Abstract
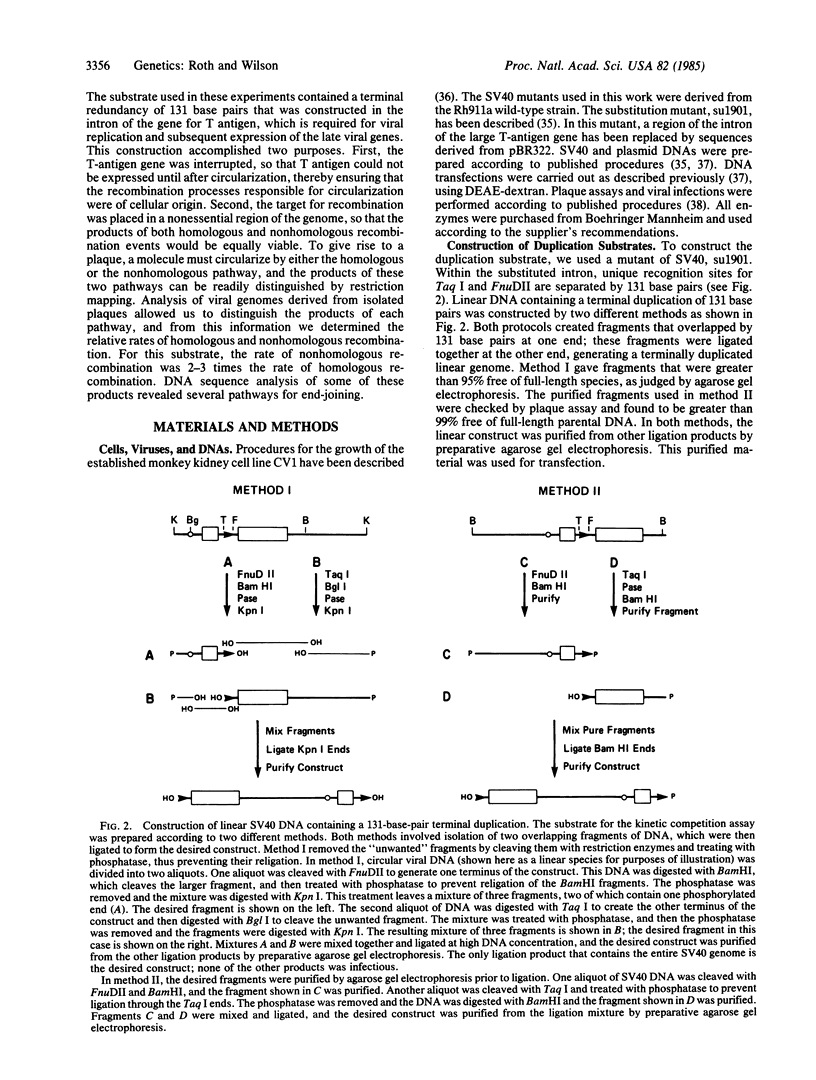
Both homologous and nonhomologous recombination events occur at high efficiency in DNA molecules transfected into mammalian cells. Both types of recombination occur with similar overall efficiencies, as measured by an endpoint assay, but their relative rates are unknown. In this communication, we measure the relative rates of homologous and nonhomologous recombination in DNA transfected into monkey cells. This measurement is made by using a linear simian virus 40 genome that contains a 131-base-pair duplication at its termini. Once inside the cell, this molecule must circularize to initiate lytic infection. Circularization can occur either by direct, nonhomologous end-joining or by homologous recombination within the duplicated region. Although the products of the two recombination pathways are different, they are equally infectious. Since homologous and nonhomologous recombination processes are competing for the same substrate, the relative amounts of the products of each pathway should reflect the relative rates of homologous and nonhomologous recombination. Analysis of individual recombinant genomes from 164 plaques indicates that the rate of circularization by nonhomologous recombination is 2- to 3-fold higher than the rate of homologous recombination. The assay system described here may prove to be useful for testing procedures designed to influence the relative rates of homologous and nonhomologous recombination.
