ABSTRACT
The cytoplasmic helicase RIG-I is an established sensor for viral 5′-triphosphorylated RNA species. Recently, RIG-I was also implicated in the detection of intracellular bacteria. However, little is known about the host cell specificity of this process and the bacterial pathogen-associated molecular pattern (PAMP) that activates RIG-I. Here we show that RNA of Salmonella enterica serovar Typhimurium activates production of beta interferon in a RIG-I-dependent fashion only in nonphagocytic cells. In phagocytic cells, RIG-I is obsolete for detection of Salmonella infection. We further demonstrate that Salmonella mRNA reaches the cytoplasm during infection and is thus accessible for RIG-I. The results from next-generation sequencing analysis of RIG-I-associated RNA suggest that coding bacterial mRNAs represent the activating PAMP.
IMPORTANCE
S. Typhimurium is a major food-borne pathogen. After fecal-oral transmission, it can infect epithelial cells in the gut as well as immune cells (mainly macrophages, dendritic cells, and M cells). The innate host immune system relies on a growing number of sensors that detect pathogen-associated molecular patterns (PAMPs) to launch a first broad-spectrum response to invading pathogens. Successful detection of a given pathogen depends on colocalization of host sensors and PAMPs as well as potential countermeasures of the pathogen during infection. RIG-I-like helicases were mainly associated with detection of RNA viruses. Our work shows that S. Typhimurium is detected by RIG-I during infection specifically in nonimmune cells.
INTRODUCTION
Pattern recognition receptors (PRRs) recognize broadly shared molecular structures known as pathogen-associated molecular patterns (PAMPs). Five main families of PRRs are known: Toll-like receptors (TLR), RIG-I-like receptors (RLR), NOD-like receptors (NLR), cytoplasmic DNA receptors, and C-type lectin-like receptors (reviewed in references 1, 2, 3, 4, and 5). Recognition of a microbial PAMPs results in the activation of PRR-specific downstream-signaling cascades and expression of a variety of antimicrobial and proinflammatory cytokines and chemokines.
In contrast to the use of purified artificial PAMPs, successful induction of an immune response during infection depends on a number of factors, including expression of the PRR in the infected cell type, colocalization of the PRR and PAMP in the same subcellular compartment, and the ability of the cell to overcome pathogen evasion strategies that serve to block innate immune recognition and signaling. Depending on the cell type and the pathogen studied, these factors may differ.
RIG-I is a member of the RLR family (4). Upon binding of 5′-triphosphorylated RNA, RIG-I undergoes conformational changes and posttranslational modifications that allow multimerization and interaction with the mitochondrial antiviral signaling protein (MAVS) (6). Subsequent signaling events ultimately result in formation of the beta interferon (IFN-β) enhanceosome and IFN-β expression. IFN-β is a key cytokine in innate antiviral immune responses, mediating expression of hundreds of IFN-stimulated genes (ISGs) that are responsible for the establishment of an antimicrobial state in the infected tissue.
A role for RIG-I in bacterial sensing has recently been described (reviewed in reference 7). However, the underlying mechanisms of detection and the nature of the activating PAMP, especially under infection conditions, remain less well characterized. Here we show that RIG-I-dependent recognition of intracellular bacteria is pathogen and cell type dependent, using Salmonella Typhimurium as a model pathogen. We further demonstrate that bacterial mRNA is recognized by RIG-I during infection, leading to expression of IFN-β.
RESULTS
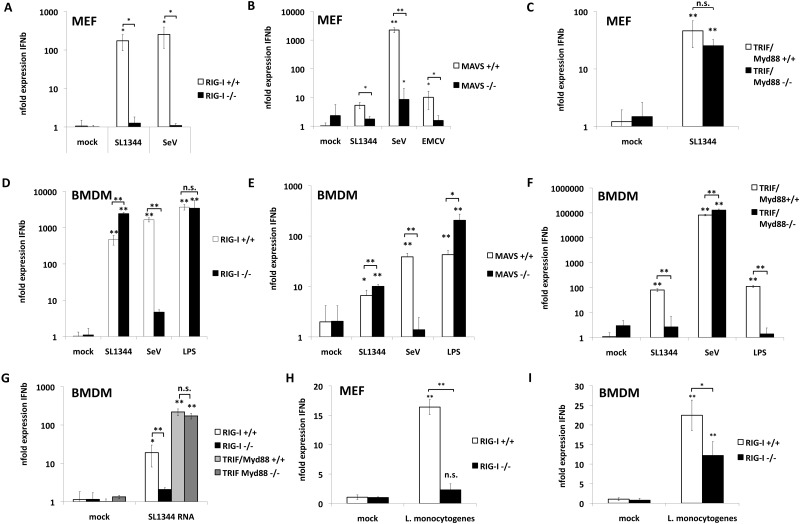
Salmonella enterica consists of a group of flagellated, Gram-negative, facultative intracellular bacteria (reviewed in reference 8). They are the leading cause for gastroenteric disease in animals, including humans. S. enterica serovar Typhimurium causes self-limiting gastroenteritis in humans. To test if RIG-I is involved in recognition of S. Typhimurium, we compared the type I interferon response in wild-type (WT) and RIG-I−/− fibroblasts upon infection. Infection of WT fibroblasts with S. Typhimurium induced expression of IFN-β mRNA (Fig. 1A). IFN-β expression was abolished in RIG-I−/− murine embryonic fibroblasts (MEFs), suggesting that RIG-I was the primary PRR leading to IFN-β expression. Reconstitution of RIG-I−/− MEFs with RIG-I restored IFN-β expression, albeit at levels lower than those seen with WT MEFs (see Fig. S1A and B in the supplemental material). We observed similar titers of intracellular bacteria in WT, RIG-I−/−, and RIG-I-reconstituted MEFs, suggesting that differences in replication did not account for the lack of IFN-β induction in RIG-I−/− MEFs (see Fig. S1C).
FIG 1 .
