Abstract
Experimental results have demonstrated that the numbers of counterions surrounding nucleic acids differ from those predicted by the nonlinear Poisson-Boltzmann equation, NLPBE. Some studies have fit these data against the ion size in the size-modified Poisson-Boltzmann equation, SMPBE, but the present study demonstrates that other parameters, such as the Stern layer thickness and the molecular surface definition, can change the number of bound ions by amounts comparable to varying the ion size. These parameters will therefore have to be fit simultaneously against experimental data. In addition, the data presented here demonstrate that the derivative, SK, of the electrostatic binding free energy, ΔGel, with respect to the logarithm of the salt concentration is sensitive to these parameters, and experimental measurements of SK could be used to parameterize the model. However, although better values for the Stern layer thickness and ion size and better molecular surface definitions could improve the model's predictions of the numbers of ions around biomolecules and SK, ΔGel itself is more sensitive to parameters, such as the interior dielectric constant, which in turn do not significantly affect the distributions of ions around biomolecules. Therefore, improved estimates of the ion size and Stern layer thickness to use in the SMPBE will not necessarily improve the model's predictions of ΔGel.
INTRODUCTION
Several recent experimental studies have tested the ability of the nonlinear Poisson-Boltzmann equation, NLPBE, to predict both the numbers and distributions of counterions around nucleic acids. Investigators have measured the numbers of bound ions around DNA and RNA with anomalous small-angle x-ray scattering experiments1, 2, 3, 4, 5, 6 and titration methods,7, 8, 9 they have compared these predictions to molecular dynamics simulations,6, 10, 11 and they have measured experimental quantities, such as changes in conformational stability upon changing the solution environment12, 13, 14 that should be sensitive to changes in the distribution of ions around the nucleic acid. There is some debate within this literature, as some studies1, 2, 3, 6 show no significant difference between the predictions of the NLPBE and experiment, while other studies claim that the NLPBE overestimates the number of monovalent cations7, 8, 10, 11, 14 and underestimates the number of multivalent cations2, 7, 8, 11, 12, 14 attracted by nucleic acids. The underestimation of the density of divalent cations has been attributed to the neglect of ion-ion correlations in the NLPBE,15 but no consensus has emerged on how the NLPBE could be modified to account for these effects. In the present study, ion-ion correlations are not considered, and therefore only 1:1 salt was considered, where ion-ion correlations are less important. That the NLPBE might overestimate of the number of monovalent ions associating with DNA is intuitively reasonable, as the NLPBE assumes that ions are pointlike (charges with no radius). Some studies have claimed that the NLPBE overestimates the experimentally measured number of bound ions as the radius of the monovalent ion increases,7, 8, 10, 14 as would be expected if the discrepancy were caused by the NLPBE's assumption of pointlike ions. This conclusion is further indicated by the observation that the dependence of the melting temperature of DNA upon cation concentration varies systematically with the size of the monovalent cation.13
To relax the assumption of pointlike ions that allow unrealistically high ion densities near highly charged interfaces, several studies have examined the size-modified Poisson-Boltzmann equation, SMPBE,6, 8, 11, 16, 17, 18, 19, 20, 21 as this mean field theory introduces an entropic penalty to tight ion packing and forbids exceeding the ion density given by close packing. Some studies of ion distributions around nucleic acids with molecular dynamics have indicated that varying the ion size in the SMPBE yields slightly better agreement between the predicted and observed ion distributions.6, 11 One study8 has attempted to determine appropriate ion sizes from measurements of the number of bound ions, but the best-fit ion sizes for Na+, and K+ ions (between 7 and 8 Å) were larger than even the size of solvated Na+ and K+ ions. Potentially, these unexpectedly large ion sizes could indicate that the dehydration of ions near the surface may be important, which would necessitate further extensions to Poisson-Boltzmann, PB, methods6, 10, 11 or that other parameters, such as the definition of the molecular surface and the thickness of the ion exclusion (Stern) layer, that also affect the number of bound ions need to be set by comparing to experimental data. For example, two studies3, 4 have attempted to account for the effect of finite ion sizes by adjusting the thickness of the Stern layer and found that 4 Å as the radius of the Rb+ ion yielded the best fit to experimental data. Apparently, the SMPBE and related theories will have to be parameterized against several different parameters simultaneously.
As has been discussed before, the NLPBE and related methods can correctly predict how the electrostatic binding free energy, ΔGel, changes when the salt concentration is changed, but they have been much less successful at predicting ΔGel itself because ΔGel is very sensitive to some details of the model.22, 23, 24 Much research has focused on better predicting the derivative,25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49SK, of ΔGel with respect to the logarithm of the concentration of 1:1 salt, log [NaCl], because it can be measured experimentally. However, over the range of concentrations considered in this study, ΔGel can be broken into a salt-dependent component of the free energy, , that is typically smaller than and uncorrelated with the salt-independent component of ΔGel, .22, 23, 24 Improved predictions of are therefore not guaranteed to improve predictions of ΔGel. As shown in the present study, SK is sufficiently sensitive to some parameters, such as the ion size, the surface definition, and the thickness of the Stern layer, that measurements of SK could be used to obtain better estimates of these parameters, but ΔGel is much more sensitive to other parameters of the model, such as the interior dielectric constant and molecular surface definition, to which SK is not sensitive. These other parameters will have to be more accurately determined before the much smaller improvements offered by varying the ion size and Stern layer thickness will produce detectable improvements in PB methods’ predictions of ΔGel.
