome (17%), with similar proportions oscillating in either phase or antiphase with pDR5 expression. Of the 16 genes encoding core carotenoid biosynthesis enzymes (35), three are present among the oscillating genes (12); however, they are not confined to a specific branch or part of the biosynthesis pathway (Table S2), and each shows a low mean expression value (<1 in all but two of 39 samples for one of the three genes). These results indicate that, overall, carotenoid metabolism or biosynthesis genes are not part of the oscillatory transcriptional output of the LR clock.
To gain additional insight into how carotenoid biosynthesis might relate to the LR clock, spatiotemporal expression of the core carotenoid biosynthesis genes was examined in the RootMap, a high-resolution compilation of transcriptional profiling data from cell lineages and developmental regions along the longitudinal axis of the root (6). Hierarchical clustering by cell types revealed that expression of the core carotenoid biosynthesis genes is high in cell types closely associated with LR formation and development (Fig. 3A). In the longitudinal dataset, a group of the core carotenoid biosynthesis genes shows higher expression in the more differentiated portions of the root (Fig. S3A). These expression data suggest that carotenoid biosynthesis preferentially occurs in the pericycle cell types and differentiated portions of the root, and thus is consistent with a role for carotenoid biosynthesis in LR formation.
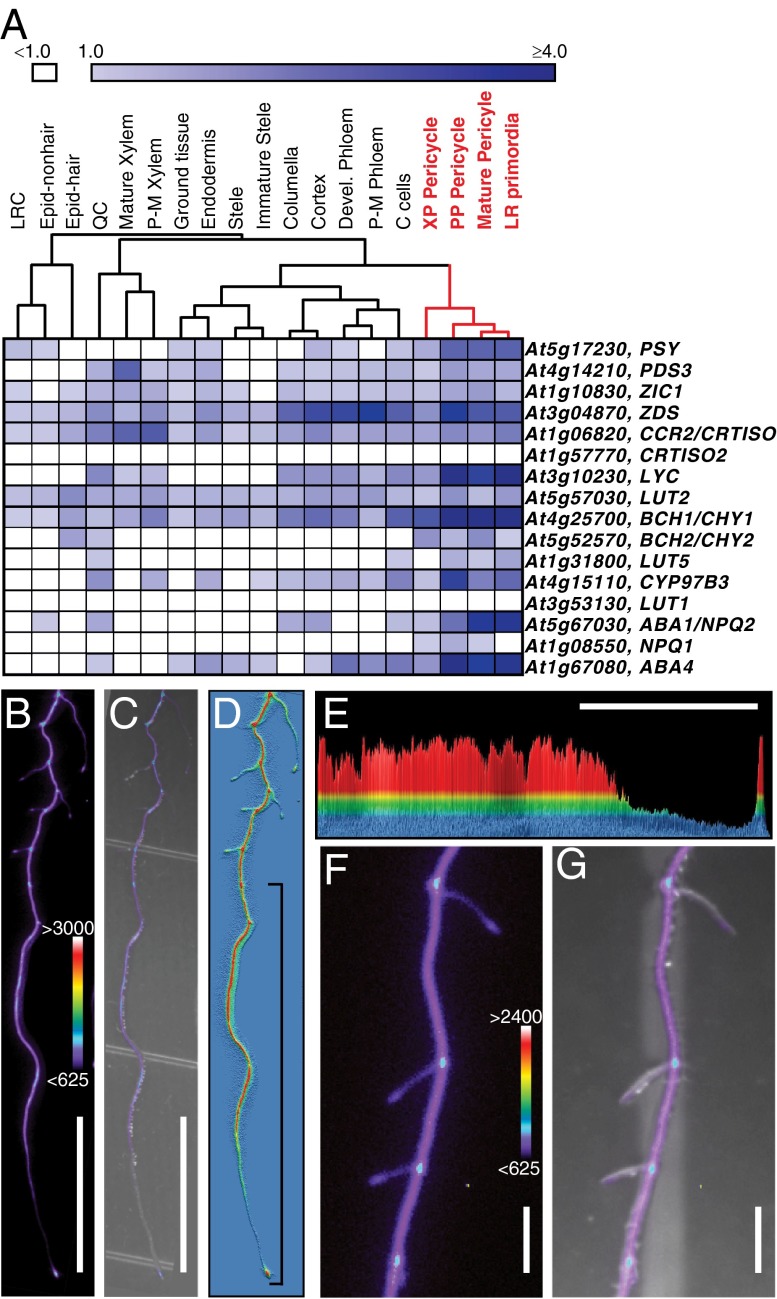
Fig. 3.
Expression of carotenoid biosynthesis genes is consistent with a role for carotenoids in LR formation. (A) Heat map of mean expression values of the core carotenoid biosynthesis genes in the root cell types (6). Core carotenoid biosynthesis gene expression is higher and most similar among cell types closely associated with LR formation (red). Note these genes are listed by their position in the pathway with PSY at the top (35). C cells, companion cells; Devel. Phloem, developing phloem; Epid-hair, epidermis hair cells; Epid-nonhair, epidermis nonhair cells; LRC, lateral root cap; QC, quiescent center; P-M phloem, proto- and metaphloem; P-M xylem, proto- and metaxylem; PP Pericycle, phloem pole pericycle; XP pericycle, xylem pole pericycle. White boxes indicate mean expression values of <1.0. (B–G) pPSY:LUC reporter gene expression in roots. (B) Luciferase activity. (C) Overlay of luciferase activity from B and bright-field image. (D and E) Histograms of luciferase activity from B; red represents high values, and blue represents no expression. The bracket in D indicates the region shown in E. (E) Two-dimensional representation of D with LUC activity on the y axis and with the root along the x axis (root tip at right). (F) Luciferase activity at the base of LRs and in later stage primordia. (G) Overlay of luciferase activity from F and bright-field image. Color bars represent the range of ADU for luciferase activity. (Scale bars: B, C, and E, 10 mm; F and G, 1 mm.)
Expression of the Rate-Limiting Carotenoid Biosynthesis Gene, PSY, Is Spatially Restricted in the Root.
PSY encodes the enzyme that catalyzes the first committed step of carotenoid synthesis. Varying expression or activity of PSY alters flux through the carotenoid biosynthesis pathway (36–38). We predicted that the expression pattern of PSY would provide functional insight into the carotenoid biosynthesis pathway in roots. However, due to low expression values (<1 in all but two of 25 samples), its expression was difficult to determine from the RootMap (Fig. S3A).
To conduct a more detailed transcriptional analysis of this key enzyme, a PSY transcriptional reporter was generated by fusing 3.5 kb upstream of the PSY start codon to the luciferase reporter gene (pPSY:LUC). In the root, pPSY:LUC showed a spatially varying expression pattern with highest activity in the more differentiated portions of the root and diminishing activity toward the OZ, where it is undetectable (Fig. 3 B–E). Unexpectedly, it appears that expression of PSY is specifically excluded from the OZ. A similar pattern of pPSY:LUC expression was also observed in longer LRs (Fig. 3 B and D). Additionally, luciferase activity was observed in the LRP, at the junction between the primary root and LRs (Fig. 3 B, D, F, and G) and at the very tip of the root, which likely corresponds to the LR cap. To examine the functional domain of the encoded protein, PSY translational fusions were generated. The pPSY:PSY:LUC transgene rescued psy mutant seedlings (Table S3), confirming that the transgenic PSY coding region was functional and that the promoter region was sufficient to recapitulate endogenous PSY expression. However, luciferase activity was weakly detectable only after very long exposure times (Fig. S3 B–E), and PSY localization could not be ascertained.
