Abstract
Cell cytotoxicity tests are among the most common bioassays using flow cytometry and fluorescence imaging analysis. The permeability of plasma membranes to charged fluorescent probes serves, in these assays, as a marker distinguishing live from dead cells. Since it is generally assumed that probes, such as propidium iodide (PI) or 7-amino-actinomycin D (7-AAD), are themselves cytotoxic, they are currently generally used only as the end-point markers of assays for live versus dead cells. In the current study, we provide novel insights into potential applications of these classical plasma membrane integrity markers in the dynamic tracking of drug-induced cytotoxicity. We show that treatment of a number of different human tumor cell lines in cultures for up to 72 h with the PI, 7-AAD, SYTOX Green (SY-G), SYTOX Red (SYR), TO-PRO, and YO-PRO had no effect on cell viability assessed by the integrity of plasma membrane, cell cycle progression, and rate of proliferation. We subsequently explore the potential of dynamic labeling with these markers in real-time analysis, by comparing results from both conventional cytometry and microfluidic chips. Considering the simplicity of the staining protocols and their low cost combined with the potential for real-time data collection, we show how that real-time fluorescent imaging and Lab-on-a-Chip platforms have the potential to be used for automated drug screening routines.
Tumor cell death is a stochastic process that often can be initiated and executed through multiple pathways, many of which are of importance in many physiological and pathological processes.1,2 Consequently, the observation of apoptosis and cell death serves as a useful end point for cell-based assays associated with the assessment of the toxic effects of pharmacological agents2,3 and is commonly implemented in flow cytometry formats using fluorescence imaging analysis.2,4 Despite the large variety of conventional fluorescence-based techniques that have been used to study the processes of cell death, there are many technological improvements that will make the implementation of these assays more effective.5 Most importantly, there is a real need for the development of new methods to provide real-time and kinetic analysis of drug-induced cytotoxicity and apoptosis.6 In such assays, the fluorescence markers, applied supravitally, should have a minimal effect on structure, function, and survival of the cells being studied.7
It is generally assumed that other commonly used fluorescent markers, such as SYTOX Green (SY-G), SYTOX Red (SY-R), TOPRO, and YO-PRO, are toxic to cells. As a consequence, these dyes are predominantly used in end-point assays8,9 to discriminate between the cells with preserved plasma membrane integrity (i.e., live cells) and those with impaired membranes (i.e., dead cells). Interestingly, no comprehensive study has been published that assesses the cytotoxic nature of these fluorescent probes.
The present study was therefore designed to investigate the cytotoxic profiles of cell impermeant fluorescent probes and subsequently to assess their potential to obtain dynamic and single-cell data on the stochastic process of cell death. We provide evidence that even when cell impermeant probes of nucleic acids are applied at concentrations higher than these used in cytotoxicity assays,3,4,9 for up to 72 h, there is no evidence of adverse effects on proliferating cells. Results indicate that plasma membrane integrity remains preserved and no significant perturbation of the cell cycle progression is apparent. These findings show that there is a possibility of using these dyes for dynamic labeling of fluorescence viability markers enabling their straightforward application in real-time single-cell analysis by both conventional flow cytometry and new microfluidic cell chips.
MATERIALS AND METHODS
Cell Culture and Labeling with Fluorescent Probes
The origin, characteristics, and culture of human tumor cell lines U937, HL60, K562, MOLT4, U2OS, and Saos2 were described previously.6 All cell cultures were maintained at 37 °C in a 5% CO2 humidified atmosphere. During experiments, cells were always in an asynchronous and exponential phase of growth. To induce apoptosis, cells were seeded on 48-well polystyrene cell culture plates (Nunc, Rochester, NY).
To explore the interaction of live cells with different fluorescent dyes, over extended periods of time, cells were continuously cultured in the presence of selected probes: SYTOX Green (SYG, 1–1000 nM), SYTOX Red (SY-R, 1–1000 nM), YO-PRO 1 (1–1000 nM), PO-PRO 1 (1–1000 nM), propidium iodide (PI, 0.1-10 μg/mL), and 7-aminoactinomycin D (7-AAD, 0.1–10 μg/ mL) (Molecular Probes, Eugene, OR) for up to 5 days.6,10
For dynamic, real-time quantification of cytotoxicity, cells were treated with staurosporine (STS; Sigma, 0.1–1 μM), cycloheximide (CHX; Sigma, 50–100 μg/mL), (camptothecin (CAM; Sigma, 1μ10 μM), or the fungal macrolide Brefeldin A (BFA; Sigma, 100μ500 nM). This was followed by staining with cell impermeable DNA stains at the end of experiment (end-point assay; 15 min at RT with 125 nM SY-G or 1 μg/mL PI). Alternatively, cells were cultured in the presence of low concentrations of selected DNA stains (25 nM SY-G or 0.25 μg/mL PI) and regularly examined in the course of the experiment (dynamic assay).
