Abstract
Methylation of cytosines is a major epigenetic modification in mammalian genomes. The levels and patterns of DNA methylation are the results of the opposing actions of methylating and demethylating machineries. Over the last two decades, great progress has been made in elucidating the methylating machinery, including the identification and functional characterization of the DNA methyltransferases (Dnmts). However, the mechanisms of demethylation and the major players involved had been elusive. A major breakthrough came in 2009, when the ten-eleven translocation (Tet) family of proteins was discovered as 5-methylcytosine (5mC) dioxygenases that convert 5mC to 5-hydroxymethylcytosine (5hmC). Studies in the past several years have established that 5hmC serves as an intermediate in DNA demethylation and that Tet proteins play important roles in epigenetic reprogramming in early embryos and primordial germ cells (PGCs). In this review, we discuss recent advances in this exciting field, focusing on the role of Tet proteins in mammalian development.
Keywords: 5-hydroxymethylcytosine, 5-methylcytosine, DNA methylation, Tet1, Tet2, Tet3
Introduction
Methylation at the 5-position of cytosine (5-methylcytosine, 5mC), which occurs predominantly in the context of CpG dinucleotides, is a common modification present in mammalian genomes. DNA methylation is essential for mammalian development and plays crucial roles in a variety of biological processes, including regulation of gene expression, genomic imprinting, X chromosome inactivation, and retrotransposon silencing1,2. Aberrant changes of genomic DNA methylation patterns and genetic mutations of components of the DNA methylation machinery are linked to numerous human diseases, including developmental syndromes, neurological diseases, immunological disorders, and various types of cancer3–6.
In mice, DNA methylation patterns are established and maintained by three active DNA methyltransferases (Dnmts) – Dnmt1, Dnmt3a, and Dnmt3b. Dnmt3a and Dnmt3b function primarily as de novo methyltransferases that set up DNA methylation patterns during early embryogenesis, whereas Dnmt1 is the major maintenance enzyme that copies the CpG methylation pattern from the parental strand onto the daughter strand during DNA replication7. Genetic studies revealed that Dnmt1 and Dnmt3b are essential for embryogenesis and Dnmt3a is required for postnatal survival8–10. Dnmt3a also cooperates with its co-factor Dnmt3L, a Dnmt3-like protein with no enzymatic activity, in mediating DNA methylation in developing germ cells, including the establishment of methylation marks at imprinting control regions (ICRs)11–13.
DNA methylation is considered to be a relatively stable modification. However, waves of global demethylation occur in two developmental stages – preimplantation embryos and developing primordial germ cells (PGCs) – through both DNA replication-independent “active” and DNA replication-dependent “passive” processes2,14. Progress in understanding the mechanisms of demethylation and the major players involved had been slow until the recent discovery that 5mC can be converted to 5-hydroxymethylcytosine (5hmC) by the ten-eleven translocation (Tet) family of dioxygenases15,16. Studies in the past several years have revealed that 5hmC is an intermediate in the process of demethylation and distinct Tet proteins appear to be involved in methylation erasure in the zygote and PGCs. Evidence has also emerged for the involvement of Tet-mediated 5mC oxidation in other biological processes.
In this review, we will discuss recent progress in our understanding of the biological functions of 5hmC and Tet proteins with an emphasis on mammalian development.
Tet proteins as 5mC dioxygenases
Although 5hmC was identified in mammalian DNA in 197217, its significance and biological function had not been explored, largely because it is present in relatively low levels in most cell types. In 2009, two research groups reported abundant 5hmC in mouse Purkinjie neurons and embryonic stem (ES) cells15,18. More importantly, Tahiliani et al. identified Tet1, Tet2, and Tet3 as mammalian homologues of the trypanosome proteins JBP1 and JBP2, which oxidize the 5-methyl group of thymine, and experimentally demonstrated that Tet1 has the capacity to catalyze the conversion of 5mC to 5hmC15. Shortly afterwards, Tet2 and Tet3 were also shown to have 5mC hydroxylase activity16,19. Subsequent studies revealed that Tet proteins can further catalyze the oxidation of 5hmC to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC), two less abundant bases20,21.
