Abstract
The validation of novel diagnostic, prognostic, and predictive biomarkers and therapeutic targets in tumor cells is of critical importance for optimizing the choice and efficacy of personalized therapies. Importantly, recent advances have led to the identification of gene-expression signatures in cancer cells, including cancer stem/progenitor cells, in the primary tumors, exosomes, circulating tumor cells (CTC), and disseminated cancer cells at distant metastatic sites. The gene-expression signatures may help to improve the accuracy of diagnosis and predict the therapeutic responses and overall survival of patients with cancer. Potential biomarkers in cancer cells include stem cell–like markers [CD133, aldehyde dehydrogenase (ALDH), CD44, and CD24], growth factors, and their cognate receptors [epidermal growth factor receptor (EGFR), EGFRvIII, and HER2], molecules associated with epithelial–mesenchymal transition (EMT; vimentin, N-cadherin, snail, twist, and Zeb1), regulators of altered metabolism (phosphatidylinositol-3′ kinase/Akt/mTOR), and drug resistance (multidrug transporters and macrophage inhibitory cytokine-1). Moreover, different pluripotency-associated transcription factors (Oct3/4, Nanog, Sox2, and Myc) and microRNAs that are involved in the epigenetic reprogramming and acquisition of stem cell–like properties by cancer cells during cancer progression may also be exploited as molecular biomarkers to predict the risk of metastases, systemic treatment resistance, and disease relapse of patients with cancer.
Introduction
Significant advancement in basic and clinical oncology during the last few years has led to earlier diagnosis and more effective therapeutic management of patients with leukemias and organ-confined tumors in the clinics (1-3). Although the surgical tumor resection may result in some cases to a complete remission, the rapid cancer progression of aggressive cancers to locally invasive and metastatic stages is generally associated with the development of resistance mechanisms by cancer cells to current antihormonal, radiation, and/or chemotherapeutic treatments and disease relapse (1-3). At the present time, the metastatic cancers remain the leading cause of the death of patients with cancer. Therefore, many research efforts have been made to identify and validate novel molecular biomarkers and therapeutic targets in cancer cells at primary and secondary tumors to prevent cancer progression and metastases and optimize the genetic- and proteomic-based individualized treatments of patients with cancer (Fig. 1; refs. 4-28).
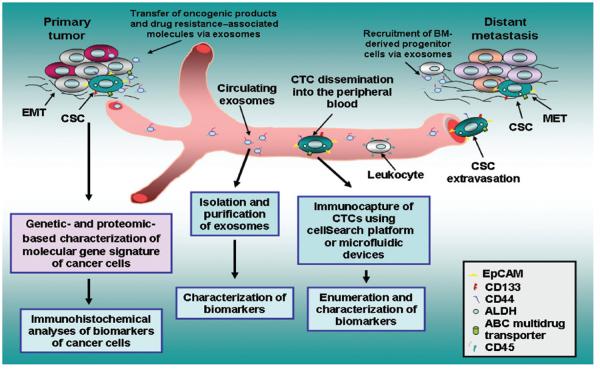
Figure 1.
Schematic representation of functions of cancer stem/progenitor cells during cancer progression and metastasis and characterization of their biomarkers. The scheme shows cancer stem/progenitor cells endowed with stem cell–like properties and which can generate the total cancer cell population at the primary and secondary tumors. Moreover, the exosomes released by cancer cells, which may contribute to the malignant transformation of other cancer cells via the transfer of oncogenic products and drug resistance–associated molecules such as EGFRvIII and P-glycoprotein, are also illustrated. The possibility to perform the characterization of molecular gene signature and biomarkers of cancer cells, exosomes, and CTCs, including cancer stem/progenitor cells expressing stem cell–like markers, is also indicated.
Importantly, accumulating lines of evidence have revealed that the shedding of cancer cells from the primary tumors into the lymphatic vessels and peripheral circulation can occur very early during the cancer development and be dependent of cellular origin, genetic alterations, and aggressiveness of cancer subtypes (16, 29-41). Hence, some patients who undergo a complete surgical tumor resection with negative margins may show the presence of circulating tumor cells (CTC) in the peripheral blood and disseminated tumor cells at the regional lymph nodes and distant tissues and organs (Fig. 1; refs. 16, 29-41). Consequently, CTCs that are able to survive in the bloodstream and spread at distant sites can persist and contribute to metastases and disease relapse even after an effective and apparently curative surgical resection of the primary tumor. In this regard, a growing body of experimental evidence has also revealed that cancer stem/progenitor cells endowed with stem cell–like properties, also designated as cancer-, tumor-, and metastasis-initiating cells, can provide critical functions for tumor growth, metastases at near and distant tissues and organs, treatment resistance, and disease relapse. In fact, it has been shown that the most cancers may originate from the malignant transformation of immature tissue-resident stem/progenitor cells or their early differentiated progenies endowed with a high self-renewal ability and aberrant differentiation potential (2, 42-44). The cancer stem/progenitor cells expressing specific stem cell–like markers such as CD133, CD44high, nestin, aldehyde dehydrogenase (ALDHhigh), and high levels of ATP-binding cassette (ABC) multidrug transporters have also been identified and isolated from primary and secondary neoplasms, including leukemias, melanomas, brain tumors, and the most epithelial cancers and cancer cell lines (9,17, 24, 44-76). It has been shown that cancer stem/progenitor cells were able to give rise to the total tumor cell mass, including differentiated cancer cells that reconstituted the histological architecture and molecular characteristics of primary and secondary tumors closely resembling to original patient’s tumors in vivo (9, 17, 45-57, 59-66, 68, 69, 71, 77). Moreover, the data from recent studies have indicated that cancer stem/progenitor cells may be more resistant than their differentiated progenies to current antihormonal, radiation and chemotherapeutic treatments, and targeted therapies (17, 22-25, 44, 52, 53, 56-64, 68, 70, 72, 77-94).
We review here recent advances on the characterization of gene products often altered in cancer stem/progenitor cells and their differentiated progenies during primary cancer progression and dissemination through the peripheral circulation and metastases. The emphasis is on molecular gene signatures, epithelial–mesenchymal transition (EMT)-like and stem cell–like biomarkers detected in cancer cells and exosomes associated with cancer progression, treatment resistance, disease relapse, and poor outcome of patients with cancer.
Heterogeneity of Cancers
The most cancers are heterogeneous diseases and include some biological subtypes characterized by specific genetic and/or epigenetic alterations occurring in cancer stem/progenitor cells and their differentiated progenies during cancer initiation and progression to locally invasive and metastatic stages (4, 6, 7, 44, 60, 78, 80, 95-109). Although the molecular events that govern the spread of cancer stem/progenitor cells and their differentiated progenies at preferential metastatic sites remain not precisely defined, some lines of evidence suggest that particular gene signatures of cancer cells at primary cancers may be associated with their propensity at metastasizing at specific tissues and organs (4, 6, 7, 95-102). More particularly, the alterations in the gene products involved in the acquisition of aggressive and migratory properties by highly tumorigenic cancer stem/progenitor cells during the EMT program at the primary tumor may result in their invasion to near lymph nodes and tissues, dissemination through the peripheral circulation, and metastases at distant sites (Fig. 1; refs. 30, 42, 90, 91, 96, 97, 110-126). Moreover, different gene products have also been identified in exosomes released by tumor cells and CTCs with stem cell–like features that may be used as diagnostic, prognostic, and/or predictive biomarkers. In this regard, we review the gene products frequently altered in cancer cells, during cancer progression to locally advanced and metastatic states, exosomes and CTCs that have been associated with treatment resistance, disease relapse, and poor overall survival of patients with cancer.
Molecular gene signatures and stem cell–like biomarkers associated with aggressive and metastatic cancers
Some tissue-specific and whole genome approaches for molecular profiling of complex gene signatures of tumors and cancer cell lines have been developed in the last few years and led to the discovery of specific gene sets related to particular cancer types and which may predict the risk of metastatic dissemination at specific tissues and organs and overall outcome of patients with cancer (Fig. 1; refs. 4-15). Moreover, the in vitro and in vivo characterization of stem cell–like markers and functional features of cancer stem/progenitor cells in tumor tissue specimens from patients with cancer and cancer cell lines has also revealed their major implications in tumor growth, metastases, treatment resistance, and disease relapse (Fig. 1; refs. 15-27). We are reporting here the gene signatures and stem cell–like biomarkers of cancer cells associated with particular subtypes of aggressive cancers, including hormone-independent prostate and breast cancers.
Prostate cancer
Prostate cancer is among the most commonly diagnosed malignancies affecting men (127-129). Although an earlier diagnosis of patients with prostate cancer has led to an improvement of their effective treatment by tumor resection surgery and/or radiation therapy, the locally advanced and metastatic castration-resistant prostate cancers (CRPC) still represent the second leading cause of cancer-related death (127-129). In fact, antihormonal treatments and first-line docetaxel-based chemotherapies of metastatic CRPCs are only palliative and result in the death of most patients after about 12 to 19 months (127-129). Therefore, this inefficacy of current treatments against CRPCs underlines the importance to identify novel biomarkers to predict the risk of propagation to CRPCs. Of clinical relevance, a 11-gene signature related to the activation of polycomb-group protein BMI-1 pathway, which provides critical functions for the self-renewal of diverse normal and cancer stem cells, has notably been identified through a comparative genomic approach using primary and metastatic tumor specimens from transgenic adenocarcinoma of the mouse prostate model and patients with prostate cancer (4). The 11-gene signature includes 8 upregulated genes (Ki67, cyclin B1, gastrulation brain homeobox 2, budding uninhibited by benzimidazoles 1 “BUB1” that acting as serine/threonine-protein kinase, kinetochore associated 2, ubiquitin-specific protease 22, host cell factor 1, and ring finger protein 2) and 3 downregulated genes (ankyrin 3, fibroblast growth factor receptor 2, and carboxylesterase 1; ref. 4). This 11-gene signature, which is associated with an upregulation of stem cell–resembling expression profile of the BMI-1–regulated pathway, has been shown to be a powerful predictor of patients with prostate cancer progression to locally invasive and metastatic stages (4). The analyses of this 11-gene signature of BMI-1–driven pathway in 1,153 clinical tumor specimens from patients with cancer diagnosed with 11 different types of cancer have also indicated that the expression of these gene products was associated with a short interval to disease recurrence, distant metastases, and death after therapy of patients with cancer (4). These cancer types comprise acute myeloid leukemia, lymphoma, mesothelioma, medulloblastoma, glioma, and prostate, breast, lung, ovarian, and bladder epithelial cancers (4). Importantly, high expression levels and/or activities of stem cell–like markers such as CD133, CD44, integrin-α2, nestin, ALDH1, and CD49f in primary prostatic adenocarcinomas and bone metastases from patients and prostate cancer cell lines have also been associated with their aggressive and invasive phenotypes and treatment resistance (17-26, 28). Particularly, the data from immunohistochemical analyses of integrin-α2, integrin-α6, and hepatocyte growth factor receptor (MET) in prostate cancer cells have revealed that an increase of expression levels of these markers in prostatic adenocarcinoma specimens from patients with prostate cancer was associated with an enhanced risk of local invasion and bone metastasis (130). It has also been reported that several ALDH isoforms were expressed in specimens of primary prostate tumors with matched bone metastases and prostate cancer cell lines, and ALDHhigh PC3M-Pro4 cells rapidly formed primary prostatic tumor and metastasized at distant sites in vivo as compared with ALDHlow PC3M-Pro4 cells (18, 19). Of therapeutic interest, the knockdown of ALDH7A1 expression in PC3M-Pro4 cells has also been observed to result in a decrease of the integrin-α2high/integrin-αvhigh/CD44+ PC3M-Pro4 stem/progenitor cell subpopulation and gene products involved in the EMT process and invasion, including snail, snail2, twist, and extracellular molecule, osteopontin (19). Moreover, the ALDH7A1 silencing in PC3M-Pro4 cells significantly inhibited their clonogenic and migratory abilities in vitro as well as their capacity to form bone metastases in vivo (19). In the same way, it has also been observed that prostate-specific antigen (PSA−/lo) prostate cancer cells from LNCaP, LAPC4, or LAPC9 cell lines or tumor specimens from patients with prostate cancer expressing stem cell–like markers such as ALDH, CD44, integrin-α2β1, and with a long-term tumor-propagating ability were able to give rise to the total mass of PSA+ prostate cancer cells in vivo (17). It has also been noted that PSA−/lo LAPC9 cells were resistant to androgen deprivation in castrated hosts, and high grade and recurrent prostatic adenocacinoma specimens of patients were enriched in PSA−/lo prostate cancer cells (17). These observations suggest that PSA−/lo prostate cancer cells may play critical functions for the tumor regrowth after antihormonal treatment initiation.