Flow Cytometry
Flow cytometry was performed using a BD FACSCalibur (BD Biosciences) analyzer, equipped with a 15 mW Argon ion and 20 mW red diode lasers, as described previously.6,10 A logarithmic amplification scale using the configuration of bandpass (BP) filters was applied: (i) 488 nm excitation line, FL1 channel (525 BP for collection of SYTOX Green and YO-PRO 1 fluorescence signals), FL2 channel (610 BP for collection of PI); (ii) 635 nm excitation line, FL4 channel (675 BP for collection of SYTOX Red fluorescence signals). Acquisition was performed in a 1024 channels resolution scale using CellQuest Pro software (Becton Dickinson). A typical run used a sample with 10 000 cells, so that a triplicate experiment involved ca. 30 000 cells. Data analysis was performed using Summit v3.1 (Dako Cytomation, Fort Collins, CO, USA) and an open access WinMDI v2.8 (http://facs.scripps.edu/software.html) software.
Microfluidic Chip-Based System
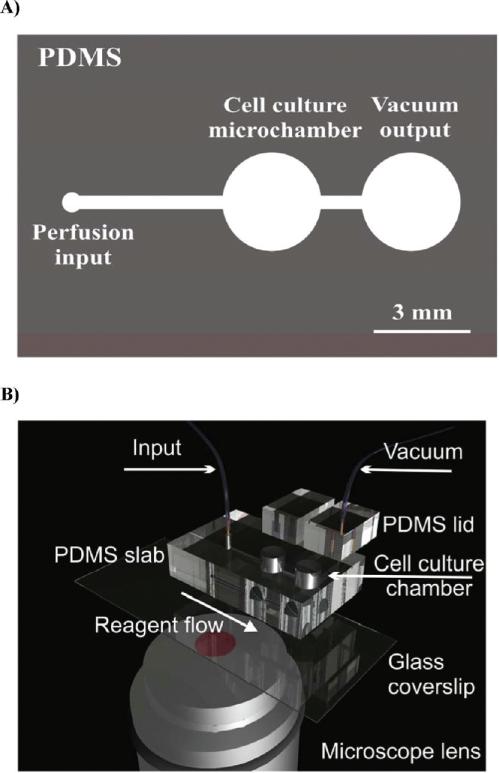
A microfluidic flow through chip-based cell culture system was fabricated in the biologically compatible elastomer polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning), as described previously.11 The device comprised two microculture chambers (3 mm in diameter) connected by a 250 μm wide perfusion channel. The device was made by casting a PDMS prepolymer (10:1 ratio w/w) against a negative relief pattern and cured thermally at 70 °C for 2 h; see Figure 1. Cell culture chambers were mechanically bored to enable microfluidic interconnects. The cast PDMS polymer microfluidic chips were sealed to quartz coverslips using conformal (reversible) bonding procedures and connected to an external syringe pump. Chip operation was described in detail elsewhere.12,13 Briefly, a vacuum driven chip allowed for a continuous perfusion of medium at a rate of up to 0.5 μL/min. Two cell culture chambers can be analyzed simultaneously on a motorized microscope stage. A 250 μm wide perfusion channel connecting two cell culture chambers permitted for a robust circulation of media and delivery of drugs while supporting long-term cell culture (Figure 1). Fluorescence images were acquired using a motorized Zeiss Axiovert 200 M epifluorescence microscope with appropriate fluorescence filter sets.
Figure 1.
Microculture device fabricated in biologically compatible elastomer. (A) Overview of the chip design. Microculture chambers are 3 mm in diameter and hold approximately 15 μL of cell culture medium. The vacuum driven chip allows for a continuous perfusion of medium at a rate of up to 0.5 μL/min. Two cell culture chambers can be analyzed simultaneously on a motorized stage. (B) Chip design explained on a 3D virtual prototype.
Data and Statistical Analysis
Data analysis and presentation was performed using ImageJ (freely available at http://rsb.info.nih.gov/ij/), Summit v3.1 (DakoCytomation, Fort Collins, CO), and open access WinMDI v2.8 (http://facs.scripps.edu/software.html) software. The student's t test was applied for comparison between groups with significance set at p < 0.05.
RESULTS
In all of our studies, we used flow cytometry and microfluidic devices to explore the interaction of live cells with different dyes, over extended periods of time. Data were collected and processed using procedures described in the Materials and Methods section.