METHODS
Ideal A-, B-, and triplex-DNA structures were created from fiber models in the X3DNA package,50, 51 the radii and charges were taken from the AMBER force field,52 and the radii, charges, and hydrogens were added to the structure files with the PDB2PQR package.53, 54 The A- and B-DNA structures had charges of −48 e, where e is the charge on a proton, and the triplex-DNA had a charge of −72 e. For comparison to the experimental data, a 24 base pair B-DNA with a sequence of 5′-GGTGACGAGTGAGCTACTGGGCGG-3′ and a −46 e net charge was built as described above. The DNA-drug complexes were taken from the RCSB Protein Data Bank55 (Protein Data Bank identifications: 1D30,56 1D86,57 1EEL,58 227D,59 261D60), and the preparation of their structure files was discussed in a previous study.22 All NLPBE and SMPBE calculations were performed with the adaptive Cartesian grid-based Poisson-Boltzmann solver (CPB),21, 61 and all atom locations were fixed, with no attempt made to model the flexibility of these molecules. Unless otherwise stated, all calculations used an interior dielectric constant of 1, an exterior dielectric constant of 80, a 0.1 M salt concentration, a temperature of 298 K, a solvent-excluded surface defined with a solvent probe radius of 1.4 Å, no Stern layer, a minimum grid spacing of 0.2 Å, and a grid that extends out to 4 times the largest dimension of the molecule.
The number of bound ions in the NLPBE was computed by integrating the excess ion density,
| (1) |
outside the molecular interior and the Stern layer, where φnl is the electrostatic potential computed from the NLPBE, is the bulk density, zi is the valence of the ith ion species, e is the fundamental charge or the charge on a proton, k is Boltzmann's constant, and T is the temperature. The number of bound ions in the SMPBE was computed by integrating a similar density,
| (2) |
where φsm is the electrostatic potential computed with the uniform ion size SMPBE,19 and ζ = ∑ζi, where ζi is the ion-exclusion factor of the ith ion species:
| (3) |
where a is the lattice spacing, and a = 2rion, where rion is the size of an ion. While a variable ion size capability is available in the CPB solver,21, 61 in the present study, only the uniform ion-size SMPBE was used. This choice was made for the following reasons: (i) we wanted to reduce the size of the parameter space considered, and (ii) the nucleic acid considered here is predominantly negatively charged. Because of the large negative charge of the nucleic acid, the concentration of anions is probably never very high, and the size of these ions therefore probably does not strongly affect the resulting predictions. By construction, Eq. 3 prevents the ion density from exceeding that given by tightly packing the ions in space.
Because charge must be conserved and only 1:1 salt was considered here, the number of counterions attracted to plus the number of coions repelled from the nucleic acid should equal its charge. This result holds for the infinite domain, but since the calculations are performed on a grid of finite extent, a correction is added that accounts for the charge contribution from outside the computational domain. Because the region outside the grid is far from the molecule, the potential in this region is very small. The ion concentration in this region can therefore be approximated by expanding either Eq. 1 or 2,
| (4) |
where ρi(r) is the local excess concentration of the ith ion species, and φ is the electrostatic potential. In the present study, the number of counterions attracted to and the number of coions repelled from the region off the grid could be approximated by qres/2, where qres is the sum of the charge on the molecule and the charge of the ions on the grid because only 1:1 salt was considered.
ΔGel was computed as described in Ref. 24, but rather than computing SK from the slope of a best-fit line of ΔGel against log [NaCl], the derivative, ∂ΔGel/∂log [NaCl] was computed at a 1:1 salt concentration of 0.1 M. This was done for the NLPBE by taking advantage of the relationship
| (5) |
where ΔΔΠ is the change in osmotic pressure upon binding. For the SMPBE, the difference between the derivative of the free energy with respect to the logarithm of the salt concentration,
| (6) |
in the bound state and that in the unbound state was taken.19
One of the parameters that has not clearly been determined in PB methods is the best way to define the boundary between the interior and exterior regions of the molecule. Most researchers use the SE surface,62, 63 which defines the interior of the molecule to be the region that cannot be accessed by a solvent probe that is constrained to lie outside all atomic spheres, but other studies have concluded that the van der Waal's surface, which defines the molecular interior simply as the union of atomic spheres, produces a better match between theoretical calculations and experimental measurements.64, 65, 66 Still other studies have proposed alternative surfaces, such as self-consistent67, 68, 69, 70, 71 and Gaussian surface definitions.72, 73 The uncertainty in the surface definition suggests computing the numbers of bound ions on different surface definitions, but doing so would yield estimates of ΔGel and SK that vary in large and unpredictable ways, whereas what we seek is an understanding of how ΔGel and SK change in response to small, continuous changes in the surface definition. Instead, rather than considering a set of such disparate surface definitions, for some of these calculations the solvent-accessible, SA, surface,74 which defines the molecular interior as the union of spheres centered at the atomic charges with radii equal to their van der Waal's radii inflated by a solvent probe radius, was used. Although this surface yielded different estimates of ΔGel than the SE surface that was used in other calculations, the SA surface could be smoothly changed by varying the solvent probe radius used to define it. In contrast, varying the probe radius in the SE surface can lead to discontinuous changes in the surface definition, and as the probe radius increases, the SE surface approaches the convex hull of the molecule, which means that the ability to change the SE surface by varying the probe radius, is limited.