The Tet proteins belong to the 2-oxoglutarate (2OG)- and Fe(II)-dependent dioxygenase (2OGFeDO) superfamily15,22. All three Tet proteins contain a C-terminal catalytic domain, which consists of a cysteine-rich region and a double-stranded β-helix (DSBH) fold characteristic of the 2OGFeDO superfamily. Tet1 and Tet3, but not Tet2, also contain an N-terminal CXXC zinc finger domain, a DNA-binding motif (Figure 1).
Figure 1.
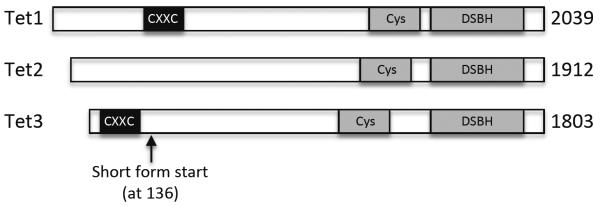
Schematic diagrams of mouse Tet proteins. There are three Tet proteins in mice: Tet1, Tet2, and Tet3. They all have a C-terminal catalytic domain, consisting of a cysteine-rich region and the double-stranded β-helix (DSBH) fold characteristic of the 2OG-dependent and Fe(II)-dependent dioxygenase family. Tet1 and Tet3, but not Tet2, have an N-terminal CXXC zinc finger domain, a DNA-binding domain.
The CXXC domain, present in multiple chromatin-interacting proteins such as CFP1, MBD1, MLL, and Dnmt1, has been shown to selectively bind unmethylated CpG dinucleotides23. Sequence alignment revealed that the CXXC domains of Tet1 and Tet3 lack a KFGG motif that is present in many other CXXC domains23,24. The DNA-binding property of the Tet1 CXXC domain is controversial: one report showed no specific DNA-binding activity24, whereas another report showed binding to unmodified, 5mC-modified, and 5hmC-modified CpG-rich DNA25. The discrepancy could be due to the different DNA-binding assays and substrates used in these studies. Recently, it was shown that the CXXC domains of Xenopus and human TET3 bind only unmodified cytosines, regardless of the sequence contexts (with a slight preference for CpG content), and that in Xenopus, the CXXC domain of Tet3 works cooperatively with the catalytic domain in targeting Tet3 to its binding sites during development26. Notably, mouse Tet3 has a shorter isoform that lacks the N-terminal 135 amino acids, including the CXXC domain (GenBank accession No. NM_183138). It would be interesting to determine the expression patterns and functional differences of the two isoforms.
Tet2 lost its CXXC domain during evolution due to a chromosomal inversion event, which split the ancestral Tet2 gene into two distinct genes, Idax, which encodes a protein containing the ancestral CXXC domain of Tet2, and Tet2, which encodes the current Tet2 protein. Interestingly, the two proteins physically interact27. IDAX, which preferentially binds DNA sequences containing unmethylated CpG via its CXXC domain, could play a role in recruiting Tet2 to its genomic targets. Strikingly, IDAX induces Tet2 degradation by caspase-mediated cleavage27.
5hmC, 5fC, and 5caC as intermediates of DNA demethylation pathways
The finding that 5mC can be converted to 5hmC by Tet proteins immediately raised the possibility that this conversion could be involved in DNA demethylation15,16. Indeed, Tet-mediated oxidation of 5mC appears to be the only source of 5hmC, as Dnmt1/Dnmt3a/Dnmt3b triple knockout ES cells lack both 5mC and 5hmC28–30 and depletion of Tet proteins substantially reduces or abolishes the production of 5hmC in various cells and tissues15,16,29,31–37. However, Tet proteins alone do not seem to be sufficient to complete DNA demethylation by converting 5hmC, 5fC, or 5caC to unmodified cytosine. Thus, it is generally accepted that 5hmC, 5fC, and 5caC are intermediates in the process of DNA demethylation.