RIG-I is essential for detection of Salmonella infection in nonphagocytic cells but is obsolete for detection in macrophages. (A) RIG-I+/+ and RIG−/− murine embryonic fibroblasts (MEFs) were infected for 8 h with S. Typhimurium SL1344 at an MOI of 10 or with SeV at an MOI of 10. (B) MAVS+/+ and MAVS−/− MEFs were infected with SL1344 at an MOI of 10, SeV at an MOI of 10, or EMCV (encephalomyocarditis virus) at an MOI of 10 for 8 h. (C) TRIF/Myd88+/+ and TRIF/Myd88−/− MEFs were infected with SL1344 at an MOI of 10 for 8 h. (D) RIG-I+/+ and RIG−/− BMDM were infected with SL1344 at an MOI of 10 or with SeV at an MOI of 10 or were treated with 2 µg/ml LPS for 4 h. Data representing average n-fold expression of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± standard deviations (SD) are depicted. (E) MAVS+/+ and MAVS−/− BMDM were infected with SL1344 at an MOI of 10 or with SeV at an MOI of 10 or were treated with 2 µg/ml LPS for 4 h. (F) TRIF/Myd88+/+ and TRIF/Myd88−/− BMDM were infected with SL1344 at an MOI of 10 or with SeV at an MOI of 10 or were treated with 2 µg/ml LPS for 4 h. (G) RIG-I+/+ and RIG−/− BMDM and TRIF/Myd88+/+ and TRIF/Myd88−/− BMDM were transfected with 1 µg SL1344 RNA for 6 h. (H) RIG-I+/+ and RIG−/− MEFs were infected for 8 h with L. monocytogenes at an MOI of 10. (G) RIG-I+/+ and RIG−/− MEFs were infected for 4 h with L. monocytogenes at an MOI of 10. Each column in this figure represents the average n-fold expression of IFN-β over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD as determined by specific qPCR.
RIG-I- and melanoma differentiation-associated protein 5 (MDA5)-dependent induction of IFN-β requires adaptor MAVS. In accordance with our data from RIG-I−/− MEFs, we detected reduced IFN-β mRNA levels in S. Typhimurium-infected MAVS−/− MEFs compared to MAVS+/+ MEFs (Fig. 1B). As expected, both Sendai virus (SeV) and encephalomyocarditis virus (EMCV), which are recognized specifically through the activity of RIG-I and MDA5, respectively, also show defective IFN-β induction in MAVS−/− MEFs. In support of the idea of primary detection of S. Typhimurium by RIG-I, fibroblasts deficient in MyD88 and Trif showed no defect in the induction of IFN-β upon S. Typhimurium infection (Fig. 1C). We confirmed RIG-I-dependent recognition of S. Typhimurium infection in two human RIG-I knockdown epithelial cells. Human A549 and HT-29 cells constitutively expressing small hairpin RNAs (shRNAs) against RIG-I show reduced IFN-β mRNA levels (see Fig. S1D and E in the supplemental material). Taken together, these data implicate primary detection of Salmonella by the RIG-I/MAVS pathway in fibroblasts in the induction of IFN-β in response to bacterial infection.
S. Typhimurium infects both phagocytic and nonphagocytic cell types in vivo. To investigate the role of RIG-I in phagocytic cells, we chose macrophages as a model system. In contrast to our results in nonphagocytic cells, S. Typhimurium infection of RIG-I−/− bone marrow-derived macrophages (BMDM) induced IFN-β responses equivalent to those seen with WT BMDM under the chosen experimental conditions (Fig. 1D), despite a significant reduction in response to Sendai virus, confirming RIG-I deficiency. In accordance, IFN-β production was also independent of the presence of MAVS in BMDM (Fig. 1E).
The lack of a role for RIG-I in IFN-β induction in BMDM suggested that other PRRs were responsible for the detection of S. Typhimurium in these cells. To test this, we infected BMDM deficient in MyD88 and TIR-domain-containing adapter-inducing IFN-β (TRIF) with S. Typhimurium. In contrast to our results with RIG-I−/− BMDM, MyD88/Trif−/− macrophages showed a significant reduction in IFN-β expression in response to S. Typhimurium and Salmonella lipopolysaccharide (LPS) but normal responses to Sendai virus (Fig. 1F). This suggests that RIG-I acts as the primary sensor for S. Typhimurium in nonphagocytic cells, whereas TLR are the primary sensors in phagocytic cells.
In contrast to the natural infection route, transfection of BMDM with Salmonella RNA led to IFN-β induction in a RIG-I-dependent and MyD88/Trif-independent fashion, confirming that BMDM are capable of recognizing bacterial RNA through RIG-I (Fig. 1G). This suggests either that bacterial RNA does not reach RIG-I in the cytoplasm during natural infection or that TLR responses mask the effect of RIG-I activation. Two recent studies showed RIG-I-dependent recognition of Listeria monocytogenes (9, 10). Abdullah and colleagues showed a 3- to 5-fold reduction in IFN-β protein levels in the supernatant of infected RIG-I−/− macrophages compared to WT cells upon infection with L. monocytogenes (9). Hagmann and colleagues demonstrated RIG-I-dependent detection of L. monocytogenes in infected nonphagocytic cells by the use of A549 cells treated with small interfering RNAs (siRNAs) against RIG-I (10). We confirmed these data using the same experimental setting as used before in the S. Typhimurium infection experiments (Fig. 1H and I), showing that accessibility of bacterial RNA for cellular sensors depends on the replication strategy of the bacterium.
Taken together, these data indicate that RIG-I is a sensor for Salmonella nonphagocytic cells in the context of intracellular infection, whereas TLR-mediated recognition of Salmonella is essential for expression of IFN-β in phagocytic cells.
Next we sought to characterize the nature of the RIG-I-detected PAMP during S. Typhimurium infection. It is well established that RIG-I detects cytoplasmic viral RNA. Total RNAs from different bacterial species were previously suggested as potential PAMPs for RIG-I (9, 11, 12). In line with this, RNA from S. Typhimurium induced IFN-β-promoter-driven expression of luciferase in 293T cells. RNase A digestion of total RNA from Salmonella confirmed that RNA and not other bacterial components conferred immunostimulatory activity (Fig. 2A).
FIG 2 .
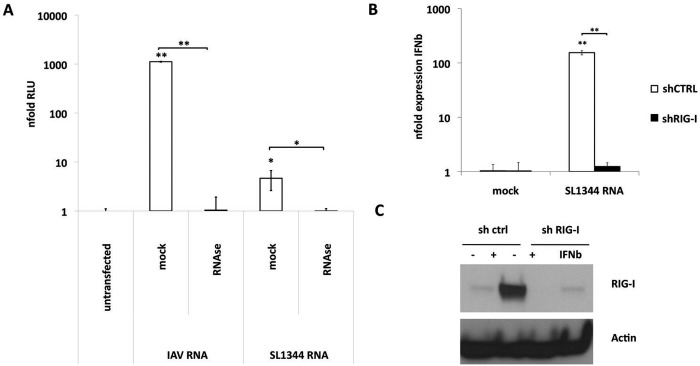
Salmonella RNA induces IFN-β expression upon transfection. (A) 293T-FF reporter cells were transfected with 1 µg of mock-treated or RNase A-treated total RNA from A549 cells infected for 8 h with influenza A/Viet Nam/1203/2004 HAlo virus (IAV) or S. Typhimurium (SL1344) at an MOI of 5. Each column represents the mean increase in n-fold relative luminescence units (RLU) compared to the average results from mock-infected cells at 24 h posttransfection of three independent biological samples each measured in technical triplicate experiments ± SD. (B) A549 control (shCTRL) and A549 shRIG-I cells were transfected with 1 µg of total RNA from S. Typhimurium (SL1344). Each column represents the average n-fold expression of IFN-β over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD. (C) Western blot analysis of total protein lysates of A549 cells stably expressing shRNAs against RIG-I (shRIG-I) or control shRNAs (sh ctrl). Cells were stimulated for 8 h with 100 U IFN-α or left untreated. Membranes were incubated with anti-RIG-I (upper panel) or anti-β-actin (lower panel).