Cell Impermeant DNA Probes Lack Short-Term and Long-Term Cytotoxicity
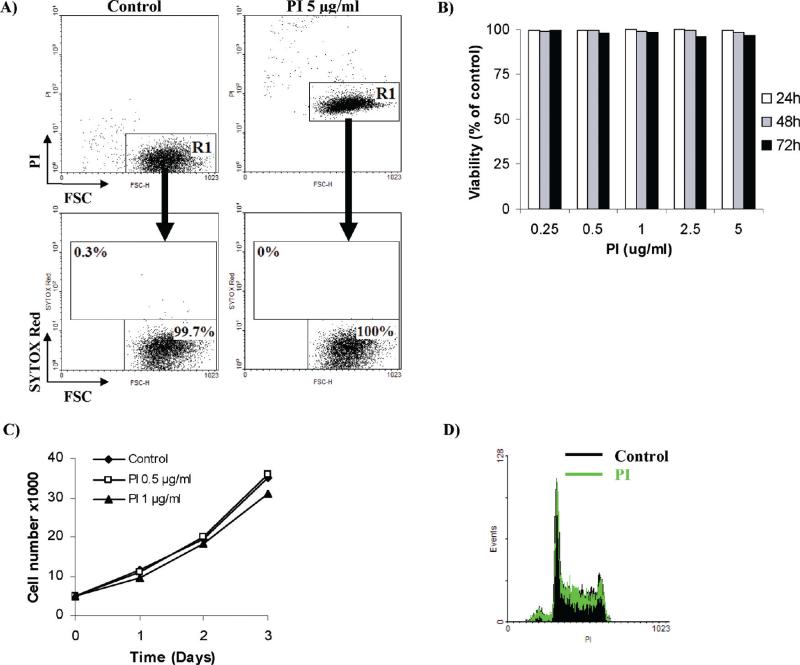
For initial screening, six cell impermeant DNA stains were chosen that differ in spectral properties, PI, 7-AAD, SY-G, SY-R, PO-PRO 1, and YO-PRO 1 (shown in Table 1). PI, the most universal probe for cytometry and imaging, was used as a reference for data presentation, throughout the study. To this end, human promyelocytic leukemia HL60 cells were treated with 5 μg/mL PI for up to 24 h to assess the impact of the dye alone upon cell viability. When analyzed by flow cytometry, all cells cultured in the presence of PI featured a profound increase in cell fluorescence (Figure 2A, upper panels). This observation was consistent with the hypothesis that PI has substantial cytotoxic properties and enters cells with permeabilized plasma membranes. We assumed, however, that if HL60 cells were indeed dead, then cell membranes should also become permeable to other cationic probes. Interestingly, when the subpopulation with elevated PI fluorescence was counterstained with a spectrally distinct (excitation 640 nm) marker of plasma membrane permeability (e.g., SY-R 5 nM), they did not exhibit elevated staining with this probe indicating that they were still viable (Figure 2A, lower panels). In fact, leukemia HL60 cells remained viable when challenged with a broad range of PI concentrations (0.25–5 μg/mL) for up to 72 h (Figure 2B). Importantly, it was found that when PI was used continuously in cell culture media, it also did not affect cell cycle and long-term cell proliferation (Figure 2C,D). Subsequent assessment of the remaining fluorescent probes on a panel of hematopoietic and epithelial tumor cell lines confirmed that these dyes affected neither cell viability nor proliferation during both short-term and long-term cell culture (Table 1). To further explore the effect of PI, we used invasive breast cancer MDA-MB-231 cells to show that the dye had no effect on directional cell motility, as shown using an established wound-closure assay (Table 2 and Supplementary Figure 1 in the Supporting Information).
Table 1.
Comparative Cytotoxicity of Six Cell Impermeant Nucleic Acid Stains
| PI | 7-AAD | SYTOX Green | SYTOX Red | YO-PRO 1 | PO-PRO 1 | |
|---|---|---|---|---|---|---|
| cell lines tested | suspension: U937, HL60, K562, MOLT-4, and Jurkat. adherent: U2OS, Saos2, MDA-MB-231, and 3T3. | |||||
| ex/em maximum (nm) | 536/617 | 546/647 | 504/523 | 640/658 | 491/509 | 435/455 |
| guide staining concentration | up to 5 μg/mL | up to 1 μg/mL | up to 1 mM | up to 10 nM | up to 1 mM | up to 1 mM |
| cytotoxic | - | - | - | - | - | - |
| phototoxica | - | N/A | - | - | - | - |
| influence on cell proliferation | - | N/A | - | - | - | - |
| influence on cell cycleb | - | N/A | - | - | - | - |
| influence on cell migrationc | - | N/A | - | - | - | - |
| dynamic viability labeling | + | + | + | + | + | + |
| photobleaching | slow (1-2 min | slow (1-2 min) | slow (1-2 min) | slow (1-2 min) | slow (1-2 min) | slow (1-2 min) |
| fluorescence signal | bright | dim | bright | bright | bright | dim |
| multiplexing | ± | ± | + | + | + | ± |
Phototoxic effects observed during kinetic cell imaging.
U937 and U2OS cell lines incubated with the probe for up to 48 h.
Directional cell motility using the would-closure assay on invasive breast cancer cells MDA-MB-23.
Figure 2.
Continuous exposure to PI has no effect on subsequent exclusion of SYTOX Red. (A) Human promyelocytic leukemia HL60 cells were exposed to increasing concentrations of propidium iodide (PI; 0.25–5 μg/mL) for up 72 h. Cell viability was estimated afterward using spectrally distinct fluorescent probe SYTOX Red. Note that no change in cell viability was assessed by exclusion of SYTOX Red following continuous exposure to PI. Similar results were achieved with SYTOX Green, SYTOX Red, PO-PRO 1, and YO-PRO 1. Results represent the mean of three independent experiments with SD values consistently lower than ± 3. (B) Reults show that the increased fluorescent signal from PI does not imply loss of cell viability. Human promyelocytic leukemia HL60 cells were incubated with or without 5 μg/mL PI for 24 h. Cells were then counterstained with 5 nM SYTOX Red and analyzed by flow cytometry. Note that the R1 subpopulation with elevated PI fluorescence (in the right panel) is not SYTOX Red+ and is thus presumed viable. Results are representative of three independent experiments. (C) Results show that PI does not disturb cell proliferation. HL60 cells were continuously challenged with low doses of PI (0.5 and 1 μg/mL) and the number of viable cells was assessed using the standard Trypan Blue assay during the 3 day study. Note the lack of cell growth retardation. The results represent the mean of at least three independent experiments. Normalized SD values were lower than ± 3. (D) Results show that exposure to PI does not disturb cell proliferation. HL60 cells were continuously challenged with 0.5 μg/mL PI for up to 48 h. Cells were then fixed, and their DNA content profile was analyzed by flow cytometry. Note the lack of any cell cycle disturbances.
