Abstract
Rationale: Biologic pathways with significant genetic conservation across human populations have been implicated in the pathogenesis of primary graft dysfunction (PGD). The evaluation of the role of recipient genetic variation in PGD has thus far been limited to single, candidate gene analyses.
Objectives: We sought to identify genetic variants in lung transplant recipients that are responsible for increased risk of PGD using a two-phase large-scale genotyping approach.
Methods: Phase 1 was a large-scale candidate gene association study of the multicenter, prospective Lung Transplant Outcomes Group cohort. Phase 2 included functional evaluation of selected variants and a bioinformatics screening of variants identified in phase 1.
Measurements and Main Results: After genetic data quality control, 680 lung transplant recipients were included in the analysis. In phase 1, a total of 17 variants were significantly associated with PGD, four of which were in the prostaglandin E2 family of genes. Among these were a coding variant in the gene encoding prostaglandin E2 synthase (PTGES2; P = 9.3 × 10−5) resulting in an arginine to histidine substitution at amino acid position 298, and three variants in a block containing the 5′ promoter and first intron of the PTGER4 gene (encoding prostaglandin E2 receptor subtype 4; all P < 5 × 10−5). Functional evaluation in regulatory T cells identified that rs4434423A in the PTGER4 gene was associated with differential suppressive function of regulatory T cells.
Conclusions: Further research aimed at replication and additional functional insight into the role played by genetic variation in prostaglandin E2 synthetic and signaling pathways in PGD is warranted.
Keywords: lung transplantation, genetics, prostaglandin, regulatory T cells
At a Glance Commentary
Scientific Knowledge on the Subject
Several biologic pathways implicated in the development of primary graft dysfunction (PGD) after lung transplantation are highly genetically conserved across human populations. Large-scale gene association studies have been useful in identifying potential mechanisms for the development of acute lung injury, a syndrome closely related to PGD after lung transplantation. The influence of genetic variation on PGD risk is incompletely understood.
What This Study Adds to the Field
We performed a large-scale candidate gene association study to detect genetic variants associated with PGD using the Lung Transplant Outcomes Group, a multicenter cohort of lung transplant recipients. We identified significant associations between polymorphisms in multiple genes in the prostaglandin E2 pathway. Furthermore, variation in the prostaglandin EP4 receptor gene (PTGER4) was associated with altered T-regulatory cell suppressor function. A more complete understanding of the role played by the prostaglandin E2 pathway family in PGD may lead to the development of new preventative therapies.
Primary graft dysfunction (PGD), a form of acute lung injury (ALI) following lung transplantation, occurs in 10–30% of cases and is a major cause of early morbidity and mortality after transplant (1–8). Recent investigations have implicated a genetic basis to ALI susceptibility and severity (9–11). The evaluation of the role of recipient genetic variation in PGD has thus far been limited to single, candidate gene analyses (12).
Several well-described biologic pathways, including airway epithelial integrity, coagulation and fibrinolytic cascades, immune activation, and endothelial cell function, have been implicated in clinical PGD pathogenesis (13–22). Many of these pathways demonstrate significant genetic conservation across human populations, between species, and also display evidence of purifying selection (23–29). These pathways may be reasonable candidates for genetic regulation of differential response to the extreme insult of the lung transplant process. Evidence exists for a contribution of genetic variation to individual risk for acute rejection and bronchiolitis obliterans syndrome, yet genetic studies of PGD have been limited to date (30–39). A large-scale genotyping approach has not been previously performed to evaluate genetic risk in PGD.
Based on the hypothesis that there is an unidentified role for recipient genetic variation in the development of PGD, we performed a large-scale candidate gene cohort study. We sought to test the association of single-nucleotide polymorphisms (SNPs) in genes with known association with inflammatory, metabolic, and vascular phenotypes with differential risk for the development of PGD after lung transplantation (phase 1). The Illumina IBC chip (Illumina, San Diego, CA) was chosen for this evaluation rather than a genome-wide array because of its inclusion of rare variants and its dense coverage of gene targets hypothesized to be involved in pulmonary and inflammatory disease states (40). We then sought to identify the functional implications of selected candidate SNPs in three ways (phase 2): (1) using cell-based assays specific to implicated variants, (2) evaluation of selected SNPs on corresponding plasma protein levels, and (3) a broad-based bioinformatics approach.
Methods
See the online supplement for more detailed methods on cohort design, subject selection, PGD definition, genotyping strategy, and T-regulatory (Treg) cell suppression assays.
Cohort Design
Subjects were enrolled from the prospective, multicenter Lung Transplant Outcomes Group cohort study (clinicaltrials.gov identifier: NCT00457847). Clinical data were prospectively collected and blood samples for DNA were collected before transplant (41). All study center institutional review boards approved this study.
PGD Definition
PGD was defined as the presence of diffuse alveolar infiltrates involving the allografts with severity determined by the Pao2/FiO2 ratio; grade 3 PGD was defined as Pao2/FiO2 less than 200 (4). The primary outcome was defined as any grade 3 PGD within 72 hours of reperfusion. Sensitivity analyses were performed with PGD defined using alternative severity definitions.
Phase 1: Study Design and Genotyping Strategy
Phase 1 was a large-scale candidate gene association study. Subjects enrolled in Lung Transplant Outcomes Group from July 2002 to July 2009 were included in the analysis. Genotyping was performed using the IBC chip (Illumina), an array designed to assay SNPs in candidate genes affecting vascular, pulmonary, and metabolic phenotypes (11, 40).
Phase 1: Statistical Analysis
Subject characteristics were compared using descriptive tests as appropriate. An odds ratio (OR) for PGD based on an additive model of genetic risk in PLINK, with significance determined by chi-square, was calculated according to genotype for all SNPs (42). Principle component analysis of the approximately 1,800 ancestry-informative markers on the chip were used to determine genetically inferred ancestry (10–12). ORs were adjusted for cardiopulmonary bypass, predisposing lung disease, and the first two principal components derived from ancestry-informative markers using logistic regression. No consensus exists for a P value for SNPs evaluated using hypothesis-driven candidate gene assays with redundant, linked SNPs. Similar to previous studies, we selected an adjusted P less than 5 × 10−4 for significance and then used functional assessments to validate the findings (43, 44).
Phase 2: Study Design
Phase 2 was a functional assessment of SNPs significantly associated with PGD in phase 1. Given the overrepresentation of prostaglandin E2 (PGE2) pathway family SNPs among all variants identified as significantly associated with PGD in phase 1, the four PGE2 pathway family SNPs were assessed functionally using Treg suppression assays. Next, we evaluated the impact of variants at selected SNPs on corresponding plasma protein PGE2 levels. To ensure generalizability of the study results, a cohort of patients was selected to include subjects with and without PGD from eight transplant centers. We also enriched the patient population to include homozygotes for both the wild-type and recessive alleles. Finally, all significant phase 1 SNPs were evaluated with a bioinformatics approach and literature searches to identify prior associations with pulmonary disease or organ transplantation (45–47).