To confirm that transfected Salmonella RNA is detected by RIG-I, we knocked down RIG-I using specific shRNAs (Fig. 2B and C). Bacterial RNA induced IFN-β expression in control cell lines but failed to induce expression in RIG-I knockdown cell lines (Fig. 2B). In summary, these data indicate that S. Typhimurium RNA is a ligand for RIG-I.
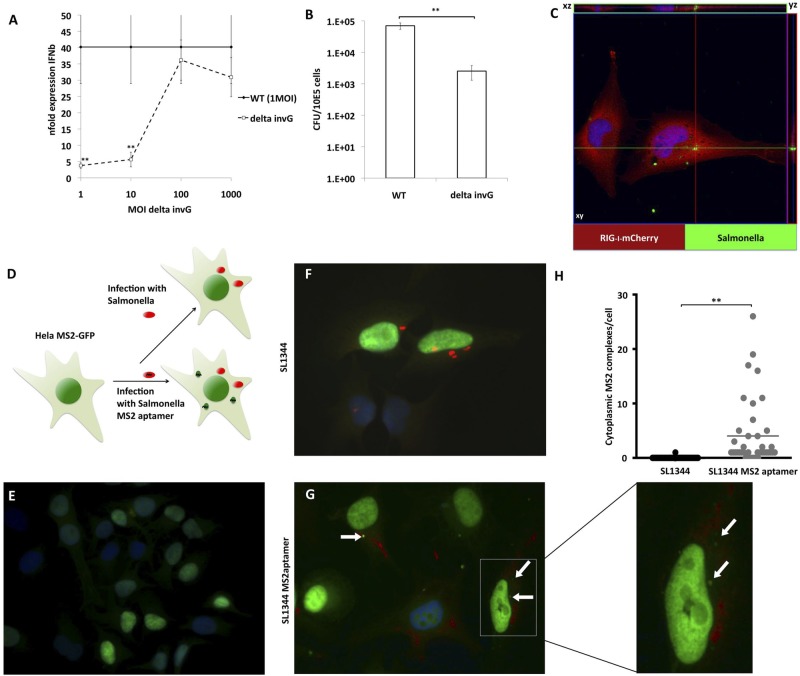
Intracellular replication of S. Typhimurium mainly takes place in vacuoles separated from the cytoplasm, termed Salmonella-containing vacuoles (SCV). It is therefore unclear how RIG-I has access to bacterial RNA. To confirm that the intracellular presence of bacteria is required for the induction of IFN-β, we infected MEFs with WT Salmonella or with an attenuated strain (SB161) lacking invG (ΔinvG), encoding a key protein of the T3SS system that shows defective invasiveness. We confirmed that the invG mutant strain showed 100-fold-reduced invasiveness in nonphagocytic cells (Fig. 3A). Interestingly, MEFs infected with WT Salmonella at a multiplicity of infection (MOI) of 1 provoked robust induction of IFN-β (Fig. 3B). The attenuated strain required 100- to 1,000-fold more bacteria to induce similar IFN-β mRNA levels, indicating that similar amounts of intracellular bacteria are required for induction of IFN-β for WT and ΔinvG Salmonella.
FIG 3 .
Salmonella Typhimurium RNA is accessible in the cytoplasm (A) MEFs were infected with S. Typhimurium WT-GFP at an MOI of 1 or with S. Typhimurium ΔinvG at the indicated MOI for 8 h. Values represent the average n-fold expression level of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD as determined by qPCR. (B) Intracellular titers of MEF infected with S. Typhimurium WT-GFP and ΔinvG at an MOI of 5, as determined by serial dilution of cell lysates. Each column represents the average titers of three independent biological samples each measured in technical triplicate experiments ± SD. (C) HeLa cells transfected with pCAGGS-RIG-I-mCherry (red) were infected 48 h posttransfection with S. Typhimurium (green) at an MOI of 10 for 8 h. Z-stacked pictures were taken at a distance of 0.5 µm, covering the whole cell body to exclude colocalization of RIG-I and S. Typhimurium signal. (D to G) HeLa cells constitutively expressing MS2-GFP (green) (D and E) were infected with WT S. Typhimurium SL1344 (F) or SL1344 expressing MS2-aptamer RNA (G) (a magnification of the indicated area is depicted on the right side). Cytoplasmic complexes of MS2-GFP are indicated with white arrows; Salmonella is indicated in red. (H) Quantification of MS2-GFP complexes in the cytoplasm of 40 randomly chosen SL1344-positive cells.
Next, we tested if RIG-I has access to SCVs, a possibility which could explain the RIG-I-dependent detection of bacterial RNA. We examined the localization of RIG-I in HeLa cells due to their large cytoplasm. By confocal laser scanning microscopy (LSM), we analyzed the localization of RIG-I-mCherry in Salmonella-infected cells. RIG-I did not colocalize with SCV (Fig. 3C), suggesting that bacterial RNA must be transferred from SCV to the cytoplasm for detection by RIG-I. To investigate whether Salmonella RNA reaches the cytoplasm during infection, we used HeLa cells constitutively expressing MS2-green fluorescent protein (MS2-GFP) with a nuclear localization signal (Fig. 3D and E). In this system, newly translated MS2-GFP translocates into the nucleus, keeping the cytoplasm free from background fluorescence. In contrast, MS2-GFP forms foci in the cytoplasm in the presence of cytoplasmic RNA containing M2 binding sites. HeLa MS2-GFP cells were infected with either S. Typhimurium SL1344 or SL1344 expressing an RNA with 24 repeats of MS2 binding sites (SL1344-MS2aptamer). SL1344-MS2aptamer-infected HeLa MS2-GFP cells showed cytoplasmic foci containing GFP at a high MOI (Fig. 3G). We did not detect these GFP accumulations in mock-infected cells (Fig. 3E). In the samples infected with WT SL1344, we found only 1 cell with a GFP complex in the cytoplasm, despite clear accumulation of intracellular bacteria (Fig. 3F). Quantification in 40 randomly chosen, SL1344-positive cells revealed an average of 4.025 MS2-GFP complexes per infected cell (Fig. 3H). Taking these findings together, this suggests that intracellular replication of Salmonella leads to RNA transfer from SCVs into the cytoplasm where RIG-I activation occurs.