Full text
PDF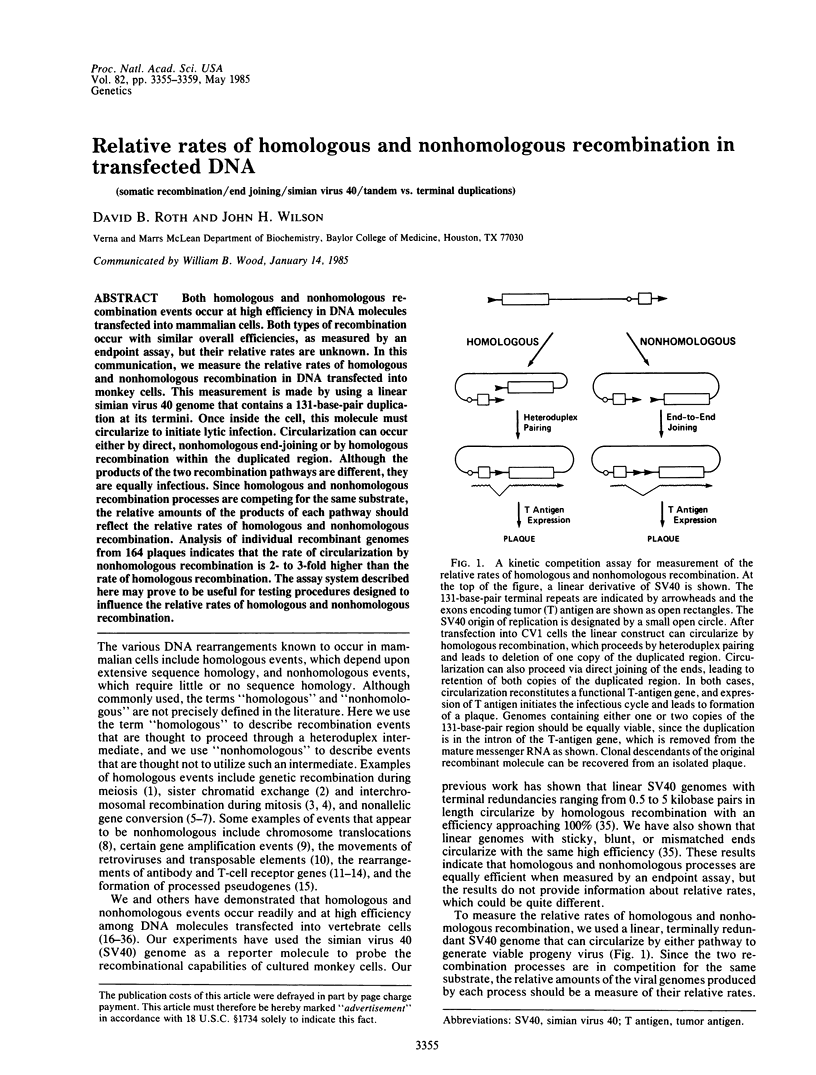



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bandyopadhyay P. K., Watanabe S., Temin H. M. Recombination of transfected DNAs in vertebrate cells in culture. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3476–3480. doi: 10.1073/pnas.81.11.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein L. B., Mount S. M., Weiner A. M. Pseudogenes for human small nuclear RNA U3 appear to arise by integration of self-primed reverse transcripts of the RNA into new chromosomal sites. Cell. 1983 Feb;32(2):461–472. doi: 10.1016/0092-8674(83)90466-x. [DOI] [PubMed] [Google Scholar]
- Brenner D. A., Kato S., Anderson R. A., Smigocki A. C., Camerini-Otero R. D. The recombination and integration of DNAs introduced into mouse L cells. Cold Spring Harb Symp Quant Biol. 1984;49:151–160. doi: 10.1101/sqb.1984.049.01.018. [DOI] [PubMed] [Google Scholar]
- Folger K. R., Wong E. A., Wahl G., Capecchi M. R. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol. 1982 Nov;2(11):1372–1387. doi: 10.1128/mcb.2.11.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger K., Thomas K., Capecchi M. R. Analysis of homologous recombination in cultured mammalian cells. Cold Spring Harb Symp Quant Biol. 1984;49:123–138. doi: 10.1101/sqb.1984.049.01.016. [DOI] [PubMed] [Google Scholar]
- Gerondakis S., Cory S., Adams J. M. Translocation of the myc cellular oncogene to the immunoglobulin heavy chain locus in murine plasmacytomas is an imprecise reciprocal exchange. Cell. 1984 Apr;36(4):973–982. doi: 10.1016/0092-8674(84)90047-3. [DOI] [PubMed] [Google Scholar]
- Hedrick S. M., Cohen D. I., Nielsen E. A., Davis M. M. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984 Mar 8;308(5955):149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- Honjo T. Immunoglobulin genes. Annu Rev Immunol. 1983;1:499–528. doi: 10.1146/annurev.iy.01.040183.002435. [DOI] [PubMed] [Google Scholar]
- Kopchick J. J., Stacey D. W. Differences in intracellular DNA ligation after microinjection and transfection. Mol Cell Biol. 1984 Feb;4(2):240–246. doi: 10.1128/mcb.4.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherlapati R. S., Eves E. M., Song K. Y., Morse B. S., Smithies O. Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3153–3157. doi: 10.1073/pnas.81.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz B. A., Haynes R. H. Phenomenology and genetic control of mitotic recombination in yeast. Annu Rev Genet. 1981;15:57–89. doi: 10.1146/annurev.ge.15.120181.000421. [DOI] [PubMed] [Google Scholar]
- Latt S. A. Sister chromatid exchange formation. Annu Rev Genet. 1981;15:11–55. doi: 10.1146/annurev.ge.15.120181.000303. [DOI] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Stachelek J. L. Evidence for intrachromosomal gene conversion in cultured mouse cells. Cell. 1983 Nov;35(1):157–165. doi: 10.1016/0092-8674(83)90218-0. [DOI] [PubMed] [Google Scholar]
- Malissen M., Minard K., Mjolsness S., Kronenberg M., Goverman J., Hunkapiller T., Prystowsky M. B., Yoshikai Y., Fitch F., Mak T. W. Mouse T cell antigen receptor: structure and organization of constant and joining gene segments encoding the beta polypeptide. Cell. 1984 Jul;37(3):1101–1110. doi: 10.1016/0092-8674(84)90444-6. [DOI] [PubMed] [Google Scholar]
- May P., Resche-Rigon M., Borde J., Breugnot C., May E. Conversion through homologous recombination of the gene encoding Simian virus 40 115,000-molecular-weight super T antigen to a gene encoding a normal-size large T antigen variant. Mol Cell Biol. 1984 Jun;4(6):1141–1151. doi: 10.1128/mcb.4.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. K., Temin H. M. High-efficiency ligation and recombination of DNA fragments by vertebrate cells. Science. 1983 May 6;220(4597):606–609. doi: 10.1126/science.6301012. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984 Nov;4(11):2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W. Direct transfer of cloned genes from bacteria to mammalian cells. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2163–2167. doi: 10.1073/pnas.77.4.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Recombination of dispersed repeated DNA sequences in yeast. Science. 1980 Sep 19;209(4463):1380–1384. doi: 10.1126/science.6251545. [DOI] [PubMed] [Google Scholar]
