Abstract
We demonstrate a microfluidic device capable of tracking the volume of individual cells by integrating an on-chip volume sensor with pressure-activated cell trapping capabilities. The device creates a dynamic trap by operating in feedback; a cell is periodically redirected back and forth through a microfluidic volume sensor (Coulter principle). Sieve valves are positioned on both ends of the sensing channel, creating a physical barrier which enables media to be quickly exchanged while keeping a cell firmly in place. The volume of individual Saccharomyces cerevisiae cells was tracked over entire growth cycles, and the ability to quickly exchange media was demonstrated.
Measuring cell growth is of primary interest to researchers who seek to study the effects of drugs, nutrients, disease, and environmental stress. This has traditionally been accomplished by monitoring the optical transmittance of large ensembles of cells and applying the Beer-Lambert Law.1,2 Such population-scale measurements provide important culture statistics, but averaging obscures the behaviour of individual cells. In addition, these techniques often require cell synchronicity in order to correlate growth with specific points in the cell cycle, but synchronicity typically decays rapidly in many cell lines including Saccharomyces cerevisiae (yeast) cultures.3 Researchers have thus adopted methods that study the growth of individual cells. Quantifying cellular growth is especially challenging since proliferating cells such as yeast or Escherichia coli are irregularly shaped, and will only increase in size by a factor of two.4 Growth will affect the mass, volume, and density of the cell; having access to each of these characteristics is important in obtaining a complete picture of this process. Time-lapse fluorescence microscopy can provide valuable information as to the cell cycle progression of individual cells,5 but 2D optics requires geometric assumptions, and, thus, can provide an incomplete picture of growth.6,7
Microfluidic lab-on-chip devices with integrated sensors can provide high-resolution growth tracking of individual cells, either through mass, volume, or density monitoring.4,7,8 Recently, a microfluidic mass sensor was used to track the buoyant mass of individual cells using a suspended microchannel resonator (SMR).4,9 Monitoring growth can also be accomplished by tracking volume using microfluidic volume sensors7 operating on the Coulter principle.10 Trapping can be achieved by either (1) cycling the target back and forth through the sensor (pressure-driven4 and electrokinetic7) or (2) holding a cell in place (posts,11 chevron structure,12 and E-Field13). The former, dynamic approach, allows a single cell to be sampled periodically by reversing flow directions after a cell is detected. Simple in its implementation, this technique also has the ability to compensate for a drifting baseline current resulting from parasitic ionic changes within the sensing channel or other sources of noise. On the other hand, static traps allow cells to be held in place while the buffer is rapidly exchanged.12 The ability to dynamically change cellular growth conditions during an experiment can lead to significant insight into the behaviour of cells in environments of varying salinity,14 oxidative,15,16 or osmotic conditions,17 as well as the effect of nutrients18 and drugs.19
In this work, we propose a device capable of tracking growth using high-resolution volume measurements, combining the best attributes of both types of measurement systems; continuous baseline correction and the ability to rapidly exchange cell media. This is accomplished by using a pressure-driven, feedback-based dynamic trap, whereby a cell is cycled back and forth through the sensor within a microfluidic channel. On-chip sieve valves positioned at both ends of the sensing channel are able to selectively capture a cell while the solution is being replaced. As proof of principle, the volume of several individual yeast cells was monitored over the course of their respective growth cycles, and the ability to quantify growth response to media exchange was demonstrated.
