Abstract
The liver kinase B1 (LKB1) tumor suppressor inhibits cell growth through its regulation of cellular metabolism and apical-basal polarity. The best understood mechanism whereby LKB1 limits cell growth is through activation of the AMP-activated-protein-kinase/mammalian-target-of-rapamycin (AMPK/mTOR) pathway to control metabolism. Since LKB1 is also required for polarized epithelial cells to resist hyperplasia it is anticipated to function through additional mechanisms. Recently, Yes-associated protein (Yap) has emerged as a transcriptional co-activator that modulates tissue homeostasis in response to cell-cell contact. Thus this study examined a possible connection between Yap and LKB1. Restoration of LKB1 expression in HeLa cells, which lack this tumor suppressor, or shRNA knockdown of LKB1 in NTERT immortalized keratinocytes, demonstrated that LKB1 promotes Yap phosphorylation, nuclear exclusion, and proteasomal degradation. The ability of phosphorylation-defective Yap mutants to rescue LKB1 phenotypes, such as reduced cell proliferation and cell size, suggest that Yap inhibition contributes to LKB1 tumor suppressor function(s). However, failure of Lats1/2 knockdown to suppress LKB1-mediated Yap regulation suggested that LKB1 signals to Yap via a non-canonical pathway. Additionally, LKB1 inhibited Yap independently of either AMPK or mTOR activation. These findings reveal a novel mechanism whereby LKB1 may restrict cancer cell growth via the inhibition of Yap.
Keywords: LKB1, Yap, Hippo, polarity, growth
Introduction
Growth and development are regulated by a balance between proliferation and apoptosis that is linked to cell polarity through poorly understood mechanisms. Disregulation of this balance results in the hyperproliferation of cancer cells as well as gross changes in their morphology and tissue organization (1). Recently, two crucial pathways have emerged that govern these events: the Hippo pathway and signals triggered by liver kinase B1 (LKB1, STK11). LKB1 is a master regulator of proliferation and apical-basal polarity, while cell structure can impact the Hippo pathway (2, 3). Despite the strong logical link between these two pathways, no evidence of their association has previously been reported.
LKB1 is a ubiquitous serine–threonine protein kinase that controls a wide range of cellular functions that include metabolism, proliferation, and cell shape (4). LKB1 heterozygous patients suffer from Peutz-Jeghers syndrome that is characterized by gastrointestinal hamartomas and increased cancer predisposition. LKB1 inactivation is also frequently observed in sporadic non-small cell lung and cervical carcinomas (4, 5). LKB1, in complex with the pseudokinase STRAD and the scaffolding protein MO25, directly phosphorylates and activates AMPK and 12 related kinases that include the microtubule affinity-regulating kinases (MARKs) family (6). In response to high levels of AMP, AMPK suppresses mTOR complex 1 (mTORC1), a central regulator of protein synthesis. LKB1 promotes efficient AMP-induced activation of AMPK, thus inducing growth arrest in response to metabolic stress (7).
LKB1 activation is also sufficient to polarize intestinal epithelial cells in the absence of cell–cell contacts (8). Reciprocally, knockdown of LKB1 results in a loss of epithelial organization and increases Myc-dependent cell proliferation (9). LKB1 and downstream MARKs regulate several conserved polarity proteins, the inactivation of which may promote tumorigenesis (2). However, it is unclear how LKB1 promotes growth arrest via its effects on cell apical-basal polarity.
The Hippo pathway regulates tissue development through a kinase cascade involving Hippo (mammalian ortholog Mst1/2) that phosphorylates ands activates Warts (Lats1/2). Active Warts then phosphorylates and inactivates the transcriptional co-activator Yki (Yap) and its binding partner Taz to both inhibit growth and promote apoptosis (3, 10, 11). During the formation of epithelial cell apical-basal polarity, mammalian Yap, or its fly ortholog Yki, is down-regulated through interaction with cell junction proteins, such as α-catenin and the atypical cadherin, fat (12, 13). Conversely, constitutive Yap expression induces epithelial-mesenchymal transition (EMT) in mammary cells grown in three-dimensional culture (14), indicating its oncogenic potential. Consistently, many studies have implicated Hippo signaling with cancer development. For instance, Yap overexpression or loss of upstream suppressors Mst, Salvador, or Merlin/NF2 (15) lead to hepatomegaly and liver carcinomas. Furthermore, multiple human cancers have elevated Yap protein and nuclear localization (16).
In mammalian cells, Lats1/2 inhibits Yap by phosphorylating five known sites. Phospho-Ser127 promotes retention of Yap in the cytosol (17) whereas phospho-Ser381 is a priming site for further phosphorylation, ubiquitylation and degradation (18). Double mutation of S127 and S381 stabilizes Yap and promotes oncogenic phenotypes, but neither mutation alone can induce cellular transformation (18).
Yap and Taz cooperate with transcription factors that are downstream of major developmental and cancer-promoting pathways such as SMADs (TGFβ and BMP pathways), β-catenin (WNT pathway), and TEAD (19). However, much of the upstream signaling that connects tissue organization to Yap regulation and the role(s) these signals play in oncogenesis remain unclear. Using HeLa cells that lack LKB1 expression, or alternatively suppressing its expression in non-transformed human keratinocytes, we now show that the regulation of cell size and proliferation by LKB1 are at least in part mediated by Yap phosphorylation, leading to nuclear exclusion and decreased stability of this transcriptional co-activator. LKB1-induced events could be partially reversed by expression of phosphorylation-defective Yap mutants. However, these phenomena occurred independently of mTOR, AMPK or Lats1/2 activation, suggesting a non-canonical pathway links LKB1 activity to Yap phosphorylation. Since LKB1-induced morphological rearrangement correlated with Yap suppression, we suggest that LKB1-induced cell polarity impacts Yap activity. This work identifies a novel link between two major tumor suppressor pathways and a potential mechanism for cell polarity to restrict cancer cell growth.