Phase 2: Treg Suppression Assay
Quantitative assays for Treg suppression function methods have been described previously (48). Aliquoted cryopreserved single healthy donor peripheral blood mononuclear cells were labeled with carboxyfluorescein diacetate succinimidyl ester, stimulated with CD3ε mAb-coated microbeads, and used as standardized responder cells in each assay. Tregs were isolated with CD4+CD25+ kit (Miltenyi Biotec, Auburn, CA) from 50 ml of blood taken from eight liver transplant recipients. Samples-in-hand from liver transplant recipients were selected because of the large volume of blood (>50 ml) required to isolate a sufficient number of Tregs from the subject and to ensure that subjects received standard immunosuppression, similar to lung transplant recipients. Four days later, the percent of dividing T cells was determined by carboxyfluorescein diacetate succinimidyl ester dilution. We controlled for the purity of Tregs and used standardized suppression assay conditions that differed only in the suppressive function of patients’ Tregs. Suppressive function was calculated as area under the standardized suppression curve (see Figure E1 in the online supplement) (48)
Phase 2: Measurement of PGE2 Plasma Concentration
Plasma PGE2 concentrations were determined using a sandwich ELISA (Cayman Chemical, Ann Arbor, MI) from pretransplant and immediate post-transplant plasma samples.
Phase 2: Statistical Analysis
Quantitative Treg suppressive function was analyzed across SNP genotypes using rank-sum tests with a P less than 0.05 considered significant. Plasma PGE2 concentrations were analyzed across PTGES2 SNP genotypes using the Kruskal-Wallis test with a P less than 0.05 considered significant. In silico gene expression results were considered significant at a P less than 0.05. Statistical analyses were performed using Stata 11.2 software (STATA Corp., College Station, TX).
Results
Patient Characteristics
After genetic quality control, 680 subjects were included in the analysis. PGD developed in 194 (29%) of 680 (95% confidence interval [CI], 25–32%) subjects. All transplant centers use a low-potassium dextran preservation solution (Perfadex; XVIVO Perfusion, Goteborg, Sweden). The characteristics of the study subjects are described in Table 1. There was no significant difference in age or sex between patients with and without PGD. Significantly more patients who developed PGD received perioperative cardiopulmonary bypass compared with those without PGD (52% vs. 29%; P < 0.001). As previously described, there were significant differences in the distribution of preoperative pulmonary diagnoses, with more idiopathic pulmonary fibrosis and less chronic obstructive pulmonary disease in the PGD group compared with the non-PGD patients (P < 0.001), and racial composition with more African American subjects in the PGD group compared with the non-PGD group (P < 0.001) (12).
Table 1:
Subject Characteristics
| Covariate | PGD (n = 194) | Non-PGD (n = 486) | P Value |
|---|---|---|---|
| Recipient variables | |||
| Age, mean | 52 | 53 | 0.6 |
| Female sex, n (%) | 93 (48) | 228 (47) | 0.9 |
| Pulmonary diagnosis, n (%) | <0.001 | ||
| Chronic obstructive pulmonary disease | 60 (31) | 219 (45) | |
| Idiopathic pulmonary fibrosis | 76 (39) | 150 (31) | |
| Cystic fibrosis | 14 (7) | 73 (15) | |
| Other | 44 (23) | 44 (9) | |
| Race, n (%) | <0.001 | ||
| White | 149 (77) | 423 (87) | |
| African American | 33 (17) | 29 (6) | |
| Other | 12 (6) | 24 (5) | |
| Operative variables | |||
| Cardiopulmonary bypass use, yes, n (%) | 101 (52) | 141 (29) | <0.001 |
Definition of abbreviation: PGD = primary graft dysfunction.
Phase 1: SNP Associations with PGD
After adjustment for preoperative pulmonary diagnosis, perioperative use of cardiopulmonary bypass, and genetically inferred race, 17 SNPs met our prespecified level of significance for PGD of P less than 5 × 10−4 (Table 2). Four of the 17 SNPs were in the PGE2 pathway genes. Of these, one SNP, rs13283456, is a coding nonsynonymous SNP in PGE synthase 2 (PTGES2, responsible for encoding a constitutive protein isoform converting PGH2 to PGE2). The minor allele was significantly associated with PGD (OR, 2.0; 95% CI, 1.4–2.9; P = 9.3 × 10−5). Three other SNPs were in the gene encoding the PGE2 receptor subtype 4 (PTGER4). Two of these SNPs, rs4434423 and rs4133101, lie in the 5′ promoter region of the gene in tight linkage disequilibrium (r2 = 0.92), and the minor alleles were associated with lower risk of PGD (OR, 0.6; 95% CI, 0.4–0.8; P = 2.8 × 10−4; and OR, 0.6; 95% CI, 0.5–0.8; P = 4.8 × 10−4). rs11957406 lies in the second intron and the minor allele was significantly associated with PGD (OR, 1.7; 95% CI, 1.3–2.3; P = 2.2 × 10−4). There were also three SNPs in TBC1D1, a gene linked with obesity and type 2 diabetes, all with intronic minor alleles associated with lower PGD risk (49–51).
Table 2:
Single-Nucleotide Polymorphism Analysis for Association with PGD
| rs Number | Gene | Gene Function/Disease Association | Location | Minor Allele | Risk Allele | MAF PGD | MAF Non-PGD | Odds Ratio (95% Confidence Interval) | P Value |
|---|---|---|---|---|---|---|---|---|---|
| rs2996044 | TBC1D1 | Cell growth and differentiation/obesity | Intron | C | T | 0.26 | 0.40 | 0.5 (0.4–0.7) | 3.0 × 10−5 |
| rs13283456 | PTGES2 | Prostaglandin E2 synthesis | Coding | T | T | 0.21 | 0.16 | 2.0 (1.4–2.9) | 9.3 × 10−5 |
| rs2925956 | TBC1D1 | Cell growth and differentiation/obesity | Intron | A | G | 0.31 | 0.41 | 0.6 (0.4–0.8) | 1.2 × 10−4 |
| rs13132184 | TBC1D1 | Cell growth and differentiation/obesity | Intron | C | T | 0.10 | 0.21 | 0.5 (0.3–0.7) | 1.6 × 10−4 |
| rs7973796 | PMCH | Hypothalamic neurotransmitter/obesity | 5′ UTR | G | A | 0.40 | 0.54 | 0.6 (0.5–0.8) | 1.7 × 10−4 |
| rs3024388 | F13A1 | Coagulation cascade/venous thrombosis | Intron | T | T | 0.30 | 0.20 | 1.8 (1.3–2.5) | 2.0 × 10−4 |
| rs12452616 | GAA | Glycogen degradation | Intron | A | A | 0.27 | 0.20 | 1.8 (1.3–2.5) | 2.1 × 10−4 |
| rs11957406 | PTGER4 | Prostaglandin E receptor (EP4) | Intron | A | A | 0.56 | 0.47 | 1.7 (1.3–2.3) | 2.2 × 10−4 |
| rs237865 | CAV3 | Muscle development | 5′ UTR | C | C | 0.51 | 0.40 | 1.7 (1.3–2.2) | 2.4 × 10−4 |
| rs17588591 | COL4A1 | Extracellular matrix/punctate palmoplantar keratoderma | Intron | G | G | 0.37 | 0.29 | 1.7 (1.3–2.3) | 2.6 × 10−4 |
| rs16836965 | CASP8 | Cell death and tumor regulation | Intron | T | T | 0.08 | 0.02 | 3.2 (1.7–6.0) | 2.7 × 10−4 |
| rs4434423 | PTGER4 | Prostaglandin E receptor (EP4) | 5′ UTR | T | A | 0.31 | 0.42 | 0.6 (0.4–0.8) | 2.8 × 10−4 |
| rs260400 | IRX4 | Ventricular differentiation and cardiac development | 5′ UTR | A | A | 0.15 | 0.09 | 2.1 (1.4–3.1) | 3.1 × 10−4 |
| rs3772843 | ITGB5 | Cell adhesion | Intron | A | A | 0.23 | 0.14 | 1.9 (1.3–2.6) | 3.2 × 10−4 |
| rs1881597 | PRKG1 | Platelet aggregation | 3′ UTR | T | C | 0.20 | 0.31 | 0.6 (0.4–0.8) | 3.8 × 10−4 |
| rs4133101 | PTGER4 | Prostaglandin E receptor (EP4) | 5′ UTR | A | G | 0.32 | 0.42 | 0.6 (0.5–0.8) | 4.8 × 10−4 |
| rs17004504 | FTCD | Folate metabolism/autoimmune hepatitis | Intron | T | T | 0.08 | 0.03 | 2.9 (1.6–5.2) | 4.8 × 10−4 |
Definition of abbreviations: MAF = minor allele frequency; PGD = primary graft dysfunction; UTR = untranslated region.