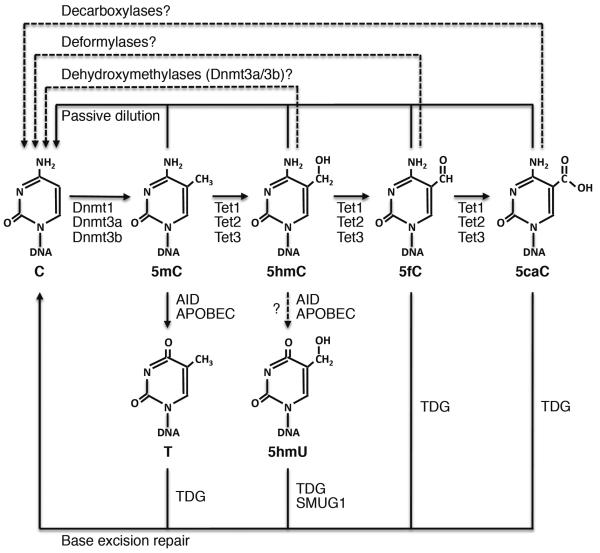
Various mechanisms of DNA demethylation involving Tet-mediated oxidation have been proposed (Figure 2). The simplest mechanism is “passive” dilution of 5hmC, 5fC, and 5caC owing to the lack of maintenance during DNA replication. In support of this mechanism, in vitro assays have revealed that Dnmt1 methylates hemi-hydroxymethylated CpG sites much more poorly than hemi-methylated CpG sites38,39.
Figure 2.
Proposed DNA demethylation pathways involving Tet proteins. Tet proteins catalyze 5mC oxidation to 5hmC, which can be further converted to 5fC and 5caC by Tet proteins or to 5hmU by AID/APOBEC deaminases (recent evidence suggests that 5hmC is an unlikely substrate for AID/APOBEC). 5mC can also be deaminated to thymine (T) by AID/APOBEC. 5fC, 5caC, 5hmU, and T can then be excised by glycosylases (TDG and SMUG1) and replaced by unmodified cytosine (C) following base excision repair. Since 5hmC, 5fC, and 5acC are poorly recognized by Dnmt1, demethylation can also be achieved by passive dilution with DNA replication. In addition, Dnmt3a and Dnmt3b have been shown to function as dehydroxymethylases that directly convert 5hmC to C in vitro, and putative deformylases and decarboxylases could directly convert 5fC and 5caC, respectively, to C. Solid lines represent processes with relatively strong evidence, and dashed lines represent processes that need to be further confirmed.
Several DNA replication-independent “active” pathways have also been suggested. First, 5hmC can be further oxidized to 5fC and 5caC, which can be recognized and excised from DNA by thymine DNA glycosylase (TDG)20,40,41. The resulting abasic site could then be repaired by the base excision repair (BER) pathway, thus generating an unmodified cytosine. Another possibility is that deformylases or decarboxylases could convert 5fC and 5caC directly to unmodified cytosine, although whether such enzymes exist remains an open question. Second, the AID/APOBEC family of deaminases has been shown to deaminate 5hmC to 5-hydroxymethyluracil (5hmU), which can then be excised by TDG and SMUG1, another DNA glycosylase, and replaced by cytosine through BER42,43. Deamination of 5mC by AID/APOBEC enzymes, resulting in a T:G mismatch leading to subsequent repair by TDG and BER, has also been implicated in DNA demethylation43–46. Third, a recent study provided in vitro evidence that Dnmt3a and Dnmt3b, in addition to their methyltransferase activity, function as dehydroxymethylases that convert 5hmC directly to cytosine. The methyltransferase and dehydroxymethylase activities seem to be regulated by the redox state of the enzymes. Reduction conditions (e.g. the presence of DTT or β-mercaptoethanol) inhibit their dehydroxymethylase activity, whereas oxidation conditions (e.g. presence of H2O2) inhibit their methyltransferase activity47. The bacterial HhaI methyltransferase (M. HhaI) has also been shown to have dehydroxymethylase activity in vitro48. Interestingly, a previous study suggests that Dnmt3a and Dnmt3b exhibit dual actions in mammalian cells, being involved in both CpG methylation and active demethylation at some loci (e.g. pS2 gene promoter), although the mechanism of demethylation was suspected to involve Dnmt3a/3b-mediated deamination of 5mC, TDG, and BER49. It would be interesting to re-visit the mechanism of demethylation and determine whether the dehydroxymethylase activities of Dnmt3a and Dnmt3b are partly responsible.