Results
LKB1 expression correlates with increased Yap phosphorylation and reduced HeLa cell proliferation
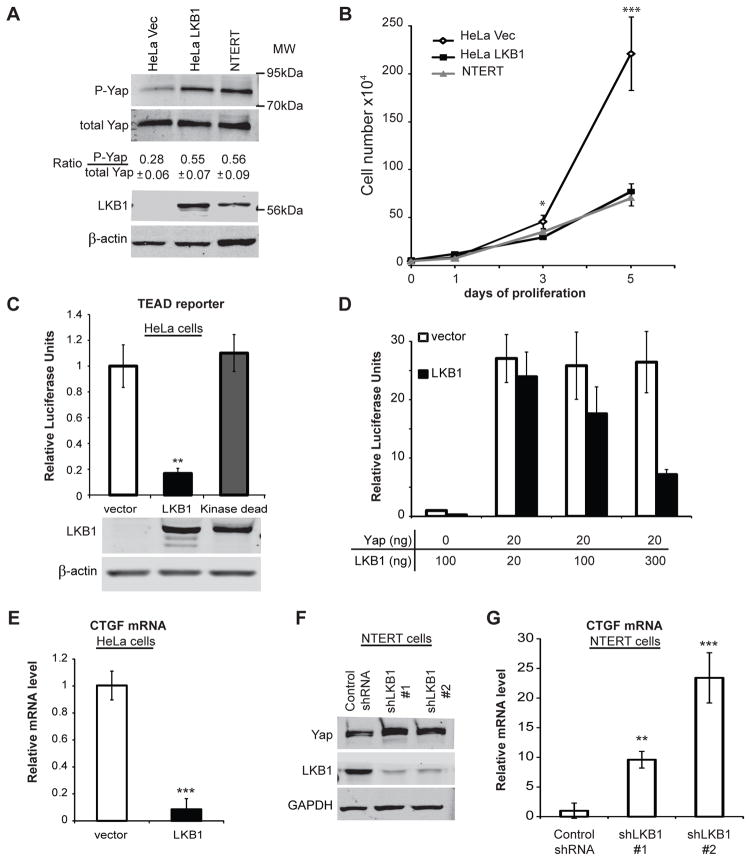
The Hippo pathway functions as part of a crowd control mechanism to limit organ size by reducing cell proliferation (3). In cultured cells, Hippo signaling is activated by high cell density, leading to phosphorylation and inhibition of the transcriptional co-activator Yap (3). Meanwhile, the tumor suppressor LKB1 provides resistance to epithelial hyperplasia (9). To investigate the potential for crosstalk between LKB1 and the Hippo pathway, the effect of LKB1 expression on the Lats1/2 phosphorylation site YapS127 (17), was measured. Using lentiviral transduction, LKB1 was stably expressed in HeLa cells (HeLa LKB1) to levels approximately twice that seen in NTERT (20) non-cancerous immortalized human keratinocytes (Figure 1a). In contrast no LKB1 was detected in HeLa cells stably expressing GFP alone (HeLa Vec). Even at low cell density, LKB1 expression raised the phospho-YapS127/total Yap ratio to a level comparable with that seen in the NTERT cell line (Figure 1a). This increase in Yap phosphorylation was correlated with over 2-fold reduction of proliferation in HeLa LKB1 cells that was similar to the growth rate of NTERT cells (Figure 1b).
Figure 1. LKB1 increased Yap phosphorylation while inhibiting Yap dependent transcription and cell growth.
A) The relative levels of Yap, phospho-YapSer127, LKB1 and β-actin were detected by immunoblot analysis of HeLa cell lysates prepared following infection with lentivirus expressing LKB1 (HeLa LKB1) or GFP control (HeLa Vec) vectors and compared to uninfected NTERT immortalized human keratinocytes. The mean ratio of phospho-YapSer127/YapTotal from three experiments is indicated below a representative immunoblot.
B) After seeding 5×104 cells/plate, trypan blue-excluded cells were counted in triplicate on the indicated days by hemocytometer. The mean number of HeLa Vec (◇) or HeLa LKB1 (■), and NTERT immortalized human keratinocytes (▲) were plotted.
C) HeLa cells were co-transfected with a luciferase reporter system containing a TEAD response element along with WT LKB1, kinase-dead LKB1or empty vector. Yap-dependent transcription was inferred from luciferase signals after 48 hr and LKB1 expression confirmed by immunoblot.
D) TEAD-driven transcription was measured as in Figure 1c using HeLa cells transfected with the indicated amount of LKB1 and Yap expressing plasmids (fixed Yap level).
E) Endogenous HeLa cell CTGF transcript was measured by RT-PCR following transfection with control or LKB1-encoding plasmids.
F) NTERT cells were infected with two independent lentiviruses encoding LKB1 shRNAs (#1 and #2) or a non-specific control sequence. After stable selection on puromycin, the relative levels of endogenous LKB1 and GAPDH from these lysates was measured by immunoblot.
G) CTGF mRNA was measured in NTERT cells as in Fig 1e following stable expression of LKB1 shRNAs #1 or #2 or a non-specific control sequence.
Luciferase data are representative of at least three independent experiments performed in triplicate. qRT-PCR is representative of two experiments performed in quadruplicate. Error was computed as standard deviation of the mean. Significance was computed by student T-test where p-values are indicated by * < 0.05; ** < 0.01; *** < 0.001; N.S., not significant.
LKB1 inhibits Yap dependent transcription
Yap binds and stimulates the transcriptional activity of Tea-domain (TEAD) transcription factors to promote cell growth (21). Thus, Yap activity was inferred by TEAD-induced gene expression from a luciferase reporter plasmid. LKB1 expression in HeLa cells reduces TEAD expression by approximately 5 fold. This inhibition required LKB1 kinase activity since a kinase-dead mutant failed to suppress luciferase activity (Figure 1c). To test the effect of LKB1 on exogenously expressed Yap, we transfected cells with various ratios of LKB1- and Yap-encoding plasmids. Remarkably, this reporter system is sensitive to as little as 20 ng of exogenous Yap. While no changes were observed upon transfection of control vector, increasing the amount of transfected LKB1-encoding plasmid resulted in dose-dependent inhibition of Yap activity (Figure 1d). Another measure of Yap activity is the transcription of connective tissue growth factor (CTGF) (21). Consistent with TEAD-luciferase data, LKB1 restoration drastically reduced CTGF mRNA level (Figure 1e). Conversely, we employed shRNAs to examine the effect of endogenous LKB1 knockdown on Yap activity in NTERT keratinocytes. Suppression of LKB1 expression by two separate shRNAs (Figure 1f) greatly enhanced CTGF transcription as determined by qRT-PCR (Figure 1g). Similar results were obtained using 293T cells (data not shown). These data indicated that LKB1 expression inhibits Yap transcriptional function.