Odds ratio and P value are based on an additive model. Analysis is corrected for first two principle components derived from ancestry informative markers, cardiopulmonary bypass use, and preoperative lung disease.
We then performed sensitivity analyses using alternative PGD outcome definitions. When comparing patients with grade 3 PGD developing any time within the first 3 days after transplant with those with no (grade 0) or mild (grade 1) PGD, the results were similar although mildly attenuated given the smaller sample size (see Table E1). The minor allele at the PTGES2 coding nonsynonomous locus, rs13283456, had an OR for PGD of 1.8 (95% CI, 1.2–2.7; P = 0.003). The associations between risk of PGD and the minor alleles of the 5′ promoter PTGER4 SNPs, rs4434423 (OR, 0.5; 95% CI, 0.4–0.7; P = 1.4 × 10−4) and rs4133101 (OR, 0.6; 95% CI, 0.4–0.8; P = 3.6 × 10−4), and the intronic PTGER4 SNP rs11957406 (OR, 1.7; 95% CI, 1.3–2.4; P = 3.7 × 10−4) were similar to the primary analysis in magnitude but slightly attenuated in significance. The results were similar when limiting the analysis to comparisons of the most extreme subjects, grade 3 versus grade 0 (see Table E2).
Phase 2: Functional Evaluation
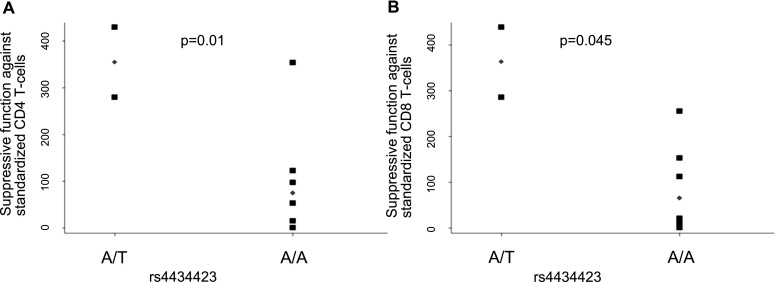
SNPs in the PGE2 pathway family were overrepresented in the phase 1 analysis (4 of 17 SNPs). SNPs from genes involved in the PGE2 signaling pathway have been previously described to be involved in immunomodulation and inflammation (52–54). Recently, the PG EP4 receptor was shown to regulate hematopoietic stem cell expansion and migration (55). Based on the immunomodulatory effect of PGE2, the three SNPs identified in the EP4 receptor were functionally evaluated in phase 2. PGE2 induces and up-regulates FOXP3 gene expression in both CD4+CD25+ Tregs and CD4+CD25−T cells (56), and PGE2 likely results in up-regulation of FOXP3 at the level of mRNA transcription (57). We therefore evaluated the suppressive function of FOXP3+ Tregs against standardized CD4 and CD8 cellular populations by PTGER4 genotype. Tregs collected from a cohort of liver transplant recipients were used for this evaluation. Tregs homozygous for the “A” risk allele at rs4434423 (n = 2) had decreased suppressor T-cell function against standardized CD4 and CD8 T-cell populations when compared with Tregs carrying the protective “T” allele (n = 6) (Figure 1); the “T” allele at the rs4434423 locus was associated with decreased risk of PGD in phase 1.
Figure 1.
T-regulatory suppressor function according to genotype at the rs4434423 locus in PTGER4. (A) Suppressive function against standardized CD4+ T cells. (B) Suppressive function against standardized CD8+ T cells. Squares represent individual patient level data. Diamonds represent the median value. Y-axis represents area under the standardized suppression curve. P value is from rank-sum test.
The effect of variation at the coding, nonsynonomous SNP, rs13283456, in PTGES2 on plasma PGE2 levels was assessed in 42 lung transplant recipients, including 18 subjects without PGD and 24 subjects with PGD. All subjects had pretransplant plasma available and 40 had corresponding post-transplant plasma available. There were nine subjects homozygous for the risk “T” allele, 17 heterozygotes (CT), and 16 wild-type “CC” homozygotes. There was no significant difference in median plasma PGE2 level measured before transplant (TT, 56.5 pg/ml; CT, 115.1 pg/ml; CC, 87.5 pg/ml; P = 0.5), immediately post-transplant (TT, 60.9 pg/ml; CT, 78.9 pg/ml; CC, 55.7 pg/ml; P = 0.6), or in the median change in PGE2 level from pretransplant to post-transplant (TT, 10.8 pg/ml; CT, −21.3 pg/ml; CC, −20.1 pg/ml; P = 0.3) (see Figure E2). There was no association between genotype and pretransplant or post-transplant PGE2 levels when evaluated within subjects with or without PGD (all P > 0.1).
Additionally, a bioinformatics evaluation of all SNPs identified as significant in phase 1 was performed. No SNPs identified in phase 1 have previously been identified to function as cis-eQTLs; genotype at the rs13283456 locus in PTGES2 was nonsignificantly associated with differential gene expression in T cells (P = 0.09) (45, 46). In silico modeling of the PTGES2 coding nonsynonomous allele, rs13283456T, leading to an arginine to histidine missense mutation at position 298, would be a benign, tolerated change, although the arginine to histidine change results in the loss of a salt bridge (47, 58–60). Three 5′ untranslated region SNPs, rs7973796 in PMCH, rs4434423 in PTGER4, and rs237865 in CAV3, and the intronic SNP rs17004504 in FTCD, are likely in promoter or regulatory regions and were predicted to have at most medium-risk effects on transcription factor binding (47).