Table 2.
Comparison between Conventional Flow Cytometry and Chip-Based Microfluidic Systems
| parameter | flow cytometry | chip-based microfluidic systems |
|---|---|---|
| sample size | 5 × 103-4 × 104 cells | 1 × 102-1 × 103 cells |
| analysis time | seconds | seconds to minutes |
| multiparametric analysis | yes | yes |
| nonoptical measurements | no | yes |
| single cell analysis | yes | yes |
| single cell real-time analysis | no | yes |
| sample manipulation during analysis | no | yes |
| integrated long-term cell culture | no | yes |
| reanalysis of the sample (e.g., fixation and secondary staining steps) | no | yes |
| cell sorting | yes | yes |
| potential for full automation | no | yes |
| manufacturing cost | high | low |
| running costs | moderate-high | low |
| operation training | complex | simple |
In a second series of experiments, we also set out to determine whether a combination of different fluorescent markers, with different plasma permeabilities, could be used during continuous monitoring in a robust cell-based assay. Using the knowledge that multiplexed exposure to cell impermeant cyanine stains has been shown to be advantageous in the discrimination and quantification of early apoptosis (using the differential uptake of these probes to explore the changes in lipid composition and/or structural relaxation of the plasma membrane8,9,14), we exposed human osteosarcoma U2OS cells to a mixture of PO-PRO 1 (250 nM), YO-PRO 1 (250 nM), and SY-R (5 nM) probes for up 5 days.
In a series of controls involving a reference chip, cells were also exposed to 1 μM Hoechst 33342 (as a positive control). No adverse effects on cell viability changes or cell growth retardation were observed following simultaneous exposure to a mixture of three cell impermeant DNA stains (Supplementary Figure 2 in the Supporting Information). Moreover, no dysfunctions in the mitochondrial function were observed during the duration of the experiments as assessed by the ΔΨm sensitive probe TMRM (Supplementary Figure 3 in the Supporting Information). In contrast, cell exposure to Hoechst 33342 led to rather rapid loss of cell viability and the capability to proliferate.
Dynamic Quantification of Drug-Induced Cytotoxicity
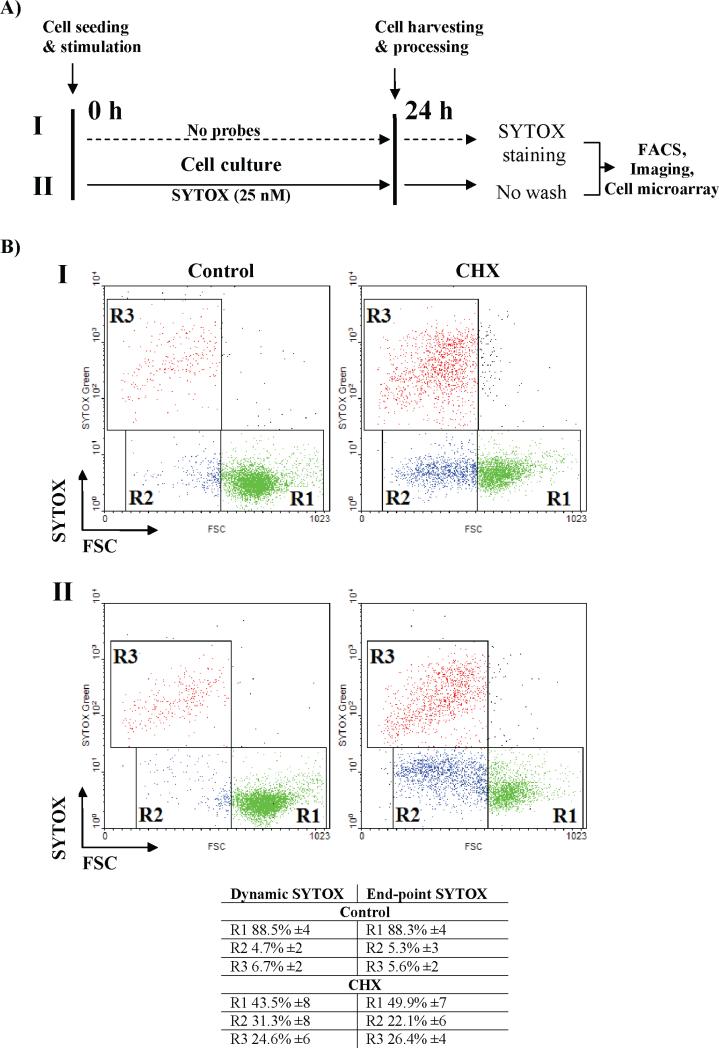
Based on these observations, we next determined the applicability of inert probes for dynamic quantification of cell death using both end-point assays and dynamic assays, based upon conventional flow cytometry. Human promyelocytic leukemia HL60 cells were exposed to a panel of apoptosis inducers (cycloheximide, STS, or CAM) for up to 24 h, at the concentrations described in the Materials and Methods. The cells were then probed using SY-G (125 nM) staining at the end of the experiment, as an end-point assay (Figure 3A, protocol I). Alternatively, cells were cultured in the presence of 25 nM SY-G with or without the indicated inducers of apoptosis (providing real-time kinetic data, as a dynamic assay). In the latter case, cells were immediately analyzed by flow cytometry at the end of the experiment without washing steps (Figure 3A, protocol II).