In addition, we have observed that sonic hedgehog (SHH/GLI-1), epidermal growth factor receptor (EGFR), pAkt, nuclear factor-κB (NF-κB), and macrophage inhibitory cytokine-1 (MIC-1) are frequently overexpressed in prostate cancer cells, including CD133+ prostate cancer stem/progenitor cells, detected in prostatic adenocarcinoma and bone metastasis specimens from patients with prostate cancer (23, 24). The high expression levels and/or activities of these oncogenic products were also detected in the side population from tumorigenic and invasive WPE1-NB26 cells and metastatic PC3M-LN4 endowed with a high prostasphere-forming ability (23-25). Moreover, the targeting of hedgehog, EGFR, pAkt, NF-κB, or MIC-1 by using cyclopamine, gefitinib, Akt inhibitor III, partenolide, or anti-MIC-1 antibody was effective at inducing the apoptotic effects on side population and non-side population cells from WPE1-NB26 and PC3M-LN4 cell lines and improving cytotoxic effects induced by docetaxel on these side population and non-side population cells (23, 24). Recent studies have also revealed that the changes in the expression levels and activities of hypoxia-inducible factors (HIF) and microRNAs (miRNA), which are short noncoding RNA molecules of about 22 nucleotides that are involved in the stringent control of normal stem cell behavior often occur in cancer cells including prostate cancer cells (27, 28, 131, 132). HIFs and miRNAs may be involved in the epigenetic reprogramming and acquisition of stem cell–like features by cancer cells (27, 28, 131, 132). More specifically, the upregulation of HIF-1α and HIF-2α has been associated with the increased expression of pluripotency-associated transcription factors (Nanog, Oct4, and Sox2) and EMT program-associated molecules in prostate cancer cells under normoxic and hypoxic conditions during prostate cancer progression to locally invasive and metastatic stages (27, 28). The downregulation of some tumor-suppressor miRNAs such as mi-R34a, let-7b, miR-106a, and miR-141 combined with an upregulation of miR-301 and miR-452 has also been observed to regulate the proliferation and tumorigenicity of CD133/CD44/integrin-α2β1 prostate cancer stem/progenitor cell population and side population cells in vitro and in vivo (131, 132).
Breast cancer
Breast cancer is among the most common cancers in women (127, 133, 134). Although a high survival rate is observed for early-stage patients with breast cancer treated by tumor resection surgery and adjuvant treatments, the propagation to locally invasive and metastatic disease state is generally associated with the development of resistance mechanisms by cancer cells to current antihormonal treatments, targeted therapies, radiation, and chemotherapies (127, 133, 134). Typically, breast cancer is a complex and highly heterogeneous disease that encompasses distinct cancer subtypes characterized by the malignant transformation of the basal and/or luminal breast epithelial cells in the mammary gland (135, 136). In this regard, the data from analyses of the expression levels of gene products in breast cancer cells, including breast cancer stem cells (BCSCs) with stem cell–like features, have indicated that breast cancer subtypes may exhibit cellular and functional heterogeneities (5-14, 137, 138). It has been observed that CD44high/CD24−/low BCSCs are frequently detected in basal-like subtypes of breast cancer whereas these immature cancer cells are very low in erbB2/HER2+ breast tumors (8). Moreover, a transcriptional analysis of basal-like breast cancer cell lines has led to the identification of 2 major cancer subclasses in which basal B seemed mesenchymal-like, whereas basal A may have either a luminal-like or basal-like morphology. It has been observed that a CD44high/CD24low epithelial basal A cell subpopulation isolated from triple-negative HMT3909S13 breast cancer cell line exhibited cancer stem cell features such as mammosphere formation and resistance to the standard chemotherapy as well as tumor-initiating capacity in vivo (6). In contrast, CD44high/CD24− mesenchymal-like basal B cell fraction does not exhibit these functional features (6). The data from comparative quantitative proteomic and gene array analyses of CD44high/CD24low BCSCs with an epithelial-like morphology have also indicated that these immature cancer cells expressed lipolysis-stimulated lipoprotein receptor, RAB25, S100A14, and mucin 1 (MUC1; ref. 6). These CD44high/CD24low BCSCs also expressed a novel 31-gene signature that was related at the presence of distant metastases in estrogen receptor negative (ER−)-human breast cancer specimens (6). Conversely, the results from another investigation have revealed that CD44high/CD24low BCSCs that were detected in patient’s primary tumors exhibited a gene signature as the claudin-low molecular subtype, which is characterized by the expression of several EMT program-associated molecules (10). It has been noted that the CD44high/CD24low BCSCs expressing mesenchymal markers were enriched in patient’s primary tumors after either endocrine letrozole-based therapy or docetaxel-based chemotherapy (10). These findings suggest that breast cancer cells endowed with an epithelial or mesenchymal phenotype can exhibit cancer stem cell properties dependent of breast cancer subtypes and be involved in the metastasis formation and treatment resistance.
On the other hand, the data from analyses of global gene expression profiles have also indicated that a common subset of 81 genes was differentially expressed between ER− and ER+ breast tumor specimens and breast cancer cell lines as well as in androgen-independent versus androgen-sensitive prostate cancer cell lines (11). It has been noted that some signaling elements of EGFR pathway were enriched in ER− breast and androgen-independent prostate cancer cell lines, suggesting that the sustained activation of this tumorigenic cascade may contribute to the growth and survival of these hormone-independent cancer cells (11). Interestingly, it has been reported that an 8-gene signature (EGFR, integrin-α3, myosin light chain kinase, retinoic acid–induced protein 14, AHNAK nucleoprotein, glutaminase, interleukin-32, and nicotinamide N-methyltransferase) was associated with the invasion and sensitivity of a panel of 78 tumor cell lines to chemotherapeutic drugs, including paclitaxel, docetaxel, erlotinib, everolimus, and dasatinib (14). This set of 8 genes also predicted the relapse-free survival for patients with breast cancer (14).
Other cancer types
Recent investigations have also led to the discovery of specific gene signatures and potential biomarkers in cancer tissue specimens for improving the personnel management of patients with other aggressive cancers such as brain, melanoma, esophageal squamous cell carcinoma (ESCC) and lung, pancreatic, colorectal, esophageal, and ovarian cancers (Table 1; refs. 3, 14, 70, 139-149). For instance, the real-time PCR data from gene expression profiles of brain tumor stem-like cells (BTSCs) from 5 human glioblastoma multiforme (GBM) and matched GBM tissue specimens have indicated that SHH and GLI1 hedgehog signaling elements were coexpressed only at high levels in BTSCs 1 to 3 and not in BTSCs 4 and 5 (141). It has also been noted that the SHH/GLI1 cascade-induced BTSC proliferation via phosphatidylinositol-3′ kinase (PI3K)/Akt/mTOR, was inhibited by small hairpin RNA (shRNA) against human GLI1 or rapamycin in vitro and in an intracranial tumor model in athymic mice (141). Of clinical interest, higher expression levels of SHH, patched receptor 1 (PTCH1), and GLI1 have also been detected in phosphatase and tensin homolog deleted on chromosome 10 (PTEN)-expressing brain tumors than in PTEN-deficient brain tumors and associated with a poorer survival time of GBM patients (141). Furthermore, it has been shown that melanoma cells from patient with melanoma specimens expressing high levels of ALDH1A1 and ALDH1A3 isoforms displayed a high self-renewal ability, and were more resistant to chemotherapeutic agents and more tumorigenic than ALDH− melanoma cells in non-obese diabetic (NOD)/severe combined immonodeficient (SCID) mice (139). It has also been noticed that the silencing of the ALDH1A gene by shRNA led to an arrest in cell cycle and apoptosis of ALDH1A+ melanoma cells, inhibition of tumor growth in vivo, and sensitized the ALDH1A+ melanoma cells to cytotoxic effects of chemotherapeutic drugs (139). These findings implicate that ALDH1A and ALDH3A isozymes are putative biomarkers for melanoma stem cells and constitute attractive therapeutic targets for treating melanoma patients. In the same way, human pancreatic ductal adenorcinoma (PDAC) specimens have also been observed to contain a side population exhibiting the gene expression signatures that are associated with pancreatic cancer stem cell–like characteristics and resistance to chemotherapeutic drugs (144). Among these genes, expression levels of ABCB1 multidrug transporter and CXC chemokine receptor 4 (CXCR4) were correlated with a worse survival of patients with pancreatic cancer (144).
Table 1.
Characterization of molecular biomarkers detected in patient tissue specimens and their clinical significances in diverse aggressive and metastatic cancers
| Results and clinical significance | ||||
|---|---|---|---|---|
| Cancer type | Biomarker detection method |
Number of patients/ samples |
Biomarkers and their diagnostic, prognostic, and predictive potentials |
Ref. |
| 11 different cancer types including hematologic, brain, and epithelial cancers |
Microarray and qRT-PCR analyses |
1,153 clinical tumor specimens from patients with cancer diagnosed with acute myeloid leukemia, lymphoma, mesothelioma, medulloblastoma, glioma, and prostate, breast, lung, ovarian, or bladder epithelial cancers |
An 11-gene signature that was associated with an upregulation of stem cell–resembling expression profile of the BMI-1–regulated pathway has been shown to be a powerful predictor of prostate cancer progression and metastases. The expression of this 11-gene signature has also been associated with a short interval to disease recurrence, distant metastases and death after therapy of patients with 11 different cancer types. |
(4) |
| Prostate cancer | Immunhistochemical staining |
Tissue microarray (TMA) containing primary prostate tumor sections (n = 30) as well as 10 primary prostate tumors and their 10 matched bone metastases from patients with prostate cancer. |
ALDH7A1 isoform was highly expressed in 83% clinical specimens of primary prostate tumors (TMA) as well as in 80% and 90% of prostate cancers and their matched bone metastases, respectively. These data suggest that ALDH7A1 could constitute a potential biomarker for the stratification of patients with prostate cancer at risk of developing metastatic disease. |
(18) |
| Prostate cancer | Immunhistochemical staining of PSA+ and PSA−/lo cells in prostate cancer sections |
Primary tumor specimens from untreated patients with Gleason score 7 (n = 10) and 9 or 10 (n = 10), and treatment-failed patients (n = 23) |
High grade and recurrent prostatic adenocacinoma specimens of patients were enriched in PSA−/lo prostate cancer cells suggesting that PSA−/lo prostate cancer cells with stem cell–like properties may play critical functions in the tumor regrowth after treatment initiation. |
(17) |
| Prostate cancer | Immunhistochemical staining |
76 primary tumor specimens from patients with prostate cancer with Gleason scores 6–10 plus 30 bone metastasis specimens from patients with prostate cancer. |
The expression levels of EGFR, Ser473-pAkt, NF-κB p65, and MIC-1 proteins were significantly enhanced in the same subset of 76 cases of prostate cancer specimens during the disease progression and detected in a small subpopulation of CD133+ prostate cancer cells and the bulk tumor mass of CD133− prostate cancer cells. All of these biomarkers were also overexpressed in 80% to 100% of 30 prostate cancer metastasis bone tissue specimens indicating their potential use as diagnostic and prognostic biomarkers. |
(24) |
| Breast cancer | Analysis of a gene expression signature from CD44+/CD24−/low cells and mammosphere (MS)- forming cancer cells in breast cancer specimens |
The gene expression signature was analyzed in 36 tumors (18 luminal A/B, 13 basal-like, and 5 ErbB2-enriched). For therapeutic significance of signature, 18 patients with breast cancer pairs before and after therapy with letrozole or 12 patient pairs before and after docetaxel therapy were used. |
A gene expression signature common to both CD44+/CD24−/low BCSCs and MS-forming breast cancer cells was mainly detected in patient’s primary tumors as the claudin-low molecular subtype expressing several EMT program-associated molecules. The CD44+/CD24−/low- MS and claudin-low signatures were also enriched in patient’s primary tumors after either endocrine letrozole-based therapy or docetaxel-based chemotherapy. These results support the potential application of the CD44+/ CD24−/low-MS signature to predict the treatment resistance and relapse. |
(10) |
| Breast and prostate cancers |
Comparative analyses of profiles of hormone- sensitive and -independent breast cancers and prostate cancer cell lines with gene expression data sets from patient’s breast cancers. |
Breast ER-status signature was established from ER− (hormone- independent) and ER+ (hormone-dependent) invasive breast cancer (n = 295) profile datasets and 18 different breast cancer cell lines (10 of them ER−). The gene signature data sets from 8 androgen-sensitive and androgen-independent prostate cancer cell lines was also used. |
A common subset of 81 genes was differentially expressed between ER− and ER+ breast tumor specimens and breast cancer cell lines as well as in androgen-independent versus androgen-sensitive prostate cancer cell lines. EGFR signaling elements were also enriched in ER− breast and androgen-independent prostate cancer cell lines suggesting that the EGFR tumorigenic cascade may contribute to hormone-independent phenotypes of the breast cancer and prostate cancer cells. |
(11) |
| Breast and lung cancers | Comparative analyses of invasion assay and Affymetrix gene expression data on a panel of cancer cell lines with chemo- sensitivity data. |
Invasion-associated genes established from 60 human cancer cell lines developed by the National Cancer Institute were compared with chemo-sensitivity data of 99 anticancer drugs and clinical data from adjuvant chemotherapy cohorts in 508 breast and 71 patients with lung cancer and a cohort of untreated patients (controls). |