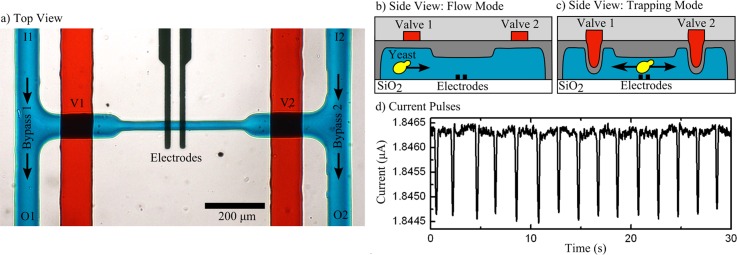
Devices were fabricated using multilayered soft lithography with polydimethylsiloxane (PDMS) molding.20 The completed device is pictured in Figure 1(a); full fabrication protocols are presented as supplementary material.21 To maximize measurement sensitivity, it is optimal to choose a channel width and height slightly larger than the dimensions of the target cell.22 However, yeast cells are asymmetrically shaped and tend to tumble as they traverse the sensor. Preliminary testing suggested this effect could be mitigated by having cells flow along trajectories far from the electrodes (through buoyancy), where electric field is more uniform. Thus, a channel height of 20 μm was chosen as a compromise. Channel height increases to 28 μm in the wider part of the central and bypass channels, a result of using a mold made out of reflowed photoresist.23 Channel width was set at 25 μm through the sensor, and widens to 80 μm at the sieve valves to facilitate valve actuation, which requires a high width to height ratio.20 The fluidic layer is integrated in a 35 μm thick PDMS spin-coated layer, above which sits a 50 μm tall valve channel in a 4 mm PDMS layer. Tubing connects I1 and I2 to a common inlet vial, V1 and V2 to vials filled with deionised water and O1 and O2 connect to empty vials (not pictured). Inlet pressures I1 and I2, and valve pressures V1 and V2 are controlled with manual regulators (SMC IR2000-N02-R and SMC IR2010-N02-R); outlet pressures are computer-controlled (SMC ITV-1011). This pressure scheme is detailed elsewhere.24 Current pulses caused by transiting particles/cells (Figure 1(d)) were acquired by applying a 50 kHz, 220 mV AC voltage between a pair of electrodes and measuring the drawn current. This frequency is sufficiently elevated to avoid the electrical double layer capacitance at the electrode-electrolyte interface,25 but low enough to avoid sensitivity to cell impedance or substrate.26 The electrical setup used for these experiments has been described previously.24,27 A temperature controller maintains the device at 30 °C.
FIG. 1.
(a) Micrograph of the microfluidic device. Two parallel bypass channels are connected by a sensing channel with sensing electrodes. Pressure is applied at inlets (I1, I2) and outlets (O1, O2) to control flow conditions. Valves (V1, V2) are positioned over each end of the sensing channel. Food coloring is used to highlight the valve (red) and fluidic layers (blue). (b) Flow mode: valves are unpressurized, and cells flow freely through the device. (c) Trapping mode: valves are pressurized to capture a cell within the central channel. Pressure-driven flow cycles the cell back and forth across the sensor. (d)Typical current pulses measured for a yeast cell.
The cell capture, media exchange, and detection process occurs as follows. A cell suspension is loaded into the bypass channel and made to flow through the central sensing channel by imposing a pressure gradient (Figure 1(b)). Cells flowing through the sensor are observed optically; once a cell of interest is observed (a cell without a bud), valves are sealed (V1 = V2 = 35 psi). This stops all flow through the sensor, and enables bypass channels to be flushed and replaced with fresh media. After 2 min, valve channels are pressurized to 24 psi where they compress the channel to a sufficient height to physically restrict the passage of yeast cells, while allowing the media to flow through the central channel (Figure 1(c)). The pressure gradient between bypasses causes the media in the central channel to be flushed out, while the target cell is physically trapped. Replacing the media in the central channel takes 2 min. At this stage, a pressure-driven feedback-based dynamic trap can be initiated. In this dynamic trap mode, the pressure settings at O1 and O2 are adjusted to redirect the cell back and forth through the sensor, based on current pulses measured from cells transiting through the sensor. Through custom LabView® software, these outlet pressure settings are feedback-adjusted to maintain a speed of 250 μm/s in both directions at a detection frequency of 30 cells/min (Figure 1(d)). To minimize the effects of channel stretching/shrinking, the sum of pressures at O1 and O2 is held constant. This precaution was taken since the sensing channel structured within the flexible PDMS polymer will alter its geometry based on internal pressure.28 The short central channel ensures steady nutrient replenishment from the bypasses. For example, a glucose molecule takes ∼4 min to diffuse from the bypass to the electrodes. In practice, Taylor-Aris dispersion will reduce this replenishment time considerably. Based on video analysis, 25% of the central channel's media is replenished every pressure reversal (video presented as supplementary material21). Polystyrene microspheres of 3.9 ± 0.3 μm, 5.6 ± 0.2 μm, and 8.3 ± 0.7 μm (NIST size standards) were used to calibrate the sensor, and obtain the current pulse-to-volume calibration for every solution (supplementary material21). The validity of this calibration method is discussed elsewhere.29 Care was taken to limit trajectory-based variations in signal: the device is positioned with electrodes at the top of the sensing channel, and with the negatively buoyant cells/particles flowing along the bottom. Based on previous experimental and theory work, we found that signal amplitude can vary as much as 3.5 fold for different heights.27 The effect of trajectory on current pulse amplitude has also been reported elsewhere.30,31 In this work, buoyancy is used to ensure that the cell flows along a trajectory at the same distance from the electrodes for every measurement.
