Abstract
Messenger RNA precursors (pre-mRNAs) are produced as the nascent transcripts of RNA polymerase II (Pol II) in eukaryotes and must undergo extensive maturational processing, including 5′-end capping, splicing, and 3′-end cleavage and polyadenylation. This review will summarize the structural and functional information reported over the past few years on the large machinery required for the 3′-end processing of most pre-mRNAs, as well as the distinct machinery for the 3′-end processing of replication-dependent histone pre-mRNAs, which have provided great insights into the proteins and their subcomplexes in these machineries. Structural and biochemical studies have also led to the identification of a new class of enzymes (the DXO family enzymes) with activity toward intermediates of the 5′-end capping pathway. Functional studies demonstrate that these enzymes are part of a novel quality surveillance mechanism for pre-mRNA 5′-end capping. Incompletely capped pre-mRNAs are produced in yeast and human cells, in contrast to the general belief in the field that capping always proceeds to completion, and incomplete capping leads to defects in splicing and 3′-end cleavage in human cells. The DXO family enzymes are required for the detection and degradation of these defective RNAs.
In eukaryotes, mRNA precursors (pre-mRNAs) are transcribed by RNA polymerase II (Pol II) from the genome and must undergo extensive cotranscriptional processing to become mature mRNAs. The typical progression of pre-mRNA maturation involves 5′-end capping, splicing, and 3′-end cleavage and polyadenylation. The integrity and precision of each of these steps are critical for generating stable, functional mRNAs. Moreover, recent studies have demonstrated the importance of alternative splicing, alternative polyadenylation (APA), and RNA editing in producing an incredibly diverse, often cell-specific mRNA library that contributes to the biological complexity of higher eukaryotes.
5′-end capping occurs very early during Pol II transcription, typically after the synthesis of ∼20 nucleotides of the pre-mRNA. Capping has been linked to splicing and 3′-end processing of the pre-mRNA, and the export of the mature mRNA. In addition, the 5′-end cap is directly recognized by the eukaryotic translation initiation factor eIF-4E, which is essential for mRNA translation by the ribosome.
A majority of pre-mRNAs acquire a poly(A) tail after 3′-end processing, which is important for the export of the mature mRNAs from the nucleus to the cytoplasm. The poly(A) tail also promotes the translation of the mRNAs and protects them from degradation. In comparison, 3′-end processing of replication-dependent histone pre-mRNAs involves only the cleavage reaction, and these mRNAs do not carry a poly(A) tail. Instead, a conserved stem–loop structure at their 3′-end supports many of the functions that are associated with the poly(A) tail.
This review will focus on recent advances (within the past ∼5 years) in structural and functional studies of pre-mRNA 3′-end processing, and the newly reported structures are summarized in Table 1. There are also many other excellent reviews on these topics, some of which are listed here.1−8 In addition, a novel quality surveillance mechanism for 5′-end capping was discovered recently and will be reviewed here, as well. Other aspects of pre-mRNA processing, such as splicing, APA,9−11 and poly(A) length regulation,12,13 and other mechanisms of mRNA quality control and decay, such as nonsense-mediated decay and no-go decay, will not be covered here because of space limitations.
Table 1. Recently Published Structures of Protein Factors Involved in Pre-mRNA 3′-End Processing or 5′-End Capping Quality Surveillance.
| protein factor | subdomains | Protein Data Bank entries | refs |
|---|---|---|---|
| CPSF | |||
| archaeal CPSF-73 | metallo-β-lactamase, β-CASP | 2YCB, 2XR1 | (21), (22) |
| archaeal CPSF-73–RNA complex | metallo-β-lactamase, β-CASP | 3AF6 | (20) |
| CPSF-30–NS1A complex | CPSF-30: zinc fingers 2 and 3; NS1A: effector | 2RHK | (29) |
| CstF | |||
| Rna14–Rna15 complex | Rna14: full length; Rna15: hinge | 4EBA, 4E85, 4E6H | (37) |
| Rna14–Rna15 complex | Rna14: monkeytail; Rna15: hinge | 2L9B | (40) |
| CstF-50 | homodimerization | 2XZ2 | (39) |
| Rna15 | RRM | 2X1B | (44) |
| Rna15–RNA complex | RRM | 2X1A, 2X1F | (44) |
| Rna15–Hrp1–RNA complex | Rna15: RRM; Hrp1: RRM | 2KM8 | (43) |
| CF Im | |||
| CF Im25 | full length | 3BAP, 2CL3, 3BHO, 2J8Q | (50), (51) |
| CF Im25–RNA complex | full length | 3MDG, 3MDI | (52) |
| CF Im25–CF Im68 complex | CF Im25: full length; CF Im68: RRM | 3Q2S | (53) |
| CF Im25–CF Im68–RNA complex | CF Im25: full length; CF Im68: RRM | 3Q2T | (53) |
| CF Im25–CF Im59 complex | CF Im25: full length; CF Im59: RRM | 3N9U | unpublished (2010) |
| PAP | |||
| PAPγ | core | 4LT6 | (74) |
| PAP–Fip1 complex | PAP: full length; Fip1: NTD fragment | 3C66 | (23) |
| PAPD1 | 3PQ1 | (77) | |
| cytoplasmic polyadenylation | |||
| CPE-binding protein (CPEB) | ZZ domain | 2M13 | (86) |
| symplekin-Ssu72 | |||
| symplekin | NTD | 3ODR, 3ODS, 3GS3, 3O2T | (88), (146) |
| Ssu72 | full length | 3OMW, 3OMX, 3FDF | (147) |
| Ssu72–Pol II CTD pSer5 complex | full length | 3P9Y | (98) |
| symplekin–Ssu72 complex | Symplekin: NTD; Ssu72: full length | 3O2S | (88) |
| symplekin–Ssu72–Pol II CTD pSer5 complexes | Symplekin: NTD; Ssu72: full length | 3O2Q, 4IMJ, 4IMI | (88), (97) |
| symplekin–Ssu72–Pol II CTD pSer7 complexes | Symplekin: NTD; Ssu72: full length | 4H3H, 4H3K | (99) |
| histone mRNA 3′-end processing | |||
| SLBP–RNA complex | RBD fragment | 2KJM | (148) |
| SLBP–3′hExo–RNA complex | SLBP: RBD; 3′hExo: full length | 4L8R | (112) |
| SLBP–SLIP1 complex | SLBP: fragment; SLIP1: full length | 4JHK | (106) |
| mRNA 5′-end capping quality surveillance | |||
| DXO–RNA complexes | full length | 4J7L, 4J7M | (143) |
| DXO–m7GpppG complex | full length | 4J7N | (143) |
| Dxo1 | full length | 4GPU, 4GPS | (142) |
| Dom3Z (DXO) | full length | 3FQI | (140) |
| Dom3Z–GDP complex | full length | 3FQJ | (140) |
| Rai1 | full length | 3FQG | (140) |
| Rat1–Rai1 complex | 3FQD | (140) |
Canonical Pre-mRNA 3′-End Processing
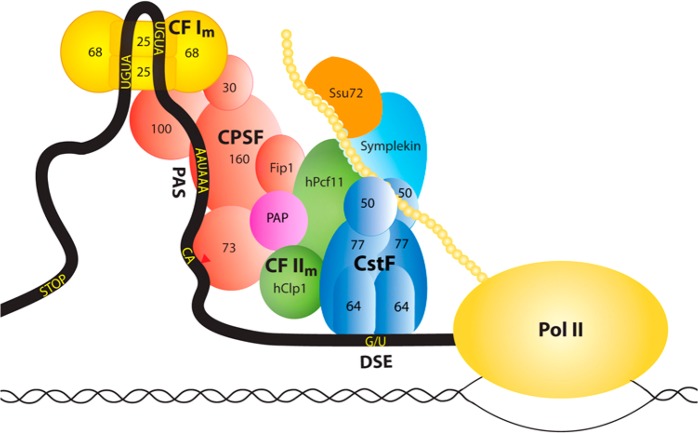
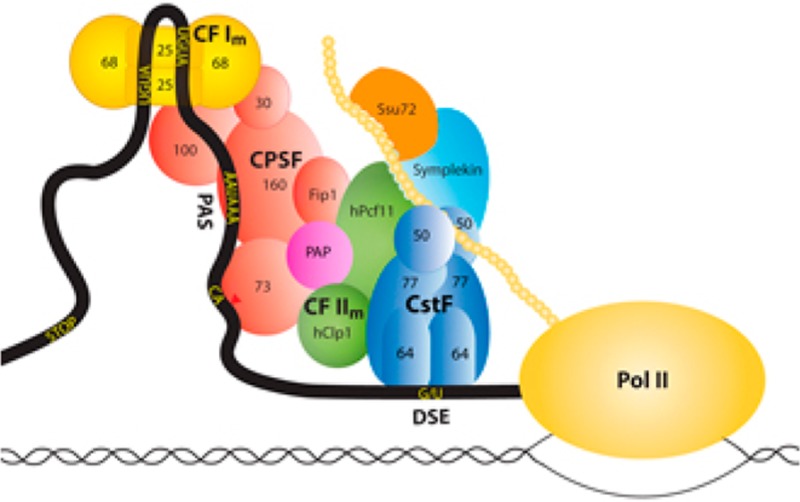
Key sequence elements in the 3′-untranslated regions (UTRs) of pre-mRNAs are recognized for 3′-end processing. In mammals, the major sequence elements include a hexanucleotide poly(A) signal (PAS, oftentimes AAUAAA) 10–30 nucleotides upstream of the cleavage site,14 the cleavage site itself (oftentimes after a CA dinucleotide), and a U- or G/U-rich downstream sequence element (DSE) (Figure 1). In addition, auxiliary sequence elements can be recognized, which may also help alter the site of 3′-end processing by APA.
Figure 1.
Schematic drawing of the canonical mammalian pre-mRNA 3′-end processing machinery showing the various protein factors and their subcomplexes. Many additional protein factors are involved in 3′-end processing but are not shown.
A large protein machinery is responsible for 3′-end processing in mammals, which consists of several subcomplexes such as cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor I (CF Im), CF IIm, and other protein factors such as poly(A) polymerase (PAP), symplekin, and Ssu72 (Figure 1). CPSF-73 (the 73 kDa subunit of CPSF) is the endoribonuclease for the cleavage reaction. CPSF-160 recognizes the PAS, and CstF-64 recognizes the DSE. This large machinery ensures the fidelity of 3′-end processing, supports APA in response to specific molecular and cellular environments, and is also connected to the DNA damage response.15 Moreover, many protein factors in the machinery communicate with other transcription processes, such as Pol II initiation and termination. A proteomic study identified more than 90 protein factors that may be associated with the pre-mRNA, although the exact roles of many of these proteins in 3′-end processing remain to be established.16
Similarly, pre-mRNA 3′-end processing in yeast also involves a large protein machinery. Many of the protein factors have homologues in the mammalian machinery, although the subcomplexes in yeast can have compositions and functions different from those of the subcomplexes in mammals. For example, yeast CF IA contains Rna14 and Rna15, the homologues of mammalian CstF-77 and CstF-64, respectively. CF IA also contains Clp1 and Pcf11, which belong to CF IIm in mammals (Figure 1). CF II in yeast contains Ysh1, Ydh1, and Yhh1, which are homologues of CPSF-73, CPSF-100, and CPSF-160, respectively. CF II also contains Pta1, the homologue of symplekin, while the other two subunits of CPSF, CPSF-30 (Yth1) and hFip1 (Fip1), belong to polyadenylation factor I (PF I). CF II, PF I, and many other protein factors comprise the cleavage and polyadenylation factor (CPF) in yeast.
The machinery for 3′-end processing of most eukaryotic pre-mRNAs will be termed the canonical machinery, to distinguish it from the machinery required for 3′-end processing of replication-dependent histone pre-mRNAs (see below). We will describe below the various subcomplexes and protein factors of the mammalian machinery, together with the equivalent proteins in the yeast machinery.
CPSF
CPSF has five subunits: CPSF-160, CPSF-100, CPSF-73, CPSF-30, and hFip1. CPSF-160 contains three β-propeller domains. CPSF-73 and CPSF-100 contain a metallo-β-lactamase and a β-CASP domain in the N-terminal region. CPSF-30 has five zinc fingers and one zinc knuckle. hFip1 does not contain any recognizable domains and is likely disordered on its own.