An 8-gene signature (EGFR, integrin-α3, myosin light chain kinase, retinoic acid induced protein 14, AHNAK nucleoprotein, glutaminase, interleukin-32, and nicotinamide N-methyltransferase) was associated with the invasion and sensitivity of a panel of 60 tumor cell lines to paclitaxel, docetaxel, erlotinib, everolimus, and dasatinib. The 8-gene signature also predicted the relapse-free survival for patients with breast and lung cancer. |
(14) |
| Non–small cell lung cancer (NSCLC) |
Immunohistochemical analysis |
Various histological tissue specimens of lung squamous cell carcinoma (SCC) and lung adenocarcinomas in 2 independent large TMA sets (n1 = 287 and n2 = 511) were used. |
A marked nuclear expression of SOX2 embryonic stem cell transcriptional factor was detected in all normal bronchial epithelia, alveolar bronchiolization structures, and premalignant lesions in SCC development (hyperplasia, dysplasia, and carcinoma in situ). In contrast, SOX2 protein expression was not seen in all normal alveoli and atypical adenomatous hyperplasias. Moreover, SOX2 expression was greatly higher in lung SCCs compared with lung adenocarcinomas. |
(143) |
| Pancreatic cancer | Whole-genome expression analyses of side population cells and main non-side (non-side population) cell fraction isolated from human pancreatic cancer resections specimens by FACS |
Side population cells were obtained from human PDAC specimens (n = 32) and the prognostic value of the side population gene signature was validated in a large independent series of PDAC patients (n = 78). |
32 PDAC specimens contained a side population cell subpopulation exhibiting an upregulation of genes associated with markers of putative pancreatic cancer stem and therapy resistance. The validation of side population cell–derived gene signatures of 32 or 10 up- or downregulated genes in a different set of 78 PDAC samples has also indicated that the expression levels of ABCB1 multidrug transporter and CXCR4 were correlated with worse patient survival suggesting their potential used as prognostic biomarkers and therapeutic targets. |
(144) |
| Brain cancer | The mRNA expression levels of PTEN, SHH, PTCH1, and GLI1 were determined by real- time PCR assays. |
55 GBM tissue specimens of patients (age range = 29–75) who were diagnosed with de novo glioblastoma and treated with external beam radiation therapy to 60 Gy and chemo-therapy were used. The GBM samples were separated into a PTEN-expressing group and a PTEN-deficient group on the basisof their PTEN expression. |
The expression levels of SHH and GLI1 were significantly higher in PTEN- expressing GBM specimens than in PTEN-deficient GBMs. The higher expression levels of SHH, PTCH1, and GLI1 in PTEN-coexpressing GBM specimens were also associated with reduced survival of patients. These data indicate that hedgehog signaling elements may constitute potential prognostic biomarkers and molecular targets for individualized treatment of GBM patients overexpressing SHH/ GLI1. |
(141) |
| Melanoma | Analyses of ALDH activity by Aldefluor assay and gene expression by microarray analyses, qRT-PCR and immunohischemistry |
3 primary and 6 metastatic tumors from melanoma patients |
8 patient’s tumors harbored small ALDH+ subpopulations, ranging from 0.08% to 1.15%. The gene expression signatures of melanoma cells expressing high levels of ALDH1A and ALDH3A from patient’s melanoma specimens were characterized by expression of some genes associated with stem cell functions. |
(139) |
Altogether, these studies have led to the identification and validation of gene expression signatures and potential diagnostic, prognostic, and predictive biomarkers related to stem cell–like cancer cells, EMT program, treatment resistance, and relapse of different aggressive cancers. Future investigations are however necessary to validate the expression of these gene signatures and their implications in the treatment resistance on larger cohorts of tumor tissue specimens from patients with cancer. In this regard, recent studies have also revealed the possibility to analyze the phenotypic features of exosomes secreted by cancer cells and CTCs, including circulating cancer stem/progenitor cells, to assess the tumor stage and risk of cancer progression, metastases, and disease relapse of patients with cancer.
Molecular biomarkers of exosomes
Accumulating lines of evidence have indicated that exosomes that are secreted by normal and cancer cells can act as key mediators in signal transduction mechanisms that are involved in short- and long-distance cell-to-cell communication (150-157). In fact, the exosomes are small microvesicles (sized about 50–100 nm) derived from cellular endocytosis that may contain specific lipids, proteins, mRNAs, and miRNAs encapsulated in a cholesterol-rich phospholipid membrane (150-157). More particularly, the exosomes that are released by cancer cells in their surrounding microenvironment and peripheral circulation may adhere via their antigenic surface molecules or transfer their cytoplasmic contents through membrane fusion to their targeted host cells, including other cancer cells, stromal cells, endothelial cells, and immune cells (Fig. 1; refs. 150-157). Hence, the shedding of exosomes by cancer cells under normoxic and hypoxic conditions may modulate immune system and promote the malignant behavior of recipient cells in their tumor microenvironment and angiogenesis as well as the formation of premetastatic niches, metastases at distant tissues and organs, and treatment resistance (Fig. 1 and Table 2; refs. 150-160). Importantly, recent advances in the development of isolation techniques of exosomes from some types of body fluids or cancer cell culture supernatants using centrifugation, ultrafiltration, immunoprecipitation, magnetic activated cell sorting(MACS), and immunoaffinity capture methods have led to an improvement of the purity of exosomal preparations (150-159, 161-164). Hence, several studies carried with these isolation methods have indicated the presence of exosomes in blood, plasma, serum, lymph, urine, cerebrospinal fluid, or ascites from patients with melanoma and brain, ovarian, breast, pancreatic, colon, bladder, prostate, and lung cancers as well as conditioned medium of different cancer cell types (Fig. 1 and Table 2; refs. 150-159, 161-164). It has also been observed by proteomic profiling that different exosomal preparations of different cancer types frequently expressed a set of biomarkers. The common biomarkers of most exosomes include tubulin, actin, tetraspanins (CD9, CD63, CD81, and CD82), Alix, TSG101, clathrin, small GTPases (Rab proteins), annexins, heat-shock proteins (Hsp70 and Hsp90), 14-3-3 proteins, and MHC class I molecules that are involved in the biosynthesis, cytoskeletal structure, and trafficking of exosomes (159, 162-164). Furthermore, tumor cell–derived exosomes also harbored specific RNAs, miRNAs, and proteins, including stem cell–like markers (CD133, CD44, and/or CD105) and oncogenic products (EGFR, EGFRvIII mutant, erbB2/HER2/Neu, MET, claudins, and/or FASN) that are related to cancer progression, metastases, and treatment resistance (150-159, 161-166). For instance, the data from proteomic analyses of highly purified exosomes derived from 2 ovarian cancer cell lines, OVCAR-3 and IGROV1, have indicated that several proteins of tumorigenic pathways that are overexpressed in ovarian cancer tissues and associated with migration, invasion, immune modulation, angiogenesis, and metastasis were also detected in exosomes (162). These oncogenic products comprise EGFR, epithelial cell surface antigen (EpCAM), proliferation cell nuclear antigen (PCNA), tubulin β-3 chain (TUBB3), apolipoprotein E (APOE), and claudin 3 (CLDN3; ref. 162). Moreover, the exosomes expressing T-cell receptor (TCR), CD20, HLA-DR, B7-2, HER2, CA125, and histone H2A have been detected in 85.4% (35/41) of ascites samples from patients with ovarian cancer (167). It has also been shown that the exosomes induced the apoptosis of precursor and mature dendritic cells and peripheral blood mononuclear cells (PBMC) and impaired the cytotoxic activity of PBMCs in the presence of dendritic cells suggesting their implication in the modulation of antitumor immunity (167).
Table 2.
Potential applications of molecular biomarkers of circulating exosomes for the diagnosis, prognosis, and prediction of treatment responses of diverse aggressive and metastatic cancers
| Potential application of exosomes |
Cancer type | Exosome source and purification and detection methods |
Results and their significances | Ref. |
|---|---|---|---|---|
| Diagnostic biomarkers |
Ovarian cancer | Tumor-derived exosomes were isolated from peripheral blood of 50 women diagnosed with serous papillary adenocarcinoma of the ovary (stages I–IV) using a MACS procedure with anti-EpCAM coupled to magnetic microbeads. |
An 8 diagnostic miRNA signature (miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205, and miR-214) was detected in ovarian tumor-derived EpCAM-exosomes and ovarian tumor specimens isolated from same patients with cancer. In contrast, these exosomal miRNAs were not be detected in normal controls. Moreover, the levels of circulating exosomes detected in women increased with the stages I–IV of ovaian cancer. |
(166) |
| Diagnostic biomarkers |
Prostate cancer | Exosomes were prepared by differential centrifugation from urine samples of 11 patients with prostate cancer. |
The expression of prostate cancer biomarkers, including prostate cancer gene-3 (PCA-3) and TMPRSS2; ERG gene fusion, was detected in urinary exosomes suggesting their potential use for the diagnosis and monitoring of patients with prostate cancer. |
(161) |
| Diagnostic biomarker |
Brain tumor | Exosomes were purified by differential centrifugation from serum samples from 25 GBM patients and 30 healthy individuals. |
Tumor-specific and constitutively active EGFRvIII mutant was detected by RT-PCR in serum exosomes from 7 of 25 (28%) GBM patients whereas EGFRvIII mRNA was not detected in serum exosomes from 30 normal individuals used as controls. |
(152) |
| Diagnostic and prognostic biomarker |
Esophageal squamous cell cancer (ESCC) |
Exosomes were extracted from the serum samples of patients with ESCC and benign diseases without systemic inflammation. |
The exosomal miR-21 expression was significantly upregulated in serum samples from patients with ESCC versus benign diseases without systemic inflammation. Exosomal miR-21 positively correlated with tumor progression and aggressiveness, lymph node status, and the presence of metastasis with inflammation and clinical stage without inflammation. |
(165) |
| Diagnostic and prognostic biomarkers |
Melanoma | Exosomes were purified by centrifugation from peripheral blood samples from stage I–IV melanoma subjects. |
An exosome-specific melanoma signature comprised of MET, tyrosinase-related protein- 2, very late antigen-4, HSP70, and HSP90 has been associated with stage and outcome of patients with melanoma. |
(169). |
| Predictive biomarker | Breast cancer | Exosomes were isolated from HER2- overexpressing breast carcinoma cell lines SKBR3 and BT474 or breast cancer patient’s serum samples. |
Exosomes from HER2+ SKBR3 and BT474 tumor cell-conditioned supernatants or breast cancer patients’ serum expressing HER-2 have been observed to interact with humanized antibody trastuzumab. Moreover, the data from functional assays have also revealed that exosomes suppressed the inhibitory activity of trastuzumab on the SKBR3 cell proliferation whereas lapatinib had no effects. These findings suggest that HER2+ exosomes can modulate the sensitivity of breast cancer cells to trastuzumab, and consequently HER2-driven tumor aggressiveness. |
(172) |
| Diagnostic and predictive biomarker |
Prostate cancer | Exosomes were purified by ultracentrifugation from plasma and serum samples from 39 patients with prostate cancer, 20 benign prostatic hyperplasia (BPH) patients, 8 recurrent prostate cancers and 16 healthy controls. |
Survivin level was significantly upregulated in the tumor-derived exosomes compared with those from BPH and controls. Exosomal survivin level was also higher in patients that had relapsed after chemotherapeutic treatment compared with controls. |
(171) |
| Diagnostic, prognostic and predictive biomarkers |
Brain cancer | Profiling of circulating exosomes labeled with target-specific magnetic nanoparticles was directly performed in blood samples of 24 GBM patients and 8 healthy volunteers using microfluidic chip. Exosomes were detected with miniaturized nuclear magnetic resonance system |
Data of protein profiling of circulating exosomes have indicated that the expression of a combination of 4 markers (EGFR, EGFRvIII mutant, podoplanin, and cytosolic isocitrate dehydrogenase 1) considerably improved the detection accuracy (>90%) as compared with a single marker (<76). Circulating exosome profiling also predicted treatment outcomes of GBM patients and differentiated between responders and nonresponders to a treatment with alkylating agent, temozolomide plus radiation. |
(170) |
In addition, the tumor-specific and constitutively active form of EGFR designated as EGFRvIII mutant has also been detected in serum exosomes from 7 of 25 patients with glioblastoma (152). It has been noted that EGFRvIII protein expressed by glioma cells may be transferred via exosomes to other glioma cells lacking EGFRvIII, and thereby promoted their oncogenic transformation via the activation of PI3K/Akt and mitogen-activated protein kinase (MAPK) pathways (Figs. 1 and 2A and Table 2; ref. 153). These molecular events induced the changes in the expression of EGFRvIII-regulated genes such as vascular growth factor “VEGF,” Bcl-xL, and p27 in host glioma cells and increased their anchorage-independent growth capacity (153). It has also been observed that exosomes released from glioblastoma tumor cells also contained mRNA, miRNA, and angiogenic proteins that could be transferred to host cells, including normal brain microvascular endothelial cells, and thereby stimulate the tubule formation (153). In the same way, cancer stem cells expressing the mesenchymal stem cell marker CD105 from human renal cell carcinoma also released exosomes that contained proangiogenic mRNAs and miRNA (168). It has also been noted that the exosomal mRNAs and miRNA in turn activated the growth of normal endothelial cells and induced the vessel formation and angiogenesis after implantation in SCID mice (168). Moreover, the treatment of SCID mice with exosomes from CD105+ cells was effective at promoting the formation of lung metastases induced by intravenous injection of renal carcinoma cells (168).