Tet proteins in demethylation in preimplantation embryos
Genome-wide analysis reveals that the male and female gametes have different levels of CpG methylation, with ~90% in sperm and ~40% in oocytes50,51. After fertilization, most DNA methylation marks inherited from gametes are erased during preimplantation development, exceptions including those associated with ICRs and intracisternal-A particles (IAPs) that resist this wave of global demethylation50–53. The mechanisms by which the paternal and maternal genomes undergo demethylation are distinct. In the zygote, the male pronucleus, but not its female counterpart, undergoes rapid global loss of 5mC before the onset of DNA replication, suggesting an active mechanism54–56. In contrast, the maternal genome is passively demethylated during cleavage divisions, presumably due to the exclusion of Dnmt1, the maintenance DNA methyltransferase, from the nucleus57.
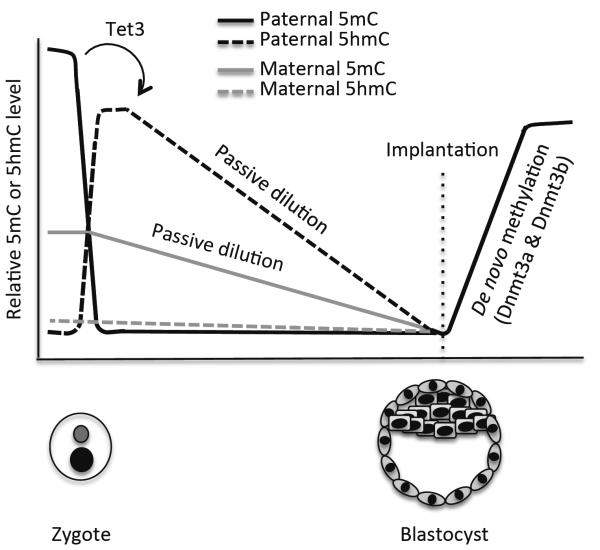
Recent studies using immunofluorescence revealed that, in the zygote, concomitant with the loss of the 5mC signal in the male pronucleus, there is a dramatic increase in 5hmC, as well as 5fC and 5caC, thus suggesting Tet-mediated 5mC oxidation58–61. Tet3, but not Tet1 and Tet2, is highly expressed in oocytes and zygotes36,58,59. Indeed, depletion of maternal Tet3 blocks the conversion of 5mC to 5hmC in the male pronucleus in the zygote36,58. 5mC oxidation seems to be a key step in the erasure of paternal methylation marks, as Tet3 deficiency inhibits demethylation of paternal genes36. Although BER has been proposed to be involved in active demethylation in preimplantation embryos62,63, 5hmC, 5fC, and 5caC do not appear to be rapidly replaced by unmodified cytosine. Instead, they persist in the paternal genome and gradually decline during cleavage divisions36,59–61. These results suggest that, while 5hmC, 5fC, and 5caC are generated in the zygote by an enzyme-catalyzed process, their loss during preimplantation development is primarily through a DNA replication-coupled passive process (Figure 3).
Figure 3.
DNA demethylation during preimplantation development. Shortly after fertilization, paternal 5mC is rapidly oxidized by Tet3. The resulting 5hmC, as well as maternal 5mC, gradually declines during subsequent cleavage divisions primarily through passive dilution. After implantation, Dnmt3a/3b-mediated de novo methylation occurs to establish lineage-specific methylation patterns.
Although the maternal and paternal genomes are exposed to an identical environment in the zygote, the maternal genome is protected from Tet3-mediated 5mC oxidation. PGC7 (also known as Stella and Dppa3), a maternal factor, has recently been shown to be required for this protection. Depletion of maternal PGC7 results in conversion of 5mC to 5hmC in both the male and female pronuclei58. Consistent with this finding, a previous study showed that PGC7 protects the maternal genome from demethylation in early embryos64. In normal zygotes, Tet3 is enriched and preferentially associated with the male pronucleus36,65. PGC7 seems to bind histone H3K9me2, which is abundant in the maternal chromatin but absent in the paternal chromatin with the exception of some imprinted loci, and inhibit Tet3 binding to the maternal chromatin and paternally imprinted loci65.
