Abstract
Hadrontherapy is a form of external radiation therapy, which uses beams of charged particles such as carbon ions. Compared to conventional radiotherapy with photons, the main advantage of carbon ion therapy is the precise dose localization along with an increased biological effectiveness. The first results obtained from prostate cancer patients treated with carbon ion therapy showed good local tumor control and survival rates. In view of this advanced treatment modality we investigated the effects of irradiation with different beam qualities on gene expression changes in the PC3 prostate adenocarcinoma cell line. For this purpose, PC3 cells were irradiated with various doses (0.0, 0.5 and 2.0 Gy) of carbon ions (LET=33.7 keV/μm) at the beam of the Grand Accélérateur National d’Ions Lourds (Caen, France). Comparative experiments with X-rays were performed at the Belgian Nuclear Research Centre. Genome-wide gene expression was analyzed using microarrays. Our results show a downregulation in many genes involved in cell cycle and cell organization processes after 2.0 Gy irradiation. This effect was more pronounced after carbon ion irradiation compared with X-rays. Furthermore, we found a significant downregulation of many genes related to cell motility. Several of these changes were confirmed using qPCR. In addition, recurrence-free survival analysis of prostate cancer patients based on one of these motility genes (FN1) revealed that patients with low expression levels had a prolonged recurrence-free survival time, indicating that this gene may be a potential prognostic biomarker for prostate cancer. Understanding how different radiation qualities affect the cellular behavior of prostate cancer cells is important to improve the clinical outcome of cancer radiation therapy.
Keywords: microarray, carbon ion therapy, motility genes, biomarkers, PC3 prostate adenocarcinoma
Introduction
Recent advances in radiotherapy, such as hadrontherapy, have been added as a radiation treatment choice for specific types of cancer. The inverted depth-dose profile and the sharp dose fall-off after the Bragg peak offered by charged particle beams allow for a more precise localization of the radiation dosage to the tumor (1). Due to this greater precision the surrounding healthy tissue receives a much lower dose compared to conventional radiotherapy with photons. The use of carbon ion beams offers, besides this ballistic advantage, also a biological advantage. High-linear energy transfer (LET) carbon ion radiation has been shown to have a higher relative biological effectiveness (RBE) compared to conventional low-LET photon therapy (2), and is therefore more effective in inducing DNA damage, cell cycle arrest and cell death in tumor cells (3–5). This accounts for the highly lethal effects of carbon ions, even on radioresistant (with respect to X-rays) tumors. Currently, carbon ion radiotherapy has been approved for treatment of specific types of cancer, including prostate cancer (6–8).
Prostate cancer is the second most frequently diagnosed cancer and the sixth leading cause of cancer mortality in males worldwide (9). Most prostate cancer-related deaths are due to metastasis (9–11). So far, first results obtained from prostate cancer patients treated with carbon ion therapy, showed good local control and high local and biochemical relapse-free rates (8,12–15). However, an increased survival rate may be accompanied by potential long-term biological consequences after carbon ion radiotherapy, including metastasis (16,17).
Metastases occur when cancerous cells acquire properties which allow them to detach from the original cancer site and adhere to a target organ to form a new tumor (10). Changes in gene expression can dysregulate cell signaling pathways, thereby leading to changes in functional cell behavior which can ultimately result in cancer metastasis (18,19). Acquiring these properties is a multi-step process starting with tumor growth, followed by detachment of cancer cells, migration and invasion in the surrounding tissue, circulation into blood vessels and implantation to a distant organ (20).