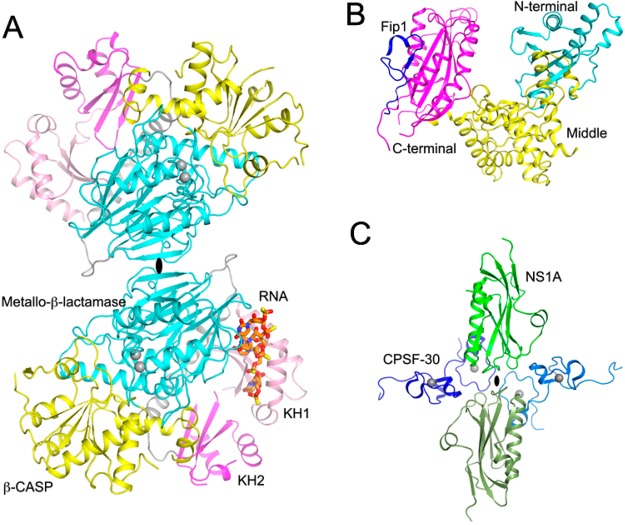
The structure of human CPSF-73 showed that its active site is located at the interface of the metallo-β-lactamase and β-CASP domains.17 CPSF-73 homologues are found in all three domains of life, with important functions in RNA processing and/or decay.18,19 Recently published structures of CPSF-73 homologues from two different archaeal species revealed the presence of two type II K homology (KH) RNA-binding motifs at the N-terminus, as well as the formation of a homodimer via the C-terminal region of the metallo-β-lactamase domain (Figure 2A). The RNA is likely recognized by the KH domains in one monomer and cleaved by the active site in the other monomer.20−22 This mechanism may be unique to archaea as mammalian CPSF-73 and its yeast homologue Ysh1 do not contain KH domains at the N-terminus.
Figure 2.
Recently published structures of CPSF subunits. (A) Structure of the Methanosarcina mazei CPSF-73 homologue dimer [Protein Data Bank (PDB) entry 2XR1].21 The bound position of the RNA analogue is modeled from the structure of the Pyrococcus horikoshii CPSF-73 homologue (PDB entry 3AF6).20 The 2-fold axis of the dimer is depicted as a black oval. (B) Structure of yeast PAP in complex with Fip1 (PDB entry 3C66).23 (C) Structure of human CPSF-30 (second and third zinc fingers) in complex with the influenza virus NS1A effector domain (PDB entry 2RHK).29 All the structures were produced with PyMOL (http://www.pymol.org).
Fip1, the yeast homologue of hFip1, tethers PAP to the processing machinery, which recognizes an intrinsically unstructured segment in Fip1 near its N-terminus (Figure 2B).23 PAP mutants that retain polymerase activity but cannot bind Fip1 are nonetheless lethal, indicating that the Fip1–PAP interaction serves an essential function in yeast. An N-terminal deletion mutant of Fip1 in which this binding site is disrupted cannot complement the loss of wild-type Fip1, but the mutant is fully functional if it is fused directly to PAP.24
CPSF-30 is targeted by the C-terminal effector domain of the nonstructural protein (NS1A) from the influenza A family of viruses,25−27 and the viral polymerase stabilizes this complex.28 NS1A binding inhibits host antiviral responses such as production of type I interferon and activation of dendritic cells. The effector domain of NS1A is recognized by the second and third zinc fingers of CPSF-30, in a 2:2 heterotetrameric complex (Figure 2C).29 Single-site mutations of NS1A residues in the interface prevent binding to CPSF-30, and an influenza virus carrying such a mutation in NS1A cannot inhibit interferon-β pre-mRNA processing and is attenuated in cells.
Arabidopsis thaliana CPSF-30 (AtCPSF-30) binds the A-rich near upstream element (NUE, which contains the AAUAAA motif) that is present in a subset of pre-mRNAs, located 10–30 nucleotides upstream of the cleavage site.30 Binding of RNA by AtCPSF-30 is mostly mediated through the first of its three zinc fingers.31 AtCPSF-30 also possesses endonuclease activity, which is mediated by its third zinc finger and inhibited by the N-terminal region of AtFip1(V), a plant homologue of Fip1.31 Loss of AtCPSF-30 results in an enhanced tolerance to oxidative stress because of the overexpression of proteins with thioredoxin- and glutaredoxin-like domains.32 The nuclease activity of AtCPSF-30 itself is redox-sensitive, as the third zinc finger contains a disulfide bond that stabilizes the overall structure of the protein.33 Some of these properties may be unique to AtCPSF-30 as it is localized in the cytoplasm in the absence of other CPSF subunits.34
CstF
CstF contains three subunits: CstF-50, CstF-64, and CstF-77. CstF-50 has a WD40 domain in the C-terminal region. CstF-64 has a RNA recognition module (RRM) at the N-terminus, followed by a hinge region, a Pro/Gly-rich region, and a small C-terminal domain (CTD). CstF-77 contains a HAT domain in the N-terminal region, followed by a Pro-rich region.
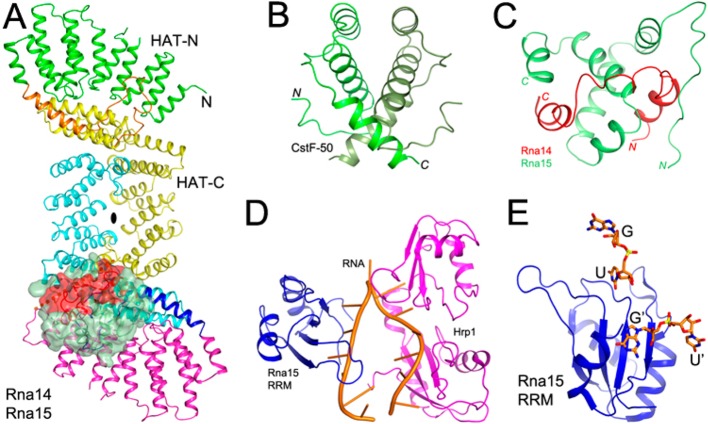
The crystal structure of the HAT domain of CstF-77 revealed a dimeric association, providing the first evidence that CstF may function as a dimer.35,36 The recently published crystal structure of the Kluyveromyces lactis Rna14–Rna15 complex also showed a dimeric association of this heterodimer into a heterotetramer, mediated by the HAT domain of Rna14 (Figure 3A).37 Mutation of two residues in the HAT domain dimer interface caused a temperature-sensitive phenotype in yeast, and the cell extract was defective in cleavage and polyadenylation.38 The structure of the N-terminal segment of CstF-50 is also a dimer (Figure 3B).39 Overall, the structures as well as biochemical studies support a stable, dimeric association of the CstF complex. While Rna14 and Rna15 are dimeric in the CF IA complex in yeast, Clp1 and Pcf11 may actually be monomeric, giving an overall 2:2:1:1 stoichiometry for the complex.38
Figure 3.
Recently published structures of the CstF subunits and their homologues. (A) Structure of the K. lactis Rna14–Rna15 dimer (PDB entry 4EBA).37 Only one copy of the complex between the Rna14 C-terminal Pro-rich segment (red) and the Rna15 hinge region (pale green) is ordered (shown as a molecular surface). (B) Structure of the Drosophila CstF-50 N-terminal domain dimer (PDB entry 2XZ2).39 (C) Structure of the heterodimer of the Pro-rich segment of Rna14 (red) with the hinge region of Rna15 (pale green) (PDB entry 2L9B).40 (D) Structure of the yeast Rna15 RRM (blue)–Hrp1 (magenta)–RNA (orange) complex (PDB entry 2KM8).43 (E) Structure of the yeast Rna15 RRM–RNA complex (PDB entry 2X1F).44 The binding site at the top (RNA labeled GU) mediates specific recognition. The RNA in front of the β-sheet (labeled G′U′) is related by crystal symmetry to the GU RNA and is not specifically recognized.
The interactions between Rna14 and Rna15 are mediated by the C-terminal Pro-rich region of Rna14 and the hinge region of Rna15 (Figure 3C).37,40 The formation of the CstF-64–CstF-77 complex is also important for the nuclear localization of CstF.41 CstF-77 contains a poly(A) site within its intron 3, and the usage of this site depends on the expression levels of CstF-77, resulting in a negative feedback mechanism.42
The amino acid sequences of the RRMs of CstF-64 and Rna15 are ∼50% identical, but the RRMs display distinct sequence preferences for RNA. CstF-64 recognizes the G/U-rich DSE, while Rna15 recognizes the A-rich positioning element (PE) in yeast. This distinct preference for Rna15 may be due to Hrp1, which constitutes CF IB but does not have a counterpart in the mammalian machinery. A crystal structure of the Rna15 RRM–Hrp1–RNA complex showed that the A-rich RNA interacts with the surface of the RRM β-sheet (Figure 3D),43 which is the canonical mode of RNA recognition for RRMs. On the other hand, a crystal structure of Rna15 RRM in complex with U-rich RNA, in the absence of Hrp1, shows that the RRM has a second, noncanonical RNA binding surface, which involves conserved loops above the β-sheet of the RRM (Figure 3E).44 The interaction between Hrp1 and the Rna14–Rna15 dimer has also been studied by NMR, and a model for the Rna14–Rna15–Hrp1–RNA complex has been proposed.45
The regulation of the RNA preference of Rna15 by Hrp1 may have important functional relevance. If there are two copies of Rna15 and only one copy of Hrp1 in the yeast 3′-end processing machinery, the two copies of Rna15 may bind to two different sequence elements in the transcript, one being A-rich and the other U- or G/U-rich. This may explain why 3′-end processing is enhanced by the addition of U-rich sequences between the PE and the cleavage site in the absence of Hrp1.44 Whether CstF-64 has a binding partner that is functionally homologous to Hrp1 to facilitate an A-rich sequence preference has not been determined. Musashi1, a mammalian homologue of Hrp1, with a similar RNA binding mode,46 is known to bind to the 3′-UTR of mRNAs but exerts its control at the translation level rather than the transcription level. Such regulation of the RNA preference of CstF-64 could also be important for its function in APA.
A second isoform of CstF-64 in mammals, CstF-64τ, was originally thought to be restricted to the testis and brain, although recent studies suggest that it is more widely expressed.47 CstF-64τ may complement the function of CstF-64. Moreover, a dimeric CstF complex could include one copy each of CstF-64 and CstF-64τ, which could be another mechanism for regulating 3′-end processing and APA. In addition, a family of splicing variants of CstF-64 has been identified, known as βCstF-64, which may have roles in APA in neuronal cells.48
CF Im
CF Im is comprised of two subunits: CF Im25 and CF Im68. CF Im25 has a Nudix nucleotide hydrolase fold but lacks hydrolase activity. CF Im68 is the most common second subunit, but there are alternative 59 and 72 kDa subunits. These three proteins contain an N-terminal RRM, a central Pro/Gly-rich region, and a C-terminal Arg/Ser-, Arg/Asp-, and Arg/Glu-rich segment. CF Im binds UGUA elements and is typically positioned 40–50 nucleotides upstream of the cleavage site.49
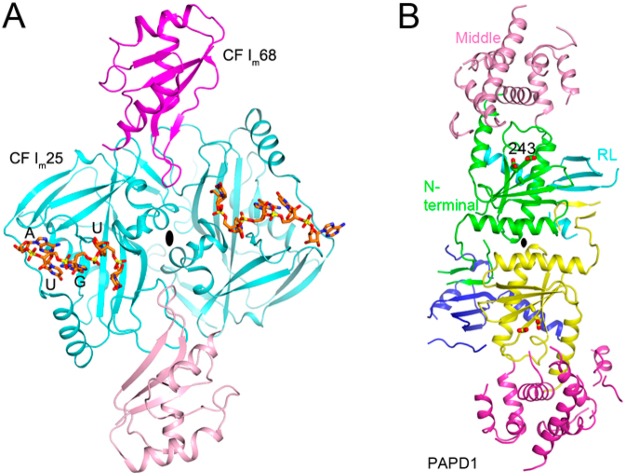
Several crystal structures have been reported for this complex over the past few years, which have greatly enhanced our understanding of its molecular mechanism.50−54 The structures show that CF Im is a heterotetramer, with a central CF Im25 dimer and two CF Im68 monomers bound to opposite sides of the CF Im25 dimer (Figure 4A).53,54 The two Nudix domains in the CF Im25 dimer are arranged antiparallel to each other, which would require the two UGUA cis elements of the pre-mRNA to make a 180° turn to bind to them simultaneously (Figure 1). The two RRMs of CF Im68 enhance RNA binding by ∼3-fold and promote RNA loop formation but are dispensable.