Discussion
Using a large, multicentered cohort study of lung transplant recipients, we identified 17 SNPs in 13 genes to be prioritized for research on PGD after lung transplantation. Of these, there were four SNPs in two genes in the PGE2 family, PTGES2 and PTGER4, which were significantly associated with PGD. The A risk allele in PTGER4 rs4434423 was associated with decreased Treg suppressor cell function, indicating a potential functional consequence of the observed genetic variation, consistent with a pathogenic consequence of this variation.
The PG EP4 receptor plays a central role in immunomodulation and control of inflammation mediated by PGE2 (52–54). Antagonism of the EP4 receptor results in impaired immunosuppressive responses to ultraviolet irradiation (53). Activation of the EP4 receptor inhibits the activation and proliferation of T cells (54). The documented immunosuppressive role of the EP4 receptor is consistent with our demonstration of increased Treg suppressor function in cells possessing the rs4434423T allele, which was associated with lower PGD risk. Although our bioinformatics evaluation of the identified PTGER4 SNPs only identified the rs443443 to have a very low to medium risk of altering transcription factor binding, the recent ENCODE project highlights the tremendous diversity in genetic regulatory elements, outside of traditional transcription factor binding (61). The molecular mechanism linking the genetic variation with altered Treg cell function needs to be further elucidated.
There is no previous literature evaluating the role played by Tregs in the development of PGD. However, Treg cells play a central role in the resolution of ALI (62, 63). The adoptive transfer of Tregs into lymphocyte-deficient mice results in the resolution of fibroproliferation through reduced fibrocyte recruitment (64). In an endotracheal lipopolysaccharide-induced model of ALI, transplantation of human umbilical cord mesenchymal stem cells results in a significant increase in the number of CD4+CD25+Foxp3+ Tregs, an increase in the IL-10 levels, and a decrease in proinflammatory cytokines (65). A detailed evaluation of the role of Tregs in the development of PGD after lung transplantation is the focus of ongoing investigation.
Given the central role of PGE2 in cancer immunology, inflammation, and autoimmunity, there is significant interest in developing therapies targeting PGE2-associated pathways. Synthetic EP4 receptor agonists inhibit proinflammatory mediators and decreased mortality in a mouse liver ischemia reperfusion model (66). EP4 agonists also diminished infarction area in a rat myocardial ischemia reperfusion model (67). Pharmacologic activation of the EP4 receptor prolonged cardiac allograft survival in mouse models (68). In addition to its immunomodulatory effects, PGE2 has potent vasodilatory properties, likely mediated by the EP4 receptor (69). Our findings of a genetic association between PGE-associated genetic variation and PGD, combined with the exiting knowledge on PGE2 and EP4 agonism, indicate that evaluating the role of PGE2 and EP4 receptor agonists in the treatment and prevention of PGD is an area ripe for future investigation.
PTGES2 encodes a constitutively expressed PG synthase distinct from cyclooxygenase-2. Although the protein crystal structure of PGES2 indicates the altered amino acid moiety encoded, by rs13283456T, is located in the protein binding pocket and may therefore alter protein synthetic function, the function of this protein alteration is unknown at this time (70). This SNP variant has previously been associated with altered risk of type II diabetes, (71) and it is postulated that replacement of arginine, with a pKa of 12.5, to histidine, with a pKa of 6.0, may alter protein function and decrease PGE2 production. The addition of PGE2 results in activation of FOXP3+ Treg cells and suppression of effecter T cells (56, 72). This is consistent with our finding that a variant of PTGES2 with hypothesized decreased synthetic function is associated with the proinflammatory state leading to severe PGD after transplant. We demonstrated no association between variation in PTGES2 and altered PGE2 protein plasma levels in an enrichment sampled cohort of lung transplant recipients. PGE2 is rapidly metabolized in vivo, with an estimated half-life of less than 15 seconds (73, 74). Rapid degradation of PGE2 in blood may hamper our ability to adequately assess protein levels in response to genetic variation in PTGES2. Alternatively, this may indicate that variation in the receptors is more significant in altering the activity of this pathway, or that other PGE2 synthase isoforms may have influence. Further evaluation of alterations in the synthetic function of PGE synthase 2 (e.g., through measurement of longer-lasting urinary metabolites or in lung tissue) is needed to further refine the role played by the rs13283456T variant.
We limited our functional evaluation to a small segment of the 17 SNPs identified in our initial screen given the abundance of PGE-associated SNPs identified in phase 1. Future investigations should focus on evaluating the potential implications of the other genes associated with PGD. The 11 genes tagged by the other 13 SNPs identified in phase 1 were not further functionally evaluated because they have not previously been associated with lung disease, organ transplantation, or ischemia-reperfusion injury (Table 2). However, one can hypothesize merit for future functional evaluation of several genes identified in phase 1. Given the newly identified association between obesity and PGD, two genes, TBC1D1 and PMCH, which have been associated with metabolism and weight, are intriguing targets for future characterization (51, 75, 76). We identified an intronic variant locus, rs3024388, in F13A1, the gene encoding the factor XIII A1 subunit, to be associated with PGD (Table 2). Genetic variation at the rs5985 locus in F13A1 has previously been associated with protection against myocardial infarction (77). We also demonstrated differences at the rs1881597 locus in PRKG1, encoding protein kinase, cGMP-dependent, regulatory type 1, to be associated with PGD (Table 2). This protein plays an important role in inhibiting platelet aggregation. Previous studies have shown that plasma differences in plasminogen activator inhibitor-1, an inhibitor of fibrinolysis, are associated with altered risk of PGD after lung transplantation (78, 79). The association of variant F13A1 and PRKG1 with PGD provides support for further investigation into the possible mechanistic role of coagulation, fibrinolysis, and clot formation in the pathogenesis of PGD.
There are several limitations to our study. First, there is no available replication cohort to validate our genotyping findings. However, our cohort is multicentered and broadly generalizable. Additionally, although there is no population-based replication, we validated the findings using biologic mechanisms (80). We identified multiple SNPs in two genes in the same pathway and then demonstrated a potential functional effect of the variants in the EP4 receptor. The Treg suppressor function was evaluated in a population of liver transplant subjects, not lung transplant recipients. The genotype-specific differential Treg suppressor function identified in this population was therefore independent of PGD. The prospective assessment of Treg function in the development of PGD is an area of active research. Although the functional assessment should be interpreted cautiously because of the relatively small sample size of patients used for Treg assays, taken together the functional implications of the identified genetic differences enhance the validity of our findings. Our study does not take into account the potential role played by donor genetic variation and the interaction between donor and recipient genotypes. We focused on the effect of recipient genetic variation and the risk of PGD. Separate evaluation of the impact of donor gene expression on the risk of PGD is ongoing (81). Next, our functional assessment focused on a single cell type, Tregs. Future studies will assess the impact of PGE2 pathway genetics variants on other cell types, including alveolar macrophages. Finally, we did not perform a full genome-wide association study and thus may have missed the potentially important association of other genetic variation in PGD. However, the overall population of lung transplant recipients is small, thus limiting the power necessary for performing a genome-wide association study evaluation. Additionally, because of the limited study population, our study was only powered to detect relatively common variants with modest effect sizes and may therefore have missed rare variants with less substantial effect sizes. Importantly, however, our candidate gene approach focused on genes with hypothesized roles in metabolic pathways implicated in PGD, therefore increasing the potential validity of our findings.