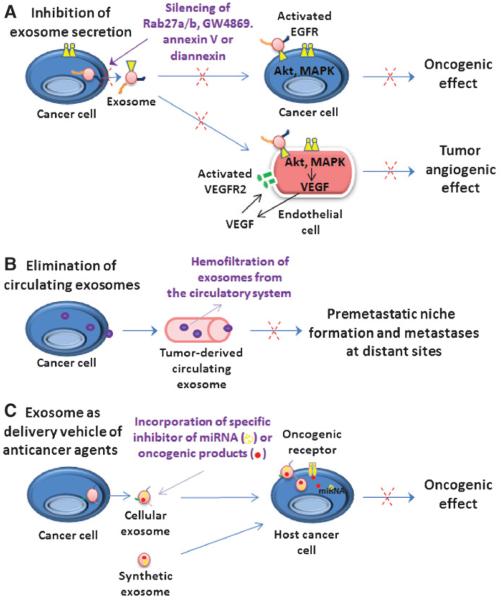
Figure 2.
Schematic representation of functions of exosomes released by cancer cells in cancer progression, metastasis, angiogenesis, and treatment resistance and their potential therapeutic applications. A, the oncogenic molecules such as EGFR or EGFRvIII, which can be shed from cancer cells via exosome secretion and transferred to other adjacent cancer cells and/or endothelial cells. Hence, the activated EGFR or EGFRvIII may stimulate MAPK and Akt pathways and expression of their targeted genes, including VEGF, in host cancer cells and endothelial cells in their surrounding tumor microenvironment. VEGF released by endothelial cells in turn can activate in an autocrine manner the VEGFR2 cascade and tumor angiogenesis. Of therapeutic interest, the inhibition of exosome secretion from cancer cells carried by shRNA silencing of Rab27a/b, using an inhibitor of sphingomyelinase (GW4869) or annexin/diannexin and which represent potential strategies to prevent the promoting effects of exosomes on tumor progression, angiogenesis, and metastasis is indicated. B, the elimination of tumor cell–derived exosomes from the entire circulatory system, which can be performed by extracorporeal hemofiltration using novel medical devices such as an affinity plasmapheresis platform designated as Aethlon ADAPT system is illustrated. C, the systemic delivery of therapeutic agents incorporated in cellular or synthetic exosomes and which are able to inhibit the functions of tumor-specific mRNAs, miRNAs such as spherical nucleic acid constructs, and/or oncogenic products in target cancer cells is also illustrated.
Importantly, several lines of experimental evidence have also indicated that the exosomes may be involved in the formation of premetastatic niches and transfer of more aggressive and drug-resistance phenotypes to cancer cells and modulation of treatment responses of patients with cancer (Table 2; refs. 150, 169-175). For instance, CD133-expressing exosomes purified from FEMX-I cell line, which was derived from a lymph node metastasis of a patient with malignant melanoma, have been observed to express several prometastatic proteins, including CD44, MAPK4K, GTP-binding proteins, ADAM10, and annexin A2 and high levels of miRNAs with cancer-related functions (173). It has been shown that primary melanoma-derived exosomes can induce the recruitment and growth of melanoma cells to sentinel lymph nodes in vivo by inducing extracellular matrix deposition and vascular proliferation in the lymph nodes (174). Moreover, the exosomes from highly metastatic melanomas also contributed to the premetastatic niche formation through the upregulation of the MET oncoprotein in CD45−/KITlow/+/tyrosine kinase with immunoglobulin-like and EGF-like domains-2 (TIE2+) bone marrow-derived progenitor cells (Fig. 1; ref. 169). The enhanced expression of MET by bone marrow progenitor cells resulted in their reprogramming toward provasculogenic and prometastatic phenotypes (169). The gene signature of exosomes released by melanoma, including MET, tyrosinase-related protein-2, very late antigen-4, HSP70, and HSP90, has also been associated with a poor prognosis of patients with melanoma (Table 2; ref. 169). Moreover, it has been reported that the exosomes isolated from serum samples of patients with prostate cancer promoted the proliferation and invasion of prostate cancer cell lines whereas the exosomes from age-matched healthy controls have not significant effects (150). The exposure of DU145 and 22Rv1 prostate cancer cells to exosomes from serum samples of patients with prostate cancer treated with docetaxel was also effective at enhancing their docetaxel resistance as compared with exosomes from untreated prostate cancer patient’s sera (150). It has also been noticed that docetaxel-resistant DU145RD and 22Rv1RD cell lines secreted the exosomes expressing P-glycoprotein, which could be transferred and conferred docetaxel resistance to parental DU145 and 22Rv1 cell lines (Fig. 1; ref. 150). The PCR-based analyses of urine samples from patients with prostate cancer have also indicated that the transcripts of prostate cancer gene-3 (PCA-3) and TMPRSS2;ERG gene fusion were detected in urinary exosomes suggesting their potential use as a noninvasive method for the diagnosis and follow-up of patients with prostate cancer (161). Moreover, it has also been shown that the exosomes released by SKBR3 breast cancer cells overexpressing HER2 oncoprotein interfered with the antitumoral activity of the anti-HER2 antibody, trastuzumab (Herceptin; ref. 172).
Therapeutic applications of exosomes
The molecular targeting of tumor cell–derived exosomes by interfering with their biosynthesis, secretion, and/or interactions with cell antigens or receptors also represents a promising approach to prevent the cancer progression, premetastatic niche formation, and metastases at distant sites (Fig. 2; refs. 160, 169, 176-179). For example, the targeting of neutral sphingomyelinase 2, which is involved in the ceramide biosynthesis and upregulation of exosome secretion, with chemical drug GW4869 has been shown to inhibit the lung metastases in lung cancer cell-bearing mice in vivo (176). Moreover, the interference with molecular pathways of exosome secretion by shRNA silencing of small GTPase Rab27a or Rab27b or 2 Rab27 effectors, known as Slp4 and Slac2b, also inhibited the exosome secretion by cervical adenocarcinoma HeLa cells, 4T1 and TS/A mammary carcinoma cells, and melanoma cells (169, 177, 178). It has also been noted that Rab27a blockade resulted in decreased primary tumor growth and lung dissemination of a metastatic carcinoma (4T1), but not of a nonmetastatic carcinoma (TS/A; ref. 177). In A431 human tumor xenografts in mice, angiogenic endothelial cells stained positively for human EGFR and phospho-EGFR, whereas treatment with annexin homodimer, diannexin which cloak phosphatidylserine residues on the surfaces of exosomes led to a reduction of tumor growth rate and microvascular density (Fig. 2A; ref. 160). Of particular clinical interest, a novel medical device using an affinity plasmapheresis platform termed as Aethlon ADAPT system was also effective to perform extracorporeal hemofiltration and elimination of tumor cell–derived exosomes from the entire circulatory system (Fig. 2B; ref. 179).
Recent investigations have also revealed that exosomes could be used as natural cell–derived membrane vehicles for systemic delivery of therapeutic agents interfering with the functions of tumor-specific mRNAs, miRNAs, and oncogenic products in target cells as well as immune regulatory tools in cancer immunotherapy (Fig. 2C; refs. 180-183). As a matter of fact, exosomes engineered to deliver spherical nucleic acid constructs into PC3 prostate cancer cells downregulated miR-21 target by approximately 50% (182). Furthermore, it has been shown exosomes that are released by tumor cells and dendritic cells may be used to elicit-specific immune responses against established tumors (180, 181). For instance, the data from a phase I clinical trial have indicated that a combination of autologous ascites-derived exosomes with granulocyte macrophage colony-stimulating factor (GM-CSF) was safe and well tolerated and induced tumor antigen-specific cytotoxic T-cell responses without side effects in patients with advanced colorectal cancer (180).
Together these data suggest important roles of exosomes secreted by cancer cells in their surrounding microenvironment in the acquisition of a more malignant behavior by host cancer cells and promoting immune escape and tumor neovascularization during cancer progression. Moreover, the exosomes can play critical functions in the establishment of premetastatic niches, recruitment, and homing of cancer cells at distant tissues and organs, metastases, and treatment resistance. Hence, these results support the interest to use the exosome-specific biomarkers as new diagnostic, prognostic, and predictive indicators and to develop new exosome-based-targeted therapies for treating aggressive and metastatic cancers.
Molecular biomarkers of CTCs
The enumeration of CTCs isolated from whole blood samples of patients with metastatic cancers before or posttreatment initiation and characterization of their molecular biomarkers may constitute a noninvasive and real-time method for improving accuracy of diagnosis and prognosis and choice of their therapeutic management, including use of appropriate targeted therapy (16, 31-35, 184). Several strategies, including CellSearch platform, microfluidic devices, and cell size–based separation methods, have been developed to detect, enrich, and isolate rare CTCs from peripheral blood samples (Fig. 1 and Table 3; refs. 16, 31-34). These detection techniques involve the CTC immunocapture from peripheral blood samples using different specific antibodies or size-based filters (Fig. 1 and Table 3; refs. 16, 31-34). More particularly, the CellSearch technology, which consists to use immunomagnetic beads coated with an antibody directed against EpCAM molecules, can allow researchers to isolate epithelial cancer cells expressing EpCAM surface marker (16, 31, 33-35, 184, 185). In a second step, the analyses of different biomarkers of the interest expressed by captured CTCs can be performed by quantitative real-time reverse transcription PCR (qRT-PCR) using DNA extracted from CTCs or immunofluorescence staining of CTCs (Fig. 1; refs. 16, 31, 34, 184). The biomarkers of CTCs include stem cell–like markers (CD133, CD44, ALDH, and ABC multidrug transporters), cytokeratins, EGFR, EGFRvIII, HER2, and K-Ras as well as the absence of the leukocyte-specific marker CD45 (Table 3; refs. 16, 31, 34, 184). Hence, it has been observed that the number and phenotypic features of CTCs captured using CellSearch assay, microfluidic CTCships, and other detection techniques given significant information on the treatment benefit and helped to predict the overall outcome of patients with locally advanced and metastatic breast, prostate, ovarian, pancreatic, lung, colorectal, and pancreatic cancers (Table 3; refs. 16, 31-35, 37, 184, 186-189). For instance, the characterization of CTCs from patients with metastatic CRPC has indicated that the majority of CTCs coexpressed epithelial proteins (EpCAM, cytokeratins, and E-cadherin), mesenchymal markers (vimentin, N-cadherin, and O-cadherin), and the stem cell–like marker CD133 (34).