Figure 4.
Structures of CF Im and PAPD1. (A) Structure of the human CF Im25 (cyan)–CF Im68 (magenta)–UGUA RNA (orange) complex dimer (PDB entry 3Q2T).53 (B) Structure of the human mitochondrial PAPD1 D325A mutant dimer (PDB entry 3PQ1).77 The domains are shown in different colors. The catalytic residues are shown as stick models.
CF Im has a key role in APA49,55,56 and in the export of mRNA from the nucleus.57 Knockdown of the 25 and 68 kDa subunits in HEK293 cells increased the extent of global use of proximal poly(A) sites. On the other hand, knockdown of CF Im59 has no effect on poly(A) site choice. Therefore, the CF Im25–CF Im68 complex promotes the selection of distal poly(A) sites, producing mRNAs with an extended 3′-UTR that may be subject to specific 3′-UTR-mediated regulation. CF Im68 interacts with the nuclear export machinery through the Thoc5 protein of the TREX complex and the nuclear export receptor NXF1/TAP and shuttles between the nucleus and the cytoplasm.58 Knockdown of Thoc5 also promotes the usage of proximal poly(A) sites.59
CF Im68 is recruited by the capsid protein of HIV60 and helps the virus to evade host innate immune recognition.61 A C-terminal deletion of CF Im68 promotes HIV-1 capsid disassembly.62
CF IIm
CF IIm comprises two subunits: hClp1 and hPcf11. hClp1 contains three domains: N-terminal, central, and C-terminal domains. The central domain contains a Walker A P-loop motif and can bind ATP. hPcf11 contains a Pol II C-terminal domain (CTD) interaction domain (CID) at the N-terminus, two zinc fingers, a short sequence between the two zinc fingers that interacts with Clp1,63 and other sequence motifs. Their homologues in yeast, Clp1 and Pcf11, belong to CF IA and interact with Rna14 and Rna15. An equivalent interaction between CF IIm and CstF in mammalian cells has not been demonstrated. In addition to the Clp1–Pcf11 interface identified from earlier studies,63 there may be a “distant” binding site for Pcf11 on Clp1.64,65
hClp1 exhibits ATP hydrolase activity and plays an important role in pre-tRNA splicing66−68 as well as pre-mRNA 3′-end processing. Yeast Clp1 does not have ATP hydrolase activity but still requires ATP binding for its function. Yeast cells carrying Clp1 mutations in the ATP binding pocket, which induce a conformational change but do not occlude ATP binding, are not viable.65,69 The correct conformation of Clp1 induced by ATP binding may be essential for interactions with other protein factors in the machinery, including Ssu72, Ysh1, Pta1, and Rna14. Clp1 may therefore be an important structural protein in the machinery, which is supported by the observation that reconstitution of CF IA from individual components requires Clp1.64 Clp1 may contribute to gene looping and transcriptional directionality at bidirectional promoters, possibly through its interaction with Ssu72.
hClp1 may also compete with the mRNA nuclear export factor Aly (Yra1 in yeast) for binding to hPcf11, and yeast Clp1 can displace Yra1 from Pcf11 in affinity experiments.70 Recombinant Yra1 inhibits in vitro CF IA-mediated cleavage and polyadenylation reactions.71
PAP
CPSF stimulates the activity of PAP so that it processively extends the poly(A) tail, the length of which is regulated by the nuclear poly(A)-binding protein (PABPN1).72 Once the tail reaches ∼250 nucleotides, PABPN1 interferes with this stimulation. PABPN1 can also stimulate PAP and cause hyperadenylation, which can mediate RNA degradation by the exosome.73
A recently published structure of the human PAPγ core,74 from the γ clade of mammalian PAPs, confirms the three-domain core structure shared among the canonical PAPs, with N-terminal, middle, and C-terminal domains (Figure 2B).
Noncanonical PAPs have very weak conservation of sequence with respect to that of canonical PAPs and lack the C-terminal domain in the core.75,76 Some of these enzymes catalyze the oligo- or polyuridylation of their substrates and are also known as poly(U) polymerases (PUPs) or terminal uridylate transferases (TUTs or TUTases). The structure of human mitochondrial PAPD1 reveals a dimer of this enzyme, involving a RL (RNA-binding domain-like) domain unique to this enzyme (Figure 4B), and biochemical studies suggest that dimerization is required for PAPD1 activity.77 PAPD1 can use all four nucleotides as substrates in vitro, and how it achieves nucleotide specificity in the mitochondria is currently not known. Structures of other noncanonical PAPs (TUTs) have also been reported.76
Cytoplasmic Polyadenylation
Cytoplasmic polyadenylation is important for the post-transcriptional control of gene expression through the reactivation of deadenylated and dormant but otherwise intact cytoplasmic mRNAs, which is directed by the presence of a cytoplasmic polyadenylation element (CPE) in the 3′-UTR of the mRNAs.78,79 The CPE is bound by the regulatory cytoplasmic element binding protein (CPEB), which in turn interacts with many other proteins to regulate cytoplasmic polyadenylation and mRNA translation. This CPEB complex contains competing deadenylase (PARN) and PAP (Gld2) enzymes to regulate the length of the poly(A) tail.80 CPEB-dependent protein synthesis plays a key role in synapse formation and long-term memory persistence in sensory neuron–motor neuron cultures,81 and CPEB prion-like multimerization is associated with changes in synapse persistence.82
Many protein factors can affect the expression of their target mRNA by binding to CPEB and freeing the mRNA for polyadenylation and translation.79 For example, translation of the mRNA for tumor suppressor p53 is promoted by the interaction between CPEB and noncanonical PAP Gld483 and inhibited by overexpression of CPEB in the absence of Gld2.84 Another tumor suppressor protein, parafibromin, also exerts control over cell fate through its interaction with CPEB85 and affects the translation of multiple genes.
CPEB also recruits several protein factors of the canonical, nuclear 3′-end processing machinery, including CPSF and symplekin, for cytoplasmic polyadenylation. A recent solution structure of the C-terminal zinc-binding domain of CPEB reveals a structural similarity to ZZ-type zinc fingers, which are known to facilitate protein–protein interactions with sumoylated proteins.86 Both symplekin and CPSF are known to be sumoylated, suggesting a possible mechanism for how CPEB recognizes these factors.
Symplekin–Ssu72–Pol II CTD Complex
Symplekin is a scaffold protein and mediates interactions among many proteins in the 3′-end processing machinery. The yeast homologue, Pta1, shares very weak sequence conservation with symplekin but has generally equivalent protein partners. The N-terminal domain (NTD) of symplekin/Pta1 interacts with Ssu72, a central region with CstF-64/Pti1 (a yeast homologue of Rna15), and a C-terminal domain with CPSF-73/Ysh1.87−89
Ssu72 is a Pol II CTD phosphatase and has functions in 3′-end processing as well as gene looping,90 which helps to maintain correct transcription directionality and prevents transcription of certain noncoding RNAs from bidirectional promoters.91 The Pol II CTD contains heptapeptide repeats (26 in yeast and 52 in humans) with the Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 consensus sequence, and the phosphorylation state of the CTD regulates the function of Pol II.92−94 Ssu72 is a well-characterized pSer5 phosphatase and has recently been reported to have pSer7 phosphatase activity, as well.95,96
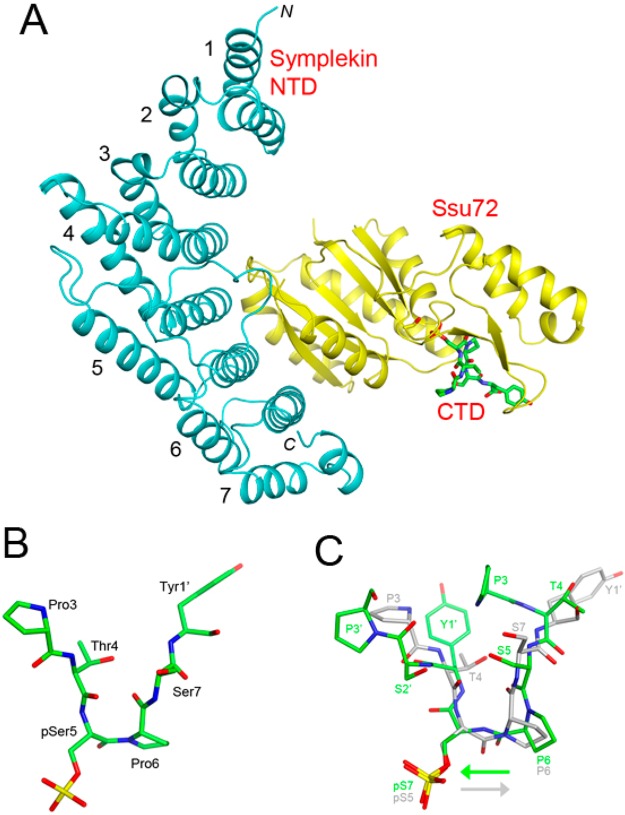
The recent crystal structure of the symplekin NTD in complex with Ssu72 and a Pol II CTD pSer5 peptide defines the detailed interactions in this ternary complex (Figure 5A).88 Although the NTD–Ssu72 interface is ∼25 Å from the active site of Ssu72, the NTD can stimulate the phosphatase activity of Ssu72, indicating that symplekin is not just a passive scaffold in the 3′-end processing machinery.
Figure 5.
Structures of human symplekin NTD–Ssu72–Pol II CTD phosphopeptide complexes. (A) Structure of the human symplekin NTD (cyan)–Ssu72 (yellow)–Pol II CTD pSer5 phosphopeptide (green) complex (PDB entry 3O2Q).88 The seven pairs of antiparallel helices are labeled. (B) Mode of binding of the Pol II CTD pSer5 peptide in the active site of Ssu72. (C) Overlay of the modes of binding of the Pol II CTD pSer7 peptide (green) and the pSer5 peptide (gray) in the active site of Ssu72 (PDB entry 4H3H).99 The directions of the polypeptide backbones are denoted by the arrows. The primed numbers indicate residues in the second CTD repeat.
The structure also reveals that the pSer5–Pro6 peptide bond of the Pol II CTD assumes the cis configuration in the active site of Ssu72 (Figure 5B), the first time that a protein phosphatase (or protein kinase) has been shown to recognize the cis configuration of the substrate. Subsequent structures of another ternary complex (with pThr4 and pSer5)97 as well as an Ssu72–phosphopeptide binary complex98 confirmed this finding. The peptidyl-prolyl isomerase Pin1 enhances the dephosphorylation activity of Ssu72.88 The symplekin NTD can regulate the function of Ssu72 in transcription-coupled 3′-end processing with the HeLa cell nuclear extract.88 Studies in yeast show that the Pta1 NTD can inhibit 3′-end processing, and binding of Ssu72 to the NTD relieves this inhibition.87
The binding mode of the pSer5 peptide in the active site of Ssu72 contrasts with its reported pSer7 phosphatase activity, as it could require that the pSer7–Tyr1 peptide bond be in the cis configuration. The crystal structure of the symplekin NTD–Ssu72–Pol II CTD pSer7 peptide ternary complex showed that the peptide, with all its amide bonds in the trans configuration, is bound in the reverse orientation compared to that of the pSer5 peptide in the active site of Ssu72 (Figure 5C).99 The substrate and the general acid for catalysis are misaligned in this complex. In vitro assays using peptide substrates indicate that the pSer7 phosphatase activity is ∼4000-fold weaker than the pSer5 phosphatase activity. On the other hand, assays with the entire CTD as the substrate, using antibodies to monitor dephosphorylation, showed no differences between the two activities. The reason behind this discrepancy is currently not known. A CTD peptide phosphorylated at Ser2, Ser5, and Ser7 is bound exclusively with pSer5 in the active site, suggesting that Ssu72 has a higher affinity for pSer5 than for pSer7.99
Replication-Dependent Histone Pre-mRNA 3′-End Processing
Metazoan replication-dependent (also known as canonical) histone proteins H1, H2A, H2B, H3, and H4 are involved in de novo chromatin packaging during DNA replication, while variant histones, notably, H3.3, H2A.Z, CENP-A, macroH2A, and H1.0, are important for chromatin remodeling, centromere function, and epigenetic silencing.100−102 Variant histone pre-mRNAs carry introns and are cleaved and polyadenylated. In comparison, the replication-dependent histone pre-mRNAs do not have introns and are cleaved but not polyadenylated. Their 3′-end processing is conducted by a different machinery, which will be the focus of this section.