Saccharomyces cerevisiae (BY4743 Mat a/alpha, genotype: his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 ade2::LEU2/ade2::URA3) was cultured to exponential phase at 30 °C in an incubator/shaker in yeast bacto-peptone (YPD) with 2% w/v glucose, supplemented with 0.2 M NaCl, 0.05% bovine serum albumin (BSA) and 42 mg/l adenine. Sodium chloride was added to enable the current pulse measurement, at a concentration where cells are viable;32 BSA was used to prevent cell agglomeration; adenine was supplemented since this particular yeast mutant does not produce its own supply. A cell suspension was introduced into the device, from which a cell at the early stages of its cell cycle was captured, and dynamically trapped for 100 min. Three typical cell growth results are shown in Figure 2(a). Since the culture was not synchronized, this leads to variability between “initial” cell volumes: there is a 27% difference in initial volume between the cells identified by red squares and green triangles. This is caused by (1) optical limits, whereby cells chosen for study are not all at the exact same cell cycle stage and (2) differences in the age of the mother cell: the more buds a mother cell has produced, the larger it becomes.33 On average, captured yeast cell demonstrated a doubling time consistent with growth rates under ideal incubator/shaker conditions; nutrient depletion, electric field, and shear stresses are not affecting growth. Optical inspection of budding cells confirms that most growth is occurring at the daughter cell, as expected.33 An elevated signal-to-noise ratio allows for high resolution volumetric measurements (4 μm3); cell asymmetry7 and trajectory variability27,30,31 lead to a relative standard deviation of 6% for cells and 4% for microspheres of similar size. While mass or protein synthesis methods have indicated linear34 or exponential4,6,35,36 growth curves, volume-based methods have suggested sigmoidal patterns.7,37 Prior to daughter cell emergence, and later in the cycle as the daughter cell emerges, volumetric growth rate declines.38 In this work, it is difficult to ascertain with mathematical rigor the shape of the growth profile; however, for each cell, volume increases steadily throughout the growth cycle before declining near the end of the cycle.
FIG. 2.
(a) Growth curves for 3 cells trapped in succession. Simultaneous optical and electrical measurements allow cell cycle stage to be correlated with volume. Pictures of cell corresponding to the red squares are presented in 15 min increments. A cell is cycled through the sensor every 2 s. For clarity, each data point for yeast volume represents the average of data points over a period of 5 min, with standard deviation. (b) Demonstration of an interrupted growth cycle, where YPD + 0.2 M NaCl was replaced with 0.2 M NaCl at 40 min, and then again returned to YPD + 0.2 M NaCl at 80 min. The media exchange process takes 4 min.
To demonstrate our ability to easily exchange media while maintaining a trap, the solution was exchanged 40 min into a yeast growth cycle; culture media was replaced with a pure saline solution 0.2 M NaCl + 0.05% BSA, and then replaced again with culture media at 80 min (Figure 2(b)). Cell growth is halted temporarily while in saline solution, before resuming normal growth thereafter. The cell cycle time is extended by this period. The cell volume drifts downward after the initial solution change at 40 min. Though this drift lies within our uncertainty bounds, cellular responses to osmotic shock on similar timescales have been documented elsewhere.39 This result demonstrates an ability to quickly exchange cell media, and observe cellular response.