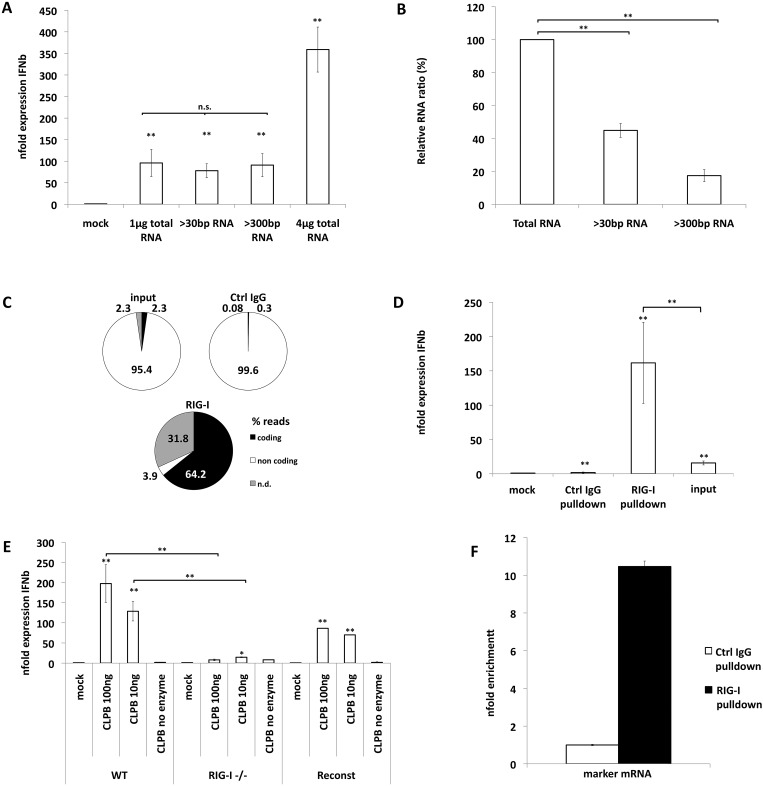
RIG-I is activated by a variety of RNA species in vitro (4). It was shown that 5′-triphosphorylated RNA is a very potent ligand of RIG-I (13–16). In addition, double-stranded RNA motifs and poly(U/UC) stretches also contribute to recognition by RIG-I (17). To understand the characteristics of the bacterial immunostimulatory RNA, we first fractionated total RNA from S. Typhimurium using size-specific-cutoff centrifugation columns (Fig. 4A; see also Fig. S2A in the supplemental material). A significant proportion of the total bacterial RNA was shorter than 500 bp (Fig. 4B). Nevertheless, removal of this 80% fraction of RNA did not affect the immunostimulatory capacity of the remaining RNA (Fig. 4A). This suggests that small bacterial RNAs do not play a major role in activating RIG-I during bacterial infection.
FIG 4 .
Salmonella coding RNAs bind to RIG-I and activate IFN-β expression in a 5′-triphosphate-dependent fashion (A) Column fractionation was used to eliminate RNAs smaller than 30 or smaller than 500 bp. A549 cells were transfected with 1 µg or 4 µg of total RNA and equal volumes of fractionated RNAs. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD are depicted. n.s., not significant. (B) Concentrations of RNAs after column fractionation were compared to total RNA as determined by spectrometry. (C) Relative representations (%) of reads of coding (black) and noncoding (white) RNAs from total bacterial RNA (Total), GFP pulldown RNA (GFP), and RIG-I pulldown RNA groups. n.d. (gray) represents RNAs that could not be classified as coding or noncoding. (D) A549 cells were transfected with 40 ng of pulldown RNA for 16 h. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± are depicted. (E) In vitro-transcribed RNA was transfected into RIG-I+/+, RIG−/−, or reconstituted RIG−/− MEF. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD are depicted. Significance is indicated in reference to no-enzyme control samples. (F) Enrichment of marker mRNA from Salmonella-infected A549 cells (MOI of 10, 8 h) after pull down with RIG-I-specific antibody over pull down with control IgG antibody. Equal amounts of pulldown RNA were reverse transcribed using gene-specific primers for marker mRNA. Average n-fold levels over control IgG pull down levels from three independent biological samples each measured in technical triplicate experiments ±SD are depicted.
To identify the class of bacterial RNA responsible for RIG-I activation, lysates of epithelial cells were incubated with total RNA from S. Typhimurium. RIG-I complexed with RNA was immunoprecipitated. Next, we sequenced the RIG-I-complexed RNA by next-generation Illumina sequencing (Fig. 4C). The majority (95.4%) of the total Salmonella RNA from bacterial lysates corresponded to ribosomal and other noncoding RNAs. Only a small fraction (2.3%) of the total RNA represented coding RNAs (Fig. 4C [input]). However, the RNA specifically immunoprecipitated by RIG-I was highly (64.2%) enriched for coding bacterial RNAs without specific preference for a subset of coding RNAs (Fig. 4C; see also Table S1 in the supplemental material). Nonspecific RNA that was immunoprecipitated with control IgG had ratios of coding versus noncoding bacterial RNA similar to those seen with the input RNA, confirming the specificity of the pulldown. In order to confirm enrichment of immunostimulatory RNAs among the RIG-I-bound RNAs, we transfected input and pulldown RNAs into A549 cells and tested IFN-β mRNA expression by quantitative PCR (qPCR) (Fig. 4D). RNA pulled down with RIG-I resulted in increased IFN-β mRNA levels compared to the results seen with identical amounts of input RNA (40 ng). Transfection of the pulldown RNA into WT, RIG-I−/−, and reconstituted RIG-I−/− MEFs confirmed the immunostimulatory capacity of the RNA in a RIG-I-dependent fashion (see Fig. S2B).
Eubacteria do not encode the enzymatic machinery for synthesis of RNA caps (18); hence, intact bacterial mRNAs expose a 5′-triphosphorylated end. Removal of the first two phosphate groups initiates degradation of bacterial mRNA (19). To demonstrate that primary transcripts of coding bacterial RNAs represent a RIG-I-detectable PAMP, we utilized purified Escherichia coli RNA-polymerase holoenzyme. We generated a bacterial mRNA transcript (E. coli CLPb) in vitro (see Fig. S3A in the supplemental material). Transfection of this RNA into MEFs induces robust, RIG-I-dependent IFN-β mRNA expression in amounts as low as 10 ng/5×10E4 cells (Fig. 4E). To test if this immunostimulatory capacity is sequence specific to ClpB mRNA or a feature of all primary products of bacterial polymerase, we tested mCherry mRNA, a nonbacterial mRNA, generated with E. coli holoenzyme in vitro. As shown before for RNA generated from a bacterial template, this RNA activates IFN-β expression (data not shown). In order to validate the binding of RIG-I to coding RNAs under infection conditions, we used Salmonella expressing an exogenous marker mRNA (MS2aptamer). We infected A549 cells with these bacteria for 8 h and pulled down RIG-I-associated RNA as described before. We compared the relative amounts of MS2aptamer mRNA in isotype control and RIG-I-specific pulldowns by qPCR using identical amounts of RNA (Fig. 4F). RIG-I-specific pulldown results in a 10-fold enrichment of marker mRNA over control IgG pulldown.