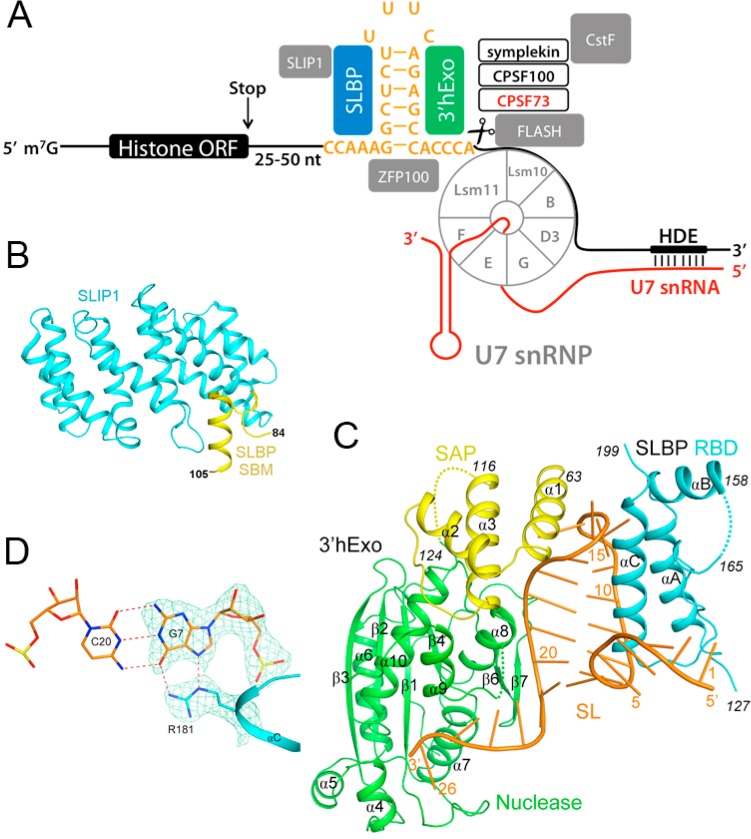
The 3′-UTRs of replication-dependent histone pre-mRNAs contain two signature cis-acting elements: a highly conserved stem–loop (SL) 25–50 nucleotides downstream of the open reading frame and a purine-rich segment further downstream (15–20 nucleotides in vertebrates) named the histone downstream element (HDE) (Figure 6A). The SL is recognized by the stem–loop-binding protein (SLBP, also known as the hairpin binding protein, HBP), which has a central role in replication-dependent histone mRNA processing and function. The SL is also bound by a 3′–5′ exoribonuclease, known as 3′hExo or Eri-1, which has an important role in histone mRNA degradation, although Drosophila lacks this nuclease. The HDE recruits the U7 snRNP, another important component for histone pre-mRNA 3′-end processing. These various factors will be described in more detail below.
Figure 6.
Structural information about histone pre-mRNA 3′-end processing. (A) Schematic drawing of the replication-dependent histone pre-mRNA 3′-end processing machinery. (B) Structure of Danio rerio SLIP1 bound to the SLIP1-binding motif (SBM) of SLBP (PDB entry 4JHK).106 (C) Structure of the human SLBP RNA binding domain (RBD)–3′hExo–stem-loop RNA complex (PDB entry 4L8R).112 (D) Specific recognition of the second guanine in the stem by Arg181 of SLBP.
SL, SLBP, and 3′hExo
The SL has a 6 bp stem and a four-nucleotide loop (Figure 6A). A G-C base pair at the second position of the stem is strictly conserved, while the loop is generally rich in pyrimidines.103 Systematic as well as focused studies on the effects of SL mutations on binding to SLBP are consistent with sequence conservation.102,104
SLBP is found in all metazoans and is a 31 kDa protein in humans.102 A highly conserved 70-residue RNA-binding domain (RBD) near the center of SLBP binds tightly to the SL, with a dissociation constant (Kd) of ∼10 nM. Phosphorylation of Thr171 in the RBD enhances the affinity, reducing the Kd by ∼7-fold. Immediately C-terminal to the RBD is a 20-residue segment that is dispensable for RNA binding but is required for efficient 3′-end processing. The N-terminal region of SLBP binds SLBP interaction protein 1 (SLIP1) (Figure 6B), which interacts with eukaryotic translation initiation factor eIF-4G and is essential for promoting the translation of histone mRNAs.105,106 Phosphorylation of several residues in this region is correlated with polyubiquitination and rapid breakdown of SLBP at the end of the S phase.107,108
3′hExo belongs to the DEDD family of 3′–5′ exonucleases and prefers single-stranded RNA as the substrate. The activity of 3′hExo requires two Mg2+ cations coordinated by four invariant acidic residues (DEDD) in its active site. In addition to the nuclease domain, 3′hExo also contains an N-terminal SAP domain, previously characterized as a nucleic acid binding motif.109 3′hExo trims up to two nucleotides from the 3′-end of histone mRNAs after cleavage, and it also participates in the rapid decay of histone mRNAs at the end of the S phase.110 In addition to its roles in histone mRNA metabolism, 3′hExo is critical for trimming the 3′-end of the 5.8S rRNAs111 and regulating microRNA homeostasis.
The recently published crystal structure of the human SLBP RBD–3′hExo–SL ternary complex provided the first molecular insights into the architecture of this complex (Figure 6B).112 The stem adopts the conformation of A-form RNA, and three of the four bases of the loop are flipped out. The SLBP RBD interacts with the 5′-flanking sequence, the 5′-arm of the stem, and the loop of SL. The SAP domain of 3′hExo interacts with the loop, and the nuclease domain with the 3′-arm and flanking sequence. In particular, the last nucleotide of the SL is located in the active site of the nuclease domain, explaining how 3′hExo can trim the last two nucleotides of the histone mRNA after cleavage.
The observed binding mode is consistent with most of the mutagenesis data on this complex. Only the guanine base in the second base pair of the stem is specifically recognized, by the side chain of Arg181 in SLBP (Figure 6D), which explains why this nucleotide is invariant in all metazoans. The structure indicates that SLBP and 3′hExo primarily recognize the shape, rather than the sequence, of the SL.
There are no direct contacts between the SLBP RBD and 3′hExo in the ternary complex (Figure 6C). The cooperative binding between the two proteins observed earlier likely results from the induced-fit behavior of the SL, as there are large conformational differences between the SL in the complex versus that free in solution.112 Therefore, binding of one protein induces a conformation of the SL that promotes the binding of the other protein.
HDE and U7 snRNP
HDE recruits the U7 snRNP through base pairing with the 5′-extension of U7 snRNA.101 The heptameric Sm ring of U7 snRNP contains Sm proteins B, D3, E, F, and G that are found in spliceosomal snRNPs as well as two unique subunits, Sm-like proteins Lsm10 and Lsm11. Lsm11 is a 40 kDa protein in humans, which is substantially larger than the typical Sm protein (∼13 kDa). It has a unique N-terminal segment that is mostly unstructured but is essential for histone pre-mRNA processing via the recruitment of other processing factors, including the zinc finger protein ZFP100 and FLASH (see below).
Other Processing Factors
Similar to polyadenylated mRNAs, CPSF-73 is the endoribonuclease for the cleavage reaction of replication-dependent histone pre-mRNAs.102 The cleavage site is also typically located after a CA dinucleotide, located five (in vertebrates) or four (in fruit fly and sea urchin) nucleotides downstream of the stem.103 The 5′-end-capped and 3′-end-cleaved histone mRNA, accompanied by SLBP and 3′hExo, is then exported into the cytoplasm by the antigen peptide transporter.
Besides CPSF-73, several other protein factors in the canonical 3′-end processing machinery are also important for histone pre-mRNA 3′-end processing, including CPSF-100, symplekin, CstF-64, and CstF-77.113,114 These proteins form the heat labile factor (HLF), discovered in the 1980s as an essential component of the histone pre-mRNA 3′-end processing machinery,115 and are recruited to the machinery through FLASH.
FLASH (FLICE-associated huge protein) was initially identified as a 220 kDa proapoptotic protein that is part of the death-inducing signaling complex (DISC).116 Only a small segment of ∼140 residues at the N-terminus of FLASH, especially a Leu-Asp-Leu-Tyr motif, is required for recruiting the various protein factors for histone pre-mRNA processing.117−121 This segment also has tight interactions with Lsm11, which brings FLASH to the 3′-end of histone pre-mRNAs.
The central region of FLASH recruits arsenite resistance protein 2 (ARS2).122 ARS2 directly binds to histone mRNAs and interacts with the nuclear cap-binding complex (CBC).123 The CBC–ARS2 complex can stimulate the 3′-end processing of histone mRNAs, presumably through CBC’s interactions with the negative elongation factor (NELF) and SLBP.123−125
The 100 kDa zinc finger protein (ZFP100) interacts with both SLBP and U7 snRNP and is crucial for efficient 3′-end processing.126 It contains a poorly conserved N-terminal domain and a C-terminal domain that is comprised of 18 C2H2-type zinc fingers. Overexpression of ZFP100 greatly enhances the 3′-end processing of a reporter RNA that mimics histone pre-mRNAs, while overexpression of the components of the U7 snRNP alone does not, indicating that ZFP100 is the limiting factor for histone pre-mRNA processing. The primary role of ZFP100 is probably to bridge the SLBP–SL complex with the U7 snRNP–HDE complex, thereby stabilizing the overall processing machinery.
Phosphorylation of Thr4 in the Pol II CTD is required for histone 3′-end processing.127 It is also required for Pol II transcription elongation.128 The kinase that phosphorylates Thr4 is CDK9, which also targets NELF.127,129 CDK9 is recruited to histone genes by the nuclear protein ataxia-telangiectasia locus (NPAT), the expression of which is negatively regulated by p53.130 It remains unclear how the Pol II CTD communicates with the integral components of the histone pre-mRNA 3′-end processing machinery.
Cell Cycle Regulation of Histone Pre-mRNA 3′-End Processing
The demand for histones during the S phase of the cell cycle is enormous, and an estimated 108 molecules of each of the five histones are synthesized within a period of several hours.131 Levels of histone mRNA increase by 35-fold at the beginning of the S phase, through transcription activation (3.5-fold increase) and enhanced pre-mRNA processing (10-fold increase). At the conclusion of the S phase, or through inhibition of DNA synthesis in the S phase, levels of histone mRNA are reduced back to the G1 phase baseline.
SLBP is a key factor in the regulation of histone mRNA levels, and the level of this protein correlates with those of histone mRNAs during the cell cycle. In late G1 phase, inhibition of protein degradation and activation of translation synergistically increase the level of SLBP.132 At the end of the S phase, several residues in the N-terminal segment of SLBP (including Ser20, Ser23, Thr60, and Thr61) are phosphorylated, which facilitates its polyubiquitination and rapid clearance by the ubiquitin–proteasome system.107,108 Pin1 may also have a role in facilitating the phosphorylation of Ser20 and Ser23, and the dephosphorylation of Thr171.108
On the other hand, artificial inhibition of DNA replication in the S phase has little effect on the cellular levels of SLBP, indicating the existence of other mechanism(s) for regulating histone levels.
A Novel Quality Surveillance Mechanism for mRNA 5′-End Capping
The 5′-end 7-methylguanosine (m7G) cap is a significant contributor to mRNA splicing, nuclear export, translation, stability, and other processes.133,134 The cap is added cotranscriptionally and attached to the terminal nucleotide of the RNA by an unusual 5′–5′ triphosphate linkage. Capping proceeds in three steps: conversion of 5′-end-triphosphorylated RNA (pppRNA, the primary transcript of Pol II) to diphosphorylated RNA (ppRNA), coupling to GMP to produce capped RNA (GpppRNA), and methylation to produce m7GpppRNA (Figure 7). A mature, methylated cap is essential for recognition by the cap-binding complex, CBC and eIF-4E, which coordinate many of the functions attributed to the cap.135,136
Figure 7.