In conclusion, we have demonstrated a microfluidic device capable of maintaining a dynamic, pressure-driven cell trap, which can monitor cellular volume over the cell cycle. Concurrent optical microscopy allows for real-time visual inspection of the cells. In addition, sieve valve integration provides for the exchange of media or the addition of drugs. Such a platform could also be key in cancer cell cytotoxicity assays,40 where growth response to anticancer drugs could be monitored.
Acknowledgments
The authors thank Hilary Phenix and Mads Kaern for useful discussions and for supplying initial yeast cultures, Lukasz Andrzejewski for assistance in scripting, and Benjamin Watts for microfabrication help. This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Canadian Foundation for Innovation (CFI).
REFERENCES
- 1.Svanberg S., Atomic and Molecular Spectroscopy: Basic Aspects and Practical Applications; with 14 Tables (Springer, 2004). [Google Scholar]
- 2.Myers F. B. and Lee L. P., Lab Chip 8, 2015 (2008). 10.1039/b812343h [DOI] [PubMed] [Google Scholar]
- 3.Walker G. M., Methods Cell Sci. 21, 87 (1999). 10.1023/A:1009824520278 [DOI] [PubMed] [Google Scholar]
- 4.Godin M., Delgado F. F., Son S., Grover W. H., Bryan A. K., Tzur A., Jorgensen P., Payer K., Grossman A. D., Kirschner M. W., and Manalis S. R., Nat. Methods 7, 387 (2010). 10.1038/nmeth.1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn A. T., Jones J. T., and Meyer T., Cell Cycle 8, 1044 (2009). 10.4161/cc.8.7.8042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woldringh C. L., Huls P. G., and Vischer N. O., J. Bacteriol. 175, 3174 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J., Stowers C. C., Boczko E. M., and Li D., Lab Chip 10, 2986 (2010). 10.1039/c005029f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burg T. P., Godin M., Knudsen S. M., Shen W., Carlson G., Foster J. S., Babcock K., and Manalis S. R., Nature 446, 1066 (2007). 10.1038/nature05741 [DOI] [PubMed] [Google Scholar]
- 9.Bryan A. K., Goranov A., Amon A., and Manalis S. R., Proc. Natl. Acad. Sci. U. S. A. 107, 999 (2010). 10.1073/pnas.0901851107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coulter W. H., Proc. Natl. Electron. Conf. 12, 1034 (1956). [Google Scholar]
- 11.Luongo K., Holton A., Kaushik A., Spence P., Ng B., Deschenes R., Sundaram S., and Bhansali S., Biomicrofluidics 7, 034108 (2013). 10.1063/1.4809590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua S. Z. and Pennell T., Lab Chip 9, 251 (2009). 10.1039/b806003g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park H., Sens. Actuators, B 150, 167 (2010). 10.1016/j.snb.2010.07.020 [DOI] [Google Scholar]
- 14.Melamed D., Pnueli L., and Arava Y., RNA 14, 1337 (2008). 10.1261/rna.864908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castells-Roca L., Mühlenhoff U., Lill R., Herrero E., and Bellí G., Mol. Microbiol. 81, 232 (2011). 10.1111/j.1365-2958.2011.07689.x [DOI] [PubMed] [Google Scholar]
- 16.Demasi A. P. D., Pereira G. A. G., and Netto L. E. S., FEBS J. 273, 805 (2006). 10.1111/j.1742-4658.2006.05116.x [DOI] [PubMed] [Google Scholar]
- 17.Hohmann S., Microbiol. Mol. Biol. Rev. 66, 300 (2002). 10.1128/MMBR.66.2.300-372.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cullen P. J. and Sprague G. F., Proc. Natl. Acad. Sci. 97, 13619 (2000). 10.1073/pnas.240345197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beaumont T. E., Shekhar T. M., Kaur L., Pantaki-Eimany D., Kvansakul M., and Hawkins C. J., Cell Death Dis. 4, e619 (2013). 10.1038/cddis.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Unger M. A., Chou H.-P., Thorsen T., Scherer A., and Quake S. R., Science 288, 113 (2000). 10.1126/science.288.5463.113 [DOI] [PubMed] [Google Scholar]
- 21. See supplementary material at http://dx.doi.org/10.1063/1.4867035E-BIOMGB-8-001402 for fabrication protocol, cell trap video, and pulse-to-volume calibration.