RESULTS
Bound ions
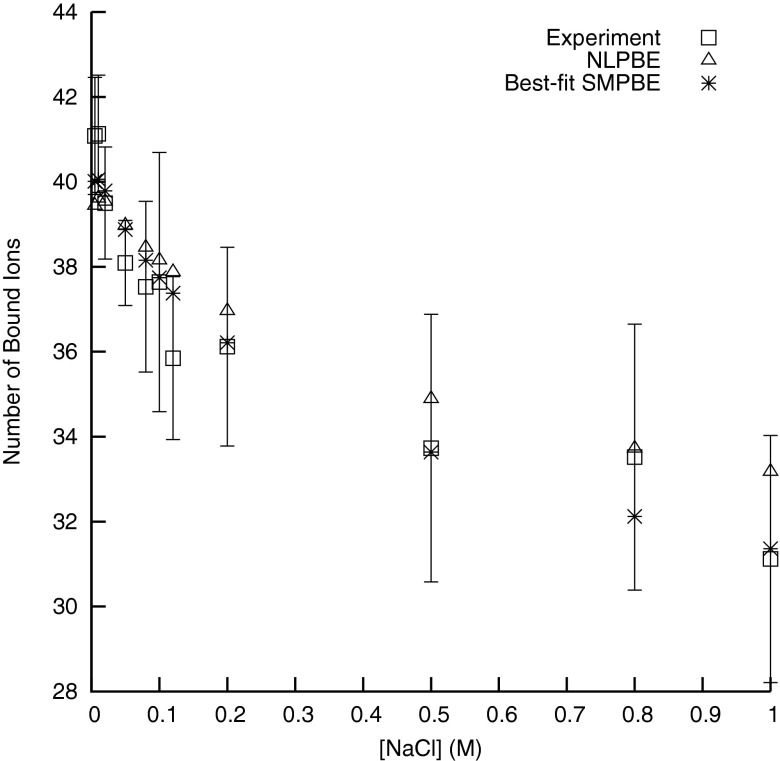
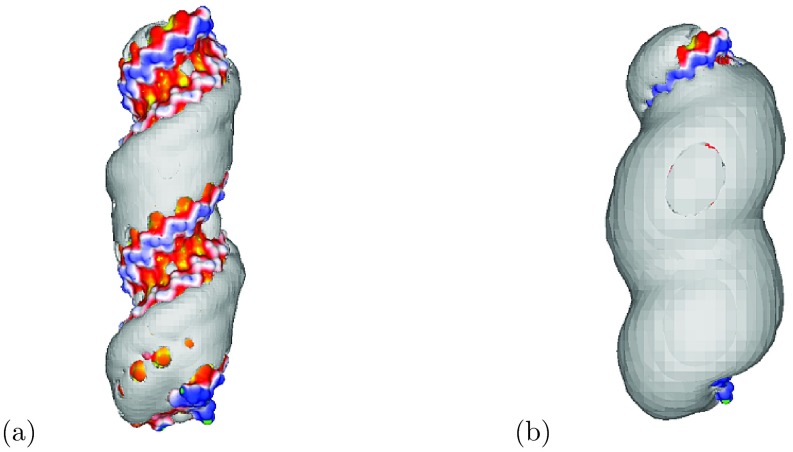
That the NLPBE overestimates the number of bound ions around the 24 base pair B-DNA considered here for concentrations above 0.01 M can be seen from Figure 1, where the predictions of the NLPBE and the experimental measurements taken from a data set considered in the literature8 are shown. As described above, using the SMPBE with a finite ion size offers one method of reducing this discrepancy between theoretical predictions and experimental measurements. Figure 2 shows isocontours where the predictions of the local ion concentrations given by the SMPBE with ion sizes of 2 and 4 Å were 0.1 M less than those given by the NLPBE. Because the SMPBE predicts that these regions have lower ion concentrations than those predicted by the NLPBE, its predictions of the numbers of bound counterions will be smaller than those of the NLPBE. These regions are localized near the molecular surface, where φ is large, and inside these isocontours the differences between the two theoretical predictions are larger.
Figure 1.
The numbers of bound ions around B-DNA from experiment, predicted by the NLPBE, and predicted by the SMPBE with a best-fit combination of a 4 Å Stern layer and a 2 Å ion size at the experimental salt concentrations, [NaCl].
Figure 2.
Isocountour plots of the differences between the ion distributions predicted by the NLPBE and those predicted by the uniform ion size SMPBE with ion radii of 2 (a) and 4 (b) Å. The isocontour corresponds to a concentration difference of 0.1 M. The surfaces of the molecules are colored by the electrostatic potentials computed with the NLPBE. Yellow regions correspond to −8 kT/e, red regions correspond to −5.75 kT/e, white corresponds to −3.5 kT/e, blue regions correspond to −1.25 kT/e, and green regions correspond to 1 kT/e.
As discussed above, other researchers have attempted to use the experimental data in Figure 1 to determine what ion size should be used for Na+ ions, but the resulting best-fit ion sizes were larger (between 7 and 8 Å) than expected. As described above, these findings could indicate that current PB models neglect important physics, but they could also simply indicate that other parameters that affect the number of bound ions will also need to be fit against experimental data. As can be seen in Figure 3, the number of bound ions can be changed not just by changing the size of the ion in the SMPBE but also by changing the thickness of the Stern layer or the solvent probe radius used to define the SAS. Unfortunately, the experimental data considered here do not strongly constrain these parameters. Because the number of bound ions changes slowly with respect to the parameters, small errors in the experimental measurements lead to large changes in the resulting best-fit estimates of the parameters. Additionally, the predictions of the NLPBE without a Stern layer are already so close to the experimental predictions (Figure 1) that one runs the risk of merely fitting experimental error.
Figure 3.
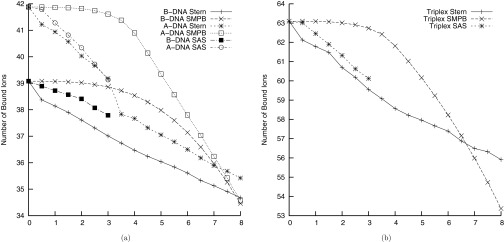
The number of bound counterions predicted by the nonlinear Poisson-Boltzmann equation as a function of the thickness of the Stern layer and the solvent probe radius of the solvent accessible surface (SAS), and the number of bound counterions predicted by the uniform ion size size-modified Poisson-Boltzmann equation, SMPBE, as a function of the ionic radius for (a) the B-DNA and A-DNA fiber models and (b) the triplex-DNA fiber model. The thickness of the Stern layer, the solvent probe radius in the SAS surface, and the ionic radius in the SMPBE were measured in angstroms. All curves except those showing the effect of changing the probe radius in the SAS used the solvent-excluded surface.