PSY was overexpressed by driving expression with the UBIQUITIN 10 promoter (pUBQ10) in the presence and absence of visual markers (pUBQ10:PSY:GFP, pUBQ10:PSY:LUC, and pUBQ10:PSY). All of these overexpression constructs complemented psy mutant seedlings (Table S3). The PSY:GFP fusion protein was detectable in plastids when overexpressed (Fig. S4 A–F), but it was not detectable and failed to rescue psy mutants when expressed under the endogenous promoter (Table S3). Additionally, despite overexpression and very high expression of the pUBQ10:LUC control (Fig. S3 H–J), PSY:LUC activity was weakly detectable only after long exposure times (Fig. S3 F and G). The most parsimonious explanation for these results is based on the localization of PSY to the plastoglobules (39). We propose that the suborganellar localization of PSY may render PSY:GFP fusions only partially functional and C-terminally fused LUC inaccessible to its substrate, luciferin, which is required for visualization of the marker. These observations suggest that although C-terminal reporter constructs are biologically functional in planta, they are not useful for analysis of endogenous PSY expression in planta. Nevertheless, expression of pPSY:LUC is consistent with spatial restriction of PSY activity and, therefore, carotenoid biosynthesis. This restriction to more differentiated regions of the root and apparent preferential exclusion from the OZ suggest a non–cell-autonomous function for a carotenoid derivative along the longitudinal axis.
Plants expressing PSY under the constitutive 35S (35S:PSY) (36) or UBQ10 promoter did not show alterations in LR capacity compared with control seedlings (Fig. S4G). Because plants overexpressing PSY (35S:PSY) have increased levels of carotenes, particularly in the roots (36), these results indicate that LR clock function is not disrupted by a general increase in carotenoid content. Additionally, they suggest that synthesis of the carotenoid derivative (apocarotenoid) that functions in LR formation is controlled downstream of precursor availability. This type of control would be similar to the apocarotenoid ABA, which is not produced in excess in leaves upon overexpression of PSY (40) and whose rate-limiting step typically occurs at the enzymatic cleavage of its carotenoid precursor when precursors are not limiting (41–44).
Genetic Analyses Implicate a β-Carotene Derivative Distinct from ABA or Strigolactone in LR Formation.
The cyclization of lycopene is a key branch point in the carotenoid biosynthesis pathway, leading to α- and β-carotene and their respective derivatives. Because CPTA blocks the cyclization of lycopene and reduces LR capacity, we hypothesized that a downstream carotenoid is required for LR formation. To narrow down the list of candidate carotenoids further, plants with mutations in key carotenoid metabolism genes were examined for changes in LR capacity.
The α-carotene branch of the pathway was examined beginning with the carotenoid chloroplast regulatory 1 (ccr1)/set domain group 8 (sdg8) and ccr2 [carotenoid isomerase (CRTISO)] mutants. Mutation of either ccr1 or ccr2 results in limited flux through the α-carotene branch, and these genes are required for normal lutein accumulation (45, 46) (Fig. 1A and Fig. S1C). In ccr1 seedlings, LR capacity was not significantly different from WT. The ccr2 seedlings showed a decrease in LR capacity compared with WT, although this decrease was modest, particularly compared with seedlings in which carotenoid biosynthesis was inhibited (Fig. 4 A and B). Although CCR2 encodes a CRTISO and isomerization of lycopene occurs upstream of cyclization (Fig. S1C), nonenzymatic isomerization of lycopene occurs in light (photoisomerization), leading to suppression of the biochemical phenotype (47). Depending on growth conditions and tissue or plastid type, cis-carotenoids accumulate and xanthophyll levels are reduced in ccr2 mutants (45, 46). Therefore, the decrease in LR capacity in ccr2 may indicate that LR formation is sensitive to even minor perturbations in flux through the carotenoid biosynthesis pathway. To address the requirement for α-carotene or its derivatives in LR formation more specifically, we next examined LR capacity in seedlings with mutations in the LUTEIN 2 (LUT2, EPSILON CYCLASE) or LUTEIN 1 (LUT1, EPSILON HYDROXYLASE) gene, both of which encode enzymes with direct roles in synthesis of α-carotene and lutein, respectively (48–50). Seedlings with mutations in either the LUT2 or LUT1 gene showed no change in LR capacity (Fig. 4A). Altogether, these results indicate that the α-carotene branch is dispensable for LR formation under our conditions.
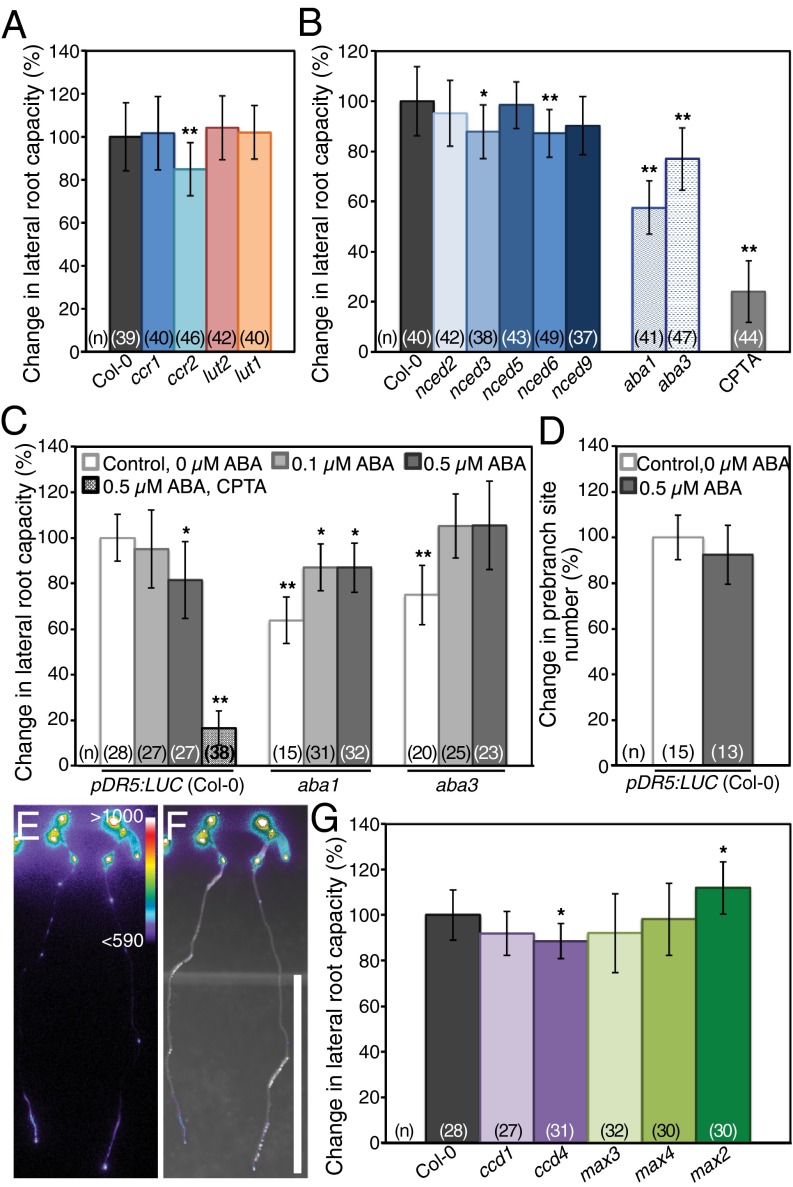
Fig. 4.