Figure 3.
(A) Dynamic tracking of cytotoxicity using simplified no-wash protocols: workflow of modified no-wash protocol. Note that viability marker SYTOX Green (100 nM) is continuously present in the culture medium (lower panel, II) as opposed to standard, end-point staining procedures (upper panel, I). (B) Human promyelocytic leukemia HL60 cells were exposed to a pro-apoptotic drug cycloheximide (CHX, 50 μg/mL) for 24 h. After stimulation, cells were collected and stained according to two protocols described in (A). Data were acquired using a FACS Calibur flow cytometer equipped with a 488 nm excitation line and a standard optical filter configuration. Note the excellent agreement between results obtained with end-point (I) vs dynamic no-wash (II) protocols. The results represent the mean of at least three independent experiments.
Under normal culture conditions, a small fraction of necrotic cells could be detected, with both assays showing comparable sensitivity (5.6 ± 2% end-point SY-G vs 6.7 ± 2% dynamic SY-G; Figure 3B). Upon treatment with the apoptosis inducer cycloheximide, end-point and dynamic SY-G assays again gave similar estimates of the three cell populations: live, apoptotic, and late apoptotic/necrotic (Figure 3B). Furthermore, both assays had comparable sensitivity over a broad range of stimuli and exposure times (R2 ≥ 0.97 for p < 0.05 in a Pearson and Lee linear correlation test) (not shown).
The low-dosage, continuous labeling procedure (protocol II) not only provided similar results to the standard end-point staining protocol (protocol I) but also provided a proof of principle that these assays could be adapted for a high-throughput screening protocol (HTS).6 For example, cells incubated in the presence of SY-G were viable even after 72 h, providing the potential for a noninvasive and long-term tracking of cell viability, in either a flow cytometry or a microfluidic format.
When the assay was implemented in a no-wash HTS protocol, in order to circumvent the elevated fluorescence signal, a 5-fold reduced concentration of SY-G was used without compromising the assay's sensitivity. Importantly, similar results were also obtained with the remaining fluorescent probes such as PI, SY-R, and YO-PRO 1 (Table 1). This opens up a broad spectrum of new opportunities for dynamic multispectral flow cytometry and time-lapse imaging, detailed here for the first time.
Kinetic Cytotoxicity Assays for On-Chip Analysis on a Single Cell Level
Microfluidic devices are considered as an emerging technology in the field of high-throughput screening.15–17 However, widespread application of on-chip cell-based assays still is primarily used in research-based applications. This is in part due to the slow implementation of new bioassays that can fully leverage the advantages of microchip devices.6 For our screening, we adapted a simple and self-contained microculture chip-based system, which supports a continuous perfusion of cells over extended periods of time (Figure 1) and which can be easily expanded to conduct high-throughput analyses.
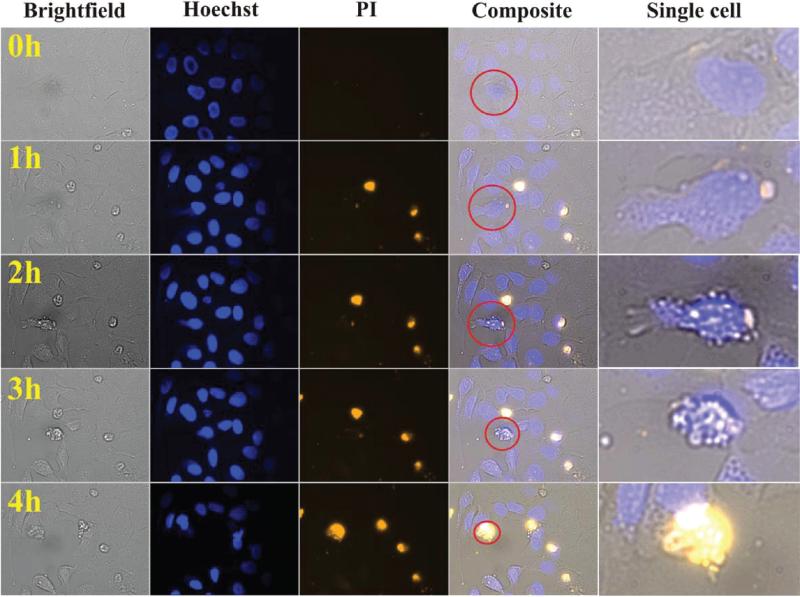
First, we performed a qualitative on-chip analysis of tumor cell death using automated time-resolved imaging. Human osteosarcoma U2OS cells were loaded onto a chip and allowed to attach to the glass surface overnight. Cells were then continuously perfused with 2 μM staurosporine in the presence of 0.25 μg/mL PI. The chips were placed inside a temperature-controlled chamber surrounding the microscope and imaged every 15 min over a period of 4 h (Figure 4). The first morphologically recognizable event of apoptosis was seen at approximately 1 h and involved loss of cell–cell contacts (Figure 4, right panels). Subsequent stages of apoptosis proceeded with (i) cell dehydration, shrinkage, and plasma membrane blebbing (2 h), (ii) rounding up and loss of attachment with the substrate (3 h), and (iii) increased plasma membrane permeability to PI (4 h). The latter events represent a final destabilization of plasma membrane structure during apoptosis (secondary necrosis; Figure 4, right panels). In contrast to a standard end-point analysis, on-chip kinetic imaging allowed for a dynamic and noninvasive observation of apoptotic cell death on a single-cell level.