Table 3.
Characterization of molecular biomarkers of CTCs and their clinical significances in diverse aggressive and metastatic cancers
| Results and clinical significance | ||||
|---|---|---|---|---|
| Cancer type | CTC detection method | Number of CTCs detected by patients/samples |
Biomarkers and their diagnostic, prognostic, and predictive potentials |
Ref. |
| Breast cancer | AdnaTest Breast Cancer Select for detection of EpCAM, MUC1, HER2 and β-actin transcripts |
CTCs were detected by AdnaTest in 19% of 502 blood samples from patients with breast cancer. Blood samples from 502 primary patients with breast cancer versus 30 healthy donor samples |
29% samples positive for CTCs were characterized by RT-PCR by the expression of at least one EMT marker (twist, Akt2, and PI3Kα) whereas 97% of 30 healthy donor samples investigated were negative. Moreover, 14% samples positive for CTCs expressed ALDH1 stem cell–like marker whereas 95% of 30 healthy donor samples were negative. |
(38) |
| Breast cancer | AdnaTest Breast Cancer Select for detection of EpCAM, MUC1, and HER2 transcripts |
226 blood samples from 39 patients with metastatic breast cancer obtained during their follow-up of chemotherapy, hormonal, or trastuzumab antibody- based treatment |
CTCs were detected by AdnaTest in 31% of 226 blood samples. The samples positive for CTCs were characterized by RT-PCR by the expression of at least one EMT marker (twist, Akt2, and PI3Kα) and ALDH1 in 62% and 69% cases as compared with 7% and 14% in blood samples negative for CTCs, respectively. The EMT and ALDH1 expression levels in blood samples from patients unresponsive to therapy were of 62% and 44% relative to only 10% and 5% in responders. |
(39) |
| Breast cancer | CellSearch EpCaM-based immunocapture method |
Blood samples from 16 women with metastatic breast cancer with disease progression |
Data from analyses performing by CellSearch system and immunostaining have revealed that over 75% of CTCs detected in blood samples from patients with metastatic cancers breast cancer coexpressed epithelial markers (EpCAM and E-cadherin) and mesenchymal proteins (vimentin and N-cadherin). |
(34) |
| Breast cancer | Microfluidic capture of CTCs with epithelial- and tumor-specific antibodies directed against EpCAM, EGFR, and HER2 |
Blood samples from 41 patients with metastatic breast cancer at various stages of treatment. |
CTCs were detected in 41% of blood samples from patients with breast cancer. CTCs expressed epithelial (cytokeratins, EpCAM, and cadherin 1) and/or mesenchymal markers (fibronectin, cadherin 2, and serpin peptidase inhibitor, clade E). The enrichment of mesenchymal CTCs was associated with disease progression and expression of TGF-β signaling pathway components. |
(37) |
| Prostate cancer | CellSearch EpCaM-based immunocapture method |
Blood samples from 41 men with metastatic castration-resistant prostate cancer with disease progression |
Data from analyses performing by Cell Search System and immunostaining have indicated that over 80% of CTCs detected in blood samples from metastatic CRPC patients coexpressed epithelial markers (EpCAM, cytokeratin, and E-cadherin), mesenchymal proteins (vimentin, N-cadherin, and O-cadherin), and CD133 stem cell–like marker. |
(34) |
| Ovarian cancer | The monocyte blood fraction containing CTCs was enriched by 2-layer density gradient and RNA extracted from the enriched cell fraction |
Blood samples from 216 patients with epithelial ovarian cancer obtained before primary treatment and 6 months after adjuvant platinium- based chemotherapy and 39 cases of healthy women |
CTCs were detected in 24.5% of the baseline and 20.4% of the follow-up samples. The detection of CTCs correlated with the presence of ascites, suboptimal debulking and elevated CA- 125, and human epididymis protein 4 (HE-4) levels. Moreover, the CTC detection was more frequent in platinium resistant patients than in the group sensitive to platinium treatment and indicative of poor outcome of patients. |
(187) |
| Pancreatic cancer | CellSearch system | Blood samples from 26 patients with pancreatic cancer |
CTCs expressing cytokeratins were detected in 42% of blood samples and patients with pancreatic cancer with CTCs exhibited significantly shorter overall survival. |
(188) |
| Pancreatic cancer | CellSearch system or isolation by size of epithelial tumor cells (ISET) |
Blood samples from 54 patients with pancreatic cancer |
CTCs were detected by CellSearch EpCaM-based immunocapture method and ISET in 40% versus 93% of patients with pancreatic cancer, respectively. The immunostaining analyses have also indicated that CTCs expressed variable levels of EpCaM, pan-cytokeratin, E-cadherin, vimentin, and cytokeratin 7. |
(186) |
| Colorectal cancer | Pre- and postoperative EpCAM based-flow cytometry analyses |
Blood samples from 76 patients with colorectal cancer undergoing surgical resection |
CTCs were detected in 71% patients preoperatively, and all metastatic patients showed high levels of CTCs. Colorectal tumor surgical resection was associated with a significant decrease in CTCs. A high postoperative level of CTCs was also related to cancer relapse. The progression-free survival rate of patients with colorectal cancer without CTCs was increased from 16% to 86%, with a reduction in the risk of disease relapse greater than 90%. |
(185) |
| Lung cancer | Centrifugal force-based separation technique termed dean flow fractionation (DFF) |
Blood samples from 20 patients with metastatic lung cancer at different stages of treatment versus 20 blood samples from healthy individuals |
5 to 88 CTCs were detected by DFF in all blood samples analyzed from patients with metastatic lung cancer whereas healthy subjects had a negligible number of CTCs. Moreover, the data from immunostaining have revealed that CTCs detected in blood samples from patients with advanced lung cancer coexpressed cytokeratin and CD133 stem cell–like marker. |
(40) |
| Non–small cell lung cancer (NSCLC) |
CellSearch system or mutation detection in circulating tumor DNA |
Blood samples from 41 patients with relapsed or refractory NSCLC enrolled in a single-arm phase II multicenter trial with erlotinib and pertuzumab |
CTCs were detected using CellSearch system in 78% of patients with NSCLC and a greater baseline of CTC counts was associated with a response to treatment by response evaluation criteria in solid tumor (RECIST). A decrease of CTC counts upon treatment of patients was also related with FDG-PET and RECIST response and longer progression-free survival. Data from RT-PCR analyses have also indicated that the detection of mutation in EGFR in ctDNA and DNA isolated from CTCs was associated with a most substantial decrease in CTC counts over the course of treatment of NSCLC patients with erlotinib and pertuzumab. |
(184) |
| Epithelial malignancies |
Multimarker quad- μ-nuclear magnetic resonance (μ-NMR) |
Biopsies of 58 patients with epithelial malignancies including breast (n = 4), gastrointestinal (n = 17), genitourinary (n = 4), gynecologic (n = 7), pancreatic (n = 10), lung (n = 9), and poorly differentiated adenocarcinoma (n = 7) and 6 patients with benign diagnosis |
Data from quad μ-NMR expression profiles of the biopsies obtained by the fine needle aspiration of 58 patients with different epithelial malignancies have revealed that the combined analyses of EpCAM, HER2, EGFR, and MUC1 markers may be used to achieve the diagnosis of patients with cancer in 99.2% of samples. |
(189) |
| Melanoma | RNA isolation from whole blood samples |
Blood sample from 230 melanoma patients, including 154 patients at early stages I–II and 76 patients at late stages III–IV versus 152 cases of healthy individuals |
Data from quantitative RT-PCR analyses have indicated that 92% of melanoma patients expressed MLANA, TGF-β2, PAX3d, MCAM, and ABCG5 multidrug transporter. Importantly, the expression of MLANA and ABCG5 could predict disease recurrence whereas MLANA expression was associated with a poor outcome of patients with melanoma after treatment. |
(41) |
In addition, the results of qRT-PCR analyses of CTCs from patients with breast cancer have also indicated a direct relationship between the expression of stem cell–like markers such as Nanog, Oct3/4, Sox-2, nestin, and CD34 on CTCs and the stage of the disease progression (36). Moreover, the analyses of blood samples from 502 patients with breast cancer with the AdnaTest BreastCancer Select for the detection of EpCAM, MUC1, HER2, and β-actin transcripts have revealed that CTCs were detected in 19% of samples, and 29% samples expressed at least one of EMT markers (twist1, Akt2, and PI3Kα) whereas ALDH1 stem cell–like marker was detected in 14% of the samples (38). In another study, CTCs were also detected in 31% of 226 blood samples from 39 patients with metastatic breast cancer and 62% were positive for at least one of EMT markers and 69% for ALDH1 (39). It has also been noticed that the expression of EMT markers and ALDH1 was detected in 62% and 44% of patients with breast cancer that were nonresponders to the therapy whereas only 10% and 5% of the responders expressed these markers, respectively suggesting their potential use as predictive biomarkers (39).
More recently, single CTCs and multicellular clusters in a viable state have also been isolated from bloodstream of patients with metastatic lung cancers by dean flow fractionation (40). This size-based separation method may overcome the problems associated with the immunocapture with antibodies that is dependent of antigens expressed by CTCs (40). The number of CTCs detected by dean flow fractionation in whole blood samples from 20 patients with metastatic lung cancer was 39.1 ± 24.8 CTCs/mL whereas CTCs in blood samples from 20 healthy individuals was negligible (0.79 ± 0.42 CTCs/mL; ref. 40). It has also been observed that the CTC population detected in blood samples from patients with advanced lung cancer contained cells coexpressing cytokeratin and CD133 stem cell–like marker (40). Moreover, the qRT-PCR analyses of peripheral blood samples from patients with primary melanoma (early stages 0–II, n = 154) and metastatic melanoma (late stages III–IV, n = 76) have indicated that all samples contained CTCs (41). It has also been noted that melan-A known as melanoma antigen and stem cell–like marker, ABCB5 multidrug transporter were expressed in 45% and 49% of patients showing melanoma recurrence after treatment initiation, respectively (41).
The use of specific biomarkers of CTCs, including stem cell–like markers, represents a promising strategy for improving the diagnosis, prognosis, and therapeutic management of patients with aggressive and metastatic cancers.
Conclusions and Perspectives
The characterization of gene products expressed in tumor tissue specimens and CTCs from patients with cancer and exosomes released by cancer cells has led to the identification and validation of some gene signatures and potential stem cell–like markers. These altered gene products could be used as noninvasive diagnostic, prognostic, and predictive tools in conjunction for monitoring the risk of tumor progression and metastases and determining the choice of therapeutic treatments of patients with cancer. Furthermore, the molecular targeting of specific markers of cancer stem/progenitor cells, CTCs, and exosomes also represents a potential strategy could prevent cancer progression and metastases and overcome the treatment resistance and disease relapse of patients with cancer.
Future investigations are however required to optimize the sensitivity and reproducibility of immunocapture methods used for the enrichment and enumeration of CTCs and circulating exosomes from whole blood samples to consider their low number, potential heterogeneity, and specific biomarkers. Indeed, the detection and purification of rare CTCs, including circulating cancer stem cells, and circulating exosomes may be obscured by the nonspecific background of other hematopoietic cell types and overwhelming material in whole blood samples. Therefore, the optimization of in vivo detection platforms suitable for a continuous monitoring of whole blood volume could markedly improve the probability to detect rare CTCs, including circulating cancer cells with stem cell–like features. Moreover, the use of additional antibodies directed against epithelial, mesenchymal, and/or stem cell–like surface markers of CTCs could be included in immunoaffinity capture methods, such as CellSearch platform and microfluidic ships, to detect the EpCAM− CTCs or exosomes. It will also be of interest to further establish the relationship between the expression levels of EMT process- and stem cell–like markers detected on captured CTCs and exosomes and the stage of tumor progression, treatment response, and survival rate using a large cohort of patients with cancer. Hence, the combined use of these new biomarkers detected in primary and metastatic tumor tissues, CTCs, and exosomes should lead to the development of multiplex biomarker approaches for improving the accuracy of current diagnostic and prognostic tests and efficacy of individualized treatments for patients with cancer. Of clinical relevance, the development of new nanotheranostic platforms for the combined in vivo detection by imaging and nanobubble-based targeted therapies of CTCs and circulating exosomes also may constitute a promising approach for real-time diagnosis and treatment of patients with aggressive and metastatic cancers.