In summary, using a targeted, large-scale candidate gene approach, we identified 17 SNPs in lung transplant recipients associated with risk of PGD after lung transplantation. These SNPs were enriched in the PGE2 synthetic and metabolic genes PTGES2 and PTGER4. Furthermore, we identified a correlation between PTGER4 genetic variation and suppressor function of Treg cells. Taken together, our results indicate that PGE2 pathways provide a novel target for preventing and treating severe PGD after lung transplantation.
Acknowledgments
Acknowledgment
The authors acknowledge the contributions of the participating centers and investigators in the Lung Transplant Outcomes Group. For a full list of centers and investigators, see the online supplement.
Footnotes
Supported by National Institutes of Health grants R01 HL087115, R01 HL081619, R01 HL096845, R01 HL114468, K12 HL090021, K24 HL115354, K24 HL103844, and K24 HL103836.
Author Contributions: Conception and design: J.M.D., R.J.S., E.C., N.J.M., R.F., S.M.K., D.J.L., S.B., V.N.L., A.W., J.O., K.W., M.C., S.A., S.M.P., L.B.W., and J.D.C. Acquisition of data: J.M.D., T.A., A.K., R.J.S., M.H.L., E.C., R.F., S.M.K., J.C.L., L.B.W., S.M.P., S.B., V.N.L., A.W., J.O., K.W., M.C., D.J.L., E.D., and J.D.C. Analysis and interpretation of data: J.M.D., T.A., A.K., R.J.S., M.H.L., E.C., R.F., S.M.K., N.J.M., E.D., and J.D.C. Drafting or revising the manuscript for important intellectual content: J.M.D., T.A., R.J.S., E.C., R.F., S.M.K., N.J.M., J.C.L., W.W.H., R.A., L.B.W., S.M.P., S.B., V.N.L., A.W., J.O., K.W., M.C., D.J.L., S.A., E.D., and J.D.C. Final approval of the version to be published: J.M.D., T.A., A.K., R.J.S., M.H.L., E.C., R.F., S.M.K., N.J.M., J.C.L., W.W.H., R.A., L.B.W., S.M.P., S.B., V.N.L., A.W., J.O., K.W., M.C., D.J.L., S.A., E.D., and J.D.C.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201307-1283OC on January 27, 2014
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Arcasoy SM, Fisher A, Hachem RR, Scavuzzo M, Ware LB ISHLT Working Group on Primary Lung Graft Dysfunction. Report of the ISHLT working group on primary lung graft dysfunction. Part V: Predictors and outcomes. J Heart Lung Transplant. 2005;24:1483–1488. doi: 10.1016/j.healun.2004.11.314. [DOI] [PubMed] [Google Scholar]
- 2.Bharat A, Kuo E, Steward N, Aloush A, Hachem R, Trulock EP, Patterson GA, Meyers BF, Mohanakumar T.Immunological link between primary graft dysfunction and chronic lung allograft rejection Ann Thorac Surg 200886189–195.discussion 196–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christie JD, Bellamy S, Ware LB, Lederer D, Hadjiliadis D, Lee J, Robinson N, Localio AR, Wille K, Lama V, et al. Construct validity of the definition of primary graft dysfunction after lung transplantation. J Heart Lung Transplant. 2010;29:1231–1239. doi: 10.1016/j.healun.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie JD, Carby M, Bag R, Corris P, Hertz M, Weill D ISHLT Working Group on Primary Lung Graft Dysfunction. Report of the ISHLT working group on primary lung graft dysfunction. Part II: definition. A consensus statement of the International Society for Heart and Lung Transplantation. J Heart Lung Transplant. 2005;24:1454–1459. doi: 10.1016/j.healun.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 5.Christie JD, Kotloff RM, Ahya VN, Tino G, Pochettino A, Gaughan C, DeMissie E, Kimmel SE. The effect of primary graft dysfunction on survival after lung transplantation. Am J Respir Crit Care Med. 2005;171:1312–1316. doi: 10.1164/rccm.200409-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christie JD, Van Raemdonck D, de Perrot M, Barr M, Keshavjee S, Arcasoy S, Orens J ISHLT Working Group on Primary Lung Graft Dysfunction. Report of the ISHLT working group on primary lung graft dysfunction. Part I: Introduction and methods. J Heart Lung Transplant. 2005;24:1451–1453. doi: 10.1016/j.healun.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Daud SA, Yusen RD, Meyers BF, Chakinala MM, Walter MJ, Aloush AA, Patterson GA, Trulock EP, Hachem RR. Impact of immediate primary lung allograft dysfunction on bronchiolitis obliterans syndrome. Am J Respir Crit Care Med. 2007;175:507–513. doi: 10.1164/rccm.200608-1079OC. [DOI] [PubMed] [Google Scholar]
- 8.Whitson BA, Prekker ME, Herrington CS, Whelan TP, Radosevich DM, Hertz MI, Dahlberg PS. Primary graft dysfunction and long-term pulmonary function after lung transplantation. J Heart Lung Transplant. 2007;26:1004–1011. doi: 10.1016/j.healun.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Christie JD, Ma SF, Aplenc R, Li M, Lanken PN, Shah CV, Fuchs B, Albelda SM, Flores C, Garcia JG. Variation in the myosin light chain kinase gene is associated with development of acute lung injury after major trauma. Crit Care Med. 2008;36:2794–2800. doi: 10.1097/ccm.0b013e318186b843. [DOI] [PubMed] [Google Scholar]
- 10.Christie JD, Wurfel MM, Feng R, O’Keefe GE, Bradfield J, Ware LB, Christiani DC, Calfee CS, Cohen MJ, Matthay M, et al. Trauma ALI SNP Consortium (TASC) investigators. Genome wide association identifies PPFIA1 as a candidate gene for acute lung injury risk following major trauma. PLoS ONE. 2012;7:e28268. doi: 10.1371/journal.pone.0028268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer NJ, Li M, Feng R, Bradfield J, Gallop R, Bellamy S, Fuchs BD, Lanken PN, Albelda SM, Rushefski M, et al. ANGPT2 genetic variant is associated with trauma-associated acute lung injury and altered plasma angiopoietin-2 isoform ratio. Am J Respir Crit Care Med. 2011;183:1344–1353. doi: 10.1164/rccm.201005-0701OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond JM, Meyer NJ, Feng R, Rushefski M, Lederer DJ, Kawut SM, Lee JC, Cantu E, Shah RJ, Lama VN, et al. Lung Transplant Outcomes Group. Variation in PTX3 is associated with primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med. 2012;186:546–552. doi: 10.1164/rccm.201204-0692OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bobadilla JL, Love RB, Jankowska-Gan E, Xu Q, Haynes LD, Braun RK, Hayney MS, Munoz del Rio A, Meyer K, Greenspan DS, et al. Th-17, monokines, collagen type V, and primary graft dysfunction in lung transplantation. Am J Respir Crit Care Med. 2008;177:660–668. doi: 10.1164/rccm.200612-1901OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christie JD, Robinson N, Ware LB, Plotnick M, De Andrade J, Lama V, Milstone A, Orens J, Weinacker A, Demissie E, et al. Association of protein C and type 1 plasminogen activator inhibitor with primary graft dysfunction. Am J Respir Crit Care Med. 2007;175:69–74. doi: 10.1164/rccm.200606-827OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Covarrubias M, Ware LB, Kawut SM, De Andrade J, Milstone A, Weinacker A, Orens J, Lama V, Wille K, Bellamy S, et al. Lung Transplant Outcomes Group. Plasma intercellular adhesion molecule-1 and von Willebrand factor in primary graft dysfunction after lung transplantation. Am J Transplant. 2007;7:2573–2578. doi: 10.1111/j.1600-6143.2007.01981.x. [DOI] [PubMed] [Google Scholar]