LKB1 restoration alters Yap subcellular localization
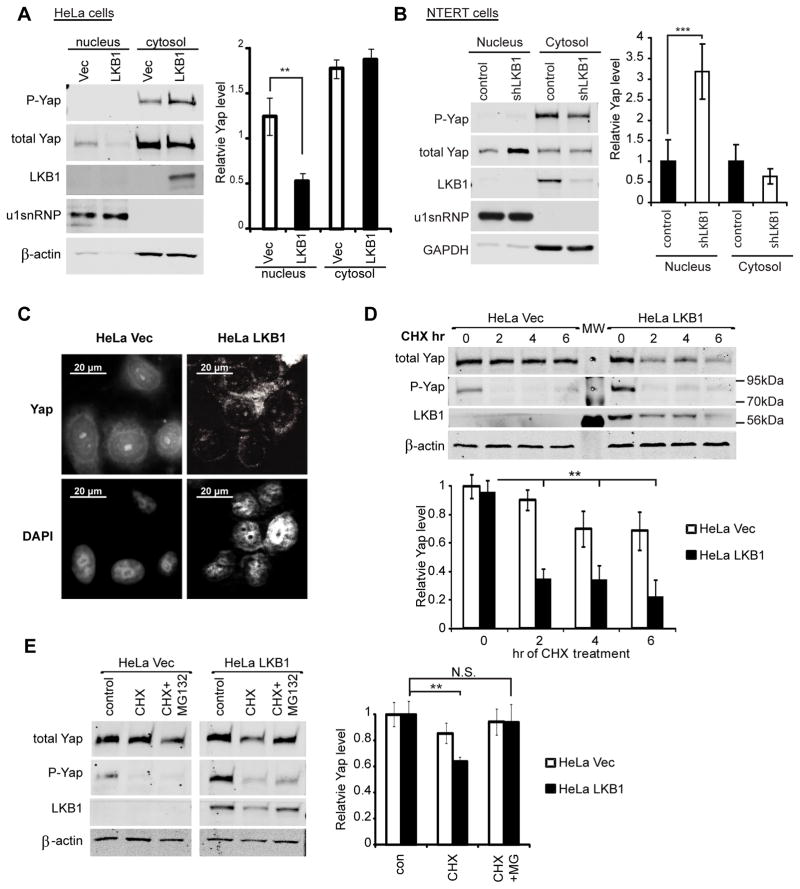
Yap dependent transcription is inhibited via the hyperphosphorylation of Yap on multiple sites. Currently, there are 5 known Lats1/2 phosphorylation sites on Yap (18). Among these, phosphoserine 127 allows for 14-3-3 binding and Yap cytoplasmic retention (22). In order to evaluate whether this spatial regulation occurs in LKB1 cells, we employed both nuclear fractionation and fluorescence microscopy. Firstly, hypotonic lysis was used to separate nucleic and cytoplasmic fractions. Endogenous Yap from both fractions was then detected by western blotting. We independently fixed cells and visualized the subcellular localization of endogenous Yap protein by immunofluorescence microscopy. Consistent with the increased phospho-YapS127 level in LKB1 expressing cells, both localization methods showed a significant decrease in nuclear Yap protein in HeLa LKB1 cells comparing to HeLa Vec cells of the same plating density (Figures 2a and c). Consistently, knockdown of LKB1 in NTERT cells caused a significant increase in nuclear Yap (Figure 2b). We concluded that LKB1 restoration increases S127 phosphorylation, promoting Yap nuclear exclusion.
Figure 2. LKB1 expression decreases Yap nuclear localization and stability in HeLa cells.
A) The relative levels of Yap, phospho-YapSer127, LKB1, U1snRNP and β-actin were measured by immunoblot from nucleus- and cytosol-enriched fractions prepared from HeLa cells stably expressing LKB1 (HeLa LKB1) or GFP control vector (HeLa Vec).
B) NTERT cells were treated as in Fig 2A.
C) HeLa LKB1 and HeLa Vec cells were fixed and immunostained for Yap. Nuclei were counterstained with DAPI.
D) HeLa LKB1 and HeLa Vec cells grown at low density were treated with 100 μg/ml cycloheximide (CHX) for the indicated times. The levels of Yap, phospho-YapSer127, LKB1 and β-actin were measured by immunoblot analysis from cell lysates (upper panel). The average Yap levels from three independent experiments were also computed (lower panel).
E) HeLa LKB1 and HeLa Vec cells were treated with CHX and 10 μM MG132 for 2 hr. Protein levels were then measured by immunoblot analysis (left panel). The average levels of Yap from three independent experiments are also shown (right panel).
Errors and significances are represented as in Figure 1.
LKB1 expression increases Yap degradation by the proteasome
Since proteasomal degradation is another important Yap regulating mechanism (3), we examined whether LKB1 add-back decreases Yap protein stability. To monitor the stability of Yap, HeLa LKB1 and HeLa Vec cells were treated with cycloheximide (CHX) to inhibit new protein synthesis. Plated at low density, HeLa Vec exhibited insignificant decrease in Yap protein level over 6 hours of CHX treatment. Meanwhile, Yap level rapidly decreased in LKB1 cells (Figure 2d). To confirm that the decrease in Yap level was due to proteasomal degradation, we used the proteasome inhibitor, MG132. As previously observed, we saw a significant decrease in Yap level in LKB1 cells treated with CHX alone compared to DMSO control. However, addition of MG132 along with CHX returned Yap protein to control level (Figure 2e). Consistently, LKB1 knockdown in NTERT cells caused an increase in total cellular Yap protein (Figure 1f). Altogether, these results indicated that LKB1 decreases Yap stability by promoting its proteasomal degradation.
LKB1-mediated inhibition of Yap functions requires Yap phosphorylation
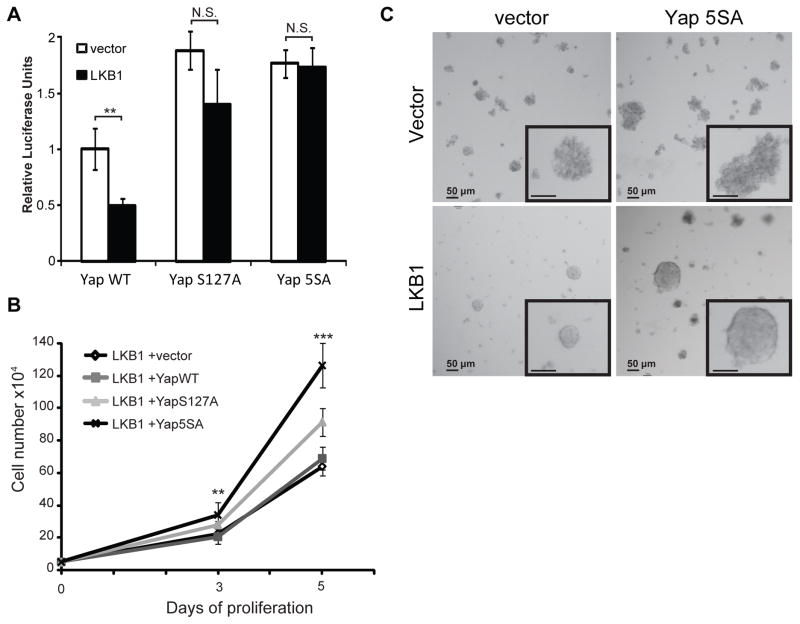
Figure 1 demonstrated that LKB1 expression induces phosphorylation of YapSer127. This correlated with decreased Yap nuclear localization and transcriptional co-activation. We hypothesized that if Yap phosphorylation was the key event triggered by LKB1 expression to regulate gene expression and cell growth, then blocking such phosphorylation event(s) would rescue LKB1 tumor suppressor phenotypes. Two phosphorylation-defective Yap mutants were used in addition to a wild type (YapWT) construct to test this hypothesis: YapS127A has Ser 127 mutated to Ala while the Yap5SA mutant lacks all five reported Lats phosphorylation sites (S61, S109, S127, S164, and S381) (18). The TEAD-luciferase reporter assay was used as above to monitor Yap activity following transient transfection. While YapWT activity was inhibited by LKB1 (Figures 3a), the S5A mutant was insensitive to LKB1 action (Figure 3a). A statistically insignificant but reproducible inhibition of YapS127A activity by LKB1 suggested that additional Yap phosphorylation sites might also play a role in suppressing its activity. These results suggested that Yap phosphorylation is required for LKB1 to suppress gene expression.