Mutant analyses and ABA supplementation indicate that a β-carotene derivative distinct from ABA or strigolactone participates in LR formation. Quantification of LR capacity in mutants required for α-carotene biosynthesis (A) and mutations in each NCED and two ABA biosynthesis genes compared with their respective controls (B). For comparison, the change in LR capacity of CTPA-treated seedlings is included. (C) Quantification of LR capacity in control, 0.1 μM ABA, 0.5 μM ABA or CPTA, and 0.5 μM ABA-treated seedlings. (D) Quantification of prebranch site number in control and 0.5 μM ABA-treated seedlings [this difference is not statistically significant (Student t test, P > 0.05)]. (E and F) pDR5:LUC expression in seedlings grown on CPTA and 0.5 μM ABA. Luciferase activity (E) and overlay of bright-field and luciferase activity (F) are shown. Color bar in E represents range of ADU for luciferase activity. (Scale bar: 1 cm.) (G) Quantification of LR capacity in seedlings with mutations in CCD genes and MAX2 compared with their respective controls. Asterisks indicate statistically significant differences compared with controls (Student t test: *P < 1 × 10−3; **P < 1 × 10−6). Error bars represent SD.
Next, we examined the β-carotene branch of the pathway, from which ABA and strigolactones are derived. These apocarotenoids are phytohormones with roles in environmental response and development (21–25, 51–61). Production of apocarotenoids, including ABA and strigolactone, requires oxidative cleavage of carotenoid precursors. In Arabidopsis, a nine-member family of enzymes with two subgroups [four carotenoid cleavage dioxygenases (CCDs) and five 9-cis-epoxycarotenoid dioxygenases (NCEDs)] carries out these cleavage reactions (62). We first examined the NCEDs, which cleave 9′-cis-neoxanthin and/or 9-cis-violaxanthin as the first step toward ABA biosynthesis (Fig. S1C). The five NCED genes show specificity for the same substrate in vitro but have differential expression patterns and distinct suborganellar localization within the plastid (43, 63, 64). Seedlings with mutations in nced2 and nced5 each had an LR capacity similar to WT controls (Fig. 4B). The LR capacity of nced3, nced6, and nced9 seedlings was slightly decreased compared with WT, which may reflect either functional redundancy among the NCEDs or an inherent sensitivity of LR formation to alterations in carotenoid catabolism.
With functional redundancy a concern among the NCEDs, we next examined other ABA-deficient mutants. The ABSCISIC ACID DEFICIENT 1 (ABA1) gene is upstream of the NCEDs and encodes zeaxanthin epoxidase, which functions to convert zeaxanthin sequentially into antheraxanthin and all trans-violaxanthin (Fig. S1C). Plants with mutations in aba1 accumulate high levels of zeaxanthin and are deficient in downstream carotenoids and ABA (65, 66). The ABA3 gene is downstream of the NCEDs and encodes a molybdenum cofactor sulfurase required at the final step of ABA biosynthesis (67–69). Seedlings with mutations in each of these genes showed a reduction in LR capacity compared with WT (Fig. 4B); however, neither mutation reduced LR capacity to the same degree as chemical or genetic inhibition of carotenoid biosynthesis (Fig. 1 D and F).
We hypothesized that if the reduction in LR and prebranch site number under carotenoid-deficient conditions were primarily due to the absence of ABA, then carotenoid- and ABA-deficient seedlings supplied with exogenous ABA would resume LR and prebranch site formation. Exogenous ABA has been shown to rescue other ABA-deficient phenotypes, such as wilting (66, 69). However, exogenous ABA has also been shown to arrest LRP development postemergence without altering LRP number, suggesting that ABA function is restricted to the later stages of LR development (70). To determine if ABA has a role in the establishment of competence to form an LRP, we examined LR capacity in WT seedlings treated with 0.1 μM and 0.5 μM ABA. For seedlings treated with 0.1 μM ABA, we observed no difference in LR capacity with the control, although for seedlings treated with 0.5 μM ABA, we observed a modest but significant reduction in LR capacity (Fig. 4C). To determine whether 0.5 μM ABA reduced the number of sites competent to form an LRP, prebranch site number was examined; however, no significant difference was observed (Fig. 4D). These ABA treatments were previously shown to reduce the number of visible LRs to approximately one-third of the control or to zero, respectively (70). Our results indicate that the LR capacity assay largely overrides the ABA-sensitive checkpoint predicted to operate in LRP development postemergence and that it is highly unlikely that ABA has an impact on the establishment of prebranch sites.
Next, we tested if exogenous ABA could rescue LR capacity in ABA- or carotenoid-deficient seedlings. We found that ABA supplementation of aba biosynthetic mutants increased LR capacity (Fig. 4C). Seedlings with mutations in ABA3 are defective in the final step of ABA biosynthesis, and exogenous ABA rescued LR capacity in aba3 mutants to WT levels. The complete rescue of LR capacity by both concentrations of ABA suggests that there is some optimum range of ABA levels for normal LR capacity but that modest changes are observed on either side of this range. Alternatively, it may suggest that altered ABA levels result in changes in carotenoid precursor availability to different signaling pathways. Mutations in the upstream gene ABA1 result in decreases in carotenoid levels downstream of zeaxanthin, including reduction in ABA synthesis, and in aba1 mutants, exogenous ABA also increased LR capacity but unexpectedly failed to rescue it to WT levels. In contrast to the aba mutants, seedlings treated simultaneously with CPTA and ABA showed no increase in prebranch site or LR capacity (Fig. 4 C, E, and F). Although these results support some role for ABA in LR formation, they indicate that upon inhibition of carotenoid biosynthesis, ABA deficiency is not sufficient to account for the reduction in LR capacity and prebranch site number observed.
The four CCDs have diverse substrates, cleavage activities, and subcellular localization, and they also show functional redundancy (62). Two of these enzymes, CCD7/MORE AXILLARY GROWTH 3 and CCD8/MAX4, are known to be required for strigolactone biosynthesis (54, 55, 71). Seedlings with a mutation in strigolactone biosynthesis genes showed similar LR capacity as WT (Fig. 4G). Additionally, seedlings with a mutation in the MAX2 gene, which encodes an F-box protein that functions in perception of strigolactones (61, 72), showed a slight increase in LR capacity compared with WT (Fig. 4G). A modest decrease in LR capacity was observed in seedlings with mutations in the CCD1 and CCD4 genes. These genetic analyses indicate that no single CCD is required for LR formation and that strigolactone is not involved in the early steps of this process. In summary, our analyses do not support a primary role for either of the two known β-carotene–derived apocarotenoid signaling molecules (ABA or strigolactone) in LR formation. Instead, an uncharacterized apocarotenoid, likely derived from the β-carotene branch of the pathway, is implicated in LR formation.
Carotenoid Cleavage Inhibitor, D15, Functions as an Inhibitor of CCD Activity in Vitro.
Synthesis of distinct biologically active apocarotenoids requires specific oxidative cleavage of precursors. For example, ABA synthesis requires cleavage of 9′-cis-neoxanthin and/or 9-cis-violaxanthin at the 11,12 position (73), whereas strigolactone synthesis requires cleavage of 9-cis-β-carotene at the 9,10 position (71). Because the CCDs and NCEDs show functional redundancy among family members and have multiple cleavage activities and substrate promiscuity, eliminating a role for any one enzyme in LR formation based on genetic analysis is difficult. An approach to addressing these challenges is to design selective inhibitors against specific carotenoid cleavage activities. Several of these inhibitors have been shown to induce shoot branching phenotypes in WT consistent with disruption of the carotenoid cleavage that is required for strigolactone synthesis (74).