- 22.Golden J. P., Justin G. A., Nasir M., and Ligler F. S., Anal. Bioanal. Chem. 402, 325 (2012). 10.1007/s00216-011-5415-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fordyce P. M., Diaz-Botia C. A., DeRisi J. L., and Gomez-Sjoberg R., Lab Chip 12, 4287 (2012). 10.1039/c2lc40414a [DOI] [PubMed] [Google Scholar]
- 24.Riordon J., Mirzaei M., and Godin M., Lab Chip 12, 3016 (2012). 10.1039/c2lc40357a [DOI] [PubMed] [Google Scholar]
- 25.Nieuwenhuis J. H., Kohl F., Bastemeijer J., Sarro P. M., and Vellekoop M. J., Sens. Actuators, B 102, 44 (2004). 10.1016/j.snb.2003.10.017 [DOI] [Google Scholar]
- 26.Gawad S., Cheung K., Seger U., Bertsch A., and Renaud P., Lab Chip 4, 241 (2004). 10.1039/b313761a [DOI] [PubMed] [Google Scholar]
- 27.Riordon J., Catafard N. M., and Godin M., Appl. Phys. Lett. 101, 154105 (2012). 10.1063/1.4759033 [DOI] [Google Scholar]
- 28.Hardy B. S., Uechi K., Zhen J., and Pirouz Kavehpour H., Lab Chip 9, 935 (2009). 10.1039/b813061b [DOI] [PubMed] [Google Scholar]
- 29.Sun J., Gao Y., Isaacs R. J., Boelte K. C., Lin P. C., Boczko E. M., and Li D., Anal. Chem. 84, 2017 (2012). 10.1021/ac203212g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gawad S., Schild L., and Renaud P., Lab Chip 1, 76 (2001). 10.1039/b103933b [DOI] [PubMed] [Google Scholar]
- 31.Sun T., Green N. G., Gawad S., and Morgan H., Nanobiotechnology IET 1, 69 (2007). 10.1049/iet-nbt:20070019 [DOI] [PubMed] [Google Scholar]
- 32.Logothetis S., Walker G., and Nerantzis E., Zb. Matice Srp. Za Prir. Nauke Issue 113, 271 (2007). 10.2298/ZMSPN0713271L [DOI] [Google Scholar]
- 33.Hartwell L. H. and Unger M. W., J. Cell Biol. 75, 422 (1977). 10.1083/jcb.75.2.422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitchison J. M., Exp. Cell Res. 15, 214 (1958). 10.1016/0014-4827(58)90077-6 [DOI] [PubMed] [Google Scholar]
- 35.Elliott S. G. and McLaughlin C. S., Proc. Natl. Acad. Sci. 75, 4384 (1978). 10.1073/pnas.75.9.4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Talia S. D., Skotheim J. M., Bean J. M., Siggia E. D., and Cross F. R., Nature 448, 947 (2007). 10.1038/nature06072 [DOI] [PubMed] [Google Scholar]
- 37.Scopes A. W. and Williamson D. H., Exp. Cell Res. 35, 361 (1964). 10.1016/0014-4827(64)90102-8 [DOI] [PubMed] [Google Scholar]
- 38.Mitchison J. M., in International Review of Cytology (Academic Press, 2003), pp. 165–258. [DOI] [PubMed] [Google Scholar]
- 39.Knudsen S. M., von Muhlen M. G., Schauer D. B., and Manalis S. R., Anal. Chem. 81, 7087 (2009). 10.1021/ac900968r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugiura S., Edahiro J., Kikuchi K., Sumaru K., and Kanamori T., Biotechnol. Bioeng. 100, 1156 (2008). 10.1002/bit.21836 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4867035E-BIOMGB-8-001402 for fabrication protocol, cell trap video, and pulse-to-volume calibration.