Figure 3. The ability of LKB1 to suppress Yap dependent transcription and cell growth is partially reversed by phosphorylation-deficient Yap mutants.
A) HeLa cells were transfected with Yap- wild type (WT), S127A, or 5SA in combination with vector control or LKB1. TEAD dependent transcription was measured as described in Figure 1c.
B) HeLa cells were co-infected with LKB1 and vector control (◇), YapWT (■), YapS127A (▲), and Yap5SA (X) mutants. Following puromycin selection, cell accumulation was measured as in Figure 1b.
Error and significance are represented as described in Figure 1.
C) HeLa cells stably expressing the indicated cDNAs were imaged after 10 days culture in Matrigel. Representative images are shown. Insets are 2-fold increase magnification of colonies.
As shown in Figure 1b, LKB1 strongly reduced HeLa cells proliferation. Since the transcriptional activity of Yap phospho-mutants was not inhibited by LKB1, we next addressed whether these mutants could rescue the suppression of proliferation seen following LKB1 expression. Lentiviruses encoding LKB1 and various Yap transcripts were used to co-infect HeLa cells. After puromycin selection, cell accumulation was monitored as in Figure 1b. Despite LKB1 suppressing endogenous Yap activity (Figure 1b), expression of the Yap5SA mutant significantly promoted cell proliferation. Consistent with the transcriptional activity data (Figure 3a), the YapS127A mutant had a weaker effect than Yap5SA on rescuing cell proliferation. Meanwhile, YapWT expression did not significantly change cell growth (Figure 3b). The overexpression of Yap mutants did not significantly change cell accumulation in the absence of LKB1 (data not shown). We also examined the effect of LKB1 and Yap5SA expression on HeLa cell growth in a 3-dimensional Matrigel matrix. Consistent with our previous data, LKB1 significantly reduced cell growth in Matrigel (Figure 3C, bottom left panel). Interestingly, Yap5SA expression greatly increased cell proliferation both in the presence and absence of LKB1 but did not disrupt LKB1-induced cell-cell compaction. The expression of either YapWT or YapS127A did not altered cell growth pattern (data not shown). We concluded that the hyper-phosphorylation of Yap contributes to LKB1-mediated suppression of cell proliferation.
Phosphorylation-defective Yap mutants partially reversed cell size reduction by LKB1
Cell size is a complex phenotype that is often influences by proliferative state and could be the outcome of various distinct pathways (23, 24). Various studies have reported the regulation of cell and organ size by both LKB1 and the Hippo pathway. For example, conditional knockout of murine LKB1 resulted in increased islet beta cell volume (25). Meanwhile, Yap conditional overexpression in other tissues was reported to cause liver enlargement and thickening of epidermal layers (26, 27). Therefore, we examine whether LKB1 regulates HeLa cell size by inhibiting Yap.
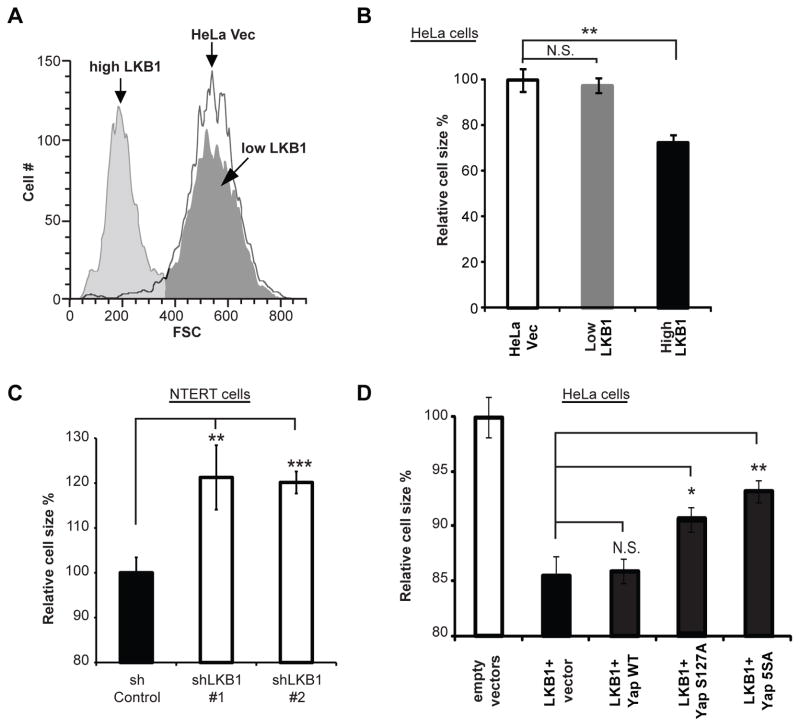
Utilizing the HeLa LKB1 and HeLa Vec cells described above, we first confirmed the ability of LKB1 to reduce cell size. The lentiviral pCDH plasmid encodes for both GFP and LKB1, thus LKB1 levels could be correlated with GFP fluorescence. Two distinct populations were observed in GFP/LKB1 add-back cells; those that expressed high levels of recombinant proteins and those that had much lower levels (Figure 4a). While the relative volume of low GFP/LKB1-expressing cells was similar to HeLa Vec cells, consistent with previous reports (5, 25), the cells highly expressing LKB1 had significantly smaller average size (Figure 4b). Meanwhile, knockdown of LKB1 in NTERT cells increased cell volume (Figure 4c) confirming LKB1 function in cell size reduction.
Figure 4. LKB1-induced cell shrinkage is partially reversed by expression of YapS127A or Yap5SA.
A) HeLa cells infected with a lentivirus carrying LKB1 and GFP (separated by a T2A self-cleaving peptide) or GFP alone were analyzed by flow cytometry forward scatter (FSC). Two sub-populations were observed in GFP/LKB1 add-back cells: one with high and one with low GFP signal.
B) The mean FSC of each population described in Figure 4a was calculated as an arbitrary unit of cell size, which was then normalized to vector control to obtain relative cell size. Three samples of each population were analyzed and the average relative sizes presented.
C) NTERT cells were treated as in 4A/B.
D) Stable cell lines generated from co-infection with LKB1 and Yap transcripts as described in Figure 3b were sorted by GFP signals/LKB1 expression. Cell sizes were then analyzed. The mean FSCs were normalized to vector control, averaged from three samples, and presented as mean ± S.D.