To illustrate that these experimental data do not strongly constrain the choice of parameters in the SMPBE, the numbers of bound ions predicted by the SMPBE at each experimental salt concentration were computed for all combinations of Stern layer thicknesses and ion sizes between 0 and 5 Å in increments of 0.5 Å. The weighted least squares residuals between the experimental data and the predictions of the SMPBE for each parameter set were then computed
| (7) |
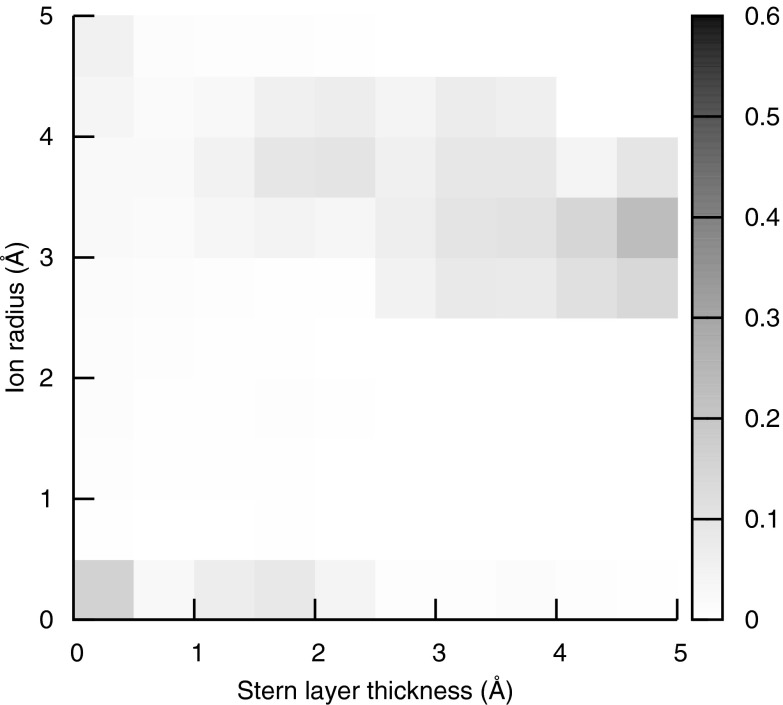
where the summation is carried out over the concentrations of NaCl used in the experimental data (0.005, 0.01, 0.02, 0.05, 0.08, 0.1, 0.12, 0.2, 0.5, 0.8, and 1.0 M), ηi(τ, rion) is the number of bound counterions at the ith salt concentration predicted with the uniform ion size SMPBE, a Stern layer thickness of τ and an ion radius of rion, is the experimental number of bound counterions at the ith salt concentration, and σi is the reported error in each experimental measurement. The parameter set corresponding to the smallest R2 was a Stern layer thickness of 4 Å and an ion size of 2 Å, and the resulting predictions of the numbers of bound ions can be seen in Figure 1. However, the error in the experimental data generates uncertainty in the best choice of these parameters, as can be seen from Figure 4. To create this figure 1 000 000 artificial data sets were generated, each of which were created by adding a random error to each number of bound ions in the experimental data set taken from a normal distribution with an average of zero and a standard deviation equal to the reported experimental error. The weighted least squares calculation described above was then repeated for each of these artificial data sets and the set of parameters that gave the smallest R2 was found. The probability density of the best-fit parameters for these artificial data sets is plotted in Figure 4. Clearly, the experimental error in the measurements of the numbers of bound ions leads to large uncertainties in the parameters, and more experimental data will be needed before truly optimal values of these parameters can be determined.
Figure 4.
The probability density of each combination of parameters created by generating 1 000 000 artificial data sets and choosing the parameter combination that generated the smallest R2 for each data set.
The electrostatic binding free energy and its salt dependence
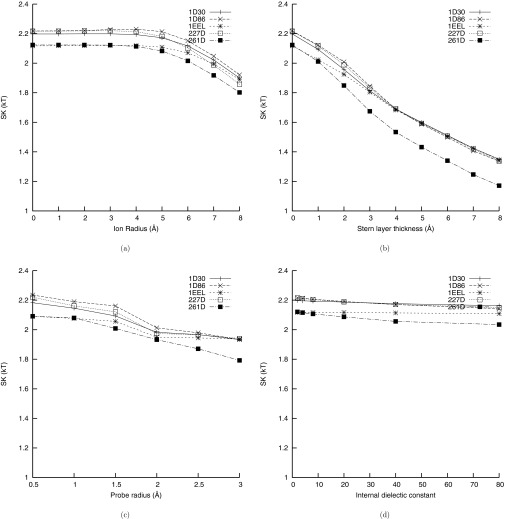
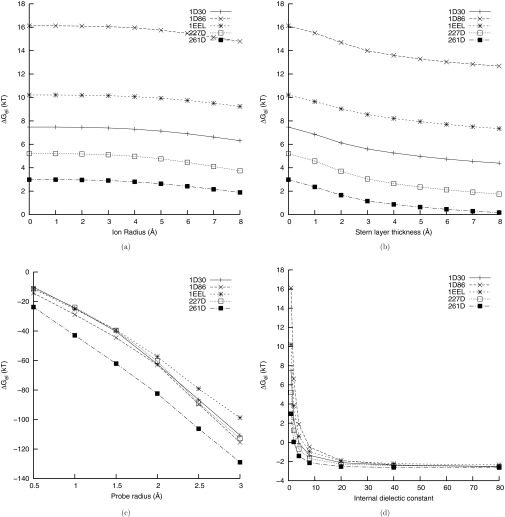
Several studies have demonstrated that while the NLPBE and allied methods describe the ion atmosphere sufficiently well to reproduce experimental measurements of SK,22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49 they have been less successful at predicting ΔGel itself. Perhaps part of the explanation for this discrepancy is the different sensitivities of these two quantities to the parameters in the calculations. In Figure 5, the SMPBE's predictions of SK as functions of the ion size, the thickness of the Stern layer, the radius of the probe used to define the SA surface, and the interior dielectric constants, are shown. In Figure 6 the same plots are shown for ΔGel. From these two figures, it is evident that ΔGel is extremely sensitive to the interior dielectric constant and the size of the solvent probe radius in the SA surface, while SK is much less sensitive to these quantities. This may help explain why the NLPBE and related methods have been more successful at predicting SK than at predicting ΔGel. At the same time, SK is sensitive to some parameters, such as the thickness of the Stern layer and the ion size, and could therefore be used to better determine these parameters, but it is much less sensitive to other parameters, such as the interior dielectric constant and surface definition, to which ΔGel is sensitive. This observation implies that using experimental measurements of SK to improve predictions of ΔGel is unlikely to work unless supplemented by more precisely determined internal dielectric constants and surface definitions.