Acknowledgments
Grant Support
The authors were supported by the grants from the NIH(R01CA138791, EDRN U01, and SPORE P50 CA127297).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Mimeault M, Batra SK. New advances on critical implications of tumor- and metastasis-initiating cells in cancer progression, treatment resistance and disease recurrence. Histol Histopathol. 2010;25:1057–73. doi: 10.14670/hh-25.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mimeault M, Batra SK. Novel biomarkers and therapeutic targets for optimizing the therapeutic management of melanomas. World J Clin Oncol. 2012;3:32–42. doi: 10.5306/wjco.v3.i3.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–21. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kok M, Koornstra RH, Mook S, Hauptmann M, Fles R, Jansen SP, et al. Additional value of the 70-gene signature and levels of ER and PR for the prediction of outcome in tamoxifen-treated ER-positive breast cancer. Breast. 2012;21:769–78. doi: 10.1016/j.breast.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Leth-Larsen R, Terp MG, Christensen AG, Elias D, Kuhlwein T, Jensen ON, et al. Functional heterogeneity within the CD44 high human breast cancer stem cell-like compartment reveals a gene signature predictive of distant metastasis. Mol Med. 2012;18:1109–21. doi: 10.2119/molmed.2012.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu JC, Voisin V, Bader GD, Deng T, Pusztai L, Symmans WF, et al. Seventeen-gene signature from enriched Her2/Neu mammary tumor-initiating cells predicts clinical outcome for human HER2+: ERα- breast cancer. Proc Natl Acad Sci U S A. 2012;109:5832–37. doi: 10.1073/pnas.1201105109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K, et al. The CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10:R53. doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Hajj M, Wicha MS, Ito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–15. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Creighton CJ. A gene transcription signature associated with hormone independence in a subset of both breast and prostate cancers. BMC Genom. 2007;8:199. doi: 10.1186/1471-2164-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R, et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet. 2003;362:362–9. doi: 10.1016/S0140-6736(03)14023-8. [DOI] [PubMed] [Google Scholar]
- 13.Rodenhiser DI, Andrews JD, Vandenberg TA, Chambers AF, Souter LH, Andrews JD, et al. Gene signatures of breast cancer progression and metastasis. Breast Cancer Res. 2011;13:201–58. doi: 10.1186/bcr2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu YC, Chen HY, Yuan S, Yu SL, Lin CH, Wu G, et al. Genome-wide analysis of three-way interplay among gene expression, cancer cell invasion and anti-cancer compound sensitivity. BMC Med. 2013;11:106. doi: 10.1186/1741-7015-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glinsky GV. “Stemness” genomics law governs clinical behavior of human cancer: implications for decision making in disease management. J Clin Oncol. 2008;26:2846–53. doi: 10.1200/JCO.2008.17.0266. [DOI] [PubMed] [Google Scholar]
- 16.Toloudi M, Apostolou P, Chatziioannou M, Papasotiriou I. Correlation between cancer stem cells and circulating tumor cells and their value. Case Rep Oncol. 2011;4:44–54. doi: 10.1159/000324403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin J, Liu X, Laffin B, Chen X, Choy G, Jeter CR, et al. The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell. 2012;10:556–69. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70:5163–73. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- 19.van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Pelger RC, van der Pluijm G. The aldehyde dehydrogenase enzyme 7A1 is functionally involved in prostate cancer bone metastasis. Clin Exp Metastasis. 2011;28:615–25. doi: 10.1007/s10585-011-9395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Pelger RC, van der Pluijm G. Integrin αV expression is required for the acquisition of a metastatic stem/progenitor cell phenotype in human prostate cancer. Am J Pathol. 2011;179:2559–68. doi: 10.1016/j.ajpath.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Horst G, van den Hoogen C, Buijs JT, Cheung H, Bloys H, Pelger RC, et al. Targeting of alpha(v)-integrins in stem/progenitor cells and supportive microenvironment impairs bone metastasis in human prostate cancer. Neoplasia. 2011;13:516–25. doi: 10.1593/neo.11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L, Huang X, Zheng X, Wang X, Li S, Zhang L, et al. Enrichment of prostate cancer stem-like cells from human prostate cancer cell lines by culture in serum-free medium and chemoradiotherapy. Int J Biol Sci. 2013;9:472–9. doi: 10.7150/ijbs.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mimeault M, Johansson SL, Henichart JP, Depreux P, Batra SK. Cytotoxic effects induced by docetaxel, gefitinib, and cyclopamine on side population and non-side population cell fractions from human invasive prostate cancer cells. Mol Cancer Ther. 2010;9:17–630. doi: 10.1158/1535-7163.MCT-09-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mimeault M, Johansson SL, Batra SK. Pathobiological implications of the expression of EGFR, pAkt, NF-κB and MIC-1 in prostate cancer stem cells and their progenies. PLoS One. 2012;7:e31919. doi: 10.1371/journal.pone.0031919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mimeault M, Johansson SL, Batra SK. Marked improvement of cytotoxic effects induced by docetaxel on highly metastatic and androgen-independent prostate cancer cells by downregulating macrophage inhibitory cytokine-1. Br J Cancer. 2013;108:1079–91. doi: 10.1038/bjc.2012.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castellon EA, Valenzuela R, Lillo J, Castillo V, Contreras HR, Gallegos I, et al. Molecular signature of cancer stem cells isolated from prostate carcinoma and expression of stem markers in different Gleason grades and metastasis. Biol Res. 2012;45:297–305. doi: 10.4067/S0716-97602012000300011. [DOI] [PubMed] [Google Scholar]
- 27.Salnikov AV, Liu L, Platen M, Gladkich J, Salnikova O, Ryschich E, et al. Hypoxia induces EMT in low and highly aggressive pancreatic tumor cells but only cells with cancer stem cell characteristics acquire pronounced migratory potential. PLoS ONE. 2012;7:e46391. doi: 10.1371/journal.pone.0046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–52. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melchior SW, Corey E, Ellis WJ, Ross AA, Layton TJ, Oswin MM, et al. Early tumor cell dissemination in patients with clinically localized carcinoma of the prostate. Clin Cancer Res. 1997;3:249–56. [PubMed] [Google Scholar]
- 30.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–61. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Punnoose EA, Atwal SK, Spoerke JM, Savage H, Pandita A, Yeh RF, et al. Molecular biomarker analyses using circulating tumor cells. PLoS One. 2010;5:e12517. doi: 10.1371/journal.pone.0012517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saad F, Pantel K. The current role of circulating tumor cells in the diagnosis and management of bone metastases in advanced prostate cancer. Future Oncol. 2012;8:321–31. doi: 10.2217/fon.12.3. [DOI] [PubMed] [Google Scholar]
- 33.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14:6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 34.Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, et al. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res. 2011;9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paterlini-Brechot P. Organ-specific markers in circulating tumor cell screening: an early indicator of metastasis-capable malignancy. Future Oncol. 2011;7:849–71. doi: 10.2217/fon.11.32. [DOI] [PubMed] [Google Scholar]
- 36.Apostolou P, Toloudi M, Chatziioannou M, Ioannou E, Papasotiriou I. Cancer stem cells stemness transcription factors expression correlates with breast cancer disease stage. Curr Stem Cell Res Ther. 2012;7:415–9. doi: 10.2174/157488812804484639. [DOI] [PubMed] [Google Scholar]
- 37.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kasimir-Bauer S, Hoffmann O, Wallwiener D, Kimmig R, Fehm T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012;14:R15. doi: 10.1186/bcr3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep. 2013;3:1259. doi: 10.1038/srep01259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reid AL, Millward M, Pearce R, Lee M, Frank MH, Ireland A, et al. Markers of circulating tumour cells in the peripheral blood of patients with melanoma correlate with disease recurrence and progression. Br J Dermatol. 2013;168:85–92. doi: 10.1111/bjd.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mimeault M, Batra SK. New promising drug targets in cancer- and metastasis-initiating cells. Drug Discov Today. 2010;15:354–64. doi: 10.1016/j.drudis.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mimeault M, Batra SK. Novel therapies against aggressive and recurrent epithelial cancers by molecular targeting tumor- and metastasis-initiating cells and their progenies. Anticancer Agents Med Chem. 2010;10:137–51. doi: 10.2174/187152010790909353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mimeault M, Batra SK. Hypoxia-inducing factors as master regulators of stemness properties and altered metabolism of cancer- and metastasis-initiating cells. J Cell Mol Med. 2013;17:30–54. doi: 10.1111/jcmm.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 46.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 47.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Gesch-wind DH, Bronner-Fraser M, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100:15178–83. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan X, Curtin J, Xiong Y, Liu G, Waschsmann-Hogiu S, Farkas DL, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23:9392–400. doi: 10.1038/sj.onc.1208311. [DOI] [PubMed] [Google Scholar]
- 49.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–21. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 50.Yu SC, Ping YF, Yi L, Zhou ZH, Chen JH, Yao XH, et al. Isolation and characterization of cancer stem cells from a human glioblastoma cell line U87. Cancer Lett. 2008;265:124–34. doi: 10.1016/j.canlet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 51.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–11. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 52.Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. Brca1 breast tumors contain distinct CD44+/CD24− and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008;10:R10. doi: 10.1186/bcr1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. doi: 10.1186/bcr1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–14. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- 55.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90(+) cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–66. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 56.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, et al. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311–20. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mimeault M, Hauke R, Mehta PP, Batra SK. Recent advances on cancer stem/progenitor cell research: therapeutic implications for overcoming resistance to the most aggressive cancers. J Cell Mol Med. 2007;11:981–1011. doi: 10.1111/j.1582-4934.2007.00088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.She JJ, Zhang PG, Wang ZM, Gan WM, Che XM. Identification of side population cells from bladder cancer cells by dyecycle violet staining. Cancer Biol Ther. 2008;7:1663–8. doi: 10.4161/cbt.7.10.6637. [DOI] [PubMed] [Google Scholar]
- 60.Huang D, Gao Q, Guo L, Zhang C, Wei J, Li H, et al. Isolation and identification of cancer stem-like cells in esophageal carcinoma cell lines. Stem Cells Dev. 2009;18:465–73. doi: 10.1089/scd.2008.0033. [DOI] [PubMed] [Google Scholar]
- 61.Chiba T, Miyagi S, Saraya A, Aoki R, Seki A, Morita Y, et al. The polycomb gene product BMI1 contributes to the maintenance of tumor-initiating side population cells in hepatocellular carcinoma. Cancer Res. 2008;68:7742–9. doi: 10.1158/0008-5472.CAN-07-5882. [DOI] [PubMed] [Google Scholar]
- 62.Shi GM, Xu Y, Fan J, Zhou J, Yang XR, Qiu SJ, et al. Identification of side population cells in human hepatocellular carcinoma cell lines with stepwise metastatic potentials. J Cancer Res Clin Oncol. 2008;134:1155–63. doi: 10.1007/s00432-008-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sung JM, Cho HJ, Yi H, Lee CH, Kim HS, Kim DK, et al. Characterization of a stem cell population in lung cancer A549 cells. Biochem Biophys Res Commun. 2008;371:163–7. doi: 10.1016/j.bbrc.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 64.Friel AM, Sergent PA, Patnaude C, Szotek PP, Oliva E, Scadden DT, et al. Functional analyses of the cancer stem cell-like properties of human endometrial tumor initiating cells. Cell Cycle. 2008;7:242–9. doi: 10.4161/cc.7.2.5207. [DOI] [PubMed] [Google Scholar]