All results are representative of at least two independent experiments performed in triplicate. * p < 0.05; ** p < 0.01; N.S. not significant
To address whether the inhibition of Yap contributed to this effect, lentiviruses carrying YapWT or the phosphorylation-defective S127A or 5SA mutants were co-transduced with LKB1-encoding virus into HeLa cells. While YapWT had no effect, YapS127A and Yap5SA mutants significantly rescued LKB1-induced cell size reduction (Figure 4d). The Yap5SA mutant was most effective, again suggesting contribution of phospho-sites other than S127 in mediating LKB1’s effects. However, even Yap5SA expression did not completely return HeLa cells to control size (Figure 4d). Thus, inhibition of Yap may be just one factor contributing to suppression of cell volume by LKB1.
Neither AMPK nor mTOR activity mediated LKB1’s regulation of Yap
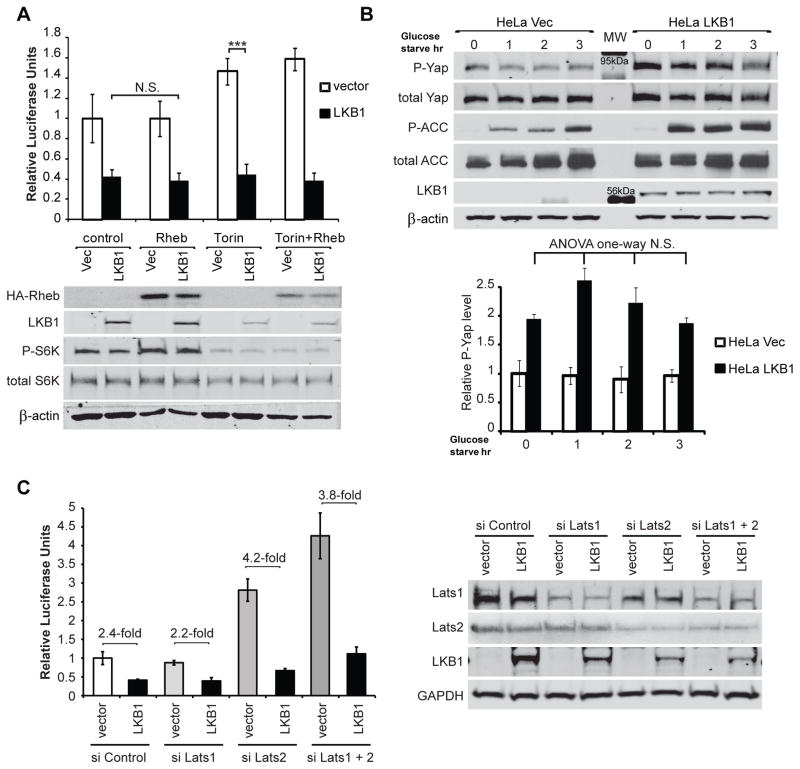
Numerous studies have reported that LKB1 regulates cell size through activation of AMPK and subsequent inhibition of mTORC1-induced protein synthesis via phosphorylation of TSC2 and/or raptor (2). Since Yap phosphorylation-defective mutants were found above to reverse LKB1-induced cell size reduction, we next addressed whether LKB1 regulates Yap via mTOR activation. The ATP analogue Torin1 (28) was used to effectively block both mTORC1 and C2 activities. To activate mTORC1, we overexpressed its upstream regulator, Rheb (29). mTORC1 activates S6 kinase (S6K), promoting ribosomal S6 protein phosphorylation to control protein synthesis and cell size (30). Thus we used phospho-S6K as a read-out for mTORC1 activity. As expected, Torin1 reduced and Rheb increased phospho-S6K level (Figure 5a). However, changes in mTORC1 activity did not influence the ability of LKB1 to inhibit Yap activity (Figure 5a). This suggested that LKB1 inhibits Yap independently of mTOR. Since Yap mutants only partially rescued LKB1 effects on cell size and proliferation (Figures 3b and 4c), we speculate that Yap and mTOR mediate independent aspects of signaling downstream of LKB1. Surprisingly, the inhibition of mTORC1 by the drugs Torin1 (Figure 4d) and rapamycin, or by glutamine starvation (data not shown) caused a small but significant increase in Yap activity (ANOVA one-way p<0.01). Since mTORC1 is a major metabolic sensor, this enhancement implies a possible negative feedback regulation between these two pathways downstream of LKB1.
Figure 5. LKB1 regulates Yap independently of mTOR or AMPK activation.
A) HeLa cells were co-transfected with LKB1, Rheb and/or control vector as indicated. After 24 hr, cells were treated with the mTOR kinase inhibitor Torin1 (250 nM) for 16 hr. TEAD-luciferase activity was then measured as in Figure 1c. HA-Rheb, LKB1, phospho-S6K, total S6K and β-actin were measured as controls for protein expression and mTOR activity.
B) HeLa Vec and HeLa LKB1 cells were glucose starved for up to 3 hr as indicated. Lysates were then immunoblotted to measure phospho-YapSer127, Yap, phospho- and total acetyl CoA carboxylase (ACC), LKB1 and β-actin. AMPK activation was confirmed by increased ACC phosphorylation. The average levels of phospho-YapSer127 from three experiments are indicated in lower panel.
C. HeLa cells were transiently transfected with indicated siRNAs and TEAD luciferase plasmids. TEAD-luciferase activity as well as Lats 1, Lats2, LKB1 and GAPDH were then measured.
Figures A and B are representative of three experiments. Errors and significance are as described in Figure 1. C is representative of 2 independent experiments. ANOVA one way was performed to compare groups of four data points. N.S. not significant
LKB1 directly phosphorylates AMPK, enabling efficient activation upon AMP elevation, thus allowing cells to quickly response to metabolic stress (2). In order to examine possible mTOR-independent effects of AMPK on Yap, we induced metabolic stress via glucose starvation. AMPK phosphorylates acetyl-CoA carboxylase (ACC) to reduce glucose metabolism (31). Therefore we measured phospho-ACC as a positive control for AMPK activation. In this experiment, YapS127 phosphorylation was measured instead of Yap transcriptional activity due to the short duration of the experiment precluding significant changes in luciferase expression. Although AMPK was strongly activated by 1–3 hr of glucose starvation (large increase in ACC phosphorylation), Yap phosphorylation remained unchanged (Figure 5b). Thus we concluded that LKB1 inhibits Yap independently of AMPK activation.
LKB1 suppresses Yap function independently of stress fiber formation or Lats1/2 kinases
Two recent studies reported that stress fibers, induced by cell spreading, control Yap nuclear retention and thus its transcriptional activity (32, 33), although stress fiber disruption may preferentially affect the phosphorylation and stability of the Yap-related Taz protein (33). Since the Rho GTPase plays a role in promoting stress fiber formation, we overexpressed a constitutively active RhoAQ63L mutant (34) to examine if HeLa cell stress fiber disruption couples LKB1 to suppression of Yap activity. Consistent with published studies (33), RhoAQ63L strongly increased TEAD luciferase activity. However, LKB1 expression promoted a similar fold suppression of TEAD activity in vector control and RhoAQ63L cells (Supplemental Figure 1a). Therefore, it is likely that the mechano-response is mediated by a separate pathway from polarity/crowd control, with both signals converging on Yap/Taz phosphorylation.