An aryl-C3N hydroxamic acid analog, D15 (Fig. S5A), was designed and synthesized based on previously reported carotenoid cleavage inhibitors (74). D15 is a candidate inhibitor for carotenoid cleavage at the 9,10 position and was found to elicit an LR capacity phenotype in the absence of an albino phenotype (Fig. 5 A–C). To determine its inhibitory function against carotenoid cleavage enzymes, we examined the activity of D15 against recombinant proteins from tomato (LeCCD1a and LeNCED1) in vitro. D15 was found to be a stronger inhibitor of LeCCD1a activity than of LeNCED1 activity (Table S4), suggesting that it is a more potent inhibitor of 9,10 cleavage enzymes (CCD) than 11,12 cleavage enzymes (NCED).
Fig. 5.
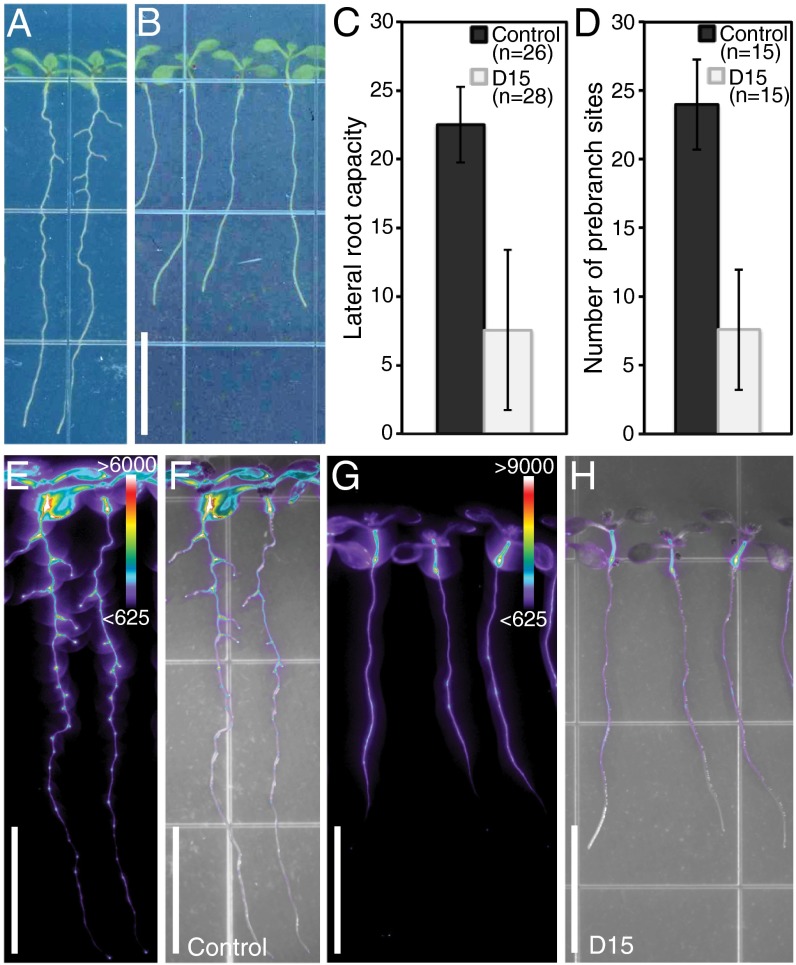
Treatment with the carotenoid cleavage inhibitor, D15, reduces LR capacity and prebranch site number without inducing an albino phenotype. pDR5:LUC expression in control (A, E, and F) and D15-treated (B, G, and H) seedlings. Quantification of LR capacity (C) and prebranch site number (D) are shown; these differences are statistically significant (Student t test, P < 1 × 10−6). Error bars represent SD. Luciferase activity (E and G) and luciferase activity overlaid with bright-field images (F and H) are shown. Color bars in E and G represent the range of ADU for luciferase activity. (Scale bar: 1 cm.)
D15 Treatment Indicates an Uncharacterized Apocarotenoid Is Involved in LR Formation.
Seedlings treated with D15 showed a modest reduction in primary root length but a highly significant decrease in LR capacity (Fig. 5 A–C and Fig. S5B). This decrease in LR capacity was similar to that observed in seedlings in which carotenoid biosynthesis was inhibited either pharmacologically or genetically (compare Fig. 5C with Fig. 1 D and F). Additionally, D15-treated seedlings have a clear reduction in prebranch site number (Fig. 5 D–H). Unlike treatment with the carotenoid biosynthesis inhibitors, which resulted in a small, albino shoot phenotype (Fig. 1C), D15-treated seedlings exhibited green shoots of comparable size to controls (Fig. 5B). This observation clearly demonstrates a function for carotenoids in LR formation that can be distinguished from possible secondary impacts of carotenoid deficiency and can unequivocally be uncoupled from their role as photoprotective pigments.
Additionally, we measured ABA content in CPTA- and D15-treated roots and shoots and found that in contrast to CPTA-treated tissues, D15-treated tissues do not have reduced ABA content (Fig. S5 D and E). These results are consistent with the inhibitory activity of D15 in vitro and with our phenotypic data excluding ABA deficiency as the primary basis for reduced LR capacity under carotenoid-deficient conditions. Furthermore, they indicate that a possible increase in ABA content, perhaps due to changes in metabolic flux through the pathway upon D15 treatment, also cannot explain the reduction in LR capacity. These results provide further evidence that a previously uncharacterized apocarotenoid is required for LR formation.
The D15-induced decrease in LR capacity and preferential inhibition of LeCCD1 activity in vitro suggest that cleavage of a carotenoid at the 9,10 position is required for LR formation. To assess whether the reduction in LR capacity is specific to D15 or is a common feature of inhibitors that exhibit preferential inhibition of LeCCD1 activity, we tested previously characterized inhibitors that were shown to be active in planta (74). Seedlings treated with D1, D5, and D6 (Fig. S5A and Table S4) had no detectable changes in shoot pigmentation and exhibited normal or near-normal LR capacity (Fig. S5C). Despite preferential inhibition of LeCCD1 activity in vitro, these four carotenoid cleavage inhibitors do not elicit the same LR capacity phenotype, suggesting that they may have distinct target specificity or inhibitory activities in planta.
The effect of D15's inhibitory activity on carotenoid content in planta was unknown. Additionally, because D15 specifically reduces LR capacity without having obvious effects on photoprotective pigments, we reasoned that analysis of carotenoid content in D15-treated seedlings might provide insight into the synthesis of the apocarotenoid(s) that function in LR formation. To determine how D15 affects carotenoid content in planta, we conducted HPLC analysis on tissue from the shoots and roots of D15-treated seedlings. D15-treated shoots exhibited little change in carotenoid content, with only zeaxanthin levels showing a modest decrease, and no change in chlorophyll levels compared with controls (Table 1 and Fig. S5F). Strikingly, carotenoid content was significantly increased by approximately twofold in D15-treated roots compared with control roots (Table 1). These results indicate that D15 treatment has an impact on carotenoid cleavage activity downstream of the photoprotective function of carotenoids and suggests that D15-inhibited carotenoid cleavage activity leads to altered carotenoid levels specifically in the root. Taken together, our results implicate an uncharacterized apocarotenoid, likely derived from carotenoid cleavage at the 9,10 position, in the earliest steps of LR formation.