To determine if RIG-I activation by S. Typhimurium RNA is dependent on a 5′-triphosphate, we treated total RNA of S. Typhimurium, in vitro-transcribed (IVT) bacterial RNA, influenza virus A (IAV) RNA, and long poly(I·C) with calf intestinal phosphatase (CIP) for 5 to 60 min and transfected these RNAs in A549 cells (see Fig. S3B in the supplemental material). Long poly(I·C) is recognized in a 5′ triphosphate- and RIG-I-independent manner by MDA-5, resulting in induction of IFN. CIP treatment of poly(I·C) did not affect IFN-β induction, confirming the absence of RNase contamination of the CIP enzyme. Dephosphorylation abolished the IFN-β-inducing potential of IAV RNA within 5 min. In contrast, the immunostimulatory capacity of S. Typhimurium RNA and IVT bacterial RNA decreased more slowly, reaching ~1% of the initial IFN-β-inducing capacity after 40 to 60 min of CIP treatment (see Fig. S3B in the supplemental material).
RIG-I binds synthetic 5′-triphosphorylated RNAs with its regulatory domain (RD) (13). To validate whether immunostimulatory bacterial RNA is recognized by a similar mechanism, we performed pulldown experiments with different domains of RIG-I. Equal amounts of pulldown RNA were subsequently transfected into A549 cells. As shown for viral RNA ligands, we found pulldown of IFN-β inducing RNA only when using either full-length RIG-I or the RIG-I regulatory domain (see Fig. S3C in the supplemental material).
Our data suggest that RIG-I preferentially binds to coding RNA from S. Typhimurium during bacterial infection. Recognition occurs through the RD of RIG-I, and immunostimulatory activity depends on 5′ triphosphorylation.
DISCUSSION
Recently, multiple studies investigated the role of RLR signaling in antibacterial responses (reviewed in reference 7). However, the underlying mechanisms of detection and the nature of the activating PAMP remain less well understood.
S. Typhimurium invades nonphagocytic as well as phagocytic cells in vivo. The composition of the intracellular compartment that Salmonella establishes for replication depends on the host cell type (reviewed in reference 20) and the mechanism of bacterial entry (21). It was therefore unclear whether the innate immune system provides PRR in nonphagocytic cells to detect this invasion and whether the mechanisms of detection are similar in phagocytic and nonphagocytic cells.
Here we demonstrate that recognition of S. Typhimurium requires different subsets of PRR in phagocytic and nonphagocytic cells. The reasons for this are most likely multifactorial. Macrophages rely highly on TLR- and NLR-dependent recognition of phagocytized bacterial pathogens (22). This broad panel of PRRs allows the detection of multiple easily accessible bacterial PAMPs, such as components of the bacterial cell wall or flagella. Moreover, the potential routes of infection of S. Typhimurium differ between phagocytic and nonphagocytic cells, allowing the presented PAMPs to be located in different subcellular compartments, with different sets of PRRs.
We show that Salmonella RNA is transferred into cytoplasm in nonphagocytic cells during infection. It binds to RIG-I and activates IFN-β production. These data are in accordance with an earlier study showing that transfection of Helicobacter pylori RNA into RIG-I-overexpressing 293T cells induces production of IFN-β mRNA (23). Our RIG-I pulldown experiments implicate bacterial mRNA as the immunostimulatory species among the total RNA. By column fractionation, we could rule out an impact of small RNA molecules on the immunostimulatory capacity of total bacterial RNA. A recent study showed that bacterial tRNA can activate TLR7, when lacking a posttranscriptional 2′-O-methylated G(m) 18 modification (24). It is well established that RIG-I preferentially binds to 5′-triphosphorylated RNA species (4, 13). Bacterial mRNA is uncapped and 5′-triphosphorylated. Accordingly, dephosphorylation of bacterial RNA abolished its immunostimulatory capacity in our hands. For in vitro-synthesized bacterial mRNA, it was previously shown that the addition of a 3′ poly(A) tail reduces the stimulatory capacity of this RNA in dendritic cells, suggesting the presence of immunostimulatory structures at the 3′ end of bacterial mRNA (25). However, detection of bacterial RNAs through RIG-I might play a minor role due to the dominance of TLR signaling in these cells.
In a recent study, Abdullah and colleagues demonstrated that macrophages partially depend on RIG-I for production of interferon upon L. monocytogenes infection (9). The same group very recently published data revealing the dependence on RIG-I of the type I interferon response to Listeria infection in nonimmune cells (10). In conjunction with our data from S. Typhimurium infections, this implies that different intracellular bacterial pathogens are sensed by different mechanisms, depending on the biology of the invading pathogen. This highlights the importance of accessibility of bacterial PAMPs to cellular PRRs for the innate host response during infection.
In summary, we demonstrate RIG-I-mediated innate immune recognition of S. Typhimurium mRNA in nonphagocytic cells under infection conditions. This is in contrast to IFN-β production in phagocytic cells in response to S. Typhimurium infection, which relies on TLR signaling. Our data implicate a role for RIG-I in detection of mRNA of invading bacterial pathogens that complements the orchestra of well-studied PRRs known to detect other bacterial PAMPs.
MATERIALS AND METHODS
Reagents.
Sendai virus RNA was isolated from SeV Cantell-infected embryonated chicken eggs. TLR-grade LPS solution from Salmonella Minnesota R594 was purchased from Alexis.
Bacteria.
Salmonella Typhimurium WT SL1344 (ATCC), SL1344 transformed with pM973, encoding GFP under the control of the SPI-2 promoter PssaG (WT-GFP) (26), and SB161 (ΔinvG) (27) cells were grown in standard LB liquid culture. For infection of cell cultures, Salmonella was freshly inoculated from glycerol stocks and grown in LB medium for 16 h at 37°C and 300 rpm. Cultures were diluted 1:500 with fresh medium and grown to an optical density at 420 nm (OD420) of 0.6 to 0.8. CFU levels were estimated by optical density at 420 nm and confirmed by plating serial dilutions on MacConkey’s agar. Intracellular bacterial titers were determined by permeabilization with phosphate-buffered saline (PBS) containing 0.1% (vol/vol) Triton X-100 for 15 min. Bacterial supernatants were diluted and spotted on MacConkey’s agar containing suitable antibiotics. MS2-aptamer RNAs expressing S. Typhimurium SL1344 were generated by electroporation of S. Typhimurium SL1344 with pCR4-24XMS2SL-stable (Addgene plasmid 31865 [28]) and selection with 100 µg/ml ampicillin.