Figure 5.
The sensitivity of the derivative, SK, of ΔGel with respect to the logarithm of the 1:1 salt concentration computed at 0.1 M salt concentration as a function of the different parameters of the model for the DNA-drug complexes in this study. (a) SK computed with the uniform ion size SMPBE as a function of the ion size. (b) SK computed with the NLPBE as a function of the thickness of the Stern layer. (c) SK computed with the NLPBE and the solvent accessible surface (SAS) as a function of the size of the probe used to define the SAS. (d) SK computed with the NLPBE as a function of the internal dielectric constant. All figures except (c) were created using the solvent-excluded surface rather than the SAS.
Figure 6.
The sensitivity of the electrostatic binding free energy, ΔGel to various parameters for the DNA-drug complexes in this study. (a) ΔGel computed with the uniform ion size SMPBE as a function of the ion size. (b) ΔGel computed with the NLPBE as a function of the thickness of the Stern layer. (c) ΔGel computed with the NLPBE and the solvent accessible surface (SAS) as a function of the size of the probe used to define the SAS. (d) ΔGel computed with the NLPBE as a function of the internal dielectric constant. All figures except (c) were created using the solvent-excluded surface rather than the SAS.
CONCLUSIONS
Recent research has shown that the NLPBE underestimates the numbers of monovalent cations attracted by nucleic acids, and because these discrepancies increase with the radius of the cations, it has been attributed to the assumption of pointlike ions in the NLPBE.2, 7, 8, 11, 12, 14 The SMPBE6, 8, 11, 16, 17, 18, 19, 20, 21 provides a straightforward theoretical modification to the NLPBE that may be able to capture these effects, but previous work that attempted to fit the ion size against experimental data found unexpectedly large radii.8 Potentially, these unusually large radii indicate the need to parameterize the SMPBE not only against the ion size in the theory but also against the other parameters, such as the Stern layer thickness and molecular surface definition. Indeed, previous studies that attempted to account for the effects of ion size by varying the thickness of the Stern layer found reasonable values.3, 4 Predicting these parameters with experimental measurements of the numbers of bound ions will require more accurate experimental measurements than the ones used here because changes in these parameters lead to small changes in the predicted numbers of ions and therefore small uncertainties in the numbers of bound ions can lead to large uncertainties in the underlying parameters. The values of SK predicted by the SMPBE are also sensitive to these parameters, so experimental measurements of SK could be used to help fix these parameters.48, 49
In the present study, a set of DNA-drug complexes was chosen that were closely spaced in ΔGel but also with significant SK, as the rank ordering of such complexes would be expected to be as sensitive as possible to changes in the ion atmosphere. However, although the predictions of SK did change with the ion size and the thickness of the Stern layer, and even the ordering of complexes by SK changed, the changes in the spacing in SK were small compared to the differences in ΔGel. These differences therefore changed little with either changes in the ion size or the thickness of the Stern layer, and the ranking of complexes by ΔGel did not change. Perhaps for other systems better values for the thickness of the Stern layer and the ion size would lead to improved estimates of ΔGel, but such systems would either have to be more closely-spaced in ΔGel or have SK that are more sensitive to changes in the ion size and Stern layer thickness. Given the sensitivity of ΔGel to the other parameters, better predictions of the thickness of the Stern layer and the ion size would not produce detectable improvements in the predictions of ΔGel by the NLPBE and related methods for the systems considered here.
ACKNOWLEDGMENTS
This publication was made possible by Grant No. 5R44GM073391-03 from the National Institute of General Medical Sciences of the National Institute of Health. Two of the authors (R.C.H. and M.O.F.) would like to thank Mr. Travis Mackoy for his help in preparing Figure 2.
References
- Lipfert J., Chu V. B., Bai Y., Herschlag D., and Doniach S., “Low-resolution models for nucleic acids from small-angle X-ray scattering with applications to electrostatic modeling,” J. Appl. Crystallogr. 40, s229–s234 (2007). 10.1107/S0021889807001707 [DOI] [Google Scholar]
- Andresen K., Qiu X., Pabit S. A., Lamb J. S., Park H. Y., Kwok L. W., and Pollack L., “Mono- and trivalent ions around DNA: A small-angle scattering study of competition and interactions,” Biophys. J. 95, 287–295 (2008). 10.1529/biophysj.107.123174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabit S. A., Qiu X., Lamb J. S., Li L., Meisburger S. P., and Pollack L., “Both helix topology and counterion distribution contribute to the more effective charge screening in dsRNA compared with dsDNA,” Nucl. Acids Res. 37, 3887–3896 (2009). 10.1093/nar/gkp257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabit S. A., Meisburger S. P., Lamb J. S., Li L., Blose J. M., Jones C. D., and Pollack L., “Counting ions around DNA with anomalous small-angle X-ray scattering,” J. Am. Chem. Soc. 132, 16334–16336 (2010). 10.1021/ja107259y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack L., “SAXS studies of ion-nucleic acid interactions,” Ann. Rev. Biophys. 40, 225–242 (2011). 10.1146/annurev-biophys-042910-155349 [DOI] [PubMed] [Google Scholar]