The ability of Yap5SA to revert the effects of LKB1 expression additionally suggested Lats1/2 kinases may couple LKB1 to Yap phosphorylation. To confirm this we transiently knocked down Lats1 and/or 2 using siRNA and measured Lats protein levels and TEAD-driven luciferase activity. Surprisingly, suppression of Lats1/2 expression did not impact the ability of LKB1 to suppress Yap activity (Figure 5c), suggesting that LKB1 regulated Yap activity via a non-canonical pathway.
Disruption of LKB1-induced morphological changes decreases Yap phosphorylation
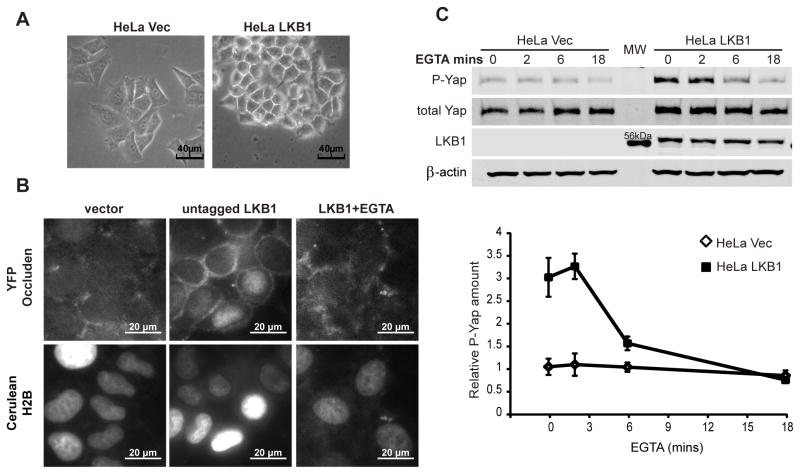
Changes in epithelial cell morphology or structure, such as adherens junction formation, have been reported to affect Yap cellular localization (13). Furthermore, LKB1 restoration promoted morphological changes and HeLa cell-cell contacts even at low culture density (Figure 6a). Those cells not adjacent to others also established a distinct non-spread morphology. This was consistent with Clevers’ report that LKB1 activation alone is sufficient to polarize dissociated intestinal epithelial cells (8). Therefore, we examined whether cellular structure induced by LKB1 might mediate Yap phosphorylation. We first confirmed that LKB1 promoted epithelial architecture by imaging live cells expressing fluorescently-tagged proteins. pYFP-occluden (a component of tight junctions) and pCFP-histone 2B (a nuclear marker) were co-transfected with vector or untagged LKB1 into HeLa cells. Live cell fluorescence images showed a polarized distribution of occluden at cell junctions in LKB1-expressing cells (Figure 6b). Such peripheral distribution was also observed in the absence of cell contact (data not shown).
Figure 6. Cellular structure promoted by LKB1 plays a role in creating and maintaining elevated phospho-Yap levels.
A) Phase-contrast images suggested HeLa LKB1 cells have increase cell-cell attachment.
B) YFP-tagged occluden (a component of tight junction) and cerulean histone 2B (a nuclear marker) were co-transfected with vector or untagged LKB1 into HeLa cells. Fluorescent live cell images were taken from vector control, LKB1 cells, and LKB1 cells after 6 min EGTA treatment.
C) HeLa Vec and HeLa LKB1 cells were treated with EGTA to disrupt adherens junctions for indicated times. Lysates from these cells were immunoblotted for phospho-YapSer127, total Yap, LKB1 and β-actin (upper panel). The average levels of phospho-YapSer127 of Vec (◇) and LKB1 (■) cells from three experiments were also computed (lower panel).
Blot and fluorescence images are representative. Error and significance are as described in Figure 1.
Since adherens junctions are a major contributor to epithelial cell polarity that rely on Ca2+-dependent homotypic interaction of E-cadherin molecules, we disrupted cell contacts by chelating extracellular calcium and examined the effect on Yap phosphorylation (35). Upon chelation of Ca2+ with EGTA, the peripheral localization of occluden in LKB1-expressing cells was lost within 6 minutes (Figure 6b). Ca2+-chelation also drastically decreased YapSer127 phosphorylation in a time dependent manner in LKB1 cells, while no significant change was observed in the already low Yap level of vector control cells (Figure 6c). Surprisingly, the total Yap level in LKB1 cells was also slightly decreased (Figure 6c). This suggested that cell polarity, induced by LKB1, impacts Yap phosphorylation and disruption of such structure lead to Yap degradation.
Discussion
The architecture of a cell contributes to its survival, proliferation and function. A key component of this architecture is the establishment of apical-basal polarity whereby certain organelles and proteins are asymmetrically distributed. Polarity is established though a variety of signaling complexes that are integrated into the control of proliferation by poorly defined mechanisms. Recently, the Hippo (Mst-Lats-Yap) signaling pathway has emerged as a major growth regulator with its endpoint, phosphorylation of the Yap transcriptional co-activator, being regulated by cell polarity/structure (13, 32). Interestingly, the LKB1 tumor suppressor is currently the only protein reported to cause cellular polarization in a cell autonomous fashion (8). The data reported here reveal a novel polarity-growth connection in which LKB1 activity regulates the subcellular distribution and proteasomal degradation of Yap leading to decreased cell proliferation and cell size.
The master kinase LKB1 is required for epithelial structure-related resistance to hyperplasia (9) at least in part by regulating the Par-3/Par-6/atypical protein kinase C tight junction complex (36). Consistent with a previous report in intestinal epithelial cells (8), expression of LKB1 autonomously promotes the polarization of HeLa cells in 2- and 3-D cultures and relocalization of the tight junction protein occluden to the cell periphery. LKB1 also suppresses cell proliferation and its re-expression in HeLa cells reduced their growth rate to that of non-transformed human keratinocytes. Since the YAP transcriptional co-activator is a key regulator of cell growth in response to cell polarity, we examined the effect of LKB1 by its re-expression in HeLa cells and suppression of endogenous LKB1 gene expression in NTERT or 293T cells. LKB1 expression inhibited the ability of both endogenous and exogenously expressed Yap to induce the activation of a well-characterized luciferase-coupled TEAD response element as well as endogenous CTGF transcription. These data demonstrated for the first time that Yap activity is closely associated with LKB1 status of tumor cells. Since Yap directly promotes the expression of proliferative genes, these findings indicate an alternative mechanism for LKB1-mediated suppression of cancer cell growth.
In many developmental systems and malignancies, the localization, integrity, and function of Yap are tightly regulated by its phosphorylation status. Consistently, we found that each of these parameters was closely linked to LKB1 expression in HeLa and NTERT cells. Specifically, LKB1 promoted phosphorylation of Ser127 that correlated with nucleus to cytoplasmic localization and enhanced Yap degradation. The decrease in nuclear Yap levels in LKB1 expressing cells also correlated with reduced proliferation of 2 and 3 dimensional cell cultures as well as cell size. The impacts of LKB1 expression on gene expression, cell growth and cell size were all suppressed by the expression of phosphorylation-resistant Yap mutants but not the overexpression of WT Yap. These data support the notion that LKB1 impacts cell growth via Yap phosphorylation. Consistent with previous reports (18), the Yap5SA mutant was more effective that Yap127A, suggesting that degradation rather than mere exclusion from the nucleus plays a role in LKB1-induced events.