Listeria monocytogenes was kindly provided by Thomas Moran (Icahn School of Medicine at Mount, Sinai). L. monocytogenes was grown in brain heart infusion broth.
Cells.
Stimulation of 293T-FF cells was described previously (16). RIG-I+/+, RIG-I−/−, reconstituted RIG-I−/− MEF, MAVS−/− MEF, and TRIF/MyD88 double-knockout (DKO) fibroblasts were described earlier (29–31). All MEFs were cultivated in Dulbecco’s modified Eagle’s medium (DMEM)–10% fetal calf serum (FCS) containing penicillin-streptomycin. Mouse bone marrow-derived macrophages (BMDM) were generated and cultured as described earlier (32).
HeLa, HT29, and A549 cells were purchased from ATCC and cultured in DMEM–10% FCS containing penicillin-streptomycin. MEFs were kindly provided by Shizuo Akira’s group (30, 33, 34). BMDM were isolated as described before (32).
HeLa cells stably expressing MS2-GFP were generated by lentiviral transduction and subsequent puromycin selection. The MS2-GFP open reading frame (ORF) was cloned from pMS2-GFP (Addgene plasmid 27121 [35]) into pLVX-IRES (internal ribosome entry site)-puro (Clontech). Cells were selected with 2 µg/ml puromycin for 2 weeks.
A549 and HT-29 cell lines stably expressing shRNAs (GAGGTGCAGTATATTCAGGTTCAAGAGACCTGAATATACTGCACCTCTTTTTT) against RIG-I were generated by lentiviral transduction using pLKOshRNA RIG-I (kindly provided by R. E. Randall, University of St. Andrews, United Kingdom). Cells were selected with 2 µg/ml puromycin for 2 weeks.
Mice.
Generation of RIG-I+/+ and RIG-I−/− mice was described earlier (33). MAVS−/− mice were described earlier (30). TRIF/MyD88 DKO mice were described earlier (34). Bacterial infection was performed similarly to a previously published protocol (36).
Ethics statement.
All research studies involving the use of animals were reviewed and approved by the Institutional Review Boards (IRB) of Mount Sinai School of Medicine (MSSM). All human cell lines used were purchased from ATCC. All animal procedures performed in this study were in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines and have been approved by the IACUC of Mount Sinai School of Medicine (protocol 020274).
RNA extraction and qPCR.
Total RNAs from cultured cells and bacteria were isolated with a Qiagen RNeasy Minikit (Qiagen) and TRIzol (Invitrogen), respectively.
Mean n-fold expression levels of cDNA from three individual biological samples, each measured in triplicate, were normalized to 18S rRNA levels and calibrated to mock-treated samples according to the 2−ΔΔCT method (37). The primer sequences were as follows: for the murine gene, the IFN-β forward primer was 5′-CAGCTCCAAGAAAGGACGAAC-3′, the IFN-β reverse primer was 5′-GGCAGTGTAACTCTTCTGCAT-3′, the 18S forward primer was 5′-GTAACCCGTTGAACCCCATT-3′, and the 18S reverse primer was 5′-CCATCCAATCGGTAGTAGCG-3′; for the human gene, the IFN-β forward primer was 5′-TCTGGCACAACAGGTAGTAGGC-3′ and the IFN-β reverse primer was 5′-GAGAAGCACAACAGGAGAGCAA-3′; and for the other primers, the MS2aptamer forward primer was 5′-ACGGTACTTATTGCCAAGAAAGC-3′ and the MS2aptamer reverse primer was 5′-GATGAACCCTGGAATACTGGAGC-3′.
Western blot analyses.
SDS-PAGE and Western blot analyses were performed as described previously (38).
RNA.
For fractionation, total RNAs of S. Typhimurium were depleted of small RNA fractions using ChromaSpin columns (Clontech).
For dephosphorylation, RNAs were treated with 10 U of RNase A/µg of RNA for 1 h at 37°C or with 10 U of calf intestinal alkaline phosphatase (CIAP)/µg of RNA for 5 min to 1 h at 37°C. Treated RNAs were purified using Qiagen RNeasy Mini columns.
For in vitro transcription, in vitro-transcribed bacterial RNA was generated using purified, sigma-saturated E. coli holoenzyme (EpiBio) and linearized pClpB (Addgene plasmid 1235; kindly provided by Susan Lindquist [39]) or pFPVmCherry (Addgene plasmid 20956; kindly provided by Olivia Steele-Mortimer, NIH, Washington, DC [40]) as a template. RNAs were purified using Qiagen RNeasyMini columns. Template DNA was removed by on-column digestion with RNase-free DNase (Qiagen).
Denaturing RNA gel electrophoresis.
Denaturing agarose gels (1% agarose, 6.5% formaldehyde, 2.5 mM sodium acetate, 1.25 mM EDTA, 20 mM MOPS [morpholinepropanesulfonic acid], pH 7.0) were prepared under RNase-free conditions. RNA was heated to 65°C for 5 min in 5 mM EDTA–50% formamide–0.0125% bromophenol blue–0.0125% xylene cyanol. Gels were run in 2.5 mM sodium acetate–1.25 mM EDTA–20 mM MOPS (pH 7.0) containing 8.7% formaldehyde.
RIG-I pulldown and deep sequencing.
Immunoprecipitation of RIG-I and recovery of associated RNA were described earlier (16). For RNA sequencing, cDNA libraries for RNA samples were prepared using an Illumina sample preparation kit (Illumina) and RNA sequencing was performed on an Illumina platform at the Genomics Sequencing Core Facility of the Mount Sinai School of Medicine as described previously (16). Sequences were matched to the genome of Salmonella Typhimurium LT2 (41). RIG-I-specific pulldown reads were defined as 5-fold enriched over total RNA and 1.5-fold enriched over pulldown with the isotype control (anti-GFP). Control pulldown reads were defined as 5-fold enriched over total RNA and 1.5-fold enriched over RIG-I pulldown.
Microscopy.
Immunofluorescence microscopy was performed using a Zeiss Axioplan fluorescence microscope and Axiovision image acquisition. Confocal laser scanning microscopy was performed using a Zeiss 510 laser scanning microscope (LSM) at the MSSM Microscopy Shared Resource Facility.
Statistical analysis.
Experimental variation was calculated using Excel software and is depicted as standard variations of the results of independent biological replicate experiments. Statistical significance is indicated as P < 0.05 (using a single asterisk [*]) or P < 0.01 (using double asterisks [**]) and was calculated using an unpaired t test (Quick Calcs; GraphPad Software). Unless otherwise indicated, pairs of asterisks (**) over values refer to the statistical significance of comparisons to results from mock experiments and pairs of asterisks (**) over brackets refer to the statistical significance of comparisons of the values indicated by the brackets.