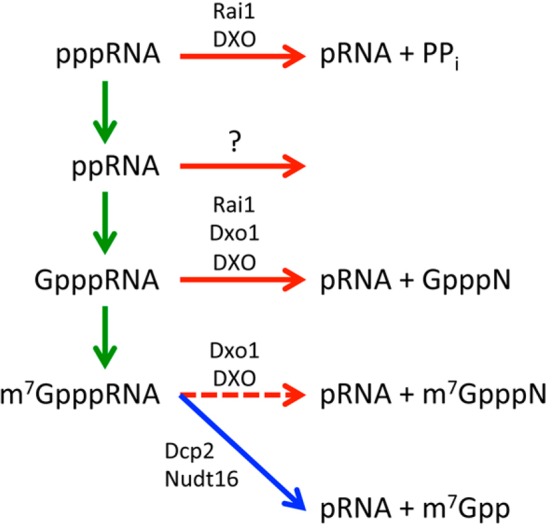
Reactions for pre-mRNA 5′-end capping and quality control. Reactions in the capping pathway are denoted by the green arrows. The intermediates in the capping pathway are recognized by the DXO family enzymes (Rai1, Dxo1, and DXO) for degradation (red arrows). The fate of the ppRNA is currently not known, although it may be possible that Rai1 and DXO also mediate its degradation. The reaction catalyzed by the classical decapping enzymes (Dcp2 and Nudt16) is denoted by the blue arrow. DXO and Dxo1 can also remove the mature cap but generate a different product (dashed red arrow).
Removal of the cap (decapping) is a regulated process catalyzed by at least two Nudix hydrolase enzymes, Dcp2 and Nudt16,137,138 which release m7GDP (m7Gpp) and 5′-end-monophosphorylated RNA (pRNA) (Figure 7). Six additional Nudix proteins possessing decapping activity in vitro have also been reported,139 although the functional role of these putative decapping enzymes in cells remains to be determined.
Until recently, it was generally accepted in the field that capping always proceeds to completion, and a quality control mechanism was not known (or deemed necessary). However, if there are defects in 5′-end capping, the intermediates of the capping pathway (pppRNA, ppRNA, and GpppRNA) could accumulate in cells, because Dcp2 and Nudt16 predominantly function on mature m7GpppRNA and have minimal activity on these intermediates.137 They are also protected against degradation by 5′–3′ exoribonucleases (XRNs), which are only active against pRNA substrates.
A novel family of enzymes that possess RNA 5′-end pyrophosphohydrolase (PPH, releasing pyrophosphate PPi), decapping, and/or distributive 5′–3′ exoribonuclease activity was recently discovered. These enzymes include Rai1140,141 and Ydr370C/Dxo1142 in yeast and Dom3Z/DXO in mammals.143 These decapping exonucleases (DXO family of enzymes) primarily act on incompletely capped mRNAs, converting them to substrates for degradation by XRNs or their own exonuclease activity (Figure 7). These biochemical activities are strongly suggestive of a hitherto unrecognized mRNA 5′-end capping quality surveillance mechanism, helping to clear transcripts with incompletely capped 5′-ends. Functional studies in yeast and mammalian cells have confirmed the presence of defects in 5′-end capping and demonstrated the importance of the DXO family enzymes in this quality control mechanism.
Biochemical Properties of the DXO Family Enzymes
The DXO activities were first identified, unexpectedly, from studies on yeast Rai1, the protein partner of the nuclear 5′–3′ exoribonuclease Rat1.140 The crystal structure of Rai1 revealed a large pocket lined with conserved residues, some of which coordinate a divalent metal ion at the bottom of the pocket. This indicated that Rai1 is an enzyme, although no catalytic activities were known for it at the time. Further studies demonstrated that Rai1 has PPH activity140 and decapping activity toward GpppRNA, while it has much weaker activity toward the mature m7GpppRNA.141 Moreover, the product of decapping is GpppN, the entire cap structure, in contrast to Dcp2 and Nudt16, which release m7GDP (m7Gpp) (Figure 7).
Rai1 has a weak sequence homologue in yeast, Dxo1 (Ydr370C). Biochemical studies showed that it has decapping activity (toward both GpppRNA and m7GpppRNA) as well as a distributive 5′–3′ exoribonuclease activity, although it lacks PPH activity.142
The mammalian homologue of Rai1, DXO (previously known as Dom3Z), has all three activities, PPH, decapping (toward both GpppRNA and m7GpppRNA), and exoribonuclease.143 These activities would allow DXO to single-handedly detect and degrade incompletely capped mRNAs. m7GpppRNAs are protected from DXO degradation by cap-binding proteins in vivo, indicating that mature mRNAs are insensitive to DXO. Therefore, DXO is expected to function preferentially on incompletely capped pre-mRNAs.
Structural Basis for the DXO Activities
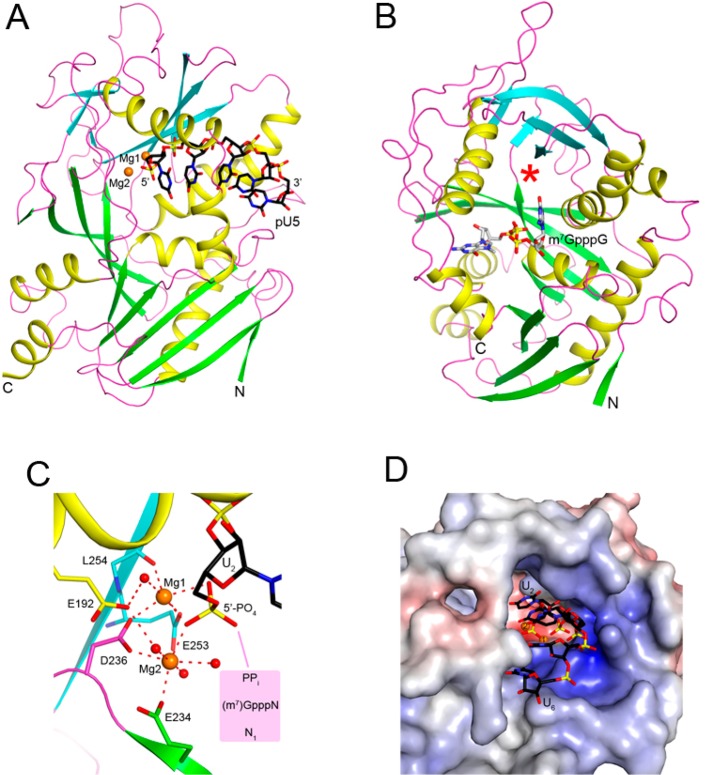
Biochemical studies showed that mammalian DXO possesses three, apparently distinct, catalytic activities: PPH, decapping, and exonuclease. On the other hand, the RNA body produced by these activities is the same, 5′-end-monophosphorylated RNA (pRNA). Crystal structures of mouse DXO in complex with 5′-end-monophosphorylated RNA oligos, 5-mer RNA (pU5) (Figure 8A), and 6-mer RNA with phosphorothioate linkages to inhibit hydrolysis [pU(S)6] as well as the m7GpppG cap analogue (Figure 8B) have defined the binding modes of the RNA substrate/product and revealed the molecular mechanism for the different activities.143
Figure 8.
Molecular mechanism for the catalytic activities of DXO family enzymes. (A) Structure of mouse DXO in complex with pU5 oligo RNA (black stick models) (PDB entry 4J7L).143 The two Mg2+ ions are shown as orange spheres. (B) Structure of mouse DXO in complex with the m7GpppG cap analogue (gray sticks) (PDB entry 4J7N). The expected location of the metal ions is indicated by the red star. The view is related to that of panel A by an ∼60° rotation around the vertical axis. (C) Binding mode of the 5′-end phosphate group of pU5. This RNA is bound in the active site as the product. Binding of the pyrophosphate (PPi), the cap structure [(m7)GpppN], or the first nucleotide (N1) on the other side of the catalytic machinery explains the three catalytic activities. (D) The active site of DXO is located at the bottom of a deep pocket, which is large enough to accommodate only ssRNA.
The pU5 oligo is bound in the DXO active site as a product, with its 5′-end phosphate group mimicking the scissile phosphate of the substrate. A second metal ion is bound in the active site in the presence of this oligo, and a terminal oxygen atom of the 5′-end phosphate group is a bridging ligand to both metal ions (Figure 8C). The pU(S)6 oligo is bound in the active site as a substrate, revealing the recognition pocket for the first nucleotide (especially its 5′-phosphate) for the 5′–3′ exonuclease activity. However, there are disruptions to the conformation of this oligo at the scissile bond caused by the incorporation of the phosphorothioate linkages and the fact that only one metal ion (Ca2+, to prevent hydrolysis) is present in the active site.
The structures demonstrate that the same active site machinery supports the three activities, and it is the distinct binding modes of the substrates that determine the outcome of the reaction. The 5′-end PPi pyrophosphate, (m7)GpppN cap, or the first nucleotide is bound on the other side of the catalytic machinery from the RNA body (Figure 8C). An attack on the scissile phosphate group, likely by a water/hydroxide coordinated to one of the metal ions, then leads to the hydrolysis.
At the same time, different DXO family enzymes have distinct biochemical activity profiles. For example, Rai1 has PPH and GpppRNA decapping activities but no exonuclease activity, while DXO has all the activities (Figure 7). Further studies are needed to elucidate the molecular mechanisms of these differences.
The DXO family enzymes share four conserved sequence motifs.142 Motif I is an Arg residue and recognizes the 5′-phosphate group of the pRNA substrate and pppRNA. Motif II, GΦXΦE (where Φ is an aromatic or hydrophobic residue and X any residue), provides a ligand to the metal ion [Glu192 in mouse DXO (Figure 8C)]. Motif III, EhD (where h is a hydrophobic residue), is ligated to both metal ions in the pU5 complex [Glu234 and Asp236 (Figure 8C)]. Motif IV, EhK, provides a ligand to the metal ion (Glu253), and the Lys residue is likely to stabilize the transition state of the reaction.
The structures of the DXO family enzymes have a remote relationship to that of D-(D/E)XK nucleases,142,144,145 which include some viral and phage nucleases. However, there is little sequence conservation with these enzymes, and only the Asp residue of motif III (EhD) and motif IV (EhK) [the D-(D/E)XK motif] are shared among them. The level of structural conservation outside of these two motifs is much lower among these enzymes. The D-(D/E)XK enzymes also include some type II restriction endonucleases, such as HincII, EcoRV, EcoRI, BamHI, and BglI,142 but the level of structural conservation with the DXO family enzymes is much lower. For example, the active site of the DXO family enzymes is located at the bottom of a deep pocket (Figure 8D), which is consistent with their exonuclease, decapping, and PPH activity. In comparison, the active site of the type II enzymes is much more open, in line with their endonuclease activity.
Functions of DXO Family Enzymes in 5′-End Capping Quality Control
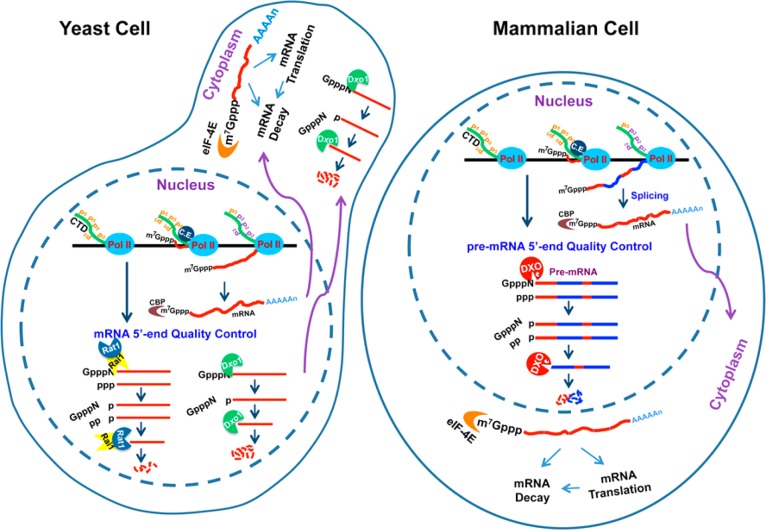
Functional studies in yeast cells harboring a deletion of Rai1 and/or Dxo1 reveal a role for these proteins in ensuring the integrity of mRNA 5′-end caps. Incompletely capped mRNAs were observed in rai1Δ cells following nutritional stress (glucose or amino acid starvation), suggesting that Rai1 is necessary for their detection and degradation.141 Moreover, incompletely capped mRNAs are detected under normal, nonstress growth conditions in rai1Δdxo1Δ doubly disrupted yeast strains.142 Collectively, these findings demonstrate that incompletely capped transcripts are normally generated in yeast cells (Figure 9), providing direct evidence that the capping process is less efficient than initially envisioned.