- Kirmizialtin S., Pabit S. A., Meisburger S. P., Pollack L., and Elber R., “RNA and its ionic cloud: Solution scattering experiments and atomically detailed simulations,” Biophys. J. 102, 819–828 (2012). 10.1016/j.bpj.2012.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Greenfeld M., Travers K. J., Chu V. B., Lipfert J., Doniach S., and Herschlag D., “Quantitative and comprehensive decomposition of the ion atmosphere around nucleic acids,” J. Am. Chem. Soc. 129, 14981–14988 (2007). 10.1021/ja075020g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V. B., Bai Y., Lipfert J., Herschlag D., and Doniach S., “Evaluation of ion binding to DNA duplexes using a size-modified Poisson-Boltzmann theory,” Biophys. J. 93, 3202–3209 (2007). 10.1529/biophysj.106.099168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V. B., Bai Y., Lipfert J., Herschlag D., and Doniach S., “A repulsive field: Advances in the electrostatics of the ion atmosphere,” Curr. Opin. Chem. Biol. 12, 619–625 (2008). 10.1016/j.cbpa.2008.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A. A., Draper D. E., and Pappu R. V., “Molecular simulation studies of monovalent counterion-mediated interactions in a model RNA kissing loop,” J. Mol. Biol. 390, 805–819 (2009). 10.1016/j.jmb.2009.05.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizialtin S., Silalahi A. R. J., Elber R., and Fenley M. O., “The ionic atmosphere around A-RNA: Poisson-Boltzmann and molecular dynamics simulations,” Biophys. J. 102, 829–838 (2012). 10.1016/j.bpj.2011.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert D., Leipply D., Shiman R., and Draper D. E., “The influence of monovalent cation size on the stability of RNA tertiary structures,” J. Mol. Biol. 390, 791–804 (2009). 10.1016/j.jmb.2009.04.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen E., Muse J. M., and Stellwagen N. C., “Monovalent cation size and DNA conformational stability,” Biochemistry 50, 3084–3094 (2011). 10.1021/bi1015524 [DOI] [PubMed] [Google Scholar]
- Anthony P. C., Sim A. Y. L., Chu V. B., Doniach S., Block S. M., and Herschlag D., “Electrostatics of nucleic acid folding under conformational constraint,” J. Am. Chem. Soc. 134, 4607–4614 (2012). 10.1021/ja208466h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj-Iglic V. and Iglic A., “A simple statistical mechanical approach to the free energy of the electric double layer including the excluded volume effect,” J. Phys. II 6, 477–491 (1996). 10.1051/jp2:1996193 [DOI] [Google Scholar]
- Borukhov I., Andelman D., and Orland H., “Steric effects in electrolytes: A modified Poisson-Boltzmann equation,” Phys. Rev. Lett. 79, 435–438 (1997). 10.1103/PhysRevLett.79.435 [DOI] [Google Scholar]
- Manciu M. and Ruckenstein E., “Lattice site exclusion effect on the double layer interaction,” Langmuir 18, 5178–5185 (2002). 10.1021/la020194r [DOI] [Google Scholar]
- Netz R. R. and Orland H., “Beyond Poisson-Boltzmann: Fluctuation effects and correlation functions,” Eur. Phys. J. E 1, 203–214 (2000). 10.1007/s101890050023 [DOI] [Google Scholar]
- Silalahi A. R. J., Boschitsch A. H., Harris R. C., and Fenley M. O., “Comparing the predictions of the nonlinear Poisson-Boltzmann equation and the ion size-modified Poisson-Boltzmann equation for a low-dielectric charged spherical cavity in an aqueous salt solution,” J. Chem. Theory Comput. 6, 3631–3639 (2010). 10.1021/ct1002785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Wong Z., and Li B., “Mean-field description of ionic size effects with nonuniform ionic sizes: A numerical approach,” Phys. Rev. E 84, 021901 (2011). 10.1103/PhysRevE.84.021901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boschitsch A. H. and Danilov P. V., “Formulation of a new and simple nonuniform size-modified Poisson-Boltzmann description,” J. Comput. Chem. 33, 1152–1164 (2012). 10.1002/jcc.22946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenley M. O., Harris R. C., Jayaram B., and Boschitsch A. H., “Revisiting the association of cationic groove-binding drugs to DNA using a Poisson-Boltzmann approach,” Biophys. J. 99, 879–886 (2010). 10.1016/j.bpj.2010.04.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenley M. O., Russo C., and Manning G. S., “Theoretical assessment of the oligolysine model for ionic interactions in protein DNA complexes,” J. Phys. Chem. B 115, 9864–9872 (2011). 10.1021/jp204915y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. C., Bredenberg J. H., Silalahi A. R. J., Boschitsch A. H., and Fenley M. O., “Understanding the physical basis of the salt dependence of the electrostatic binding free energy of mutated charged ligand–nucleic acid complexes,” Biophys. Chem. 156, 79–87 (2011). 10.1016/j.bpc.2011.02.010 [DOI] [PubMed] [Google Scholar]
- Mascotti D. P. and Lohmann T. M., “Thermodynamics of single-stranded RNA binding to oligolysines containing tryptophan,” Biochemistry 31, 8932–8946 (1992). 10.1021/bi00152a033 [DOI] [PubMed] [Google Scholar]
- Zacharias M., Luty B. A., Davis M. E., and McCammon J. A., “Poisson Boltzmann analysis of the λ repressor operator interaction,” Biophys. J. 63, 1280–1285 (1992). 10.1016/S0006-3495(92)81723-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascotti D. P. and Lohmann T. M., “Thermodynamics of single-stranded RNA and DNA interactions with oligolysines containing tryptophan,” Biochemistry 32, 10568–10579 (1993). 10.1021/bi00091a006 [DOI] [PubMed] [Google Scholar]