SUPPLEMENTAL MATERIAL
(A and B) RIG-I+/+ and RIG−/− MEFs and RIG-I-reconstituted RIG−/− MEFs were infected with S. Typhimurium at an MOI of 1 (A) or an MOI of 10 (B) for 8 h. Values represent the average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD as determined by qPCR. (C) Intracellular titers of RIG-I+/+ and RIG−/− MEFs and RIG-I-reconstituted RIG−/− MEFs infected with S. Typhimurium SL1344 at an MOI of 1, as determined by serial dilution of cell lysates. Each column represents the average titers of three independent biological samples each measured in technical triplicate experiments ± SD. Download
Characterization of bacterial mRNA pathogen-associated molecular patterns (PAMPs). (A) Denaturing RNA agarose gel. A 1-µg volume of total RNA and corresponding volumes of RNA after fractionation were loaded and stained with ethidium bromide. (B) RNA from full-length RIG-I pull down was transfected in wt, RIG-I−/−, and reconstituted RIG-I−/− MEFs. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD are depicted. (C) Total RNA of Salmonella was incubated with the indicated fragments of RIG-I expressed in 293T cells. Coprecipitated RNA was transfected into RIG-I+/+ MEFs. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD are depicted. Download
(A) Bacterial mRNA (ClpB) was generated in vitro using E. coli holoenzyme saturated with sigma factor. The purity and correct size of in vitro-transcribed RNA were confirmed by denaturing RNA agarose gel. (B) Total RNA from IAV, S. Typhimurium, IVT RNA, and poly(I·C) were treated with 10 U of CIAP/µg of RNA for indicated time points. Purified RNAs were transfected into A549 cells. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels 16 h posttransfection of three independent biological samples each measured in technical triplicate experiments ± SD are depicted. Significance is indicated in reference to influenza A virus samples. (C) Total RNA of Salmonella was incubated with the indicated fragments of RIG-I expressed in 293T cells. Coprecipitated RNA was transfected into RIG-I+/+ MEFs. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD are depicted. Download
Comparison of deep sequencing reads (50 bp) from total Salmonella RNA, GFP pulldown RNA, and RIG-I pulldown RNA.
ACKNOWLEDGMENTS
This study was partly funded by the NIAID Mucosal Immunity Study Team (MIST) program under grant U01AI095611 (to M.M. and A.G.-S.). J.R.P. is supported by an NIH T32 training grant (5T32AI007647-13). B.M. is supported by an NIH K99 Pathway to Independence award (1K99AI095320-01). Confocal laser scanning microscopy was performed using a Zeiss 510 laser scanning microscope at the MSSM-Microscopy Shared Resource Facility, supported with funding from an NIH-National Cancer Institute (NCI) shared resources grant (5R24 CA095823-04), a National Science Foundation (NSF) Major Research Instrumentation grant (DBI-9724504), and an NIH shared instrumentation grant (1 S10 RR0 9145-01).
We declare that we have no conflicts of interest.
We thank R. Cadagan, O. Lizardo, P. Muller, and the Mount Sinai CCMS for excellent technical assistance. We thank Julie M. Blander (Icahn School of Medicine at Mount Sinai, New York, NY), W.D. Hardt (ETH Zurich, Zürich, Switzerland), and Matthias Schnell (Thomas Jefferson University, Philadelphia, PA) for providing reagents.
Footnotes
Citation Schmolke M, Patel JR, de Castro E, Sánchez-Aparicio MT, Uccellini MB, Miller JC, Manicassamy B, Satoh T, Kawai T, Akira S, Merad M, García-Sastre A. 2014. RIG-I detects mRNA of intracellular Salmonella enterica servovar Typhimurium during bacterial infection. mBio 5(2):e01006-14. doi:10.1128/mBio.01006-14.
REFERENCES
- 1. Barber GN. 2011. Cytoplasmic DNA innate immune pathways. Immunol. Rev. 243:99–108. 10.1111/j.1600-065X.2011.01051.x [DOI] [PubMed] [Google Scholar]
- 2. Cambi A, Figdor CG. 2003. Dual function of C-type lectin-like receptors in the immune system. Curr. Opin. Cell Biol. 15:539–546. 10.1016/j.ceb.2003.08.004 [DOI] [PubMed] [Google Scholar]
- 3. Elinav E, Strowig T, Henao-Mejia J, Flavell RA. 2011. Regulation of the antimicrobial response by NLR proteins. Immunity 34:665–679. 10.1016/j.immuni.2011.05.007 [DOI] [PubMed] [Google Scholar]
- 4. Kato H, Takahasi K, Fujita T. 2011. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol. Rev. 243:91–98. 10.1111/j.1600-065X.2011.01052.x [DOI] [PubMed] [Google Scholar]
- 5. Kawai T, Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34:637–650. 10.1016/j.immuni.2011.05.006 [DOI] [PubMed] [Google Scholar]
- 6. Belgnaoui SM, Paz S, Hiscott J. 2011. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr. Opin. Immunol. 23:564–572. 10.1016/j.coi.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 7. Dixit E, Kagan JC. 2013. Intracellular pathogen detection by RIG-I-like receptors. Adv. Immunol. 117:99–125. 10.1016/B978-0-12-410524-9.00004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Broz P, Ohlson MB, Monack DM. 2011. Innate immune response to Salmonella typhimurium, a model enteric pathogen. Gut Microbes 3:62–70. 10.4161/gmic [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Böttcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, Civril F, Hopfner KP, Kurts C, Ruland J, Hartmann G, Chakraborty T, Knolle PA. 2012. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 31:4153–4164. 10.1038/emboj.2012.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hagmann CA, Herzner AM, Abdullah Z, Zillinger T, Jakobs C, Schuberth C, Coch C, Higgins PG, Wisplinghoff H, Barchet W, Hornung V, Hartmann G, Schlee M. 2013. RIG-I detects triphosphorylated RNA of Listeria monocytogenes during infection in non-immune cells. PLoS One 8:e62872. 10.1371/journal.pone.0062872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jehl SP, Nogueira CV, Zhang X, Starnbach MN. 2012. IFNgamma inhibits the cytosolic replication of Shigella flexneri via the cytoplasmic RNA sensor RIG-I. PLoS Pathog. 8:e1002809. 10.1371/journal.ppat.1002809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sander LE, Davis MJ, Boekschoten MV, Amsen D, Dascher CC, Ryffel B, Swanson JA, Müller M, Blander JM. 2011. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature 474:385–389. 10.1038/nature10072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cui S, Eisenächer K, Kirchhofer A, Brzózka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169–179. 10.1016/j.molcel.2007.10.032 [DOI] [PubMed] [Google Scholar]
- 14. Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997. 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- 15. Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997–1001. 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- 16. Baum A, Sachidanandam R, García-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:16303–16308. 10.1073/pnas.1005077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uzri D, Gehrke L. 2009. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J. Virol. 83:4174–4184. 10.1128/JVI.02449-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shuman S. 2002. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 3:619–625. 10.1038/nrm880 [DOI] [PubMed] [Google Scholar]