Figure 9.
DXO family enzymes function in 5′-end capping quality control. In eukaryotes, pre-mRNAs are transcribed in the nucleus by Pol II and processed into mature mRNAs by the addition of a 5′-end cap, intron splicing, and 3′-end cleavage and polyadenylation. The mature mRNAs are exported to the cytoplasm for protein translation. In yeast cells, incompletely capped Pol II RNA transcripts are subjected to degradation by the Rai1–Rat1 decapping–exonuclease heterodimer, which detects and degrades 5′-end uncapped RNA or 5′-end unmethylated capped RNA in the nucleus. Dxo1, which is predominantly but not exclusively in the cytoplasm, decaps and degrades unmethylated capped Pol II transcripts. In mammalian cells, the incompletely capped Pol II RNA transcripts are substrates for DXO, which decaps and exonucleolytically degrades the defectively capped pre-mRNA prior to further splicing and 3′-end processing. Collectively, the DXO enzymes can hydrolyze the 5′-end of incompletely capped RNAs to expose the 5′-end of the RNA to subsequent exonucleolytic decay (by Dxo1 and DXO directly or by the Rai1–Rat1 heterodimer) in a 5′-end capping quality control mechanism to maintain RNA fidelity. CE is the capping enzyme and CBP the cap-binding protein.
In addition, the fact that a double disruption of Rai1 and Dxo1, but not individual disruptions, is required for the accumulation of incompletely capped mRNAs during nonstress conditions indicates that the Rai1 and Dxo1 proteins function redundantly in the surveillance mechanism to detect and degrade incompletely capped transcripts. On the other hand, Dxo1 cannot complement the loss of Rai1 under nutritional stress conditions. It remains to be determined whether the incompletely capped transcripts are generated as a consequence of intrinsic stochastic inefficiency of the capping process or an indication that capping is normally a regulated process in which not all primary transcripts are destined to acquire a methylated cap.
Functional studies in human embryonic kidney 293T cells confirm the importance of DXO in ensuring mRNA 5′-end capping quality.143 A decrease in the DXO level in these cells through shRNA knockdown results in a significant accumulation of unprocessed pre-mRNAs (with splicing and polyadenylation defects) with minimal changes in mature mRNA levels. These unprocessed pre-mRNAs harbor incompletely capped 5′-ends, while the mature mRNAs contain an m7G cap. These data indicate that incompletely capped pre-mRNAs do not undergo further processing (splicing or polyadenylation), while the normally capped pre-mRNAs are licensed to undergo processing (Figure 9).
While earlier studies had demonstrated a link between capping quality and splicing of the first intron, the accumulated pre-mRNAs in DXO knockdown cells show retention of all the introns tested, irrespective of their positions.143 Therefore, the data suggest that incompletely capped pre-mRNAs are inefficiently spliced at all introns. These pre-mRNAs also have compromised 3′-end processing, consistent with earlier reports that the cleavage step is facilitated by the 5′-end cap.
The reported findings demonstrate that incompletely capped transcripts are generated in mammalian cells and define a novel link between capping and pre-mRNA processing. They also indicate that the capping process may function as a critical checkpoint that determines whether a pre-mRNA should be further processed (Figure 9). DXO serves as a surveillance protein in a 5′-end capping quality control mechanism to clear incompletely capped pre-mRNAs.
Future Perspectives
Structural, biochemical, and functional studies over the past few years have provided great new insights into pre-mRNA 3′-end processing in eukaryotes, and we are gaining a better understanding of the molecular mechanisms for the functions of various proteins in the 3′-end processing machineries in yeast and humans. However, this is still a burgeoning field of research, and there is much to learn about the architecture of these machineries and the regulation of their cellular functions, for example, in APA. It is also important to understand how the core machineries can acquire post-translational modifications and additional protein factors in response to specific cellular conditions or localizations. Further characterizations of the molecular mechanism and cellular functions of cytoplasmic polyadenylation will be another important area for research.
The discovery of the 5′-end capping quality surveillance mechanism has opened up a new field of research. For the first time, it is apparent that the capping step does not always proceed to completion. An important unanswered question is whether this is simply a consequence of the intrinsic inefficiency of the capping process or a regulated event to modulate subsequent pre-mRNA processing by controlling cap addition. If the latter is true, what are the components involved and how are the decisions about which pre-mRNAs are capped made? Regardless of how incompletely capped transcripts are generated, important future studies in this area also include defining the genomewide and cellular impacts of 5′-end capping quality surveillance, as well as the molecular mechanism of how the authenticity of the 5′-end cap influences pre-mRNA splicing and polyadenylation.
The authors declare no competing financial interest.
This research is supported by grants from the National Institutes of Health (NIH) to L.T. (GM077175 and GM090059) and M.K. (GM067005). A.R.J. was also supported by the NIH training program in Cellular and Molecular Foundations of Biomedical Science (GM008798).
Funding Statement
National Institutes of Health, United States
References
- Zhao J.; Hyman L.; Moore C. L. (1999) Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63, 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel C. R.; Bai Y.; Tong L. (2008) Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 65, 1099–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J.; Proudfoot N. J. (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700. [DOI] [PubMed] [Google Scholar]
- Millevoi S.; Vagner S. (2010) Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 38, 2757–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D. D.; Darnell R. B. (2010) RNA processing and its regulation: Global insights into biological networks. Nat. Rev. Genet. 11, 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.; Doublie S. (2011) Structural biology of poly(A) site definition. Wiley Interdiscip. Rev.: RNA 2, 732–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.; Choi E. A.; Shi Y. (2011) pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip. Rev.: RNA 2, 321–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. E. Jr. (2013) Reflection of the history of pre-mRNA processing and highlights of current knowledge: A united picture. RNA 19, 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C. S.; Moreira A. (2011) Alternative mRNA polyadenylation in eukaryotes: An effective regulator of gene expression. Wiley Interdiscip. Rev.: RNA 2, 22–31. [DOI] [PubMed] [Google Scholar]
- Shi Y. (2012) Alternative polyadenylation: New insights from global analyses. RNA 18, 2105–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B.; Manley J. L. (2013) Alternative cleavage and polyadenylation: The long and short of it. Trends Biochem. Sci. 38, 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann C. R.; Rammelt C.; Wahle E. (2011) Control of poly(A) tail length. Wiley Interdiscip. Rev.: RNA 2, 348–361. [DOI] [PubMed] [Google Scholar]
- Weill L.; Belloc E.; Bava F. A.; Mendez R. (2012) Translational control by changes in poly(A) tail length: Recycling mRNAs. Nat. Struct. Mol. Biol. 19, 577–585. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J. (2011) Ending the message: Poly(A) signals then and now. Genes Dev. 25, 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevher M. A.; Kleiman F. E. (2010) Connections between 3′-end processing and DNA damage response. Wiley Interdiscip. Rev.: RNA 1, 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; di Giammartino D. C.; Taylor D.; Sarkeshik A.; Rice W. J.; Yates J. R. III; Frank J.; Manley J. L. (2009) Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel C. R.; Kaneko S.; Zhang H.; Gebauer D.; Vethantham V.; Manley J. L.; Tong L. (2006) Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444, 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C. (2010) What is the role of RNase J in mRNA turnover?. RNA Biol. 7, 316–321. [DOI] [PubMed] [Google Scholar]
- Dominski Z.; Carpousis A. J.; Clouet-d’Orval B. (2013) Emergence of the b-CASP ribonucleases: Highly conserved and ubiquitous metallo-enzymes involved in messenger RNA maturation and degradation. Biochim. Biophys. Acta 1829, 532–551. [DOI] [PubMed] [Google Scholar]
- Nishida Y.; Ishikawa H.; Baba S.; Nakagawa N.; Kuramitsu S.; Masui R. (2010) Crystal structure of an archael cleavage and polyadenylation specificity factor subunit from Pyrococcus horikoshii. Proteins 78, 2395–2398. [DOI] [PubMed] [Google Scholar]
- Mir-Montazeri B.; Ammelburg M.; Forouzan D.; Lupas A. N.; Hartmann M. D. (2011) Crystal structure of a dimeric archaeal cleavage and polyadenylation specificity factor. J. Struct. Biol. 173, 191–195. [DOI] [PubMed] [Google Scholar]
- Silva A. P.; Chechik M.; Byrne R. T.; Waterman D. G.; Ng C. L.; Dodson E. J.; Koonin E. V.; Antson A. A.; Smits C. (2011) Structure and activity of a novel archael β-CASP protein with N-terminal KH domains. Structure 19, 622–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke G.; Ezeokonkwo C.; Balbo P. B.; Stafford W.; Moore C.; Bohm A. (2008) Structure of yeast poly(A) polymerase in complex with a peptide from Fip1, an intrinsically disordered protein. Biochemistry 47, 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezeokonkwo C.; Zhelkovsky A. M.; Lee R.; Bohm A.; Moore C. L. (2011) A flexible linker region in Fip1 is needed for efficient mRNA polyadenylation. RNA 17, 652–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twu K. Y.; Noah D. L.; Rao P.; Kuo R. L.; Krug R. M. (2006) The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J. Virol. 80, 3957–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale B. G.; Randall R. E.; Ortin J.; Jackson D. (2008) The multifunctional NS1 protein of influenza viruses. J. Gen. Virol. 89, 2359–2376. [DOI] [PubMed] [Google Scholar]
- Ramos I.; Carnero E.; Bernal-Rubio D.; Seibert C. W.; Westera L.; Garcia-Sastre A.; Fernandez-Sesma A. (2013) Contribution of double-stranded RNA and CPSF30 binding domains of influenza virus NS1 to the inhibition of type I interferon production and activation of human dendritic cells. J. Virol. 87, 2430–2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo R. L.; Krug R. M. (2009) Influenza A virus polymerase is an integral component of the CPSF30-NS1A protein complex in infected cells. J. Virol. 83, 1611–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das K.; Ma L. C.; Xiao R.; Radvansky B.; Aramini J.; Zhao L.; Marklund J.; Kuo R. L.; Twu K. Y.; Arnold E.; Krug R. M.; Montelione G. T. (2008) Structural basis for suppression of a host antiviral response by influenza virus. Proc. Natl. Acad. Sci. U.S.A. 105, 13093–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. E.; Wu X.; Liu M.; Gaffney B.; Ji G.; Li Q. Q.; Hunt A. G. (2012) Genome-wide control of polyadenylation site choice by CPSF30 in Arabidopsis. Plant Cell 24, 4376–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addepalli B.; Hunt A. G. (2007) A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-kD subunit of cleavage and polyadenylation specificity factor. Nucleic Acids Res. 35, 4453–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Addepalli B.; Yun K. Y.; Hunt A. G.; Xu R.; Rao S.; Li Q. Q.; Falcone D. L. (2008) A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS One 3, e2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addepalli B.; Limbach P. A.; Hunt A. G. (2010) A disulfide linkage in a CCCH zinc finger motif of an Arabidopsis CPSF30 ortholog. FEBS Lett. 584, 4408–4412. [DOI] [PubMed] [Google Scholar]
- Rao S.; Dinkins R. D.; Hunt A. G. (2009) Distinctive interactions of the Arabidopsis homolog of the 30 kD subunit of the cleavage and polyadenylation specificity factor (AtCPSF30) with other polyadenylation factor subunits. BMC Cell Biol. 10, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y.; Auperin T. C.; Chou C.-Y.; Chang G.-G.; Manley J. L.; Tong L. (2007) Crystal structure of murine CstF-77: Dimeric association and implications for polyadenylation of mRNA precursors. Mol. Cell 25, 863–875. [DOI] [PubMed] [Google Scholar]