Reprogramming of the parental genomes in early embryos is believed to be important for the establishment of totipotency, but its biological significance remains largely unknown. Embryos conceived from Tet3-depleted oocytes implant normally but show high frequency of degeneration and morphological abnormalities, starting from midgestation, with only ~20% surviving to term36, whereas embryos without PGC7 show preimplantation defects and rarely reach the blastocyst stage66. These findings support the notion that epigenetic reprogramming is crucial for embryonic development, although the developmental phenotypes observed may not be entirely attributable to defects in epigenetic reprogramming.
Tet proteins in demethylation in PGCs
In mice, PGCs are specified around embryonic day (E) 7.25 in the epiblast of the developing embryo, with the involvement of BMP signaling and the transcription factors BLIMP1 and PRDM1467. Shortly afterwards, PGCs begin migrating along the embryonic-extraembryonic interface and eventually arrive at the genital ridge, mostly by E11.568. PGCs initially have similar epigenetic marks as other epiblast cells, including significant levels of DNA methylation69,70, and thus need to be reprogrammed to generate an epigenome for the development of germ cells.
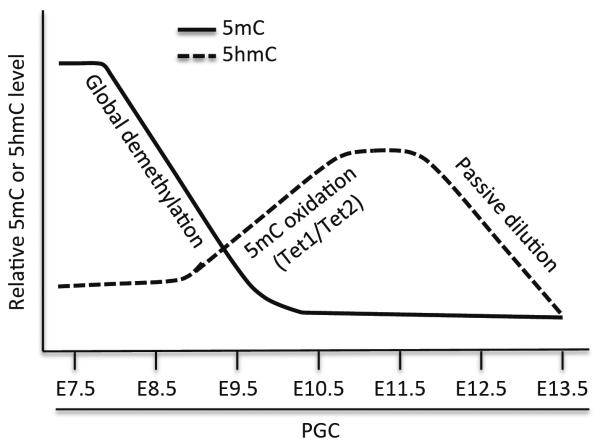
Previous studies indicated that, during their migration, PGCs undergo genome-wide demethylation, including the erasure of DNA methylation marks at ICRs71,72. Exceptions include IAPs and other active retrotransposons, which appear to be resistant to complete demethylation71–73. In the last several years, a number of groups have generated genome-wide high-resolution DNA methylation profiles of PGCs at different stages of the reprogramming process45,73–77. These studies reveal that PGCs undergo demethylation in two phases. The first phase occurs during PGC expansion and migration from ~E8.5 and involves global demethylation, affecting sequences of almost all genomic features. Passive demethylation may play a major role in this phase of genome-wide loss of methylation, as Dnmt3a and Dnmt3b, as well as Uhrf1 (also known as NP95), an essential factor for Dnmt1 function, are repressed in PGCs78,79. The second phase occurs from E9.5 to E13.5 and affects specific loci, including ICRs, CGIs on the X chromosome, and germline-specific genes73–76.
Recent studies provide evidence for the involvement of Tet-mediated 5mC oxidation in demethylation in PGCs37,75,76,80–82. Tet1 and Tet2 are expressed in PGCs between E9.25 and E11.5, but Tet3 is undetectable in PGCs75,80,81. Hackett et al. and Yamaguchi et al. used immunofluorescence to analyze PGCs at various time points and showed that both 5mC and 5hmC levels are low at E8.5, 5hmC levels begin to increase between E9.5 and E10.5, peak at ~E11.5, and then gradually decline from E11.5 to E13.575,76. Genetic studies reveal that deficiency for Tet1 or both Tet1 and Tet2 has no effect on global demethylation in PGCs, but results in defective demethylation and altered expression of specific genes, including meiotic genes and imprinted genes37,80. These results suggest that Tet1 and Tet2 are responsible for the production of 5hmC in PGCs and that 5hmC enrichment is followed by replication-coupled dilution. Using PGCs differentiated from wild-type or Tet1- and Tet2-depleted ES cells in vitro, Vincent et al. showed that, in the absence of Tet1 and Tet2, the first phase of global demethylation is unaffected, but numerous promoters and gene bodies become hypermethylated81. Taken together, these findings suggest that Tet-mediated conversion of 5mC to 5hmC is mainly involved in the second phase of demethylation in PGCs, including the erasure of imprints at ICRs (Figure 4).
Figure 4.