Therapeutic intervention may augment the metastatic potential of cancer cells, and many authors have suggested that a sub-lethal dose of photon irradiation promotes cancer cell metastasis by increasing their migration and invasion potential (21–27). In particular, Zhou et al (22) irradiated cancer cell lines from different organ sites and showed that γ-irradiation increased the capacity for migration and invasion, a finding that was also seen in glioblastoma cells (21). Interestingly, previous in vitro studies which compared the effects of particle and photon beams indicated that particle beams can decrease the migration potential of cancer cells whereas in most cases X-irradiated samples showed only a slight decrease or even an increase in their migration potential (28–32). Ogata et al (30) irradiated human fibrosarcoma cells with X-rays, protons or carbon ion beams and observed a dose-dependent decrease in cell migration and invasion caused by particle irradiation, whereas low doses of X-rays facilitated the process. Goetze et al (28) irradiated glioblastoma cells and colorectal carcinoma cells with carbon ions or X-rays and found that carbon ion irradiation suppressed the migration potential in both cell lines, while X-rays suppressed the migration potential only in the colon carcinoma cells, indicating a cell type-specific effect.
The fate of a cancer cell after radiotherapeutic intervention is believed to be controlled by a network of signaling pathways that lead to different modes of cell death or survival (33). Several studies have compared changes in gene expression of cancer cells induced by particle and photon beams (29,34–37). Particle beams were found to induce more changes in the number of genes that were differently expressed, as well as the magnitude of (dose-dependent) gene expression changes. Pathways in which these genes were involved were mostly related to cell cycle regulation, invasion and angiogenesis which may be associated with an enhanced aggressive phenotype of the cancer cells. To our knowledge, the effect of carbon ion beam radiation on gene expression of prostate cancer cells in vitro has not been verified so far.
The main aim of this study was to investigate the impact of carbon and X-irradiation on gene expression levels of the prostate adenocarcinoma cell line PC3 using whole-genome microarrays. This highly invasive cell line exhibits strong metastatic activity (38) and is widely used as an in vitro model to investigate the biological and cellular responses of human prostate cancer cells. Our results demonstrate that carbon ion irradiation induced stronger effects at the level of gene expression compared to similar doses of X-rays. Specifically after carbon irradiation, a more pronounced, dose-dependent down-regulation of genes involved in cell migration and motility was observed.
Materials and methods
Cell culture
Human PC3 prostate adenocarcinoma cells were obtained from the American Type Culture Collection (ATCC, Molsheim, France). They were cultured in F-12K medium (ATCC) supplemented with 10% fetal bovine serum (FBS) (Gibco, Life Technologies, Ghent, Belgium). Cell cultures were maintained in a humidified incubator (37°C; 5% CO2). For all irradiation experiments the same passage number of cells was used. For all conditions, we used four separate replicates.
X-irradiation
Cells were plated at a density of 3.5×105 cells in 12.5 cm2-tissue culture flasks (Falcon; VWR, Leuven, Belgium). After seeding, medium was replaced and cells were irradiated in a horizontal position with different doses of X-rays (0.0, 0.5 and 2.0 Gy) (Pantak HF420 RX machine; 250 kV, 15 mA, 1.2 mm Al equivalent and 1 mm Cu-filtered X-rays) and a calculated dose rate of 0.25 Gy/min. After irradiation, cells were further incubated for 8 h.
Carbon ion irradiation
Cells were plated in 175 cm2-tissue culture flasks (Falcon; VWR). Cells were transported by car in a transportable incubator at 37°C to the Grand Accélérateur National d’Ions Lourds (GANIL) (Caen, France). During cell transportation, culture flasks were completely filled with medium. After arrival, medium was replaced and cells were placed overnight in a humidified incubator. The following day 3.5×105 cells were plated in 12.5 cm2-tissue culture flasks (Falcon; VWR). After seeding, the flasks were completely filled with medium to allow irradiation in a vertical position. Sub-confluent cells were irradiated with a 13C beam with an initial energy of 75 MeV/u and a flux of 6.24×105 cm−2sec−1 at the D1 beam line at GANIL facility. The dosimetry was performed by physicists of CIMAP group at GANIL. It is based on the monitoring of the total ion current using the X-ray emission by a metallic thin foil inserted in the beam path. For fluxes lower than 106 cm−2 sec−1, a calibration factor between these secondary photons and the particle flux is obtained by counting the ion tracks measured on CR39 track detectors. A second step for checking the linearity is performed by a conventional ionization chamber. Taking into account the different layers before the beam arrives at the sample, the resulting LET was 33.7±1.6 keV/μm, calculated with the SRIM version 2011 code (39). To irradiate the samples at 0.0, 0.5 and 2.0 Gy absorbed doses, the requested fluences were 0, 9.3×106 cm−2 and 3.7×107 cm−2 by using the following equation:
The spot size of the beam was around 4×4 mm2. Immediately after irradiation, medium in the flasks was removed (except for 2 ml) in which the cells were further incubated for 8 h. Control samples were treated under similar conditions, including transportation and positioning identical to, and simultaneous with, that of treated samples.