- 16.Diamond JM, Kawut SM, Lederer DJ, Ahya VN, Kohl B, Sonett J, Palmer SM, Crespo M, Wille K, Lama VN, et al. Lung Transplant Outcomes Group. Elevated plasma Clara cell secretory protein concentration is associated with high-grade primary graft dysfunction. Am J Transplant. 2011;11:561–567. doi: 10.1111/j.1600-6143.2010.03431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diamond JM, Lederer DJ, Kawut SM, Lee J, Ahya VN, Bellamy S, Palmer SM, Lama VN, Bhorade S, Crespo M, et al. Lung Transplant Outcomes Group. Elevated plasma long pentraxin-3 levels and primary graft dysfunction after lung transplantation for idiopathic pulmonary fibrosis. Am J Transplant. 2011;11:2517–2522. doi: 10.1111/j.1600-6143.2011.03702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman SA, Wang L, Shah CV, Ahya VN, Pochettino A, Olthoff K, Shaked A, Wille K, Lama VN, Milstone A, et al. Lung Transplant Outcomes Group. Plasma cytokines and chemokines in primary graft dysfunction post-lung transplantation. Am J Transplant. 2009;9:389–396. doi: 10.1111/j.1600-6143.2008.02497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwata T, Philipovskiy A, Fisher AJ, Presson RG, Jr, Chiyo M, Lee J, Mickler E, Smith GN, Petrache I, Brand DB, et al. Anti-type V collagen humoral immunity in lung transplant primary graft dysfunction. J Immunol. 2008;181:5738–5747. doi: 10.4049/jimmunol.181.8.5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawut SM, Okun J, Shimbo D, Lederer DJ, De Andrade J, Lama V, Shah A, Milstone A, Ware LB, Weinacker A, et al. Lung Transplant Outcomes Group. Soluble p-selectin and the risk of primary graft dysfunction after lung transplantation. Chest. 2009;136:237–244. doi: 10.1378/chest.08-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krenn K, Klepetko W, Taghavi S, Lang G, Schneider B, Aharinejad S. Recipient vascular endothelial growth factor serum levels predict primary lung graft dysfunction. Am J Transplant. 2007;7:700–706. doi: 10.1111/j.1600-6143.2006.01673.x. [DOI] [PubMed] [Google Scholar]
- 22.Moreno I, Vicente R, Ramos F, Vicente JL, Barberá M. Determination of interleukin-6 in lung transplantation: association with primary graft dysfunction. Transplant Proc. 2007;39:2425–2426. doi: 10.1016/j.transproceed.2007.07.056. [DOI] [PubMed] [Google Scholar]
- 23.Barreiro LB, Ben-Ali M, Quach H, Laval G, Patin E, Pickrell JK, Bouchier C, Tichit M, Neyrolles O, Gicquel B, et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 2009;5:e1000562. doi: 10.1371/journal.pgen.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferwerda B, McCall MB, Alonso S, Giamarellos-Bourboulis EJ, Mouktaroudi M, Izagirre N, Syafruddin D, Kibiki G, Cristea T, Hijmans A, et al. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci USA. 2007;104:16645–16650. doi: 10.1073/pnas.0704828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, Musser JM. Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS ONE. 2007;2:e1318. doi: 10.1371/journal.pone.0001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plantinga TS, Ioana M, Alonso S, Izagirre N, Hervella M, Joosten LA, van der Meer JW, de la Rúa C, Netea MG. The evolutionary history of TLR4 polymorphisms in Europe. J Innate Immun. 2012;4:168–175. doi: 10.1159/000329492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tindall EA, Hayes VM. Comprehensive sequence analysis of the human IL23A gene defines new variation content and high rate of evolutionary conservation. DNA Res. 2010;17:117–122. doi: 10.1093/dnares/dsq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasseur E, Patin E, Laval G, Pajon S, Fornarino S, Crouau-Roy B, Quintana-Murci L. The selective footprints of viral pressures at the human RIG-I-like receptor family. Hum Mol Genet. 2011;20:4462–4474. doi: 10.1093/hmg/ddr377. [DOI] [PubMed] [Google Scholar]
- 29.Xu Z, Balsara RD, Gorlatova NV, Lawrence DA, Castellino FJ, Ploplis VA. Conservation of critical functional domains in murine plasminogen activator inhibitor-1. J Biol Chem. 2004;279:17914–17920. doi: 10.1074/jbc.M314197200. [DOI] [PubMed] [Google Scholar]
- 30.Awad M, Pravica V, Perrey C, El Gamel A, Yonan N, Sinnott PJ, Hutchinson IV. CA repeat allele polymorphism in the first intron of the human interferon-gamma gene is associated with lung allograft fibrosis. Hum Immunol. 1999;60:343–346. doi: 10.1016/s0198-8859(98)00133-5. [DOI] [PubMed] [Google Scholar]
- 31.El-Gamel A, Awad MR, Hasleton PS, Yonan NA, Hutchinson JA, Campbell CS, Rahman AH, Deiraniya AK, Sinnott PJ, Hutchinson IV. Transforming growth factor-beta (TGF-beta1) genotype and lung allograft fibrosis. J Heart Lung Transplant. 1999;18:517–523. doi: 10.1016/s1053-2498(98)00024-2. [DOI] [PubMed] [Google Scholar]
- 32.Kastelijn EA, van Moorsel CH, Rijkers GT, Ruven HJ, Karthaus V, Kwakkel-van Erp JM, van de Graaf EA, Zanen P, van Kessel DA, Grutters JC, et al. Polymorphisms in innate immunity genes associated with development of bronchiolitis obliterans after lung transplantation. J Heart Lung Transplant. 2010;29:665–671. doi: 10.1016/j.healun.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Kastelijn EA, van Moorsel CH, Ruven HJ, Karthaus V, Kwakkel-van Erp JM, van de Graaf EA, Zanen P, van Kessel DA, Grutters JC, van den Bosch JM. Genetic polymorphisms in MMP7 and reduced serum levels associate with the development of bronchiolitis obliterans syndrome after lung transplantation. J Heart Lung Transplant. 2010;29:680–686. doi: 10.1016/j.healun.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Kastelijn EA, van Moorsel CH, Ruven HJ, Lammers JW, Grutters JC. Genetic polymorphisms and bronchiolitis obliterans syndrome after lung transplantation: promising results and recommendations for the future. Transplantation. 2012;93:127–135. doi: 10.1097/TP.0b013e31823915d5. [DOI] [PubMed] [Google Scholar]