- Shapira G., Stachelek J. L., Letsou A., Soodak L. K., Liskay R. M. Novel use of synthetic oligonucleotide insertion mutants for the study of homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4827–4831. doi: 10.1073/pnas.80.15.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J., Scangos G. Recombination during gene transfer into mouse cells can restore the function of deleted genes. Science. 1983 Jan 14;219(4581):174–176. doi: 10.1126/science.6294829. [DOI] [PubMed] [Google Scholar]
- Stark G. R., Wahl G. M. Gene amplification. Annu Rev Biochem. 1984;53:447–491. doi: 10.1146/annurev.bi.53.070184.002311. [DOI] [PubMed] [Google Scholar]
- Subramani S., Berg P. Homologous and nonhomologous recombination in monkey cells. Mol Cell Biol. 1983 Jun;3(6):1040–1052. doi: 10.1128/mcb.3.6.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Upcroft P., Carter B., Kidson C. Analysis of recombination in mammalian cells using SV40 genome segments having homologous overlapping termini. Nucleic Acids Res. 1980 Jun 25;8(12):2725–2736. doi: 10.1093/nar/8.12.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upcroft P., Carter B., Kidson C. Mammalian cell function mediating recombination of genetic elements. Nucleic Acids Res. 1980 Dec 11;8(23):5835–5844. doi: 10.1093/nar/8.23.5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert F. C., Young C. S. The genetic analysis of recombination using adenovirus overlapping terminal DNA fragments. Virology. 1983 Feb;125(1):175–193. doi: 10.1016/0042-6822(83)90072-7. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Gudewicz T., Porter T., White A., Wilson J. H. How damaged is the biologically active subpopulation of transfected DNA? Mol Cell Biol. 1984 Mar;4(3):387–398. doi: 10.1128/mcb.4.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Wilson J. H. Defined oligomeric SV40 DNA: a sensitive probe of general recombination in somatic cells. Cell. 1980 Aug;21(1):141–148. doi: 10.1016/0092-8674(80)90121-x. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Wilson J. H. Simian virus 40 recombinants are produced at high frequency during infection with genetically mixed oligomeric DNA. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2876–2880. doi: 10.1073/pnas.76.6.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth J. J., Vock Hall L. Genetic demonstration of mitotic recombination in cultured Chinese hamster cell hybrids. Cell. 1984 Mar;36(3):697–707. doi: 10.1016/0092-8674(84)90350-7. [DOI] [PubMed] [Google Scholar]
- Weiss E. H., Mellor A., Golden L., Fahrner K., Simpson E., Hurst J., Flavell R. A. The structure of a mutant H-2 gene suggests that the generation of polymorphism in H-2 genes may occur by gene conversion-like events. Nature. 1983 Feb 24;301(5902):671–674. doi: 10.1038/301671a0. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H. Genetic analysis of host range mutant viruses suggests an uncoating defect in simian virus 40-resistant monkey cells. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3503–3507. doi: 10.1073/pnas.74.8.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H. Interference in SV40 DNA infections: a possible basis for cellular competence. Virology. 1978 Dec;91(2):380–388. doi: 10.1016/0042-6822(78)90385-9. [DOI] [PubMed] [Google Scholar]
- Yanagi Y., Yoshikai Y., Leggett K., Clark S. P., Aleksander I., Mak T. W. A human T cell-specific cDNA clone encodes a protein having extensive homology to immunoglobulin chains. Nature. 1984 Mar 8;308(5955):145–149. doi: 10.1038/308145a0. [DOI] [PubMed] [Google Scholar]
- de Saint Vincent B. R., Wahl G. M. Homologous recombination in mammalian cells mediates formation of a functional gene from two overlapping gene fragments. Proc Natl Acad Sci U S A. 1983 Apr;80(7):2002–2006. doi: 10.1073/pnas.80.7.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]