- Misra V. K., Sharp K. A., Friedman R. A., and Honig B., “Salt effects on ligand-DNA binding: Minor groove binding antibiotics,” J. Mol. Biol. 238, 245–263 (1994). 10.1006/jmbi.1994.1285 [DOI] [PubMed] [Google Scholar]
- Misra V. K., Hecht J. L., Sharp K. A., Friedman R. A., and Honig B., “Salt effects on protein DNA interactions: The λ cI repressor and EcoRI endonuclease,” J. Mol. Biol. 238, 264–280 (1994). 10.1006/jmbi.1994.1286 [DOI] [PubMed] [Google Scholar]
- Misra V. K. and Honig B., “On the magnitude of the electrostatic contribution to ligand DNA interactions,” Proc. Natl. Acad. Sci. U.S.A. 92, 4691–4695 (1995). 10.1073/pnas.92.10.4691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua P. C., and Cech T. R., “Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR,” Biochemistry 35, 9983–9994 (1996). 10.1021/bi9607259 [DOI] [PubMed] [Google Scholar]
- Mascotti D. P. and Lohmann T. M., “Thermodynamics of oligoarginines binding to RNA and DNA,” Biochemistry 36, 7272–7279 (1997). 10.1021/bi970272n [DOI] [PubMed] [Google Scholar]
- GuhaThakurta D. and Draper D. E., “Contributions of basic residues to ribosomal protein L11 recognition of RNA,” J. Mol. Biol. 295, 569–580 (2000). 10.1006/jmbi.1999.3372 [DOI] [PubMed] [Google Scholar]
- Kaul M. and Pilch D. S., “Thermodynamics of aminoglycoside-rRNA recognition: The binding of neomycin-class aminoglycosides to the A site of 16S rRNA,” Biochemistry 41, 7695–7706 (2002). 10.1021/bi020130f [DOI] [PubMed] [Google Scholar]
- García-García C. and Draper D. E., “Electrostatic interactions in a peptide-RNA complex,” J. Mol. Biol. 331, 75–88 (2003). 10.1016/S0022-2836(03)00615-6 [DOI] [PubMed] [Google Scholar]
- Todd B. A. and Rau D. C., “Interplay of ion binding and attraction in DNA condensed by multivalent cations,” Nucl. Acids Res. 36, 501–510 (2008). 10.1093/nar/gkm1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blose J. M., Proctor D. J., Veeraraghavan N., Misra V. K., and Bevilacqua P. C., “Contribution of the closing base pair to exceptional stability in RNA tetraloops: Roles for molecular mimicry and electrostatic factors,” J. Am. Chem. Soc. 131, 8474–8484 (2009). 10.1021/ja900065e [DOI] [PubMed] [Google Scholar]
- Islam M. M., Pandya P., Kumar S., and Kumar G. S., “RNA targeting through binding of small molecules: Studies on t-RNA binding by the cytotoxic protoberberine alkaloid coralyne,” Mol. Biosyst. 5, 244–254 (2009). 10.1039/b816480k [DOI] [PubMed] [Google Scholar]
- Islam M. M., Chowdhury S. R., and Kumar G. S., “Spectroscopic and calorimetric studies on the binding of alkaloids berberine, palmatine and coralyne to double stranded RNA polynucleotides,” J. Phys. Chem. B 113, 1210–1224 (2009). 10.1021/jp806597w [DOI] [PubMed] [Google Scholar]
- Korolev N., Berezhnoy N. V., Eom K. D., Tam J. P., and Nordenskiold L., “A universal description for the experimental behavior of salt-(in)dependent oligocation-induced DNA condensation,” Nucl. Acids Res. 37, 7137–7150 (2009). 10.1093/nar/gkp683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S. H., Spring A. M., and Germann M. W., “Thermodynamic profiling of HIV RREIIB RNA-zinc finger interactions,” J. Mol. Biol. 393, 369–382 (2009). 10.1016/j.jmb.2009.07.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athavale S. S., Ouyang W., McPike M. P., Hudson B. S., and Borer P. N., “Effects of the nature and concentration of salt on the interaction of the HIV-1 nucleocapsid protein with SL3 RNA,” Biochemistry 49, 3525–3533 (2010). 10.1021/bi901279e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paleskava A., Konevega A. L., and Rodnina M. V., “Thermodynamic and kinetic framework of selenocysteyl-tRNASec recognition by elongation factor SelB,” J. Biol. Chem. 285, 3014–3020 (2010). 10.1074/jbc.M109.081380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryawanshi H., Sabharwal H., and Maiti S., “Thermodynamics of peptide-RNA recognition: The binding of a Tat peptide to TAR RNA,” J. Phys. Chem. B 114, 11155–11163 (2010). 10.1021/jp1000545 [DOI] [PubMed] [Google Scholar]
- Weiss T. C., Zhai G. G., Bhatia S. S., and Romaniuk P. J., “An RNA aptamer with high affinity and broad specificity for zinc finger proteins,” Biochemistry 49, 2732–2740 (2010). 10.1021/bi9016654 [DOI] [PubMed] [Google Scholar]
- Williams S. G. and Hall K. B., “Coevolution of Drosophila snf protein and its snRNA targets,” Biochemistry 49, 4571–4582 (2010). 10.1021/bi100374d [DOI] [PubMed] [Google Scholar]
- Xi H., Kumar S., Dosen-Micovic L., and Arya D. P., “Calorimetric and spectroscopic studies of aminoglycoside binding to AT-rich DNA triple helices,” Biochimie 92, 514–529 (2010). 10.1016/j.biochi.2010.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- M. T.RecordJr., Lohman T. M., and de Haseth P., “Ion effects on ligand-nucleic acid interactions,” J. Mol. Biol. 107, 145–158 (1976). 10.1016/S0022-2836(76)80023-X [DOI] [PubMed] [Google Scholar]
- M. T.RecordJr., Zhang W., and Anderson C. F., “Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: A practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts,” Adv. Protein Chem. 51, 281–353 (1998). 10.1016/S0065-3233(08)60655-5 [DOI] [PubMed] [Google Scholar]
- Xiang-Jun L. and Olson W. K., “3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures,” Nucl. Acids Res. 31, 5108–5121 (2003). 10.1093/nar/gkg680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang-Jun L. and Olson W. K., “3DNA: A versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures,” Nat. Protoc. 3, 1213–1227 (2008). 10.1038/nprot.2008.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell W. D., Cieplak P., Bayly C. I., Gould I. R., Merz K. M., Ferguson D. M., Spellmeyer D. C., Fox T., Caldwell J. W., and Kollman P. A., “A second generation force field for the simulation of proteins, nucleic acids, and organic molecules,” J. Am. Chem. Soc. 117, 5179–5197 (1995). 10.1021/ja00124a002 [DOI] [Google Scholar]