Table 1.
D15 disrupts pathway catabolism and increases carotenoid content in roots
| Carotenoid levels (micrograms per gram) | ||||||||
| Tissue | Treatment | Neoxanthin | Violaxanthin | Antheraxanthin | Lutein | Zeaxanthin | β-Carotene | Total |
| Control | 38 (±2.7) | 47 (±2.2) | 11 (±0.5) | 139 (±9.1) | 2 (± 0.2) | 66 (±4.2) | 304 (±18) | |
| Shoots | D15 | 41 (±4.4) | 55 (±4.4) | 8 (±1.1) | 150 (±15.9) | 1 (± 0.3) | 69 (±7.8) | 325 (±36) |
| Fold change | 1.1 | 1.2 | 0.7 | 1.1 | 0.5* | 1.0 | 1.1 | |
| Control | 0.26 (±0.02) | 0.34 (±0.02) | 0.00 | 0.2 (±0.02) | 0.00 | 0.05 (±0.00) | 0.84 (±0.03) | |
| Roots | D15 | 0.48 (±0.02) | 0.56 (±0.04) | 0.02 (±0.01) | 0.37 (±0.04) | 0.00 | 0.10 (±0.00) | 1.53 (±0.1) |
| Fold change | 1.9** | 1.7** | — | 1.9* | — | 1.9** | 1.8** | |
P ≤ 0.05; **P ≤ 0.01.
Discussion
Time is a critical factor in establishment of the LR prepattern along the longitudinal axis of the root. The relationship between the temporal patterning mechanism and primary root growth rate permits variability in the spatial distribution of LRs (12); however, much about this LR prepatterning mechanism remains unknown. We present evidence that a carotenoid derivative functions in prebranch site formation and, therefore, LR prepatterning in Arabidopsis.
Several lines of evidence support the involvement of a carotenoid in LR formation. Inhibition of the carotenoid biosynthesis pathway, either chemically or genetically, results in seedlings with a strongly reduced LR capacity. Carotenoid-deficient seedlings also produce very few prebranch sites. Under carotenoid-deficient conditions, the close correlation between the number of prebranch sites and LR capacity suggests that there is no defect in the developmental progression of prebranch sites to LRs. Thus, carotenoid biosynthesis is tied to the earliest steps of LR formation before development of primordia.
The possibility that the reduction in LR capacity under carotenoid-deficient conditions can be attributed to an indirect, secondary consequence of the inhibitor treatment itself can be discounted based on several phenotypic observations. However, the strongest evidence against this explanation is that treatment with the carotenoid cleavage inhibitor D15 results in a similar reduction in LR capacity and prebranch site number without producing a small, albino shoot phenotype or a strong reduction in primary root length. These observations indicate that the albino and LR capacity phenotypes are separable and supports a more direct role for an apocarotenoid in LR formation.
A previous study with the carotenoid biosynthesis inhibitor fluridone (Fig. S1C) shows changes in cellular organization and reduced root meristem size as a result of ABA deficiency (75). Fluridone and NF (used in our studies) inhibit carotenoid biosynthesis at the same step yet are distinct compounds and may have unique secondary effects on the root meristem. Alternatively, our apparently contradictory observations that neither quiescent center division nor meristem size was affected by our treatments may be explained by key differences in experimental design. In our analyses, root growth and development are assessed after continuous carotenoid (and ABA) deficiency. In contrast, Zhang et al. (75) transferred older seedlings from standard media to media containing fluridone. Thus, the changes in root morphology observed by Zhang et al. (75) may be more indicative of the response to a dramatic change in carotenoid and ABA availability, suggesting that the role for ABA in the root meristem is more complex than previously reported.
We were able to exclude primary roles for the known apocarotenoid signaling molecules, ABA and strigolactone, in LR prepatterning; however, these results do not preclude ABA and strigolactone from acting later at LR emergence and meristem activation as previously described (56, 70). Additionally, we identify a carotenoid cleavage inhibitor, D15, which reduces LR capacity and prebranch site number. The use of synthetic inhibitors to dissect the specific role(s) of the CCD enzymes in plant biology can provide important insights, given the apparent functional flexibility of these enzymes (74). D15 was designed to inhibit oxidative cleavage of carotenoids; however, it is possible that D15 also inhibits the enzymatic activity of some unknown oxygenase. Because we show that D15 has an impact on carotenoid catabolism and synthesis in vitro and in vivo, respectively, and that D15- and carotenoid biosynthesis inhibitor-treated seedlings have very similar LR formation phenotypes, we believe inhibition of carotenoid cleavage by D15 is the most parsimonious explanation for our observations.
In vitro, D15 shows greater inhibition of CCD1 9,10 cleavage activity, than of NCED1 enzymatic activity, with NCED activity considered the rate-limiting step for ABA synthesis (42, 43). This result suggests that an apocarotenoid derived from a 9,10 cleavage functions in LR formation; however, it should be noted that CCD activity in vitro may not necessarily reflect the in vivo situation. Thus, it is possible that an apocarotenoid derived from cleavage at another position is required for LR formation. Despite this caveat, the in vitro activity of D15 is consistent with our results showing that exogenous ABA neither retards LR capacity in WT nor rescues the reduction in prebranch site or LR capacity upon inhibition of carotenoid biosynthesis. Additionally, we showed that changes in carotenoid content upon D15 treatment do not alter ABA content in planta. Therefore, our results are not consistent with a role for ABA as the apocarotenoid involved in establishing the LR prepattern and, instead, implicate an uncharacterized apocarotenoid in LR formation.
The other carotenoid cleavage inhibitors examined also showed preferential inhibition of CCD activity. However, the unique phenotypic consequences of D15 treatment on LR capacity suggest that there is specificity in D15 activity in planta. These inhibitors may have variable activity in planta for a variety of reasons, such as differential metabolism, uptake, or trafficking (74). Alternatively, they may exhibit variable levels of target specificity in a cellular context. Given that off-target effects were not observed for the other carotenoid cleavage inhibitors (74) and that we observed changes in carotenoid content with D15 treatment, it is unlikely that D15 reduces prebranch sites and LR capacity by some off-target effect.
The in planta impact of D15 on carotenoid cleavage enzyme activity was evidenced by enhanced carotenoid content, primarily in root tissues. These data, together with expression of core carotenoid biosynthesis genes in the root, indicate that it would not be necessary for an apocarotenoid to be transported from the shoot to participate in LR formation. Carotenoid metabolism in the shoot appeared less sensitive to the inhibitory activity of D15, with only a slight reduction in zeaxanthin levels. Interestingly, the zeaxanthin epoxidase (aba1) mutant showed a stronger reduction in LR capacity than other nonalbino mutants, and this reduction could not be fully rescued by exogenous ABA. These observations indicate that the decrease in LR capacity in aba1 mutants cannot be fully attributed to decreased ABA levels and, instead, may reflect the contribution of other carotenoids downstream of zeaxanthin to LR formation. These observations are also consistent with differences among plastid organelles, such as the leucoplast (root) and chloroplast (shoot), in regulation and accumulation of carotenoids in response to specific cellular, developmental, and environmental cues (26).