Figure 4.
Time-resolved imaging of apoptosis at a single cell level. Human osteosarcoma U2OS cells were cultured on a chip and exposed to 2 μM staurosporine for up to 4 h in the continuous presence of the plasma membrane permeability marker PI. Red circles denote the magnified cell undergoing apoptosis (right panels). Note the subsequent stages of apoptosis that involve: (i) loss of cell–cell contacts (1 h); (ii) cell dehydration, shrinkage and plasma membrane blebbing (2 h); (iii) rounding up and loss of attachment with substrate (3 h); (iv) an increase in plasma membrane permeability to PI that represents a final destabilization of plasma membrane during apoptosis (4 h).
Next, we performed a quantitative study of drug-induced tumor cell death using a dynamic imaging of U2OS cells exposed to investigational anticancer agents: STS (2 μM) and BFA (2 μM). As a positive control, U2OS cells were first challenged with 20% DMSO, while for negative controls, the cell-laden chips were perfused with fresh DMEM medium supplemented only with 0.25 μg/mL PI, Figure 5. Approximately 300 cells were analyzed for each drug and an automated image acquisition was performed at 1 h intervals. This analysis provided a stratified cell death/survival curve, as presented in Figure 5. Importantly, the stochastic nature of cell death and dissimilar kinetic profiles of STS and BFA action were easily visualized (Figure 5). STS, a pan-kinase inhibitor, exerted a rapid cytotoxic response of human osteosarcoma cells that could be clearly observed following 2 h of exposure. In contrast, BFA, a fungal 16-membered macrolide isolated from Penicillium brefeldianum, caused a delayed response observable only following 8 h of exposure, Figure 5. This observation is consistent with our earlier reports showing that the time-dependent progression of cell killing and delayed reaction was statistically significant only for relatively high doses of the BFA in follicular lymphoma cells.18
Figure 5.
Dynamic analysis of drug-induced cytotoxicity using a microculture chip-based system. Human osteosarcoma U2OS cells were exposed to 2 μM staurosporine, 2 μM Brefeldin A, and 20% DMSO for up to 24 h in the continuous presence of the plasma membrane permeability marker propidium iodide (PI). Time lapse images where acquired every hour on a motorized microscope stage. Results represent a cumulative percentage from two independent chips where a total of 300 cells for each treatment was analyzed. Note the stochastic nature of cell death and different kinetic profiles of cell death for STS and BFA.
BFA exerts endoplasmic reticulum (ER) and Golgi stress via inhibition of ADP ribosylation factor (ARF) leading to decreased coatamer protein assembly and disruption of ER–Golgi vesicular transport.19 This subsequently contributes to an unfolded protein response (UPR), activation of caspase 2, and collapse of the Golgi apparatus.18,20 The delay between initial drug perfusion and drug-induced cell killing most likely caused by the time required for the irreversible Golgi collapse to trigger the caspase cascade and proteolytic cell disassembly.18,20
We propose that this simple bioassay allows for convenient tracking of differences in stochastic cell death sensitivity that often remain undetectable with conventional end-point analysis. Such a dynamic analysis of cancer cell death may support the development and preclinical validation of combinatorial regimens to combat malignancy.6,7
DISCUSSION
Validation of potential therapeutic targets necessitates the development of new assays that provide both spatial and temporal relationships in signaling networks. Cell-based assays are therefore becoming an important part of postgenomic biomedical research. From the pharmacological perspective, the understanding of mechanistic drug action could be significantly enhanced by the kinetic analysis of cell demise.6,17,21 In this respect, supravital analysis requires the measurement of biomarkers and use of probes that do not interfere with structure or function of the cell.6,7,22 Current development of kinetic assays is, however, greatly impeded by probe cytotoxicity. Indeed nearly all cell permeable DNA fluorochromes to date interact detrimentally with cellular constituents and profoundly affect functional pathways.7,22 Interestingly, there is a widespread presumption that cell impermeant fluorescent probes such as PI, 7-AAD, and SY-G also exhibit substantial toxicity despite the lack of comprehensive experimental evidence. This is the primary reason why they are predominantly restricted to end-point DEAD/LIVE assays by both flow cytometry and fluorescence imaging analysis.