To date Lats1/2 are the only kinases proven to phosphorylate Ser127 of Yap and the ability of Yap mutants lacking Lats1/2 phosphorylation sites to partially reversed the effects of LKB1 expression were consistent such a role. Since the AMPK-related kinase, NUAK1/ARK5, is a substrate of LKB1that phosphorylates Lats (37), we first considered it as a mediator of Lats/Yap regulation. However, phosphorylation by ARK5 has been documented to destabilize Lats1 (37, 38) which would likely reduce phospho-Yap levels. While we observed a decrease in Lats1 protein level upon LKB1 expression (Supplemental Figure1b), which could be related to ARK5 activity, knockdown of Lats1/2 expression in HeLa cells did not prevent LKB1 from inhibiting Yap. Although Lats1/2 expression was not completely suppressed, this data strongly suggested that LKB1 regulates Yap independently of the Lats kinases. Conditional knock out of Mst1/2 or siRNA knock down of Mst1/2 and Lats1/2 similarly did not change Yap phosphorylation pattern or TEAD activity in keratinocytes (13). Additional reports similarly described increased Yap phosphorylation and nuclear exclusion independently of the Mst/Lats kinases (33, 39), suggesting the involvement of a non-canonical kinase.
LKB1 is known to suppress growth through the activation of AMPK and subsequently the inhibition of mTOR activity and protein synthesis. Indeed, no other signaling pathway downstream of LKB1 has been found to be activated in human tumors following LKB1 loss (2, 40). However, neither the activation of AMPK by glucose starvation nor pharmacological inhibition of mTOR activity impaired LKB1-mediated Yap phosphorylation. Therefore, the Yap transcriptional co-activator functions as an AMPK/mTOR pathway-independent mediator of LKB1’s tumor suppressor functions. None of the phosphorylation sites targeted in the Yap5SA mutant match an LKB1 consensus motif, suggesting indirect kinase or phosphatase regulation.
Destruction of LKB1-induced polarity by disrupting adherens junctions though Ca2+-chelation completely suppressed Yap phosphorylation. Since P-Yap has a short half-life (Figure 2d), the small decrease in total Yap level in LKB1 cells following EGTA treatment observed in Figure 6c suggests that P-Yap is degraded in this process. Strikingly, expression of the degradation-resistant Yap5SA mutant reverted LKB1’s growth inhibitory activity without disrupting LKB1-induced cell cohesion (most evident in Figure 3c). This both strongly implicates Yap degradation in mediating LKB1 proliferative suppression and demonstrates separation of cell polarity from proliferative influence. Taken together, these results suggest a novel mechanism where LKB1 promotes the assembly of a cellular structure that in turn suppresses the Yap oncoprotein. Identification of the novel Yap kinase and determining whether Yap must be sequestered to polarized structures prior to degradation will be the focus of future studies. In this regard, many cell junction-associated proteins (PALS1, PATJ, MUPP1, angiomotin, ZO2 and α-catenin) that in turn associate with various Ser/Thr protein kinases have been reported to interact with Yap or TAZ (reviewed in (19)).
In summary, we have identified the Yap transcriptional coactivator as a downstream target of the LKB1 tumor suppressor. LKB1 induces phosphorylation of Yap, leading to Yap nuclear exclusion and ultimately its degradation. However this was independent of the canonical Yap kinases, Lats1/2, and metabolic downstream targets of LKB1, AMPK and mTORC1. Instead, events resulting from LKB1-induced morphological transformation were responsible for Yap phosphorylation. Interfering with this inhibitory event by mutation of Yap phosphorylation sites partially reversed LKB1’s effect. Altogether, these findings point to Yap as a novel mediator of LKB1 tumor suppressive function.
Materials and methods
Reagents
LKB1 PCR products were cloned into pCDH-513B (System Biosciences) or pQCXI (Clonetech). 5xGal4-luc (41). Gal4-TEAD4, 2xFlagYap mutants (18, 42) and non-targeting shRNAs (43) were from AddGene (plasmid numbers 24640, 19045, 27370, 27371 and 1864). FlagYap cDNAs were subcloned into a modified pCDH vector using NotI/XbaI. LKB1 shRNA plasmids were from Sigma-Aldrich. siRNAs were from Thermo. Anti-phospho-YapS127, LKB1, S6K, P-S6K, ACC and P-ACC were from Cell Signaling. Anti-Yap (Abnova), β-actin and U1snRNP (Santa Cruz Biotech), anti-HA tag (Covance). Cycloheximide (Sigma-Aldrich), MG132 (CalBiochem), Torin1 (Whitehead and DFCI). HeLa and 293T cells were grown in DMEM/antibiotics/10% FBS (Atlanta Biologicals). NTERT cells were grown in Gibco keratinocyte media (Invitrogen). RIPA buffer: 50mM Tris pH 7.4, 150mM NaCl, 2mM EDTA, 1% TritonX100, 1% SDS. Fractionation buffer: 10mM HEPES pH 7.9, 1.5mM MgCl2, 10mM KCl, 0.2% IGPAL. Buffers also contained 5mM NaF, 10mM β-glycerol phosphate, 1mM PMSF and 20μg/ml aprotinin.
Transfection/infection and luciferase assays
Lentiviruses were generated in 293T cells. Target cells were plated into filtered lentiviral media containing 5ng/ml polybrene. 48 hrs later, cells were selected in 1μg/ml puromycin until mock transduced cells died (usually 2 days). Nutrient stress used glucose deficient DMEM with serum. For luciferase assays, 0.1μg Gal4-TEAD4 and 0.8μg 5xGal4-luc were transfected using Lipofectamine2000 (Invitrogen). Two days later, luciferase activity was measured using Promega reagents.
Nuclear fractionation
Cells were incubated in Fractionation buffer for 15 min, 4°C, then spun, 10 mins, 3,000 rpm and cytoplasmic fraction collected. Nuclear pellets were washed twice, resuspended in RIPA buffer and cleared by centrifugation (14,000 rpm). U1snRNP and β-actin/GAPDH were used as loading controls for the respective fractions.
Immunofluorescence staining and live cell imaging was performed as described (44).
Cell size analysis was performed using a FACSCalibur cell sorter to detect populations with high or low LKB1/GFP expression. The mean forward scatter (FSC) of each population was calculated and normalized to vector control to obtain relative cell size.
The relative quantification of mRNA levels
RNA was isolated using TRIzol (Invitrogen). First strand cDNA synthesis used reverse transcriptase (New England Biolabs) and oligo dT primer for selection of polyA-containing mRNA. qRT-PCR detection of transcripts was performed using the LightCycler-RNA master mix and Universal Probes Library (Roche). All procedures were according to manufacturers’ protocols. Primer sequences and probe numbers are available by request. CTGF mRNA levels were normalized to GAPDH expression.