- 65.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 66.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mitsutake N, Iwao A, Nagai K, Namba H, Ohtsuru A, Saenko V, et al. Characterization of side population in thyroid cancer cell lines: cancer stem-like cells are enriched partly but not exclusively. Endocrinology. 2007;148:1797–803. doi: 10.1210/en.2006-1553. [DOI] [PubMed] [Google Scholar]
- 68.Maitland NJ, Bryce SD, Stower MJ, Collins AT. Prostate cancer stem cells: a target for new therapies. Ernst Schering Found Symp Proc. 2006;5:155–79. doi: 10.1007/2789_2007_050. [DOI] [PubMed] [Google Scholar]
- 69.Bapat SA, Mali AM, Koppikar CB, Kurrey NK. Stem and progenitor-like cells contribute to the aggressive behavior of human epithelial ovarian cancer. Cancer Res. 2005;65:3025–9. doi: 10.1158/0008-5472.CAN-04-3931. [DOI] [PubMed] [Google Scholar]
- 70.Mimeault M, Batra SK. Complex oncogenic signaling networks regulate brain tumor-initiating cells and their progenies: pivotal roles of wild-type EGFR, EGFRvIII mutant and hedgehog cascades and novel multitargeted therapies. Brain Pathol. 2011;21:479–500. doi: 10.1111/j.1750-3639.2011.00505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marsden CG, Wright MJ, Carrier L, Moroz K, Pochampally R, Rowan BG. A novel in vivo model for the study of human breast cancer metastasis using primary breast tumor-initiating cells from patient biopsies. BMC Cancer. 2012;12:10. doi: 10.1186/1471-2407-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749–58. doi: 10.1038/sj.onc.1210811. [DOI] [PubMed] [Google Scholar]
- 73.Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res. 2008;6:1146–53. doi: 10.1158/1541-7786.MCR-08-0035. [DOI] [PubMed] [Google Scholar]
- 74.Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–56. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 75.Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, et al. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–51. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 76.Chiba T, Zheng YW, Kita K, Yokosuka O, Saisho H, Onodera M, et al. Enhanced self-renewal capability in hepatic stem/progenitor cells drives cancer initiation. Gastroenterology. 2007;133:937–50. doi: 10.1053/j.gastro.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 77.Frank NY, Margaryan A, Huang Y, Schatton T, Waaga-Gasser AM, Gasser M, et al. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res. 2005;65:4320–33. doi: 10.1158/0008-5472.CAN-04-3327. [DOI] [PubMed] [Google Scholar]
- 78.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salmaggi A, Boiardi A, Gelati M, Russo A, Calatozzolo C, Ciusani E, et al. Glioblastoma-derived tumorospheres identify a population of tumor stem-like cells with angiogenic potential and enhanced multidrug resistance phenotype. Glia. 2006;54:850–60. doi: 10.1002/glia.20414. [DOI] [PubMed] [Google Scholar]
- 80.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–60. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 81.Zhang X, Komaki R, Wang L, Fang B, Chang JY. Treatment of radioresistant stem-like esophageal cancer cells by an apoptotic gene-armed, telomerase-specific oncolytic adenovirus. Clin Cancer Res. 2008;14:2813–23. doi: 10.1158/1078-0432.CCR-07-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 83.Wang J, Guo LP, Chen LZ, Zeng YX, Lu SH. Identification of cancer stem cell-like side population cells in human nasopharyngeal carcinoma cell line. Cancer Res. 2007;67:3716–24. doi: 10.1158/0008-5472.CAN-06-4343. [DOI] [PubMed] [Google Scholar]
- 84.Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, et al. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101:14228–33. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haraguchi N, Utsunomiya T, Inoue H, Tanaka F, Mimori K, Barnard GF, et al. Characterization of a side population of cancer cells from human gastrointestinal system. Stem Cells. 2006;24:506–13. doi: 10.1634/stemcells.2005-0282. [DOI] [PubMed] [Google Scholar]
- 86.Loebinger MR, Giangreco A, Groot KR, Prichard L, Allen K, Simpson C, et al. Squamous cell cancers contain a side population of stem-like cells that are made chemosensitive by ABC transporter blockade. Br J Cancer. 2008;98:380–7. doi: 10.1038/sj.bjc.6604185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou J, Wang CY, Liu T, Wu B, Zhou F, Xiong JX, et al. Persistence of side population cells with high drug efflux capacity in pancreatic cancer. World J Gastroenterol. 2008;14:925–30. doi: 10.3748/wjg.14.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kurrey NK, Jalgaonkar SP, Joglekar AV, Ghanate AD, Chaskar PD, Doiphode RY, et al. Snail and slug mediate radioresistance and chemoresistance by antagonizing p53-mediated apoptosis and acquiring a stem-like phenotype in ovarian cancer cells. Stem Cells. 2009;27:2059–68. doi: 10.1002/stem.154. [DOI] [PubMed] [Google Scholar]
- 89.Mimeault M, Johansson SL, Senapati S, Momi N, Chakraborty S, Batra SK. MUC4 down-regulation reverses chemoresistance of pancreatic cancer stem/progenitor cells and their progenies. Cancer Lett. 2010;295:69–84. doi: 10.1016/j.canlet.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang L, Mezencev R, Bowen NJ, Matyunina LV, McDonald JF. Isolation and characterization of stem-like cells from a human ovarian cancer cell line. Mol Cell Biochem. 2012;363:257–68. doi: 10.1007/s11010-011-1178-6. [DOI] [PubMed] [Google Scholar]
- 91.Alvero AB, Chen R, Fu HH, Montagna M, Schwartz PE, Rutherford T, et al. Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle. 2009;8:158–66. doi: 10.4161/cc.8.1.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hamada S, Satoh K, Hirota M, Kanno A, Umino J, Ito H, et al. The homeobox gene MSX2 determines chemosensitivity of pancreatic cancer cells via the regulation of transporter gene ABCG2. J Cell Physiol. 2012;227:729–38. doi: 10.1002/jcp.22781. [DOI] [PubMed] [Google Scholar]
- 93.Steg AD, Bevis KS, Katre AA, Ziebarth A, Dobbin ZC, Alvarez RD, et al. Stem cell pathways contribute to clinical chemoresistance in ovarian cancer. Clin Cancer Res. 2012;18:869–81. doi: 10.1158/1078-0432.CCR-11-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee SO, Ma Z, Yeh CR, Luo J, Lin TH, Lai KP, et al. New therapy targeting differential androgen receptor signaling in prostate cancer stem/progenitor vs non-stem/progenitor cells. J Mol Cell Biol. 2012;424:740–6. [Google Scholar]
- 95.Ng FS, Toh TB, Ting EH, Koh GR, Sandanaraj E, Phong M, et al. Progenitor-like traits contribute to patient survival and prognosis in oligodendroglial tumors. Clin Cancer Res. 2012;18:4122–35. doi: 10.1158/1078-0432.CCR-11-3064. [DOI] [PubMed] [Google Scholar]
- 96.Bailey CM, Morrison JA, Kulesa PM. Melanoma revives an embryonic migration program to promote plasticity and invasion. Pigment Cell Melanoma Res. 2012;25:573–83. doi: 10.1111/j.1755-148X.2012.01025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng WY, Kandel JJ, Yamashiro DJ, Canoll P, Anastassiou D. A multi-cancer mesenchymal transition gene expression signature is associated with prolonged time to recurrence in glioblastoma. PLoS ONE. 2012;7:e34705. doi: 10.1371/journal.pone.0034705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsunoda Y, Sakamoto M, Sawada T, Sasaki A, Yamamoto G, Tachikawa T. Characteristic genes in luminal subtype breast tumors with CD44+CD24−/low gene expression signature. Oncology. 2011;81:336–44. doi: 10.1159/000334690. [DOI] [PubMed] [Google Scholar]
- 99.Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–93. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 100.Marquardt JU, Raggi C, Andersen JB, Seo D, Avital I, Geller D, et al. Human hepatic cancer stem cells are characterized by common stemness traits and diverse oncogenic pathways. Hepatology. 2011;54:1031–42. doi: 10.1002/hep.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yan X, Ma L, Yi D, Yoon JG, Diercks A, Foltz G, et al. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc Natl Acad Sci U S A. 2011;108:1591–6. doi: 10.1073/pnas.1018696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Marchini C, Montani M, Konstantinidou G, Orru R, Mannucci S, Ramadori G, et al. Mesenchymal/stromal gene expression signature relates to basal-like breast cancers, identifies bone metastasis and predicts resistance to therapies. PLoS ONE. 2010;5:e14131. doi: 10.1371/journal.pone.0014131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. doi: 10.1126/science.1142596. [DOI] [PubMed] [Google Scholar]
- 104.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–8. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Das B, Tsuchida R, Malkin D, Koren G, Baruchel S, Yeger H. Hypoxia enhances tumor stemness by increasing the invasive and tumorigenic side population fraction. Stem Cells. 2008;26:1818–30. doi: 10.1634/stemcells.2007-0724. [DOI] [PubMed] [Google Scholar]
- 106.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133−metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–20. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dylla SJ, Beviglia L, Park IK, Chartier C, Raval J, Ngan L, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One. 2008;3:e2428. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schmidt C. Lapatinib study supports cancer stem cell hypothesis, encourages industry research. J Natl Cancer Inst. 2008;100:694–5. doi: 10.1093/jnci/djn168. [DOI] [PubMed] [Google Scholar]
- 109.Griffero F, Daga A, Marubbi D, Capra MC, Melotti A, Pattarozzi A, et al. Different response of human glioma tumor-initiating cells to EGFR kinase inhibitors. J Biol Chem. 2009;284:7138–48. doi: 10.1074/jbc.M807111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–96. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen BY, Liu JY, Chang HH, Chang CP, Lo WY, Kuo WH, et al. Hedgehog is involved in prostate basal cell hyperplasia formation and its progressing towards tumorigenesis. Biochem Biophys Res Commun. 2007;357:1084–9. doi: 10.1016/j.bbrc.2007.04.091. [DOI] [PubMed] [Google Scholar]
- 113.Mimeault M, Batra SK. Recent progress on normal and malignant pancreatic stem/progenitor cell research: therapeutic implications for the treatment of type 1 or 2 diabetes mellitus and aggressive pancreatic cancer. Gut. 2008;57:1456–68. doi: 10.1136/gut.2008.150052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mimeault M, Batra SK. Interplay of distinct growth factors during epithelial-mesenchymal transition of cancer progenitor cells and molecular targeting as novel cancer therapies. Ann Oncol. 2007;18:1605–19. doi: 10.1093/annonc/mdm070. [DOI] [PubMed] [Google Scholar]
- 115.Cao L, Shao M, Schilder J, Guise T, Mohammad KS, Matei D. Tissue transglutaminase links TGF-β, epithelial to mesenchymal transition and a stem cell phenotype in ovarian cancer. Oncogene. 2012;31:2521–34. doi: 10.1038/onc.2011.429. [DOI] [PubMed] [Google Scholar]
- 116.Kabashima A, Higuchi H, Takaishi H, Matsuzaki Y, Suzuki S, Izumiya M, et al. Side population of pancreatic cancer cells predominates in TGF-β-mediated epithelial to mesenchymal transition and invasion. Int J Cancer. 2009;124:2771–9. doi: 10.1002/ijc.24349. [DOI] [PubMed] [Google Scholar]
- 117.Zhang Y, Wei J, Wang H, Xue X, An Y, Tang D, et al. Epithelial mesenchymal transition correlates with CD24+CD44+ and CD133+ cells in pancreatic cancer. Oncol Rep. 2012;27:1599–605. doi: 10.3892/or.2012.1681. [DOI] [PubMed] [Google Scholar]
- 118.Wang Z, Li Y, Kong D, Banerjee S, Ahmad A, Azmi AS, et al. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009;69:2400–7. doi: 10.1158/0008-5472.CAN-08-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bao B, Wang Z, Ali S, Kong D, Li Y, Ahmad A, et al. Notch-1 induces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett. 2011;307:26–36. doi: 10.1016/j.canlet.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 120.Puhr M, Hoefer J, Schafer G, Erb HH, Oh SJ, Klocker H, et al. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. Am J Pathol. 2012;181:2188–201. doi: 10.1016/j.ajpath.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 121.Shah AN, Summy JM, Zhang J, Park SI, Parikh NU, Gallick GE. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol. 2007;14:3629–37. doi: 10.1245/s10434-007-9583-5. [DOI] [PubMed] [Google Scholar]
- 122.Kikuta K, Masamune A, Watanabe T, Ariga H, Itoh H, Hamada S, et al. Pancreatic stellate cells promote epithelial-mesenchymal transition in pancreatic cancer cells. Biochem Biophys Res Commun. 2010;403:380–4. doi: 10.1016/j.bbrc.2010.11.040. [DOI] [PubMed] [Google Scholar]
- 123.Du Z, Qin R, Wei C, Wang M, Shi C, Tian R, et al. Pancreatic cancer cells resistant to chemoradiotherapy rich in “stem-cell-like” tumor cells. Dig Dis Sci. 2011;56:741–50. doi: 10.1007/s10620-010-1340-0. [DOI] [PubMed] [Google Scholar]
- 124.Giarnieri E, De Vitis C, Noto A, Roscilli G, Salerno G, Mariotta S, et al. EMT markers in lung adenocarcinoma pleural effusion spheroid cells. J Cell Physiol. 2013;228:1720–6. doi: 10.1002/jcp.24300. [DOI] [PubMed] [Google Scholar]