Here we show that cell impermeant viability probes do not adversely affect integrity of plasma membrane and cell cycle progression when used in continuous exposure experiments. An interesting question arises here, however, with regards to a potential of inducting mutations in the mitochondrial DNA by DNA-binding probes present in the medium at very low concentrations. Some authors indicated that DNA-binding dyes, such as propidium iodide, can be used to induce mutations within mitochondrial DNA or even cause loss of certain mitochondrial genes.23,24 This effect can be explained by a preferential intercalation of PI to a naked mitochondrial DNA rather than to histone-protected genomic DNA. To address this issue, we have performed assays that test mitochondrial function based on measurements of ΔΨm (not shown). Interestingly, we did not observe any dysfunctions in the mitochondrial function during the duration of experiments. From the microscopic examination of live cells cultured in the presence of DNA-binding probes, we observed, however, their intracellular accumulation pattern may suggest uptake to mitochondria, ER, lysosomes, or endosomes (Supplementary Figure 4 in the Supporting Information). As a result, one cannot exclude that preferential intercalation of PI to naked mitochondrial DNA indeed induces discrete mutational changes that do not affect mitochondrial homeostasis and cellular viability. This conundrum is currently being pursued in our laboratory further.
As a result, instead of being used solely in end-point assays they can be conveniently applied to assays for dynamic real-time analysis of drug-induced cytotoxicity. Vast reduction of sample consumption achieved with these protocols is an important consideration for restricting expenditures in high-throughput assays. Elimination of redundant centrifugation/washing/staining steps also helps the preservation of fragile apoptotic cells, while maintaining the ability to produce high data quality.2
We also demonstrate that the innovative implementation of dynamic cytotoxicity assays on microfluidic chip-based devices. Lab-on-a-Chip (LOC) microfabrication technology provides substantial technological advances over conventional analytical technologies, such as flow cytometry or microplate imaging (Table 1). It allows for robust miniaturization of various analytical components and combines small sample volumes at low Reynolds numbers with a turbulence-free flow.16,17 LOC promises both reductions in the cost of equipment and higher-throughput information.25,26 Importantly, as only low cell numbers and reagent volumes are required for such microfluidic analyzers, the ability to monitor single-cell signaling dynamics of rare subpopulations, such as those associated with cancers, provides the possibility of developing personalized therapeutics in which drug dosages and combinations of therapies can be patient-defined to treat individual disease.16,17 We show that, when used with innovative bioassays, microfluidic live-cell analysis is a practical alternative for multiparameter studies on a single-cell level. The simple PDMS-based device not only reduced the complexity of conventional cell culture protocols but also enabled time-resolved studies on apoptosis of adherent cells. The latter feature is of particular advantage for determining the kinetics of pharmaceutical efficacy and allows for sequential pharmacological stimulations and real-time analysis of cellular physiology while providing a vast reduction in the use of reagents and sample. Most importantly, the microfluidic chip can be used to analyze a greatly reduced number of cells when compared with conventional flow cytometry. We suggest that our technology is likely to become a useful adjunct technique to conventional flow cytometry especially in situations where ultralow numbers of cells need to be quantified and continuously monitored. Clearly, the adaptation of nontoxic biomarkers that can continuously circulate inside an enclosed microculture system will be beneficial for the advancement of these up and coming technologies. Our data support the hypothesis that real-time bioassays in combination with LOC devices allow for rapid and simple analysis of cell death, particularly useful if the death pattern is a stochastic rather then deterministic process. As a result, they provide sensitivity that often cannot be achieved with conventional end-point analysis.6,27
The findings show that fluoroprobes, such as PI or 7-AAD, have no distinct effect on cell viability (plasma membrane integrity) or cell cycle progression. The data (Figure 2A and Supplementary Figure 4 in the Supporting Information) indicate that, during prolonged exposure, PI is able to penetrate the plasma membrane. Upon entrance to cells, it is likely that PI binds to nucleic acids by intercalation, because the unbound (nonintercalated) PI, such as its close analogue ethidium, has minimal fluorescence whereas the fluorescence of cells exposed to PI is relatively high (Figure 2A and Supplementary Figure 4 in the Supporting Information).28 It is possible that the binding was limited to RNA, since binding of the intercalator to DNA leads to inhibition of DNA topoisomerases and induces arrest in the cell cycle, predominantly in the G2 phase (these effects were not observed during this study).29,30 It is evident that accumulation of PI in the cells and its possible binding to nucleic acids (most likely to RNA) had a negligible effect on cell cycle progression (Supplementary Figure 4 in the Supporting Information).
In summary, we show the adaptation of a series of standard fluorescence assays to dynamic, real-time bioassays that are capable of performing traditional tasks in a faster, more economical and practical manner. Such assays have the potential to be applied to a number of areas including accelerated anticancer drug discovery and therapy (particularly, high-throughput and high-content drug screening routines25,26 using patient derived cells). Further studies, however, are needed to additionally search for potential adverse effects of these probes taken up by live cells in order to exclude a possibility that in a combination and possibly in an interaction with the agents, whose cytotoxicity is being tested, the probes introduce bias in estimating the toxic effects.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by BBSRC, EPSRC, Scottish Funding Council, RASOR (D.W., S.F., and J.M.C.), and L’Oreal-UNESCO for “Women in Science” (J.S.).