DNA demethylation in PGCs. DNA demethylation in PGCs occurs in two phases. The first phase involves global demethylation. The second phase affects specific loci, including ICRs. Tet1/Tet2-mediated 5mC oxidation occurs mainly in the second phase, and 5hmC enrichment is followed by gradual decline at a rate consistent with passive dilution.
Previous studies have shown that embryonic germ cells (EGCs) have the capacity to reprogram somatic genomes, including the erasure of imprints at ICRs, in hybrid cells83. Recently, Piccolo et al. used this system to address the requirement of Tet1 and Tet2 in EGC-induced pluripotent reprogramming. Intriguingly, Tet2 induces 5mC oxidation at pluripotent genes (e.g. Oct4), as well as expression of these genes, and is thus required for the efficient reprogramming capacity of EGCs, whereas Tet1 is necessary to induce 5mC oxidation specifically at ICRs82. These results suggest that Tet1 and Tet2 may have distinct genomic targets.
Despite the participation of Tet1 and Tet2 in epigenetic reprogramming in PGCs, these enzymes do not seem to be essential for germ cell development and fertility. Mice deficient for either Tet1 or Tet2 or both are viable and fertile, although Tet1-null and Tet1/Tet2 double null female mice show reduced fertility owing to meiotic defects32,35,37,80,84.
Tet proteins in embryonic and postnatal development
Tet1, Tet2, and Tet3 show different expression patterns. Tet1 and Tet 2 are highly expressed in the inner cell mass (ICM) of mouse blastocysts, as well as in ES cells (which are derived from ICM), whereas Tet3 is highly expressed in mouse oocytes and zygotes16,58,59. Upon differentiation of mouse ES cells, Tet1 and Tet2 are rapidly downregulated and Tet3 is upregulated15,16,31. Tet2 and Tet3 also appear to be widely expressed, at various levels, in adult tissues16. Consistent with the distinct expression patterns of Tet proteins, recent genetic studies indicate that these enzymes have different functions in mammalian development.
Multiple groups have reported that depletion of Tet1 in mouse ES cells results in 5hmC reduction, alterations in gene expression, and defects in self-renewal or differentiation16,29–32. However, Tet1-null mice show no overt developmental abnormalities, though some mutant mice are slightly smaller at birth32,80. Several Tet2 mutant alleles have been generated. Tet2-null mice develop normally and are fertile35,84. However, Tet2 deletion, either systemically or in the hematopoietic compartment, results in hematological phenotypes in adult animals, characterized by progressive enlargement of the hematopoietic stem cell (HSC) pool and eventual myeloid malignancies33–35,84. Tet2 expression is ubiquitous in the hematopoietic compartment, including in the stem and progenitor subsets and in mature myeloid and lymphoid cells33,35. Although the molecular mechanisms by which Tet2 deficiency leads to the hematological phenotypes remain to be elucidated, Tet2-null mice show decreased levels of 5hmC and concurrent increased levels of 5mC in bone marrow and spleen33–35. Tet2 could regulate genes important for hematopoiesis by modulating DNA methylation. Consistent with the observed phenotypes in Tet2-deficient mice, TET2 is frequently mutated in patients with various myeloid malignancies, such as myelodysplastic syndromes (MDS), myeloproliferative neoplasms MPN), chronic myelomonocytic leukemia (CMML), acute myeloid leukemia (AML), and secondary AML (sAML)85. The fact that Tet2 deficiency in mice recapitulates the major phenotypes in human patients suggests that TET2 mutations are driver mutations in hematological malignancies.
Tet1 and Tet2 seem to have partially redundant functions in embryonic development. Although a fraction of Tet1/Tet2 double knockout (DKO) mice are viable and fertile, some DKO embryos exhibits midgestation abnormalities and most DKO animals die perinatally with a variety of malformations, such as exencephaly, hemorrhage in the head, and profound growth retardation. Tet3 is upregulated in DKO mice, suggesting that compensation by Tet3 may contribute to the viability of DKO mice37. Systemic deletion of Tet3 leads to neonatal lethality, and maternal deletion impairs reprogramming in the zygote36. Given the wide expression of Tet3 in somatic tissues16, it would be interesting to determine the function of Tet3 in adult animals.