RNA extraction
Medium was removed from the flasks 8 h after irradiation, cells were rinsed with phosphate-buffered saline (Gibco) and finally collected in 350 μl RLT buffer (Qiagen, Venlo, The Netherlands) with β-mercapto-ethanol (Sigma-Aldrich, Bornem, Belgium). Total RNA was isolated according to the manufacturer’s instructions using AllPrep DNA/RNA/protein mini kit (Qiagen). The quantity of RNA was measured with the NanoDrop Spectrophotometer (NanoDrop Products, Wilmington, DE, USA) and the quality was assessed with an Agilent Bioanalyzer (Agilent Technologies, Diegem, Belgium). RNA was stored at −80°C until further processing.
Microarrays and data analysis
Microarrays were prepared as reported previously by El-Saghire et al (40). After labeling, samples were hybridized onto Human Gene 1.0 ST Array Chips (Affymetrix, High Wycombe, UK). Raw data (.cel-files) were then imported into Partek software (Partek Genomic Suite v5.) (Partek Inc., St. Louis, MO, USA). Quality control was performed according to Partek software instructions. Based on the PCA of all 24 samples three samples were indicated as outliers and removed from the analysis. To test for differential expression two-way ANOVA analysis (with experimental conditions and dose as factors) was performed. P-values were adjusted for multiple corrections using false discovery rate (FDR) as described by the Benjamini and Hochberg (41) procedure. Genes were considered as being differentially expressed (DEX) when the fold-change (FC) was 2≤FC≤−2 and FDR ≤0.05. DEX genes were examined for functional enrichment using the ToppFun tool (http://toppgene.cchmc.org/) and applying an FDR corrected P-value of 0.05 for statistical significance. Gene Ontology lists of Molecular Function and Biological Processes were examined with the visualization tool AmiGO (v1.8) available on the Gene Ontology website. Further examination of the data set was performed with the online tool CateGOrizer in order to get a more precise view of the most important affected processes (42). Finally, DEX genes were filtered based on the cell motility gene set found on the Gene Ontology website (Table I).
Table I.
Cell motility gene set.
| Name gene set | Website | No. of genes in set | No. of genes found in data set | No. of significantly regulated genes (after 2.0 Gy C-ion) |
|---|---|---|---|---|
| GO0048870 - Cell motility | Gene Ontology | 715 | 746 | 50 |
Functional enrichment analysis
In order to determine to which extent specific gene sets were influenced by changes found in genes with Partek software, a gene set enrichment analysis (GSEA) was performed. GSEA allows calculating statistically significant enrichment of a set of DEX genes towards specific pathways or biological processes. Gene ontology databases for molecular functions, biological processes and cellular components were analyzed separately. Analysis was performed using GSEA software (v2.0.10) (43,44). Number of permutations was set at 1,000; gene sets were set as permutation type and minimum gene set size was 10. Otherwise, default settings were maintained. Gene sets were considered to be enriched with a FDR P-value ≤0.05. Enrichment maps, which indicate relationships between the enriched sets, were visualized for gene sets coding for cellular components using the Enrichment Map plug-in for the Cytoscape network visualization software (45).
cDNA synthesis
cDNA was synthesized with a GoScript™ Reverse Transcription System (Promega, Leiden, The Netherlands) on a GeneAmp PCR System 2700 (Applied Biosystems, Foster City, CA, USA). We used 0.4 μg RNA in 20 μl reactions as per the manufacturer’s instructions. cDNA samples were stored at −20°C until further reverse transcriptase PCR analysis.