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Leist M, Jaattela M. Nat. Rev. Mol. Cell Biol. 2001;2:589–598. doi: 10.1038/35085008. [DOI] [PubMed] [Google Scholar]
- 2.Darzynkiewicz Z, Juan G, Li X, Gorczyca W, Murakami T, Traganos F. Cytometry. 1997;27:1–20. [PubMed] [Google Scholar]
- 3.Ormerod MG, Sun XM, Brown D, Snowden RT, Cohen GM. Acta Oncol. 1993;32:417–424. doi: 10.3109/02841869309093620. [DOI] [PubMed] [Google Scholar]
- 4.Lecoeur H, de Oliveira-Pinto LM, Gougeon ML. J. Immunol. Methods. 2002;265:81–96. doi: 10.1016/s0022-1759(02)00072-8. [DOI] [PubMed] [Google Scholar]
- 5.Wlodkowic D, Skommer J, Darzynkiewicz Z. Cytometry, Part A. 2008;73:496–507. doi: 10.1002/cyto.a.20535. [DOI] [PubMed] [Google Scholar]
- 6.Wlodkowic D, Skommer J, Faley S, Darzynkiewicz Z, Cooper JM. Exp. Cell Res. 2009;315:1706–1714. doi: 10.1016/j.yexcr.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wlodkowic D, Darzynkiewicz Z. Cytometry, Part A. 2008;73:877–879. doi: 10.1002/cyto.a.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid I, Krall WJ, Uittenbogaart CH, Braun J, Giorgi JV. Cytometry. 1992;13:204–208. doi: 10.1002/cyto.990130216. [DOI] [PubMed] [Google Scholar]
- 9.Idziorek T, Estaquier J, De Bels F, Ameisen JC. J. Immunol. Methods. 1995;185:249–258. doi: 10.1016/0022-1759(95)00172-7. [DOI] [PubMed] [Google Scholar]
- 10.Wlodkowic D, Skommer J, Hillier C, Darzynkiewicz Z. Cytometry, Part A. 2008;73:563–569. doi: 10.1002/cyto.a.20564. [DOI] [PubMed] [Google Scholar]
- 11.Sia SK, Whitesides GM. Electrophoresis. 2003;24:3563–3576. doi: 10.1002/elps.200305584. [DOI] [PubMed] [Google Scholar]
- 12.Yin H, Pattrick N, Zhang X, Klauke N, Cordingley HC, Haswell SJ, Cooper JM. Anal. Chem. 2008;80:179–185. doi: 10.1021/ac701958z. [DOI] [PubMed] [Google Scholar]
- 13.Yin H, Zhang X, Pattrick N, Klauke N, Cordingley HC, Haswell SJ, Cooper JM. Anal. Chem. 2007;79:7139–7144. doi: 10.1021/ac071146k. [DOI] [PubMed] [Google Scholar]
- 14.Schmid I, Uittenbogaart C, Jamieson BD. Nat. Protoc. 2007;2:187–190. doi: 10.1038/nprot.2006.458. [DOI] [PubMed] [Google Scholar]
- 15.Whitesides GM. Nature. 2006;442:368–372. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 16.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 17.Sims CE, Allbritton NL. Lab Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 18.Wlodkowic D, Skommer J, Pelkonen J. Leukemia Res. 2007;31:1687–1700. doi: 10.1016/j.leukres.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Donaldson JG, Cassel D, Kahn RA, Klausner RD. Proc. Natl. Acad. Sci. U.S.A. 1992;89:6408–6412. doi: 10.1073/pnas.89.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carew JS, Nawrocki ST, Krupnik YV, Dunner K, McConkey DJ, Keating MJ. Blood. 2006;107:222–231. doi: 10.1182/blood-2005-05-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faley S, Seale K, Hughey J, Schaffer DK, VanCompernolle S, McKinney B, Baudenbacher F, Unutmaz D, Wikswo JP. Lab Chip. 2008;8:1700–12. doi: 10.1039/b719799c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wojcik K, Dobrucki JW. Cytometry, Part A. 2008;73A:555–562. doi: 10.1002/cyto.a.20573. [DOI] [PubMed] [Google Scholar]
- 23.Fukunaga M, Yielding KL. Mutat. Res. 1980;69:43–50. doi: 10.1016/0027-5107(80)90174-8. [DOI] [PubMed] [Google Scholar]
- 24.Fukunaga M, Mizuguchi Y. Chem. Pharm. Bull. (Tokyo) 1982;30:2889–2893. doi: 10.1248/cpb.30.2889. [DOI] [PubMed] [Google Scholar]
- 25.Andersson H, van den Berg A. Curr. Opin. Biotechnol. 2004;15:44–49. doi: 10.1016/j.copbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Manz A, Dittrich PS. Nature Drug Discovery. 2006;5:210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 27.Chan SD, Luedke G, Valer M, Buhlmann C, Preckel T. Cytometry, Part A. 2003;55:119–125. doi: 10.1002/cyto.a.10070. [DOI] [PubMed] [Google Scholar]
- 28.LePecq JB. Meth. Biochem. Anal. 1971;20:41–86. doi: 10.1002/9780470110393.ch2. [DOI] [PubMed] [Google Scholar]
- 29.D'Arpa P, Beardmore C, Liu LF. Cancer Res. 1990;50:6916–6924. [PubMed] [Google Scholar]
- 30.Huang X, Okafuji M, Traganos F, Luther E, Holden E, Darzynkiewicz Z. Cytometry, Part A. 2004;58A:99–110. doi: 10.1002/cyto.a.20018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.