- Dolinsky T. J., Nielsen J. E., McCammon J. A., and Baker N. A., “PDB2PQR: An automated pipeline for the setup of Poisson Boltzmann electrostatics calculations,” Nucl. Acids Res. 32, W665–W667 (2004). 10.1093/nar/gkh381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., and Baker N. A., “PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations,” Nucl. Acids Res. 35, W522–W525 (2007). 10.1093/nar/gkm276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Bhat T. N., Bourne P. E., Feng Z., Gilliland G., Weissig H., and Westbrook J., “The protein data bank and the challenge of structural genomics,” Nat. Struct. Mol. Biol. 7, 957–959 (2000). 10.1038/80734 [DOI] [PubMed] [Google Scholar]
- Larsen T. A., Goodsell D. S., Cascio D., Grzeskowiak K., and Dickerson R. E., “The structure of DAPI bound to DNA,” J. Biomol. Struct. Dyn. 7, 477–491 (1989). 10.1080/07391102.1989.10508505 [DOI] [PubMed] [Google Scholar]
- Sriram M., van der Marel G. A., Roelen H. L., van Boom J. H., and Wang A. H., “Structural consequences of a carcinogenic alkylation lesion on DNA: Effect of O6-ethylguanine on the molecular structure of the d(CGC[e6G]AATTCGCG)-netropsin complex,” Biochemistry 31, 11823–11834 (1992). 10.1021/bi00162a022 [DOI] [PubMed] [Google Scholar]
- Mazur S., Tanious F. A., Ding D., Kumar A., Boykin D. W., Simpson I. J., Neidle S., and Wilson W. D., “A thermodynamic and structural analysis of DNA minor-groove complex formation,” J. Mol. Biol. 300, 321–337 (2000). 10.1006/jmbi.2000.3869 [DOI] [PubMed] [Google Scholar]
- Laughton C. A., Tanious F., Nunn C. M., Boykin D. W., Wilson W. D., and Neidle S., “A crystallographic and spectroscopic study of the complex between d(CGCGAATTCGCG)2 and 2,5-bis(4-guanylphenyl)furan, an analogue of berenil. Structural origins of enhanced DNA-binding affinity,” Biochemistry 35, 5655–5661 (1996). 10.1021/bi952162r [DOI] [PubMed] [Google Scholar]
- Nunn C. M., Garman E., and Neidle S., “Crystal structure of the DNA decamer d(CGCAATTGCG) complexed with the minor groove binding drug netropsin,” Biochemistry 36, 4792–4799 (1997). 10.1021/bi9628228 [DOI] [PubMed] [Google Scholar]
- Boschitsch A. H. and Fenley M. O., “A fast and robust Poisson-Boltzmann solver based on adaptive Cartesian grids,” J. Chem. Theory Comput. 7, 1524–1540 (2011). 10.1021/ct1006983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly M. L., “Solvent-accessible surfaces of proteins and nucleic acids,” Science 221, 709–713 (1983). 10.1126/science.6879170 [DOI] [PubMed] [Google Scholar]
- Alexov E., “Role of the protein side-chain fluctuations on the strength of pair-wise electrostatic interactions: Comparing experimental with computed pKas,” Proteins: Struct., Funct., Bioinf. 50, 94–103 (2003). 10.1002/prot.10265 [DOI] [PubMed] [Google Scholar]
- Dong F., Vijayakumar M., and Zhou H. X., “Comparison of calculation and experiment implicates significant electrostatic contributions to the binding stability of barnase and barstar,” Biophys. J. 85, 49–60 (2003). 10.1016/S0006-3495(03)74453-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F. and Zhou H. X., “Electrostatic contribution to the binding stability of protein-protein complexes,” Proteins: Struct., Funct., Bioinf. 65, 87–102 (2006). 10.1002/prot.21070 [DOI] [PubMed] [Google Scholar]
- Alsallaq R. and Zhou H. X., “Electrostatic rate enhancement and transient complex of protein protein association,” Proteins: Struct., Funct., Bioinf. 71, 320–335 (2008). 10.1002/prot.21679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzubiella J., Swanson J. M. J., and McCammon J. A., “Coupling nonpolar and polar solvation free energies in implicit solvent models,” J. Chem. Phys. 124, 084905 (2006). 10.1063/1.2171192 [DOI] [PubMed] [Google Scholar]
- Cheng L. T., Dzubiella J., McCammon J. A., and Li B., “Application of the level-set method to the implicit solvation of nonpolar molecules,” J. Chem. Phys. 127, 084503 (2007). 10.1063/1.2757169 [DOI] [PubMed] [Google Scholar]
- Cheng L. T., Wang Z., Setny P., Dzubiella J., Li B., and McCammon J. A., “Interfaces and hydrophobic interactions in receptor-ligand systems: A level-set variational implicit solvent approach,” J. Chem. Phys. 131, 144102 (2009). 10.1063/1.3242274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Baker N. A., and Wei G. W., “Differential geometry based solvation model I: Eulerian formulation,” J. Comput. Phys. 229, 8231–8258 (2010). 10.1016/j.jcp.2010.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Baker N. A., and Wei G. W., “Differential geometry based solvation model II: Lagrangian formulation,” J. Math. Biol. 63, 1139–1200 (2011). 10.1007/s00285-011-0402-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichs M., Zhou R., Edinger S. R., and Friesner R. A., “Poisson-Boltzmann analytical gradients for molecular modeling calculations,” J. Phys. Chem. B 103, 3057–3061 (1999). 10.1021/jp982513m [DOI] [Google Scholar]
- Grant J. A., Pickup B. T., and Nicholls A., “A smooth permittivity function for Poisson Boltzmann solvation methods,” J. Comput. Chem. 22, 608–640 (2001). 10.1002/jcc.1032 [DOI] [Google Scholar]
- Lee B. and Richards F. M., “The interpretation of protein structures: Estimation of static accessibility,” J. Mol. Biol. 55, 379–400 (1971). 10.1016/0022-2836(71)90324-X [DOI] [PubMed] [Google Scholar]