There are two possible explanations for the increase in carotenoid content in D15-treated roots. First, and most directly, D15 may inhibit a root-specific carotenoid cleavage enzymatic reaction, thereby altering carotenoid catabolism and flux through the pathway. Alternatively, the increase in carotenoids may be due to metabolic feedback on the expression or activity of a key biosynthesis enzyme, such as PSY, which, if increased under D15 treatment, could lead to elevated carotenoid accumulation (76). However, in the latter case, root- and shoot-specific regulation of PSY transcription or activity would likely have to be invoked because there is but a single PSY gene in the Arabidopsis genome. Regardless, the decrease in LR capacity and prebranch site number upon D15 treatment supports the hypothesis that an as-yet unidentified carotenoid-derived molecule is specifically required for LR formation.
Apocarotenoids, other than ABA and strigolactone, serve important functions in plant biology, and it is predicted that there remain yet to be defined roles for apocarotenoids in plants. For example, β-cyclocitral, a β-carotene–derived molecule, was recently found to induce changes in nuclear gene expression and is predicted have a signaling function in response to environmental stress (77). Additionally, characterization of the BYPASS1 gene revealed a graft-transmissible signal emanating from the growing bps1 mutant root, which was sufficient to arrest WT shoot development (78, 79). This graft-transmissible signaling molecule could not be ascribed to any of the known plant hormones; however, disruption of carotenoid biosynthesis suppresses the growth arrest phenotype, suggesting that the so-called “BYPASS” (BPS) signal is carotenoid-derived (27). The BPS signal is predicted to function as a negative regulator of both root and shoot growth. Because we do not observe increased growth upon D15 treatment, we would not predict that the BPS signal and the apocarotenoid putatively involved in LR formation are the same molecule. Thus, despite decades of study, much remains to be learned about the carotenoid biosynthesis pathway as the numbers and types of molecules produced and their roles in plant biology continue to expand (71, 80).
In an effort to link carotenoid biosynthesis to the LR clock, we first examined the expression of carotenoid metabolism genes in the root. Although the tissue-specific expression of these genes was consistent with a role for carotenoids in LR formation, there was no evidence that carotenoid-related genes are part of the oscillatory transcriptional mechanism of the clock. If these genes are not part of the oscillatory mechanism, then how is carotenoid biosynthesis linked to the LR clock? Because inhibition of carotenoid biosynthesis disrupts both the output of the LR clock (LR capacity) and the oscillation in pDR5:LUC expression itself, we propose that carotenoid biosynthesis is necessary for the establishment of the LR prepattern, a process that begins in the OZ.
The root expression pattern of PSY, a key carotenoid biosynthesis gene, suggests that PSY protein is spatially restricted with a more shoot-ward maximum and preferential exclusion from a region of the root tip encompassing the OZ. Although transcript abundance does not necessarily reflect protein levels or activity, this gene expression pattern implies that PSY activity, and therefore carotenoid biosynthesis, is specifically absent from this region. This spatial restriction would necessitate a non–cell-autonomous function for carotenoids (or apocarotenoids) in both the oscillation of gene expression and the establishment of prebranch sites. Because carotenoids are large hydrophobic molecules, carotenoid cleavage would be necessary for production of a mobile apocarotenoid. We propose that this apocarotenoid may function as a positional cue along the root’s longitudinal axis, which is necessary for normal LR prepatterning. This apocarotenoid may be necessary but not sufficient, because we predict that it functions in conjunction with the oscillatory transcriptional mechanism of the LR clock to establish the LR prepattern. The expression of core carotenoid biosynthesis genes also supports a further role for carotenoids in LR formation following prebranch site formation. Thus, an alternative hypothesis is that this apocarotenoid serves as a cue from the more developed LRP, functioning to coordinate the LR clock with the development of existing primordia. Although the precise role of this carotenoid-derived signaling molecule remains to be discovered, our data are consistent with the non–cell-autonomous function of an apocarotenoid signal in establishment and/or maintenance of the LR prepattern.
Materials and Methods
Detailed information on materials and methods used in this study is provided in SI Materials and Methods.
Plant Growth and Treatment Conditions.
Seeds were surface-sterilized and plated on 1% (wt/vol) Murashige and Skoog agar media. Seeds were stratified on growth medium at 4 °C for 2–3 d. They were then placed vertically and grown under long-day conditions at 22 °C, under 85–150 μmol⋅m−2⋅s−1 illumination. Seedlings were examined at 8 das unless otherwise noted.
Plant Materials.
All seed lines are in the Columbia-0 background, except for 35S:PSY, which is in the Wassilewskija background. Transgenic reporter and overexpression lines and mutant alleles were received from the Arabidopsis Biological Resource Center and members of the Arabidopsis community.
Reporter Gene Construction and Plant Transformation.
Reporter genes were constructed by standard molecular biology methods and Invitrogen Multisite Gateway technology. Plants were transformed by the floral dip method, and transformants were identified using standard methods. Information about primers (Table S5) and genetic analyses of transformants (Table S3) is provided in SI Materials and Methods.
Quantification of Root Phenotypes.
Root, meristem, and cell lengths were measured from images of seedlings or confocal images using ImageJ software (National Institutes of Health). Roots were fixed and cleared using methods previously described (28), and LR/LRP number was examined under differential interference contrast illumination. In the LR capacity assay, the root tip is excised at 8 das and seedlings are grown for 3 more days; LRs and late-stage LRPs are then counted under a dissecting microscope.
Imaging and Confocal Microscopy.
Luciferase activity was assayed as previously described (12) with exposure times of 3–5 min or 40 min (as indicated). Bright-field and luciferase images were overlaid using Photoshop (Adobe Systems). Laser scanning confocal microscopy (Zeiss LSM 510 microscope) was used to examine roots stained with 10 μM propidium iodide to visualize cellular organization.
In Silico Analysis of Carotenoid-Related Gene Expression.
The carotenoid metabolism gene list was generated by compiling the genes associated with GO categories describing carotenoid and apocarotenoid processes (www.arabidopsis.org) and from recent publications (Table S2). This list and the 16 core carotenoid biosynthesis genes (35) were examined in the OZ dataset (12). Hierarchical clustering was conducted using the Multiple Experiment Viewer (version 4.8) program (81).
In Vitro CCD Inhibitor Assays.
The in vitro inhibitor assays were largely conducted as previously described (74). Additional details are provided in SI Materials and Methods. LeNCED1 and LeCCD1a were overexpressed in Escherichia coli as an N-terminal GST fusion protein. Inhibition assays in cell-free extracts contained 100 μM inhibitor or water.
HPLC Analyses and ABA Quantification.
Seeds were sown in two to three rows on control (ethanol) and 100 μM D15-containing plates lined with 100 μm Nitex nylon mesh (Genesee Scientific). At 7 das, tissue was collected and stored at −80 °C. HPLC-based quantification of carotenoids was performed as previously described (46, 49). ABA content was quantified using a modified ELISA-based method. Tissue was collected as described above and then ground to a fine powder from which ABA was extracted and quantified using the Phytodetek ABA Quantification Kit (Agdia). Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Miguel A. Moreno-Risueno for useful discussions, particularly in regard to the development of the LR capacity assay, and for sharing the pENTR-p2p3-LUC vector. We thank members of the laboratory of P.N.B., Ross Sozzani, Anjali Iyer-Pascuzzi, Wolfgang Busch, Terri Long, Siobhan Brady, and Miguel Moreno-Risueno for critical reading of this manuscript and helpful discussions during the course of this work. This work was supported by Grant CE140100008 of the Australian Research Council Centre of Excellence in Plant Energy Biology (to B.J.P.) and by Discovery Grant DP130102593 (to C.I.C. and B.J.P.). K.X.C. is supported by an Australian Government International Postgraduate Research Scholarship (PhD scholarship). P.J.H. is supported by a Collaborative Awards in Science and Engineering PhD studentship funded by the Biotechnology and Biological Sciences Research Council and Syngenta, Ltd. J.M.V.N. was supported by a National Institutes of Health National Research Service Award postdoctoral fellowship (GM093656-02). This work was funded by Grant D12AP0000 from the Defense Advanced Research Planning Agency (to P.N.B.) and by Grant GBMF3405 from the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (to P.N.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1403016111/-/DCSupplemental.