- 19. Celesnik H, Deana A, Belasco JG. 2007. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol. Cell 27:79–90. 10.1016/j.molcel.2007.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. 2010. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. U. S. A. 107:17733–17738. 10.1073/pnas.1006098107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drecktrah D, Knodler LA, Ireland R, Steele-Mortimer O. 2006. The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic 7:39–51. 10.1111/j.1600-0854.2005.00360.x [DOI] [PubMed] [Google Scholar]
- 22. Cai S, Batra S, Shen L, Wakamatsu N, Jeyaseelan S. 2009. Both TRIF- and MyD88-dependent signaling contribute to host defense against pulmonary Klebsiella infection. J. Immunol. 183:6629–6638. 10.4049/jimmunol.0901033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rad R, Ballhorn W, Voland P, Eisenächer K, Mages J, Rad L, Ferstl R, Lang R, Wagner H, Schmid RM, Bauer S, Prinz C, Kirschning CJ, Krug A. 2009. Extracellular and intracellular pattern recognition receptors cooperate in the recognition of Helicobacter pylori. Gastroenterology 136:2247–2257. 10.1053/j.gastro.2009.02.066 [DOI] [PubMed] [Google Scholar]
- 24. Gehrig S, Eberle ME, Botschen F, Rimbach K, Eberle F, Eigenbrod T, Kaiser S, Holmes WM, Erdmann VA, Sprinzl M, Bec G, Keith G, Dalpke AH, Helm M. 2012. Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 209:225–233. 10.1084/jem.20111044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koski GK, Karikó K, Xu S, Weissman D, Cohen PA, Czerniecki BJ. 2004. Cutting edge: innate immune system discriminates between RNA containing bacterial versus eukaryotic structural features that prime for high-level IL-12 secretion by dendritic cells. J. Immunol. 172:3989–3993 [DOI] [PubMed] [Google Scholar]
- 26. Hapfelmeier S, Stecher B, Barthel M, Kremer M, Müller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, Hardt WD. 2005. The salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J. Immunol. 174:1675–1685 [DOI] [PubMed] [Google Scholar]
- 27. Kaniga K, Bossio JC, Galán JE. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13:555–568. 10.1111/j.1365-2958.1994.tb00450.x [DOI] [PubMed] [Google Scholar]
- 28. Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2:437–445. 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- 29. Gack MU, Nistal-Villán E, Inn KS, García-Sastre A, Jung JU. 2010. Phosphorylation-mediated negative regulation of RIG-I antiviral activity. J. Virol. 84:3220–3229. 10.1128/JVI.02241-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203:1795–1803. 10.1084/jem.20060792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. 2003. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301:640–643. 10.1126/science.1087262 [DOI] [PubMed] [Google Scholar]
- 32. Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozören N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, Grant EP, Núñez G. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7:576–582. 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- 33. Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19–28. 10.1016/j.immuni.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 34. Koga R, Hamano S, Kuwata H, Atarashi K, Ogawa M, Hisaeda H, Yamamoto M, Akira S, Himeno K, Matsumoto M, Takeda K. 2006. TLR-dependent induction of IFN-beta mediates host defense against Trypanosoma cruzi. J. Immunol. 177:7059–7066 [DOI] [PubMed] [Google Scholar]
- 35. Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13:161–167. 10.1016/S0960-9822(02)01436-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt WD. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839–2858. 10.1128/IAI.71.5.2839-2858.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 38. Hale BG, Steel J, Medina RA, Manicassamy B, Ye J, Hickman D, Hai R, Schmolke M, Lowen AC, Perez DR, García-Sastre A. 2010. Inefficient control of host gene expression by the 2009 pandemic H1N1 influenza A virus NS1 protein. J. Virol. 84:6909–6922. 10.1128/JVI.00081-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shorter J, Lindquist S. 2004. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304:1793–1797. 10.1126/science.1098007 [DOI] [PubMed] [Google Scholar]
- 40. Drecktrah D, Levine-Wilkinson S, Dam T, Winfree S, Knodler LA, Schroer TA, Steele-Mortimer O. 2008. Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic 9:2117–2129. 10.1111/j.1600-0854.2008.00830.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856. 10.1038/35101614 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A and B) RIG-I+/+ and RIG−/− MEFs and RIG-I-reconstituted RIG−/− MEFs were infected with S. Typhimurium at an MOI of 1 (A) or an MOI of 10 (B) for 8 h. Values represent the average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD as determined by qPCR. (C) Intracellular titers of RIG-I+/+ and RIG−/− MEFs and RIG-I-reconstituted RIG−/− MEFs infected with S. Typhimurium SL1344 at an MOI of 1, as determined by serial dilution of cell lysates. Each column represents the average titers of three independent biological samples each measured in technical triplicate experiments ± SD. Download
Characterization of bacterial mRNA pathogen-associated molecular patterns (PAMPs). (A) Denaturing RNA agarose gel. A 1-µg volume of total RNA and corresponding volumes of RNA after fractionation were loaded and stained with ethidium bromide. (B) RNA from full-length RIG-I pull down was transfected in wt, RIG-I−/−, and reconstituted RIG-I−/− MEFs. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD are depicted. (C) Total RNA of Salmonella was incubated with the indicated fragments of RIG-I expressed in 293T cells. Coprecipitated RNA was transfected into RIG-I+/+ MEFs. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD are depicted. Download
(A) Bacterial mRNA (ClpB) was generated in vitro using E. coli holoenzyme saturated with sigma factor. The purity and correct size of in vitro-transcribed RNA were confirmed by denaturing RNA agarose gel. (B) Total RNA from IAV, S. Typhimurium, IVT RNA, and poly(I·C) were treated with 10 U of CIAP/µg of RNA for indicated time points. Purified RNAs were transfected into A549 cells. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels 16 h posttransfection of three independent biological samples each measured in technical triplicate experiments ± SD are depicted. Significance is indicated in reference to influenza A virus samples. (C) Total RNA of Salmonella was incubated with the indicated fragments of RIG-I expressed in 293T cells. Coprecipitated RNA was transfected into RIG-I+/+ MEFs. Average n-fold expression levels of IFN-β mRNA over 18S rRNA levels of three independent biological samples each measured in technical triplicate experiments ± SD are depicted. Download
Comparison of deep sequencing reads (50 bp) from total Salmonella RNA, GFP pulldown RNA, and RIG-I pulldown RNA.