- Legrand P.; Pinaud N.; Minvielle-Sebastia L.; Fribourg S. (2007) The structure of CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res. 35, 4515–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson A. R.; Tong L. (2012) Crystal structure of the Rna14-Rna15 complex. RNA 18, 1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. M. B.; Shikov S.; Kuehner J. N.; Liriano M.; Lee E.; Stafford W.; Poulsen M. B.; Harrison C.; Moore C.; Bohm A. (2011) Reconstitution of CF IA from overexpressed subunits reveals stoichiometry and provides insights into molecular topology. Biochemistry 50, 10203–10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Morcillo M.; Minvielle-Sebastia L.; Mackereth C.; Fribourg S. (2011) Hexameric architecture of CstF supported by CstF-50 homodimerization domain structure. RNA 17, 412–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Morcillo M.; Minvielle-Sebastia L.; Fribourg S.; Mackereth C. D. (2011) Locked tether formation by cooperative folding of Rna14p monkeytail and Rna15p hinge domains in the yeast CFIA complex. Structure 19, 534–545. [DOI] [PubMed] [Google Scholar]
- Hockert J. A.; Yeh H. J.; MacDonald C. C. (2010) The hinge domain of the cleavage stimulation factor protein CstF-64 is essential for CstF-77 interaction, nuclear localization, and polyadenylation. J. Biol. Chem. 285, 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W.; Ji Z.; Pan Z.; You B.; Hoque M.; Li W.; Gunderson S. I.; Tian B. (2013) The conserved intronic cleavage and polyadenylation site of CstF-77 gene imparts control of 3′ end processing activity through feedback autoregulation and by U1 snRNP. PLoS Genet. 9, e1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeper T. C.; Qu X.; Lu C.; Moore C.; Varani G. (2010) Novel protein-protein contacts faciliate mRNA 3′-processing signal recognition by Rna15 and Hrp1. J. Mol. Biol. 401, 334–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancevac C.; Goldstone D. C.; Ramos A.; Taylor I. A. (2010) Structure of the Rna15 RRM-RNA complex reveals the molecular basis of GU specificity in transcriptional 3′-end processing factors. Nucleic Acids Res. 38, 3119–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwal R. P.; Lee S. D.; Moore C.; Varani G. (2012) Structural and functional analysis of the assembly and function of the yeast pre-mRNA 3′ end processing complex CF I. Proc. Natl. Acad. Sci. U.S.A. 109, 21342–21347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T.; Nagata T.; Tsuda K.; Kobayashi N.; Imai T.; Okano H.; Yamazaki T.; Katahira M. (2012) Structure of Musashi1 in a complex with target RNA: The role of aromatic stacking interactions. Nucleic Acids Res. 40, 3218–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao C.; Choi E. A.; Weng L.; Xie X.; Wan J.; Xing Y.; Moresco J. J.; Tu P. G.; Yates J. R. III; Shi Y. (2013) Overlapping and distinct functions of CstF64 and CstF64tau in mammalian mRNA 3′ processing. RNA 19, 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarling G. S.; MacDonald C. C. (2013) Polyadenylation site-specific differences in the activity of the neuronal bCstF-64 protein in PC-12 cells. Gene 529, 220–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.; Gruber A. R.; Keller W.; Zavolan M. (2012) Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Rep. 1, 753–763. [DOI] [PubMed] [Google Scholar]
- Coseno M.; Martin G.; Berger C.; Gilmartin G. M.; Keller W.; Doublie S. (2008) Crystal structure of the 25 kDa subunit of human cleavage factor Im. Nucleic Acids Res. 36, 3474–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresaugues L.; Stenmark P.; Schuler H.; Flodin S.; Welin M.; Nyman T.; Hammarstrom M.; Moche M.; Graslund S.; Nordlund P. (2008) The crystal structure of human cleavage and polyadenylation specific factor-5 reveals a dimeric Nudix protein with a conserved catalytic site. Proteins 73, 1047–1052. [DOI] [PubMed] [Google Scholar]
- Yang Q.; Gilmartin G. M.; Doublie S. (2010) Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc. Natl. Acad. Sci. U.S.A. 107, 10062–10067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.; Coseno M.; Gilmartin G. M.; Doublie S. (2011) Crystal structure of a human cleavage factor CFI(m)25/CFI(m)68/RNA complex provides an insight into poly(A) site recognition and RNA looping. Structure 19, 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Tong S.; Li X.; Shi H.; Ying Z.; Gao Y.; Ge H.; Niu L.; Teng M. (2011) Structural basis of pre-mRNA recognition by the human cleavage factor Im complex. Cell Res. 21, 1039–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Yamamoto J.; Chen Y.; Aida M.; Wada T.; Handa H.; Yamaguchi H. (2010) Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation. Genes Cells 15, 1003–1013. [DOI] [PubMed] [Google Scholar]
- Gruber A. R.; Martin G.; Keller W.; Zavolan M. (2012) Cleavage factor Im is a key regulator of 3′ UTR length. RNA Biol. 9, 1405–1412. [DOI] [PubMed] [Google Scholar]
- Ruepp M. D.; Schumperli D.; Barabino S. M. L. (2011) mRNA 3′ end processing and more-multiple functions of mammalian cleavage factor I-68. Wiley Interdiscip. Rev.: RNA 2, 79–91. [DOI] [PubMed] [Google Scholar]
- Ruepp M. D.; Aringhieri C.; Vivarelli S.; Cardinale S.; Paro S.; Schumperli D.; Barabino S. M. L. (2009) Mammalian pre-mRNA 3′ end processing factor CF I m 68 functions in mRNA export. Mol. Biol. Cell 20, 5211–5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J.; Okuzaki D.; Inoue H.; Yoneda Y.; Maehara K.; Ohkawa Y. (2013) Human TREX component Thoc5 affects alternative polyadenylation site choice by recruiting mammalian cleavage factor I. Nucleic Acids Res. 41, 7060–7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. E.; Ambrose Z.; Martin T. D.; Oztop I.; Mulky A.; Julias J. G.; Vandegraaff N.; Baumann J. G.; Wang R.; Yuen W.; Takemura T.; Shelton K.; Taniuchi I.; Li Y.; Sodroski J.; Littman D. R.; Coffin J. M.; Hughes S. H.; Unutmaz D.; Engelman A.; KewalRamani V. N. (2010) Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasaiyaah J.; Tan C. P.; Fletcher A. J.; Price A. J.; Blondeau C.; Hilditch L.; Jacques D. A.; Selwood D. L.; James L. C.; Noursadeghi M.; Towers G. J. (2013) HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503, 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T.; Takeuchi H.; Saito H.; Sakuma R.; Inagaki Y.; Yamaoka S. (2013) A carboxy-terminally truncated human CPSF6 lacking residues encoded by exon 6 inhibits HIV-1 cDNA synthesis and promotes capsid disassembly. J. Virol. 87, 7726–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble C. G.; Beuth B.; Taylor I. A. (2007) Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res. 35, 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad R.; Maurice F.; Viphakone N.; Voisinet-Hakil F.; Fribourg S.; Minvielle-Sebastia L. (2012) An essential role for Clp1 in assembly of polyadenylation complex CF IA and Pol II transcription termination. Nucleic Acids Res. 40, 1226–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazy M. A.; Gordon J. M. B.; Lee S. D.; Singh B. N.; Bohm A.; Hampsey M.; Moore C. (2012) The interaction of Pcf11 and Clp1 is needed for mRNA 3′-end formation and is modulated by amino acids in the ATP-binding site. Nucleic Acids Res. 40, 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S.; Martinez J. (2007) The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature 447, 222–226. [DOI] [PubMed] [Google Scholar]
- Ramirez A.; Shuman S.; Schwer B. (2008) Human RNA 5′-kinase (hClp1) can function as a tRNA splicing enzyme in vivo. RNA 14, 1737–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada T.; Weitzer S.; Mair B.; Bernreuther C.; Wainger B. J.; Ichida J.; Hanada R.; Orthofer M.; Cronin S. J.; Komnenovic V.; Minis A.; Sato F.; Mimata H.; Yoshimura A.; Tamir I.; Rainer J.; Kofler R.; Yaron A.; Eggan K. C.; Woolf C. J.; Glatzel M.; Herbst R.; Martinez J.; Penninger J. M. (2013) CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 495, 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbein S.; Scola S.; Loll B.; Dichtl B. S.; Hubner W.; Meinhart A.; Dichtl B. (2011) The P-loop domain of yeast Clp1 mediates interactions between CF IA and CPF factors in pre-mRNA 3′ end formation. PLoS One 6, e29139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. A.; Cubberley G.; Bentley D. L. (2009) Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3′ end processing factor Pcf11. Mol. Cell 33, 215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. A.; Kim H.; Erickson B.; Bentley D. L. (2011) The export factor Yra1 moduates mRNA 3′ end processing. Nat. Struct. Mol. Biol. 18, 1164–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U.; Gundel M.; Knoth A.; Kerwitz Y.; Rudel S.; Wahle E. (2009) Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 284, 22803–22814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson S. M.; Conrad N. K. (2013) The human nuclear poly(A)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet. 9, e1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.; Nausch L. W.; Martin G.; Keller W.; Doublie S. (2014) Crystal structure of human poly(A) polymerase γ reveals a conserved catalytic core for canonical poly(A) polymerase. J. Mol. Biol. 426, 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.; Norbury C. J. (2010) Polyadenylation and beyond: Emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscip. Rev.: RNA 1, 142–151. [DOI] [PubMed] [Google Scholar]
- Chang J. H.; Tong L. (2012) Mitochondrial poly(A) polymerase and polyadenylation. Biochim. Biophys. Acta 1819, 992–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y.; Srivastava S. K.; Chang J. H.; Manley J. L.; Tong L. (2011) Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase. Mol. Cell 41, 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba A.; Coll O.; Gebauer F. (2011) Cytoplasmic polyadenylation and translational control. Curr. Opin. Genet. Dev. 21, 452–457. [DOI] [PubMed] [Google Scholar]
- Charlesworth A.; Meijer H. A.; de Moor C. H. (2013) Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip. Rev.: RNA 4, 437–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T.; Swanger S. A.; Takeuchi K.; Kim J. H.; Nalavadi V.; Shin J.; Lorenz L. J.; Zukin R. S.; Bassell G. J.; Richter J. D. (2012) Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol. Cell 47, 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miniaci M. C.; Kim J. H.; Puthanveettil S. V.; Si K.; Zhu H.; Kandel E. R.; Bailey C. H. (2008) Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron 59, 1024–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K.; Choi Y. B.; White-Grindley E.; Majumdar A.; Kandel E. R. (2010) Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell 140, 421–435. [DOI] [PubMed] [Google Scholar]
- Burns D. M.; D’Ambrogio A.; Nottrott S.; Richter J. D. (2011) CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 473, 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahder J. A.; Norrild B. (2011) Involvement of hGLD-2 in cytoplasmic polyadenylation of human p53 mRNA. APMIS 119, 769–775. [DOI] [PubMed] [Google Scholar]
- Zhang J. H.; Panicker L. M.; Seigneur E. M.; Lin L.; House C. D.; Morgan W.; Chen W. C.; Mehta H.; Haj-Ali M.; Yu Z. X.; Simonds W. F. (2010) Cytoplasmic polyadenylation element binding protein is a conserved target of tumor suppressor HRPT2/CDC73. Cell Death Differ. 17, 1551–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel D. J.; Wells S. B.; Hilburn B. C.; Elazzouzi F.; Perez-Alvarado G. C.; Lee B. M. (2013) The C-terminal region of cytoplasmic polyadenylation element binding protein is a ZZ domain with potential for protein-protein interactions. J. Mol. Biol. 425, 2015–2026. [DOI] [PubMed] [Google Scholar]