References
- 1.De Smet I. Lateral root initiation: One step at a time. New Phytol. 2012;193(4):867–873. doi: 10.1111/j.1469-8137.2011.03996.x. [DOI] [PubMed] [Google Scholar]
- 2.Lavenus J, et al. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013;18(8):450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Péret B, et al. Arabidopsis lateral root development: An emerging story. Trends Plant Sci. 2009;14(7):399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Van Norman JM, Xuan W, Beeckman T, Benfey PN. To branch or not to branch: The role of pre-patterning in lateral root formation. Development. 2013;140(21):4301–4310. doi: 10.1242/dev.090548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolan L, et al. Cellular organisation of the Arabidopsis thaliana root. Development. 1993;119(1):71–84. doi: 10.1242/dev.119.1.71. [DOI] [PubMed] [Google Scholar]
- 6.Brady SM, et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318(5851):801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- 7.Laskowski MJ, Williams ME, Nusbaum HC, Sussex IM. Formation of lateral root meristems is a two-stage process. Development. 1995;121(10):3303–3310. doi: 10.1242/dev.121.10.3303. [DOI] [PubMed] [Google Scholar]
- 8.Malamy JE, Benfey PN. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 1997;124(1):33–44. doi: 10.1242/dev.124.1.33. [DOI] [PubMed] [Google Scholar]
- 9.Dubrovsky JG, Rost TL, Colón-Carmona A, Doerner P. Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta. 2001;214(1):30–36. doi: 10.1007/s004250100598. [DOI] [PubMed] [Google Scholar]
- 10.Malamy JE. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 2005;28(1):67–77. doi: 10.1111/j.1365-3040.2005.01306.x. [DOI] [PubMed] [Google Scholar]
- 11.Dubrovsky JG, Soukup A, Napsucialy-Mendivil S, Jeknic Z, Ivanchenko MG. The lateral root initiation index: An integrative measure of primordium formation. Ann Bot (Lond) 2009;103(5):807–817. doi: 10.1093/aob/mcn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreno-Risueno MA, et al. Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science. 2010;329(5997):1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Smet I, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134(4):681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 15.Peñuelas J, Munné-Bosch S. Isoprenoids: An evolutionary pool for photoprotection. Trends Plant Sci. 2005;10(4):166–169. doi: 10.1016/j.tplants.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Cazzonelli CI. Carotenoids in nature: Insights from plants and beyond. Funct Plant Biol. 2011;38(11):833–847. doi: 10.1071/FP11192. [DOI] [PubMed] [Google Scholar]
- 17.DellaPenna D, Pogson BJ. Vitamin synthesis in plants: Tocopherols and carotenoids. Annu Rev Plant Biol. 2006;57:711–738. doi: 10.1146/annurev.arplant.56.032604.144301. [DOI] [PubMed] [Google Scholar]
- 18.Walter MH, Floss DS, Strack D. Apocarotenoids: Hormones, mycorrhizal metabolites and aroma volatiles. Planta. 2010;232(1):1–17. doi: 10.1007/s00425-010-1156-3. [DOI] [PubMed] [Google Scholar]
- 19.Goff SA, Klee HJ. Plant volatile compounds: Sensory cues for health and nutritional value? Science. 2006;311(5762):815–819. doi: 10.1126/science.1112614. [DOI] [PubMed] [Google Scholar]
- 20.Zhu C, et al. The regulation of carotenoid pigmentation in flowers. Arch Biochem Biophys. 2010;504(1):132–141. doi: 10.1016/j.abb.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 21.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 22.Goulet C, Klee HJ. Climbing the branches of the strigolactones pathway one discovery at a time. Plant Physiol. 2010;154(2):493–496. doi: 10.1104/pp.110.161026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SC, Luan S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012;35(1):53–60. doi: 10.1111/j.1365-3040.2011.02426.x. [DOI] [PubMed] [Google Scholar]
- 24.Matusova R, et al. The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 2005;139(2):920–934. doi: 10.1104/pp.105.061382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoneyama K, Xie X, Yoneyama K, Takeuchi Y. Strigolactones: Structures and biological activities. Pest Manag Sci. 2009;65(5):467–470. doi: 10.1002/ps.1726. [DOI] [PubMed] [Google Scholar]
- 26.Cazzonelli CI, Pogson BJ. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010;15(5):266–274. doi: 10.1016/j.tplants.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Van Norman JM, Sieburth LE. Dissecting the biosynthetic pathway for the bypass1 root-derived signal. Plant J. 2007;49(4):619–628. doi: 10.1111/j.1365-313X.2006.02982.x. [DOI] [PubMed] [Google Scholar]
- 28.Dubrovsky JG, Gambetta GA, Hernández-Barrera A, Shishkova S, González I. Lateral root initiation in Arabidopsis: Developmental window, spatial patterning, density and predictability. Ann Bot (Lond) 2006;97(5):903–915. doi: 10.1093/aob/mcj604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guevara-García A, et al. Characterization of the Arabidopsis clb6 mutant illustrates the importance of posttranscriptional regulation of the methyl-D-erythritol 4-phosphate pathway. Plant Cell. 2005;17(2):628–643. doi: 10.1105/tpc.104.028860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh MH, Goodman HM. The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol. 2005;138(2):641–653. doi: 10.1104/pp.104.058735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin G, et al. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007;17(5):471–482. doi: 10.1038/cr.2007.40. [DOI] [PubMed] [Google Scholar]
- 32.Hricová A, Quesada V, Micol JL. The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol. 2006;141(3):942–956. doi: 10.1104/pp.106.080069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Møller IM, Jensen PE, Hansson A. Oxidative modifications to cellular components in plants. Annu Rev Plant Biol. 2007;58:459–481. doi: 10.1146/annurev.arplant.58.032806.103946. [DOI] [PubMed] [Google Scholar]
- 34.Mullineaux PM, Baker NR. Oxidative stress: Antagonistic signaling for acclimation or cell death? Plant Physiol. 2010;154(2):521–525. doi: 10.1104/pp.110.161406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz-Sola MA, Rodríguez-Concepción M. Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arabidopsis Book. 2012;10:e0158. doi: 10.1199/tab.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maass D, Arango J, Wüst F, Beyer P, Welsch R. Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS ONE. 2009;4(7):e6373. doi: 10.1371/journal.pone.0006373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez-Villalón A, Gas E, Rodríguez-Concepción M. Phytoene synthase activity controls the biosynthesis of carotenoids and the supply of their metabolic precursors in dark-grown Arabidopsis seedlings. Plant J. 2009;60(3):424–435. doi: 10.1111/j.1365-313X.2009.03966.x. [DOI] [PubMed] [Google Scholar]
- 38.Meier S, Tzfadia O, Vallabhaneni R, Gehring C, Wurtzel ET. A transcriptional analysis of carotenoid, chlorophyll and plastidial isoprenoid biosynthesis genes during development and osmotic stress responses in Arabidopsis thaliana. BMC Syst Biol. 2011;5:77. doi: 10.1186/1752-0509-5-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shumskaya M, Bradbury LM, Monaco RR, Wurtzel ET. Plastid localization of the key carotenoid enzyme phytoene synthase is altered by isozyme, allelic variation, and activity. Plant Cell. 2012;24(9):3725–3741. doi: 10.1105/tpc.112.104174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fray RG, et al. Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J. 1995;8(5):693–701. [Google Scholar]
- 41.Thompson AJ, et al. Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J. 2000;23(3):363–374. doi: 10.1046/j.1365-313x.2000.00789.x. [DOI] [PubMed] [Google Scholar]