- 35.Kwakkel-van Erp JM, van de Graaf EA, Paantjens AW, van Ginkel WG, Schellekens J, van Kessel DA, van den Bosch JM, Otten HG. The killer immunoglobulin-like receptor (KIR) group A haplotype is associated with bronchiolitis obliterans syndrome after lung transplantation. J Heart Lung Transplant. 2008;27:995–1001. doi: 10.1016/j.healun.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Munster JM, van der Bij W, Breukink MB, van der Steege G, Zuurman MW, Hepkema BG, Verschuuren EA, van Son WJ, Seelen MA. Association between donor MBL promoter haplotype and graft survival and the development of BOS after lung transplantation. Transplantation. 2008;86:1857–1863. doi: 10.1097/TP.0b013e31819064b8. [DOI] [PubMed] [Google Scholar]
- 37.Palmer SM, Burch LH, Davis RD, Herczyk WF, Howell DN, Reinsmoen NL, Schwartz DA. The role of innate immunity in acute allograft rejection after lung transplantation. Am J Respir Crit Care Med. 2003;168:628–632. doi: 10.1164/rccm.200303-447OC. [DOI] [PubMed] [Google Scholar]
- 38.Palmer SM, Burch LH, Trindade AJ, Davis RD, Herczyk WF, Reinsmoen NL, Schwartz DA. Innate immunity influences long-term outcomes after human lung transplant. Am J Respir Crit Care Med. 2005;171:780–785. doi: 10.1164/rccm.200408-1129OC. [DOI] [PubMed] [Google Scholar]
- 39.Palmer SM, Klimecki W, Yu L, Reinsmoen NL, Snyder LD, Ganous TM, Burch L, Schwartz DA. Genetic regulation of rejection and survival following human lung transplantation by the innate immune receptor CD14. Am J Transplant. 2007;7:693–699. doi: 10.1111/j.1600-6143.2007.01669.x. [DOI] [PubMed] [Google Scholar]
- 40.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diamond JM, Lee JC, Kawut SM, Shah RJ, Localio AR, Bellamy SL, Lederer DJ, Cantu E, Kohl BA, Lama VN, et al. Lung Transplant Outcomes Group. Clinical risk factors for primary graft dysfunction after lung transplantation. Am J Respir Crit Care Med. 2013;187:527–534. doi: 10.1164/rccm.201210-1865OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer NJ, Feng R, Li M, Zhao Y, Sheu CC, Tejera P, Gallop R, Bellamy S, Rushefski M, Lanken PN, et al. IL1rn coding variant is associated with lower risk of acute respiratory distress syndrome and increased plasma IL1ra. Am J Respir Crit Care Med. 2013;187:950–959. doi: 10.1164/rccm.201208-1501OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tejera P, Meyer NJ, Chen F, Feng R, Zhao Y, O’Mahony DS, Li L, Sheu CC, Zhai R, Wang Z, et al. Distinct and replicable genetic risk factors for acute respiratory distress syndrome of pulmonary or extrapulmonary origin. J Med Genet. 2012;49:671–680. doi: 10.1136/jmedgenet-2012-100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Gutierrez Arcelus M, Sekowska M, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, Deloukas P, Dermitzakis ET. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, Wang HH, Yao A, Chen YT, Hsu CN. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akimova T, Kamath BM, Goebel JW, Meyers KEC, Rand EB, Hawkins A, Levine MH, Bucuvalas JC, Hancock WW. Differing effects of rapamycin or calcineurin inhibitor on T-regulatory cells in pediatric liver and kidney transplant recipients. Am J Transplant. 2012;12:3449–3461. doi: 10.1111/j.1600-6143.2012.04269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyre D, Farge M, Lecoeur C, Proenca C, Durand E, Allegaert F, Tichet J, Marre M, Balkau B, Weill J, et al. R125W coding variant in TBC1D1 confers risk for familial obesity and contributes to linkage on chromosome 4p14 in the French population. Hum Mol Genet. 2008;17:1798–1802. doi: 10.1093/hmg/ddn070. [DOI] [PubMed] [Google Scholar]
- 50.Stone S, Abkevich V, Russell DL, Riley R, Timms K, Tran T, Trem D, Frank D, Jammulapati S, Neff CD, et al. TBC1D1 is a candidate for a severe obesity gene and evidence for a gene/gene interaction in obesity predisposition. Hum Mol Genet. 2006;15:2709–2720. doi: 10.1093/hmg/ddl204. [DOI] [PubMed] [Google Scholar]
- 51.Lederer DJ, Kawut SM, Wickersham N, Winterbottom C, Bhorade S, Palmer SM, Lee J, Diamond JM, Wille KM, Weinacker A, et al. Lung Transplant Outcomes Group. Obesity and primary graft dysfunction after lung transplantation: the Lung Transplant Outcomes Group Obesity Study. Am J Respir Crit Care Med. 2011;184:1055–1061. doi: 10.1164/rccm.201104-0728OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Konya V, Ullen A, Kampitsch N, Theiler A, Philipose S, Parzmair GP, Marsche G, Peskar BA, Schuligoi R, Sattler W, et al. Endothelial e-type prostanoid 4 receptors promote barrier function and inhibit neutrophil trafficking. J Allerg Clin Immunol. 2013;131:532–540 e532. doi: 10.1016/j.jaci.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 53.Soontrapa K, Honda T, Sakata D, Yao C, Hirata T, Hori S, Matsuoka T, Kita Y, Shimizu T, Kabashima K, et al. Prostaglandin E2-prostaglandin E receptor subtype 4 (EP4) signaling mediates UV irradiation-induced systemic immunosuppression. Proc Natl Acad Sci USA. 2011;108:6668–6673. doi: 10.1073/pnas.1018625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang EH, Libby P, Vanhoutte PM, Xu A. Anti-inflammation therapy by activation of prostaglandin EP4 receptor in cardiovascular and other inflammatory diseases. J Cardiovasc Pharmacol. 2012;59:116–123. doi: 10.1097/FJC.0b013e3182244a12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoggatt J, Mohammad KS, Singh P, Hoggatt AF, Chitteti BR, Speth JM, Hu P, Poteat BA, Stilger KN, Ferraro F, et al. Differential stem- and progenitor-cell trafficking by prostaglandin E2. Nature. 2013;495:365–369. doi: 10.1038/nature11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baratelli F, Lin Y, Zhu L, Yang SC, Heuzé-Vourc’h N, Zeng G, Reckamp K, Dohadwala M, Sharma S, Dubinett SM. Prostaglandin E2 induces FOXP3 gene expression and T regulatory cell function in human CD4+ T cells. J Immunol. 2005;175:1483–1490. doi: 10.4049/jimmunol.175.3.1483. [DOI] [PubMed] [Google Scholar]