- Ghazy M. A.; He X.; Singh B. N.; Hampsey M.; Moore C. (2009) The essential N terminus of the Pta1 scaffold protein is required for snoRNA transcription termination and Ssu72 function but is dispensible for pre-mRNA 3′-end processing. Mol. Cell. Biol. 29, 2296–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K.; Nagaike T.; Xiang S.; Kilic T.; Beh M. M.; Manley J. L.; Tong L. (2010) Crystal structure of the human symplekin-Ssu72-CTD phosphopeptide complex. Nature 467, 729–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp M. D.; Schweingruber C.; Kleinschmidt N.; Schumperli D. (2011) Interactions of CstF-64, CstF-77, and symplekin: Implications on localisation and function. Mol. Biol. Cell 22, 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M.; Singh B. N.; Ansari A.; Laine J. P.; Krishnamurthy S. (2011) Control of eukaryotic gene expression: Gene loops and transcriptional memory. Adv. Enzyme Regul. 51, 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong S. M.; Zaugg J. B.; Camblong J.; Xu Z.; Zhang D. W.; Mischo H. E.; Ansari A. Z.; Luscombe N. M.; Steinmetz L. M.; Proudfoot N. J. (2012) Gene loops enhance transcriptional directionality. Science 338, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. (2009) Progression through the RNA polymerase II CTD cycle. Mol. Cell 36, 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.; Erickson B.; Luo W.; Seward D.; Graber J. H.; Pollock D. D.; Megee P. C.; Bentley D. L. (2010) Gene-specific RNA polymerase II phosphorylation and the CTD code. Nat. Struct. Mol. Biol. 17, 1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A.; Heidemann M.; Lidschreiber M.; Schreieck A.; Sun M.; Hintermair C.; Kremmer E.; Eick D.; Cramer P. (2012) CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 336, 1723–1725. [DOI] [PubMed] [Google Scholar]
- Bataille A. R.; Jeronimo C.; Jacques P.-E.; Laramee L.; Fortin M.-E.; Forest A.; Bergeron M.; Hanes S. D.; Robert F. (2012) A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol. Cell 45, 158–170. [DOI] [PubMed] [Google Scholar]
- Zhang D. W.; Mosley A. L.; Ramisetty S. R.; Rodriguez-Molina J. B.; Washburn M. P.; Ansari A. Z. (2012) Ssu72 phosphatase dependent erasure of phospho-Ser7 marks on the RNA polymerase II C-terminal domain is essential for viability and transcription termination. J. Biol. Chem. 287, 8541–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y.; Yogesha S. D.; Cannon J. R.; Yan W.; Ellington A. D.; Brodbelt J. S.; Zhang Y. (2013) Novel modifications on C-terminal domain of RNA polymerase II can fine-tune the phosphatase activity of Ssu72. ACS Chem. Biol. 8, 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Allen J. W.; Lee C.-J.; Liu P.; Nicely N. I.; Wang S.; Greenleaf A. L.; Zhou P. (2011) cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 286, 5717–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang K.; Manley J. L.; Tong L. (2012) An unexpected binding mode for a Pol II CTD peptide phosphorylated at Ser7 in the active site of the CTD phosphatase Ssu72. Genes Dev. 26, 2265–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S.; Ahmad K. (2005) Assembly of variant histones into chromatin. Annu. Rev. Cell Dev. Biol. 21, 133–153. [DOI] [PubMed] [Google Scholar]
- Dominski Z.; Marzluff W. F. (2007) Formation of the 3′ end of histone mRNA: Getting closer to the end. Gene 396, 373–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W. F.; Wagner E. J.; Duronio R. J. (2008) Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat. Rev. Genet. 9, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez M. D.; Samuelsson T. (2008) Early evolution of histone mRNA 3′ end processing. RNA 14, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L.; Meier M.; Lyons S. M.; Sit R. V.; Marzluff W. F.; Quake S. R.; Chang H. Y. (2012) Systematic reconstruction of RNA functional motifs with high-throughput microfluidics. Nat. Methods 9, 1192–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmakci N. G.; Lerner R. S.; Wagner E. J.; Zheng L.; Marzluff W. F. (2008) SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol. Cell. Biol. 28, 1182–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moeller H.; Lerner R.; Ricciardi A.; Basquin C.; Marzluff W. F.; Conti E. (2013) Structural and biochemical studies of SLIP1-SLBP identify DBP5 and eIF3g as SLIP1-binding proteins. Nucleic Acids Res. 41, 7960–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseoglu M. M.; Graves L. M.; Marzluff W. F. (2008) Phosphorylation of threonine 61 by cyclin A/Cdk1 triggers degradation of stem-loop binding protein at the end of S phase. Mol. Cell. Biol. 28, 4469–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N.; Lam T. T.; Fritz A.; Rempinski D.; O’Loughlin K.; Minderman H.; Berezney R.; Marzluff W. F.; Thapar R. (2012) The prolyl isomerase Pin1 targets stem-loop binding protein (SLBP) to dissociate the SLBP-histone mRNA complex linking histone mRNA decay with SLBP ubiquitination. Mol. Cell. Biol. 32, 4306–4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L.; Koonin E. V. (2000) SAP: A putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25, 112–114. [DOI] [PubMed] [Google Scholar]
- Hoefig K. P.; Rath N.; Heinz G. A.; Wolf C.; Dameris J.; Schepers A.; Kremmer E.; Ansel K. M.; Heissmeyer V. (2013) Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat. Struct. Mol. Biol. 20, 73–81. [DOI] [PubMed] [Google Scholar]
- Ansel K. M.; Pastor W. A.; Rath N.; Lapan A. D.; Glasmacher E.; Wolf C.; Smith L. C.; Papadopoulou N.; Lamperti E. D.; Tahiliani M.; Ellwart J. W.; Shi Y.; Kremmer E.; Rao A.; Heissmeyer V. (2008) Mouse Eri1 interacts with the ribosome and catalyzes 5.8S rRNA processing. Nat. Struct. Mol. Biol. 15, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D.; Marzluff W. F.; Dominski Z.; Tong L. (2013) Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science 339, 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev N. G.; Yario T. A.; Benson E.; Steitz J. A. (2008) Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep. 9, 1013–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. D.; Steiniger M.; Marzluff W. F. (2009) A core complex of CPSF73, CPSF100, and symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell 34, 322–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev N. G.; Steitz J. A. (2005) Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 19, 2583–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y.; Kimura T.; Murakami A.; Yajima N.; Sakamaki K.; Yonehara S. (1999) The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature 398, 777–785. [DOI] [PubMed] [Google Scholar]
- Yang X.-C.; Burch B. D.; Yan Y.; Marzluff W. F.; Dominski Z. (2009) FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell 36, 267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch B. D.; Godfrey A. C.; Gasdaska P. Y.; Salzler H. R.; Duronio R. J.; Marzluff W. F.; Dominski Z. (2011) Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA 17, 1132–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.-C.; Xu B.; Sabath I.; Kunduru L.; Burch B. D.; Marzluff W. F.; Dominski Z. (2011) FLASH is required for the endonucleolytic cleavage of histone pre-mRNAs but is dispensable for the 5′ exonucleolytic degradation of the downstream cleavage product. Mol. Cell. Biol. 31, 1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.-C.; Sabath I.; Debski J.; Kaus-Drobek M.; Dadlez M.; Marzluff W. F.; Dominski Z. (2013) A complex containing the CPSF73 endonuclease and other poyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol. Cell. Biol. 33, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath I.; Skrajna A.; Yang X.-C.; Dadlez M.; Marzluff W. F.; Dominski Z. (2013) 3′-end processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA 19, 1726–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriyama M.; Kobayashi Y.; Saito M.; Ishikawa F.; Yonehara S. (2009) Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol. Cell. Biol. 29, 4729–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber J. J.; Olejniczak S. H.; Yong J.; La Rocca G.; Dreyfuss G.; Thompson C. B. (2012) Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol. Cell 45, 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T.; Yung T. M. C.; Yamamoto J.; Tsuboi Y.; Tanabe K.; Yamaguchi Y.; Handa H. (2007) NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell 26, 349–365. [DOI] [PubMed] [Google Scholar]
- Hallais M.; Pontvianne F.; Andersen P. R.; Clerici M.; Lener D.; Benbahouche N. E.; Gostan T.; Vandermoere F.; Robert M.-C.; Cusack S.; Verheggen C.; Jensen T. H.; Bertrand E. (2013) CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat. Struct. Mol. Biol. 20, 1358–1366. [DOI] [PubMed] [Google Scholar]
- Wagner E. J.; Marzluff W. F. (2006) ZFP100, a component of the active U7 snRNP limiting for histone pre-mRNA processing, is required for entry into S phase. Mol. Cell. Biol. 26, 6702–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J.-P.; Sheth A.; Manley J. L. (2011) RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science 334, 683–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermair C.; Heidemann M.; Koch F.; Descostes N.; Gut M.; Gut I.; Fenouil R.; Ferrier P.; Flatley A.; Kremmer E.; Chapman R. D.; Andrau J. C.; Eick D. (2012) Threonine-4 of mammalian RNA polymerase II CTD is targeted by Polo-like kinase 3 and required for transcriptional elongation. EMBO J. 31, 2784–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirngruber J.; Shchebet A.; Schreiber L.; Shema E.; Minsky N.; Chapman R. D.; Eick D.; Aylon Y.; Oren M.; Johnsen S. A. (2009) CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep. 10, 894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirngruber J.; Johnsen S. A. (2010) Induced G1 cell-cycle arrest controls replication-dependent histone mRNA 3′ end processing through p21, NPAT and CDK9. Oncogene 29, 2853–2863. [DOI] [PubMed] [Google Scholar]
- Groth A.; Rocha W.; Verreault A.; Almouzni G. (2007) Chromatin challenges during DNA replication and repair. Cell 128, 721–733. [DOI] [PubMed] [Google Scholar]
- Djakbarova U.; Marzluff W. F.; Koseoglu M. M. (2014) Translation regulation and proteasome mediated degradation cooperate to keep stem-loop binding protein, a major player in histone mRNA metabolism, low in G1-phase. J. Cell. Biochem. 115, 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Kiledjian M. (2010) Regulation of mRNA decapping. Wiley Interdiscip. Rev.: RNA 1, 253–265. [DOI] [PubMed] [Google Scholar]
- Ghosh A.; Lima C. D. (2010) Enzymology of RNA cap synthesis. Wiley Interdiscip. Rev.: RNA 1, 152–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I. G.; Roberts L. O. (2008) Eukaryotic initiation factor 4E. Int. J. Biochem. Cell Biol. 40, 2675–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer P. M. (2009) Cap in hand: Targeting eIF4E. Cell Cycle 8, 2535–2541. [DOI] [PubMed] [Google Scholar]
- Song M.-G.; Li Y.; Kiledjian M. (2010) Multiple mRNA decapping enzymes in mammalian cells. Mol. Cell 40, 423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Song M.; Kiledjian M. (2011) Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA 17, 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. G.; Bail S.; Kiledjian M. (2013) Mutiple Nudix family proteins possess mRNA decapping activity. RNA 19, 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S.; Cooper-Morgan A.; Jiao X.; Kiledjian M.; Manley J. L.; Tong L. (2009) Structure and function of the 5′→3′ exoribonuclease Rat1 and its activating partner Rai1. Nature 458, 784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X.; Xiang S.; Oh C.-S.; Martin C. E.; Tong L.; Kiledjian M. (2010) Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467, 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J. H.; Jiao X.; Chiba K.; Kiledjian M.; Tong L. (2012) Dxo1 is a new type of eukaryotic enzyme with both decapping and 5′-3′ exoribonuclease activity. Nat. Struct. Mol. Biol. 19, 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X.; Chang J. H.; Kilic T.; Tong L.; Kiledjian M. (2013) A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell 50, 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L.; Makarova K. S.; Koonin E. V. (2000) Holliday junction resolvases and related nucleases: Identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28, 3417–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steczkiewicz K.; Muszewska A.; Knizewski L.; Rychlewski L.; Ginalski K. (2012) Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily. Nucleic Acids Res. 40, 7016–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S. A.; Frazier M. L.; Steiniger M.; Mast A. M.; Marzluff W. F.; Redinbo M. R. (2009) Crystal structure of the HEAT domain from the pre-mRNA processing factor symplekin. J. Mol. Biol. 392, 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Zhang M.; Zhang Y. (2011) Crystal structure of Ssu72, an essential eukaryotic phosphatase specific for the C-terminal domain of RNA polymerase II, in complex with a transition state analogue. Biochem. J. 434, 435–444. [DOI] [PubMed] [Google Scholar]
- Zhang M.; Lam T. T.; Tonelli M.; Marzluff W. F.; Thapar R. (2012) Interaction of the histone mRNA hairpin with stem-loop binding protein (SLBP) and regulation of the SLBP-RNA complex by phosphorylation and proline isomerization. Biochemistry 51, 3215–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]