Matrigel assay was performed as described (45)
Blots, densitometry, and statistical analysis
Western blots were viewed using an Odyssey infrared imaging system (Licor Biosciences) or enhanced chemiluminescence (Thermo) and X-ray film. P-values were derived using Student T-test. ANOVA analyses used Origin 8.5.1 software.
Supplementary Material
Acknowledgments
This work was supported by US Department of Defense award W81XWH1110355, the LAM Foundation and a research support funds grant from IUPUI to LAQ.
We thank Dan Spandau for NTERT cells, Jacob Adler and Bill Ranahan for experimental advice.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909–25. doi: 10.1101/gad.1211604. [DOI] [PubMed] [Google Scholar]
- 2.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–75. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–74. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hezel AF, Bardeesy N. LKB1; linking cell structure and tumor suppression. Oncogene. 2008;27:6908–19. doi: 10.1038/onc.2008.342. [DOI] [PubMed] [Google Scholar]
- 5.Gill RK, Yang SH, Meerzaman D, Mechanic LE, Bowman ED, Jeon HS, et al. Frequent homozygous deletion of the LKB1/STK11 gene in non-small cell lung cancer. Oncogene. 2011;30:3784–91. doi: 10.1038/onc.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–43. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Veelen W, Korsse SE, van de Laar L, Peppelenbosch MP. The long and winding road to rational treatment of cancer associated with LKB1/AMPK/TSC/mTORC1 signaling. Oncogene. 2011;30:2289–303. doi: 10.1038/onc.2010.630. [DOI] [PubMed] [Google Scholar]
- 8.Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, Peters PJ, et al. Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell. 2004;116:457–66. doi: 10.1016/s0092-8674(04)00114-x. [DOI] [PubMed] [Google Scholar]
- 9.Partanen JI, Nieminen AI, Makela TP, Klefstrom J. Suppression of oncogenic properties of c-Myc by LKB1-controlled epithelial organization. Proc Natl Acad Sci U S A. 2007;104:14694–9. doi: 10.1073/pnas.0704677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–34. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, et al. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr Biol. 2006;16:2090–100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–95. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, et al. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–10. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertini E, Oka T, Sudol M, Strano S, Blandino G. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8:49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
- 17.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–61. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP) Genes Dev. 2010;24:72–85. doi: 10.1101/gad.1843810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mauviel A, Nallet-Staub F, Varelas X. Integrating developmental signals: a Hippo in the (path)way. Oncogene. 2012;31:1743–56. doi: 10.1038/onc.2011.363. [DOI] [PubMed] [Google Scholar]
- 20.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–47. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–71. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basu S, Totty NF, Irwin MS, Sudol M, Downward J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell. 2003;11:11–23. doi: 10.1016/s1097-2765(02)00776-1. [DOI] [PubMed] [Google Scholar]
- 23.Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–7. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- 24.Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, Hafen E, et al. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. 1999;126:5365–72. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- 25.Granot Z, Swisa A, Magenheim J, Stolovich-Rain M, Fujimoto W, Manduchi E, et al. LKB1 regulates pancreatic beta cell size, polarity, and function. Cell Metab. 2009;10:296–308. doi: 10.1016/j.cmet.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–60. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 27.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–18. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–32. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castro AF, Rebhun JF, Clark GJ, Quilliam LA. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem. 2003;278:32493–6. doi: 10.1074/jbc.C300226200. [DOI] [PubMed] [Google Scholar]
- 30.Saucedo LJ, Edgar BA. Why size matters: altering cell size. Curr Opin Genet Dev. 2002;12:565–71. doi: 10.1016/s0959-437x(02)00341-6. [DOI] [PubMed] [Google Scholar]
- 31.Sakamoto K, McCarthy A, Smith D, Green KA, Grahame Hardie D, Ashworth A, et al. Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO J. 2005;24:1810–20. doi: 10.1038/sj.emboj.7600667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–14. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 33.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 34.Zong H, Kaibuchi K, Quilliam LA. The insert region of RhoA is essential for Rho kinase activation and cellular transformation. Mol Cell Biol. 2001;21:5287–98. doi: 10.1128/MCB.21.16.5287-5298.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Asuri S, Yan J, Paranavitana NC, Quilliam LA. E-cadherin dis-engagement activates the Rap1 GTPase. J Cell Biochem. 2008;105:1027–37. doi: 10.1002/jcb.21902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baas AF, Smit L, Clevers H. LKB1 tumor suppressor protein: PARtaker in cell polarity. Trends Cell Biol. 2004;14:312–9. doi: 10.1016/j.tcb.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Humbert N, Navaratnam N, Augert A, Da Costa M, Martien S, Wang J, et al. Regulation of ploidy and senescence by the AMPK-related kinase NUAK1. EMBO J. 2010;29:376–86. doi: 10.1038/emboj.2009.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L, Ulbrich J, Muller J, Wustefeld T, Aeberhard L, Kress TR, et al. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature. 2012;483:608–12. doi: 10.1038/nature10927. [DOI] [PubMed] [Google Scholar]
- 39.Bao Y, Nakagawa K, Yang Z, Ikeda M, Withanage K, Ishigami-Yuasa M, et al. A cell-based assay to screen stimulators of the Hippo pathway reveals the inhibitory effect of dobutamine on the YAP-dependent gene transcription. J Biochem. 2011;150:199–208. doi: 10.1093/jb/mvr063. [DOI] [PubMed] [Google Scholar]
- 40.Carretero J, Medina PP, Blanco R, Smit L, Tang M, Roncador G, et al. Dysfunctional AMPK activity, signalling through mTOR and survival in response to energetic stress in LKB1-deficient lung cancer. Oncogene. 2007;26:1616–25. doi: 10.1038/sj.onc.1209951. [DOI] [PubMed] [Google Scholar]
- 41.Hauser CA, Westwick JK, Quilliam LA. Ras-mediated transcription activation: analysis by transient cotransfection assays. Methods Enzymol. 1995;255:412–26. doi: 10.1016/s0076-6879(95)55043-7. [DOI] [PubMed] [Google Scholar]
- 42.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–41. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 43.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 44.Heller B, Adu-Gyamfi E, Smith-Kinnaman W, Babbey C, Vora M, Xue Y, et al. Amot recognizes a juxtanuclear endocytic recycling compartment via a novel lipid binding domain. J Biol Chem. 2010;285:12308–20. doi: 10.1074/jbc.M109.096230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ranahan WP, Han Z, Smith-Kinnaman W, Nabinger SC, Heller B, Herbert BS, et al. The adaptor protein AMOT promotes the proliferation of mammary epithelial cells via the prolonged activation of the extracellular signal-regulated kinases. Cancer Res. 2011;71:2203–11. doi: 10.1158/0008-5472.CAN-10-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.