Concluding remarks
Since the discovery that Tet proteins can convert 5mC to 5hmC in 200915, tremendous progress has been made in understanding the functions of these enzymes and their products (5hmC, 5fC, and 5caC). It is now widely accepted that 5hmC, 5fC, and 5caC serve as intermediates in the process of DNA demethylation. Genetic studies in mice have confirmed the involvement of distinct Tet proteins in demethylation in the zygote and PGCs. Despite the progress, complete models of DNA demethylation in various physiological contexts remain to be assembled. One of the challenges is that multiple mechanisms seem to work cooperatively to achieve demethylation, and the relative contribution of these mechanisms and how they are orchestrated need to be clarified. Some of the proposed mechanisms may not be relevant or significant. For instance, recent biochemical studies suggest that 5hmC is an unlikely substrate for the AID/APOBEC family of deaminases86,87. Emerging evidence suggests that 5hmC, in addition to its role in DNA demethylation, may function as a stable epigenetic mark. Indeed, recent studies have identified several 5hmC specific “readers”, including MBD3, MeCP2, Uhrf1, and Uhrf288–91. Further work needs to be done to determine the significance of 5hmC in regulating chromatin structure and function, including gene expression, and the mechanism by which 5hmC is maintained. Tet proteins are large molecules and may have other functions, some of which may be independent of their enzymatic activities. For example, several recent reports show that Tet proteins interact with O-GlcNAc transferase (OGT) and promote histone O-GlcNAcylation92–94. Another area of intense research is the role of Tet proteins and 5hmC in cancer. We expect to see exciting discoveries addressing these issues in the coming years.
Acknowledgements
We thank A. K. Singh for help with formatting the references. Work in our laboratory is supported by the Cancer Prevention and Research Institute of Texas (CPRIT) (R1108). T.C. is a CPRIT Scholar in Cancer Research.
References
- 1.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 2.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 3.Tomizawa S, Sasaki H. Genomic imprinting and its relevance to congenital disease, infertility, molar pregnancy and induced pluripotent stem cell. J. Hum. Genet. 2012;57:84–91. doi: 10.1038/jhg.2011.151. [DOI] [PubMed] [Google Scholar]
- 4.Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nat. Med. 2012;18:1194–1204. doi: 10.1038/nm.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballestar E. Epigenetic alterations in autoimmune rheumatic diseases. Nat. Rev. Rheumatol. 2011;7:263–271. doi: 10.1038/nrrheum.2011.16. [DOI] [PubMed] [Google Scholar]
- 6.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr. Top. Dev. Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- 8.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 9.Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, et al. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- 10.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 11.Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- 12.Hata K, Okano M, Lei H, Li E. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development. 2002;129:1983–1993. doi: 10.1242/dev.129.8.1983. [DOI] [PubMed] [Google Scholar]
- 13.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- 14.Seisenberger S, Peat JR, Reik W. Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Curr. Opin. Cell Biol. 2013 doi: 10.1016/j.ceb.2013.02.013. doi:10.1016/j.ceb.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Penn NW, Suwalski R, O'Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frauer C, Rottach A, Meilinger D, Bultmann S, Fellinger K, Hasenoder S, et al. Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS One. 2011;6:e16627. doi: 10.1371/journal.pone.0016627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Xu C, Kato A, Tempel W, Abreu JG, Bian C, et al. Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell. 2012;151:1200–1213. doi: 10.1016/j.cell.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- 29.Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- 30.Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko M, Bandukwala HS, An J, Lamperti ED, Thompson EC, Hastie R, et al. Ten-Eleven-Translocation 2 (TET2) negatively regulates homeostasis and differentiation of hematopoietic stem cells in mice. Proc. Natl. Acad. Sci. USA. 2011;108:14566–14571. doi: 10.1073/pnas.1112317108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z, Cai X, Cai CL, Wang J, Zhang W, Petersen BE, et al. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 2011;118:4509–4518. doi: 10.1182/blood-2010-12-325241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 37.Dawlaty MM, Breiling A, Le T, Raddatz G, Barrasa MI, Cheng AW, et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell. 2013;24:310–323. doi: 10.1016/j.devcel.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 39.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, et al. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen CC, Wang KY, Shen CK. The mammalian de novo DNA methyltransferases DNMT3A and DNMT3B are also DNA 5-hydroxymethylcytosine dehydroxymethylases. J. Biol. Chem. 2012;287:33116–33121. doi: 10.1074/jbc.C112.406975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liutkeviciute Z, Lukinavicius G, Masevicius V, Daujotyte D, Klimasauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nat. Chem. Biol. 2009;5:400–402. doi: 10.1038/nchembio.172. [DOI] [PubMed] [Google Scholar]