- 42.Tan BC, Schwartz SH, Zeevaart JA, McCarty DR. Genetic control of abscisic acid biosynthesis in maize. Proc Natl Acad Sci USA. 1997;94(22):12235–12240. doi: 10.1073/pnas.94.22.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iuchi S, et al. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27(4):325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 44.Li F, Vallabhaneni R, Wurtzel ET. PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic stress-induced root carotenogenesis. Plant Physiol. 2008;146(3):1333–1345. doi: 10.1104/pp.107.111120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cazzonelli CI, et al. Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell. 2009;21(1):39–53. doi: 10.1105/tpc.108.063131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ. Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell. 2002;14(2):321–332. doi: 10.1105/tpc.010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isaacson T, Ohad I, Beyer P, Hirschberg J. Analysis in vitro of the enzyme CRTISO establishes a poly-cis-carotenoid biosynthesis pathway in plants. Plant Physiol. 2004;136(4):4246–4255. doi: 10.1104/pp.104.052092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D. The Arabidopsis LUT1 locus encodes a member of the cytochrome p450 family that is required for carotenoid epsilon-ring hydroxylation activity. Proc Natl Acad Sci USA. 2004;101(1):402–407. doi: 10.1073/pnas.2237237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pogson B, McDonald KA, Truong M, Britton G, DellaPenna D. Arabidopsis carotenoid mutants demonstrate that lutein is not essential for photosynthesis in higher plants. Plant Cell. 1996;8(9):1627–1639. doi: 10.1105/tpc.8.9.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cunningham FX, Jr, et al. Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell. 1996;8(9):1613–1626. doi: 10.1105/tpc.8.9.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. Combined networks regulating seed maturation. Trends Plant Sci. 2007;12(7):294–300. doi: 10.1016/j.tplants.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Vogel JT, et al. SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J. 2010;61(2):300–311. doi: 10.1111/j.1365-313X.2009.04056.x. [DOI] [PubMed] [Google Scholar]
- 54.Gomez-Roldan V, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455(7210):189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 55.Umehara M, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455(7210):195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 56.Kapulnik Y, et al. Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta. 2011;233(1):209–216. doi: 10.1007/s00425-010-1310-y. [DOI] [PubMed] [Google Scholar]
- 57.Agusti J, et al. Strigolactone signaling is required for auxin-dependent stimulation of secondary growth in plants. Proc Natl Acad Sci USA. 2011;108(50):20242–20247. doi: 10.1073/pnas.1111902108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rasmussen A, et al. Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 2012;158(4):1976–1987. doi: 10.1104/pp.111.187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruyter-Spira C, et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 2011;155(2):721–734. doi: 10.1104/pp.110.166645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Booker J, et al. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol. 2004;14(14):1232–1238. doi: 10.1016/j.cub.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 61.Stirnberg P, van De Sande K, Leyser HM. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development. 2002;129(5):1131–1141. doi: 10.1242/dev.129.5.1131. [DOI] [PubMed] [Google Scholar]
- 62.Walter MH, Strack D. Carotenoids and their cleavage products: Biosynthesis and functions. Nat Prod Rep. 2011;28(4):663–692. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- 63.Tan BC, et al. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35(1):44–56. doi: 10.1046/j.1365-313x.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 64.Schwartz SH, Tan BC, McCarty DR, Welch W, Zeevaart JA. Substrate specificity and kinetics for VP14, a carotenoid cleavage dioxygenase in the ABA biosynthetic pathway. Biochim Biophys Acta. 2003;1619(1):9–14. doi: 10.1016/s0304-4165(02)00422-1. [DOI] [PubMed] [Google Scholar]
- 65.Niyogi KK, Grossman AR, Björkman O. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell. 1998;10(7):1121–1134. doi: 10.1105/tpc.10.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koornneef MJM, Jorna ML, Brinkhorst-van der Swan DL, Karssen CM. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) heynh. Theor Appl Genet. 1982;61(4):385–393. doi: 10.1007/BF00272861. [DOI] [PubMed] [Google Scholar]
- 67.Bittner F, Oreb M, Mendel RR. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem. 2001;276(44):40381–40384. doi: 10.1074/jbc.C100472200. [DOI] [PubMed] [Google Scholar]
- 68.Schwartz SH, Léon-Kloosterziel KM, Koornneef M, Zeevaart JA. Biochemical characterization of the aba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997;114(1):161–166. doi: 10.1104/pp.114.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Léon-Kloosterziel KM, et al. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996;10(4):655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- 70.De Smet I, et al. An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 2003;33(3):543–555. doi: 10.1046/j.1365-313x.2003.01652.x. [DOI] [PubMed] [Google Scholar]
- 71.Alder A, et al. The path from β-carotene to carlactone, a strigolactone-like plant hormone. Science. 2012;335(6074):1348–1351. doi: 10.1126/science.1218094. [DOI] [PubMed] [Google Scholar]
- 72.Stirnberg P, Furner IJ, Ottoline Leyser HM. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007;50(1):80–94. doi: 10.1111/j.1365-313X.2007.03032.x. [DOI] [PubMed] [Google Scholar]
- 73.Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. Specific oxidative cleavage of carotenoids by VP14 of maize. Science. 1997;276(5320):1872–1874. doi: 10.1126/science.276.5320.1872. [DOI] [PubMed] [Google Scholar]
- 74.Sergeant MJ, et al. Selective inhibition of carotenoid cleavage dioxygenases: Phenotypic effects on shoot branching. J Biol Chem. 2009;284(8):5257–5264. doi: 10.1074/jbc.M805453200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H, et al. ABA promotes quiescence of the quiescent centre and suppresses stem cell differentiation in the Arabidopsis primary root meristem. Plant J. 2010;64(5):764–774. doi: 10.1111/j.1365-313X.2010.04367.x. [DOI] [PubMed] [Google Scholar]
- 76.Kachanovsky DE, Filler S, Isaacson T, Hirschberg J. Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis-carotenoids. Proc Natl Acad Sci USA. 2012;109(46):19021–19026. doi: 10.1073/pnas.1214808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ramel F, et al. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA. 2012;109(14):5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Norman JM, Frederick RL, Sieburth LE. BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr Biol. 2004;14(19):1739–1746. doi: 10.1016/j.cub.2004.09.045. [DOI] [PubMed] [Google Scholar]
- 79.Van Norman JM, Murphy C, Sieburth LE. BYPASS1: Synthesis of the mobile root-derived signal requires active root growth and arrests early leaf development. BMC Plant Biol. 2011;11:28. doi: 10.1186/1471-2229-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maoka T. Recent progress in structural studies of carotenoids in animals and plants. Arch Biochem Biophys. 2009;483(2):191–195. doi: 10.1016/j.abb.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 81.Saeed AI, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.