- 125.De Bacco F, Casanova E, Medico E, Pellegatta S, Orzan F, Albano R, et al. The MET oncogene is a functional marker of a glioblastoma stem cell subtype. Cancer Res. 2012;72:4537–50. doi: 10.1158/0008-5472.CAN-11-3490. [DOI] [PubMed] [Google Scholar]
- 126.Galavotti S, Bartesaghi S, Faccenda D, Shaked-Rabi M, Sanzone S, McEvoy A, et al. The autophagy-associated factors DRAM1 and p62 regulate cell migration and invasion in glioblastoma stem cells. Oncogene. 2012;32:699–712. doi: 10.1038/onc.2012.111. [DOI] [PubMed] [Google Scholar]
- 127.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 128.Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–12. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 129.Petrylak DP, Tangen CM, Hussain MH, Lara PN, Jr., Jones JA, Taplin NE, et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–20. doi: 10.1056/NEJMoa041318. [DOI] [PubMed] [Google Scholar]
- 130.Colombel M, Eaton CL, Hamdy F, Ricci E, van der Pluijm G, Cecchini M, et al. Increased expression of putative cancer stem cell markers in primary prostate cancer is associated with progression of bone metastases. Prostate. 2012;72:713–20. doi: 10.1002/pros.21473. [DOI] [PubMed] [Google Scholar]
- 131.Liu C, Tang DG. MicroRNA regulation of cancer stem cells. Cancer Res. 2011;71:5950–4. doi: 10.1158/0008-5472.CAN-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu C, Kelnar K, Vlassov AV, Brown D, Wang J, Tang DG, et al. Distinct microRNA expression profiles in prostate cancer stem/progenitor cells and tumor-suppressive functions of let-7. Cancer Res. 2012;72:3393–404. doi: 10.1158/0008-5472.CAN-11-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Punglia RS, Morrow M, Winer EP, Harris JR. Local therapy and survival in breast cancer. N Engl J Med. 2007;356:2399–405. doi: 10.1056/NEJMra065241. [DOI] [PubMed] [Google Scholar]
- 134.Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2012;9:16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- 135.Sorlie T, Wang Y, Xiao C, Johnsen H, Naume B, Samaha RR, et al. Distinct molecular mechanisms underlying clinically relevant sub-types of breast cancer: gene expression analyses across three different platforms. BMC Genom. 2006;7:127. doi: 10.1186/1471-2164-7-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guedj M, Marisa L, de Reynies A, Orsetti B, Schiappa R, Bibeau F, et al. A refined molecular taxonomy of breast cancer. Oncogene. 2011;31:1196–206. doi: 10.1038/onc.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, et al. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:17–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 138.Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, et al. Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol. 2006;19:264–71. doi: 10.1038/modpathol.3800528. [DOI] [PubMed] [Google Scholar]
- 139.Luo Y, Dallaglio K, Chen Y, Robinson WA, Robinson SE, McCarter MD, et al. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells. 2012;30:2100–13. doi: 10.1002/stem.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gunther HS, Schmidt NO, Phillips HS, Kemming D, Kharbanda S, Soriano R, et al. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27:2897–909. doi: 10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 141.Xu Q, Yuan X, Liu G, Black KL, Yu JS. Hedgehog signaling regulates brain tumor-initiating cell proliferation and portends shorter survival for patients with PTEN-coexpressing glioblastomas. Stem Cells. 2008;26:3018–26. doi: 10.1634/stemcells.2008-0459. [DOI] [PubMed] [Google Scholar]
- 142.Tang KH, Dai YD, Tong M, Chan YP, Kwan PS, Fu L, et al. A CD90(+) tumor-initiating cell population with an aggressive signature and metastatic capacity in esophageal cancer. Cancer Res. 2013;73:2322–32. doi: 10.1158/0008-5472.CAN-12-2991. [DOI] [PubMed] [Google Scholar]
- 143.Yuan P, Kadara H, Behrens C, Tang X, Woods D, Solis LM, et al. Sex determining region Y-Box 2 (SOX2) is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung. PLoS ONE. 2010;5:e9112. doi: 10.1371/journal.pone.0009112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Van den Broeck A, Vankelecom H, Van Delm W, Gremeaux L, Wouters J, Allemeersch J, et al. Human pancreatic cancer contains a side population expressing cancer stem cell-associated and prognostic genes. PLoS ONE. 2013;8:e73968. doi: 10.1371/journal.pone.0073968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hsu HC, Liu YS, Tseng KC, Hsu CL, Liang Y, Yang TS, et al. Over-expression of Lgr5 correlates with resistance to 5-FU-based chemotherapy in colorectal cancer. Int J Colorectal Dis. 2013;28:1535–46. doi: 10.1007/s00384-013-1721-x. [DOI] [PubMed] [Google Scholar]
- 146.Mine H, Sakurai T, Kashida H, Matsui S, Nishida N, Nagai T, et al. Association of gankyrin and stemness factor expression in human colorectal cancer. Dig Dis Sci. 2013;58:2337–44. doi: 10.1007/s10620-013-2627-8. [DOI] [PubMed] [Google Scholar]
- 147.Fan F, Samuel S, Evans KW, Lu J, Xia L, Zhou Y, et al. Overexpression of snail induces epithelial-mesenchymal transition and a cancer stem cell-like phenotype in human colorectal cancer cells. Cancer Med. 2012;1:5–16. doi: 10.1002/cam4.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schreiber L, Raanan C, Amsterdam A. CD24 and Nanog identify stem cells signature of ovarian epithelium and cysts that may develop to ovarian cancer. Acta Histochem. 2013;115:330–8. doi: 10.1016/j.acthis.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 149.Amsterdam A, Raanan C, Schreiber L, Freyhan O, Schechtman L, Givol D. Localization of the stem cell markers LGR5 and Nanog in the normal and the cancerous human ovary and their inter-relationship. Acta Histochem. 2013;115:330–8. doi: 10.1016/j.acthis.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 150.Corcoran C, Rani S, O’Brien K, O’Neill A, Prencipe M, Sheikh R, et al. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS ONE. 2012;7:e50999. doi: 10.1371/journal.pone.0050999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106:768–74. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–24. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 154.Lima LG, Chammas R, Monteiro RQ, Moreira ME, Barcinski MA. Tumor-derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidylserine-dependent manner. Cancer Lett. 2009;283:168–75. doi: 10.1016/j.canlet.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 155.Jung T, Castellana D, Klingbeil P, Cuesta Hemandez I, Vitacolonna M, Orlicky DJ, et al. CD44v6 dependence of premetastatic niche preparation by exosomes. Neoplasia. 2009;11:1093–105. doi: 10.1593/neo.09822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Marhaba R, Klingbeil P, Nuebel T, Nazarenko I, Buechler MW, Zoeller M. CD44 and EpCAM: cancer-initiating cell markers. Curr Mol Med. 2008;8:784–804. doi: 10.2174/156652408786733667. [DOI] [PubMed] [Google Scholar]
- 157.Camussi G, Deregibus MC, Bruno S, Grange C, Fonsato V, Tetta C. Exosome/microvesicle-mediated epigenetic reprogramming of cells. Am J Cancer Res. 2011;1:98–110. [PMC free article] [PubMed] [Google Scholar]
- 158.King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12:421. doi: 10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cho JA, Park H, Lim EH, Kim KH, Choi JS, Lee JH, et al. Exosomes from ovarian cancer cells induce adipose tissue-derived mesenchymal stem cells to acquire the physical and functional characteristics of tumor-supporting myofibroblasts. Gynecol Oncol. 2011;123:379–86. doi: 10.1016/j.ygyno.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 160.Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci U S A. 2009;106:3794–9. doi: 10.1073/pnas.0804543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer. 2009;100:1603–7. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liang B, Peng P, Chen S, Li L, Zhang M, Cao D, et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J Proteomics. 2013;80C:171–82. doi: 10.1016/j.jprot.2012.12.029. [DOI] [PubMed] [Google Scholar]
- 163.Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Welton JL, Khanna S, Giles PJ, Brennan P, Brewis IA, Staffurth J, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics. 2010;9:1324–38. doi: 10.1074/mcp.M000063-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159–67. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 166.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 167.Peng P, Yan Y, Keng S. Exosomes in the ascites of ovarian cancer patients: origin and effects on anti-tumor immunity. Oncol Rep. 2011;25:749–62. doi: 10.3892/or.2010.1119. [DOI] [PubMed] [Google Scholar]
- 168.Grange C, Tapparo M, Collino F, Vitillo L, Damasco C, Deregibus MC, et al. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011;71:5346–56. doi: 10.1158/0008-5472.CAN-11-0241. [DOI] [PubMed] [Google Scholar]
- 169.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–91. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835–40. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Khan S, Jutzy JM, Valenzuela MM, Turay D, Aspe JR, Ashok A, et al. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS ONE. 2012;7:e46737. doi: 10.1371/journal.pone.0046737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ciravolo V, Huber V, Ghedini GC, Venturelli E, Bianchi F, Campiglio M, et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol. 2012;227:658–67. doi: 10.1002/jcp.22773. [DOI] [PubMed] [Google Scholar]
- 173.Rappa G, Mercapide J, Anzanello F, Pope RM, Lorico A. Biochemical and biological characterization of exosomes containing prominin-1/CD133. Mol Cancer. 2013;12:62. doi: 10.1186/1476-4598-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 175.Jaiswal R, Luk F, Gong J, Mathys JM, Grau GE, Bebawy M. Microparticle conferred microRNA profiles—implications in the transfer and dominance of cancer traits. Mol Cancer. 2012;11:37. doi: 10.1186/1476-4598-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–6. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, et al. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72:4920–30. doi: 10.1158/0008-5472.CAN-12-0925. [DOI] [PubMed] [Google Scholar]
- 178.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 179.Marleau AM, Chen CS, Joyce JA, Tullis RH. Exosome removal as a therapeutic adjuvant in cancer. J Transl Med. 2012;10:134. doi: 10.1186/1479-5876-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, et al. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16:782–90. doi: 10.1038/mt.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tan A, De La Pena H, Seifalian AM. The application of exosomes as a nanoscale cancer vaccine. Int J Nanomedicine. 2010;5:889–900. doi: 10.2147/IJN.S13402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Alhasan AH, Patel PC, Choi CH, Mirkin CA. Exosome encased spherical nucleic acid gold nanoparticle conjugates as potent microRNA regulation agents. Small. 2013 Sep 17; doi: 10.1002/smll.201302143. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Koppers-Lalic D, Hogenboom MM, Middeldorp JM, Pegtel DM. Virus-modified exosomes for targeted RNA delivery; a new approach in nanomedicine. Adv Drug Deliv Rev. 2013;65:348–56. doi: 10.1016/j.addr.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Punnoose EA, Atwal S, Liu W, Raja R, Fine BM, Hughes BG, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res. 2012;18:2391–401. doi: 10.1158/1078-0432.CCR-11-3148. [DOI] [PubMed] [Google Scholar]
- 185.Galizia G, Gemei M, Orditura M, Romano C, Zamboli A, Castellano P, et al. Postoperative detection of circulating tumor cells predicts tumor recurrence in colorectal cancer patients. J Gastrointest Surg. 2013;17:1809–18. doi: 10.1007/s11605-013-2258-6. [DOI] [PubMed] [Google Scholar]
- 186.Khoja L, Backen A, Sloane R, Menasce L, Ryder D, Krebs M, et al. A pilot study to explore circulating tumour cells in pancreatic cancer as a novel biomarker. Br J Cancer. 2012;106:508–16. doi: 10.1038/bjc.2011.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Obermayr E, Sanchez-Cabo F, Tea MK, Singer CF, Krainer M, Fischer MB, et al. Molecular characterization of circulating tumor cells in patients with ovarian cancer improves their prognostic significance—a study of the OVCAD consortium. Gynecol Oncol. 2013;6:42–76. doi: 10.1016/j.ygyno.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 188.Kurihara T, Itoi T, Sofuni A, Itokawa F, Tsuchiya T, Tsuji S, et al. Detection of circulating tumor cells in patients with pancreatic cancer: a preliminary result. J Hepatobiliary Pancreat Surg. 2008;15:189–95. doi: 10.1007/s00534-007-1250-5. [DOI] [PubMed] [Google Scholar]
- 189.Ghazani AA, Castro CM, Gorbatov R, Lee H, Weissleder R. Sensitive and direct detection of circulating tumor cells by multimarker micro-nuclear magnetic resonance. Neoplasia. 2012;14:388–95. doi: 10.1596/neo.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]