- 49.Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 50.Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, et al. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8:e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, et al. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 53.Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, et al. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 2011;43:811–814. doi: 10.1038/ng.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 55.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, et al. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 56.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 57.Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, et al. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 58.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 59.Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. USA. 2011;108:3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 65.Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, et al. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486:415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- 66.Payer B, Saitou M, Barton SC, Thresher R, Dixon JP, Zahn D, et al. Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 2003;13:2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 67.Saitou M. Germ cell specification in mice. Curr. Opin. Genet. Dev. 2009;19:386–395. doi: 10.1016/j.gde.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 68.Molyneaux KA, Stallock J, Schaible K, Wylie C. Time-lapse analysis of living mouse germ cell migration. Dev. Biol. 2001;240:488–498. doi: 10.1006/dbio.2001.0436. [DOI] [PubMed] [Google Scholar]
- 69.Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 70.Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 71.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 72.Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 73.Guibert S, Forne T, Weber M. Global profiling of DNA methylation erasure in mouse primordial germ cells. Genome Res. 2012;22:633–641. doi: 10.1101/gr.130997.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, et al. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science. 2013;339:448–452. doi: 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamaguchi S, Hong K, Liu R, Inoue A, Shen L, Zhang K, et al. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 2013;23:329–339. doi: 10.1038/cr.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kobayashi H, Sakurai T, Miura F, Imai M, Mochiduki K, Yanagisawa E, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 2013;23:616–627. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurimoto K, Yabuta Y, Ohinata Y, Shigeta M, Yamanaka K, Saitou M. Complex genome-wide transcription dynamics orchestrated by Blimp1 for the specification of the germ cell lineage in mice. Genes Dev. 2008;22:1617–1635. doi: 10.1101/gad.1649908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kagiwada S, Kurimoto K, Hirota T, Yamaji M, Saitou M. Replication-coupled passive DNA demethylation for the erasure of genome imprints in mice. Embo J. 2013;32:340–353. doi: 10.1038/emboj.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamaguchi S, Hong K, Liu R, Shen L, Inoue A, Diep D, et al. Tet1 controls meiosis by regulating meiotic gene expression. Nature. 2012;492:443–447. doi: 10.1038/nature11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vincent JJ, Huang Y, Chen PY, Feng S, Calvopina JH, Nee K, et al. Stage-specific roles for tet1 and tet2 in DNA demethylation in primordial germ cells. Cell Stem Cell. 2013;12:470–478. doi: 10.1016/j.stem.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piccolo FM, Bagci H, Brown KE, Landeira D, Soza-Ried J, Feytout A, et al. Different Roles for Tet1 and Tet2 Proteins in Reprogramming-Mediated Erasure of Imprints Induced by EGC Fusion. Mol. Cell. 2013;49:1023–1033. doi: 10.1016/j.molcel.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tada M, Tada T, Lefebvre L, Barton SC, Surani MA. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. Embo J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 85.Abdel-Wahab O. Genetics of the myeloproliferative neoplasms. Curr. Opin. Hematol. 2011;18:117–123. doi: 10.1097/MOH.0b013e328343998e. [DOI] [PubMed] [Google Scholar]
- 86.Nabel CS, Jia H, Ye Y, Shen L, Goldschmidt HL, Stivers JT, et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nat. Chem. Biol. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rangam G, Schmitz KM, Cobb AJ, Petersen-Mahrt SK. AID enzymatic activity is inversely proportional to the size of cytosine C5 orbital cloud. PLoS One. 2012;7:e43279. doi: 10.1371/journal.pone.0043279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frauer C, Hoffmann T, Bultmann S, Casa V, Cardoso MC, Antes I, et al. Recognition of 5-hydroxymethylcytosine by the Uhrf1 SRA domain. PLoS One. 2011;6:e21306. doi: 10.1371/journal.pone.0021306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152:1146–1159. doi: 10.1016/j.cell.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 92.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol. Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 94.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. Embo J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]