- 57.Lee BP, Juvet SC, Zhang L. Prostaglandin E2 signaling through E prostanoid receptor 2 impairs proliferative response of double negative regulatory T cells. Int Immunopharmacol. 2009;9:534–539. doi: 10.1016/j.intimp.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 58.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yue P, Melamud E, Moult J. SNPs3D: candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D’Alessio FR, Tsushima K, Aggarwal NR, West EE, Willett MH, Britos MF, Pipeling MR, Brower RG, Tuder RM, McDyer JF, et al. CD4+CD25+Foxp3+ Tregs resolve experimental lung injury in mice and are present in humans with acute lung injury. J Clin Invest. 2009;119:2898–2913. doi: 10.1172/JCI36498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pietropaoli A, Georas SN. Resolving lung injury: a new role for Tregs in controlling the innate immune response. J Clin Invest. 2009;119:2891–2894. doi: 10.1172/JCI40880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garibaldi BT, D’Alessio FR, Mock JR, Files DC, Chau E, Eto Y, Drummond MB, Aggarwal NR, Sidhaye V, King LS. Regulatory T cells reduce acute lung injury fibroproliferation by decreasing fibrocyte recruitment. Am J Respir Cell Mol Biol. 2013;48:35–43. doi: 10.1165/rcmb.2012-0198OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun J, Han ZB, Liao W, Yang SG, Yang Z, Yu J, Meng L, Wu R, Han ZC. Intrapulmonary delivery of human umbilical cord mesenchymal stem cells attenuates acute lung injury by expanding CD4+CD25+ forkhead boxp3 (Foxp3)+ regulatory T cells and balancing anti- and pro-inflammatory factors. Cell Physiol Biochem. 2011;27:587–596. doi: 10.1159/000329980. [DOI] [PubMed] [Google Scholar]
- 66.Kuzumoto Y, Sho M, Ikeda N, Hamada K, Mizuno T, Akashi S, Tsurui Y, Kashizuka H, Nomi T, Kubo A, et al. Significance and therapeutic potential of prostaglandin E2 receptor in hepatic ischemia/reperfusion injury in mice. Hepatology. 2005;42:608–617. doi: 10.1002/hep.20827. [DOI] [PubMed] [Google Scholar]
- 67.Hishikari K, Suzuki J, Ogawa M, Isobe K, Takahashi T, Onishi M, Takayama K, Isobe M. Pharmacological activation of the prostaglandin E2 receptor EP4 improves cardiac function after myocardial ischaemia/reperfusion injury. Cardiovasc Res. 2009;81:123–132. doi: 10.1093/cvr/cvn254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nomi T, Sho M, Akahori T, Kanehiro H, Nakajima Y. Protective effect of prostaglandin E2 receptors EP2 and EP4 in alloimmune response in vivo. Transplant Proc. 2006;38:3209–3210. doi: 10.1016/j.transproceed.2006.10.118. [DOI] [PubMed] [Google Scholar]
- 69.Hristovska AM, Rasmussen LE, Hansen PB, Nielsen SS, Nüsing RM, Narumiya S, Vanhoutte P, Skøtt O, Jensen BL. Prostaglandin E2 induces vascular relaxation by E-prostanoid 4 receptor-mediated activation of endothelial nitric oxide synthase. Hypertension. 2007;50:525–530. doi: 10.1161/HYPERTENSIONAHA.107.088948. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–302. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 71.Nitz I, Fisher E, Grallert H, Li Y, Gieger C, Rubin D, Boeing H, Spranger J, Lindner I, Schreiber S, et al. Association of prostaglandin E synthase 2 (PTGES2) Arg298His polymorphism with type 2 diabetes in two German study populations. J Clin Endocrinol Metab. 2007;92:3183–3188. doi: 10.1210/jc.2006-2550. [DOI] [PubMed] [Google Scholar]
- 72.Sharma S, Yang SC, Zhu L, Reckamp K, Gardner B, Baratelli F, Huang M, Batra RK, Dubinett SM. Tumor cyclooxygenase-2/prostaglandin E2-dependent promotion of FOXP3 expression and CD4+ CD25+ T regulatory cell activities in lung cancer. Cancer Res. 2005;65:5211–5220. doi: 10.1158/0008-5472.CAN-05-0141. [DOI] [PubMed] [Google Scholar]
- 73.Hamberg M, Samuelsson B. On the metabolism of prostaglandins E1 and E2 in man. J Biol Chem. 1971;246:6713–6721. [PubMed] [Google Scholar]
- 74.Samuelsson B, Granström E, Green K, Hamberg M, Hammarström S. Prostaglandins. Annu Rev Biochem. 1975;44:669–695. doi: 10.1146/annurev.bi.44.070175.003321. [DOI] [PubMed] [Google Scholar]
- 75.Chadt A, Leicht K, Deshmukh A, Jiang LQ, Scherneck S, Bernhardt U, Dreja T, Vogel H, Schmolz K, Kluge R, et al. Tbc1d1 mutation in lean mouse strain confers leanness and protects from diet-induced obesity. Nat Genet. 2008;40:1354–1359. doi: 10.1038/ng.244. [DOI] [PubMed] [Google Scholar]
- 76.Qu D, Ludwig DS, Gammeltoft S, Piper M, Pelleymounter MA, Cullen MJ, Mathes WF, Przypek R, Kanarek R, Maratos-Flier E. A role for melanin-concentrating hormone in the central regulation of feeding behaviour. Nature. 1996;380:243–247. doi: 10.1038/380243a0. [DOI] [PubMed] [Google Scholar]
- 77.Shafey M, Anderson JL, Scarvelis D, Doucette SP, Gagnon F, Wells PS. Factor XIII Val34Leu variant and the risk of myocardial infarction: a meta-analysis. Thromb Haemost. 2007;97:635–641. [PubMed] [Google Scholar]
- 78.Lau CL, Zhao Y, Kim J, Kron IL, Sharma A, Yang Z, Laubach VE, Linden J, Ailawadi G, Pinsky DJ. Enhanced fibrinolysis protects against lung ischemia-reperfusion injury. J Thorac Cardiovasc Surg. 2009;137:1241–1248. doi: 10.1016/j.jtcvs.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shah RJ, Bellamy SL, Localio AR, Wickersham N, Diamond JM, Weinacker A, Lama VN, Bhorade S, Belperio JA, Crespo M, et al. A panel of lung injury biomarkers enhances the definition of primary graft dysfunction (PGD) after lung transplantation. J Heart Lung Transplant. 2012;31:942–949. doi: 10.1016/j.healun.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J, et al. Strengthening the reporting of genetic association studies (STREGA): An extension of the strobe statement. Ann Intern Med. 2009;150:206–215. doi: 10.7326/0003-4819-150-3-200902030-00011. [DOI] [PubMed] [Google Scholar]
- 81.Cantu E, Lederer DJ, Meyer K, Milewski K, Suzuki Y, Shah RJ, Diamond JM, Meyer NJ, Tobias JW, Baldwin DA, et al. CTOT Investigators. Gene set enrichment analysis identifies key innate immune pathways in primary graft dysfunction after lung transplantation. Am J Transplant. 2013;13:1898–1904. doi: 10.1111/ajt.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]