RT-qPCR
Primers for target gene expression analysis (Table II) were purchased as pre-made assays (TaqMan Gene Expression Assay) (Applied Biosystems). Assays were performed according to the manufacturer’s instructions. Briefly, 2 μl cDNA was added to 1 μl TaqMan Gene expression Primer, 10 μl TaqMan® Fast Advanced Master Mix and 7 μl RNase free water. Assays were run on a 7500 Fast Real-Time PCR system (Applied Biosystems). First, efficiency of the primers was tested, using a five-step dilution series of an independent control sample (Table II). Expression ratios (R) were calculated using the method as described by Pfaffl (46). Finally, data were normalized by a log2 transformation and data are presented as average log2(R) ± SD.
Table II.
The Applied Biosystems assays.
| Gene symbol | Gene name | Assay ID | Ref seq | Exon boundary | Measured efficiency |
|---|---|---|---|---|---|
| FN1 | Fibronectin 1 | Hs01549967_m1 | NM_002026.2 | 3–4 | 1.98 |
| MYH9 | Myosin; heavy chain 9; non-muscle | Hs01066369_m1 | NM_002473.4 | 23–24 | 1.96 |
| NEXN | Nexilin | Hs00332124_m1 | NM_144573.3 | 10–11 | 1.91 |
| CCDC88A | Coiled-coil domain containing 88A | Hs01559766_m1 | NM_001135597.1 | 18–19 | 1.98 |
| ROCK1 | Rho-associated; coiled-coil containing protein kinase 1 | Hs01127714_mH | NM_005406.2 | 4–5 | 1.99 |
| MYH10 | Myosin; heavy chain 10; non-muscle | Hs00992050_m1 | NM_005964.1 | 21–22 | 2.01 |
| B2M | β-2-microglobulin | Hs00984230_m1 | NM_004048.2 | 3–4 | 2.05 |
Statistical analysis of RT-qPCR
Statistics were performed with GraphPad Prism 5.00. Statistical significance of differences between log2(R) of control and each experimental condition was determined using one-tailed Mann-Whitney tests. P-values ≤0.05 were considered statistically significant.
Kaplan-Meier analysis of public patient data
In order to assess the clinical relevance of our findings we cross-referenced our results with published patient data. We worked with independent, publicly accessible microarray data of prostate cancer patients (47,48) which could be imported into Partek. Our genes were cross-referenced in the patient data. Data from the study of Taylor et al (48) were imported in Partek and transformed with a two-base logarithm. Columns with ‘gene status’ were inserted, status was appointed as ‘high’ (expression data more than mean + SD); ‘intermediate’ (expression data between the mean + SD and mean − SD); and ‘low’ (expression data less than the mean − SD). Data from the study of Gulzar et al (47) were imported to Partek. Normal samples and duplicates were removed from the set. Properties, such as status and time to recurrence, were added to the data. Then, the original data were untransformed (anti-log2). Columns were filtered based on a list of our genes of interest. Columns of ‘gene status’ were inserted, status being appointed as ‘high’ (expression data >1.3 vs. reference RNA); ‘intermediate’ (expression data ≤1.3 and ≥0.77); and ‘low’ (expression data <0.77 vs. reference RNA). When status was appointed data were transferred to GraphPad Prism 5.00 for Kaplan-Meier analysis based on patient separation in high, intermediate or low gene status. Differences in recurrence-free survival were considered significant at log-rank P≤0.05.
Ethics statement
All patient data were obtained from Taylor et al (48) and Gulzar et al (47) after which they were made publicly available. Therefore, their use was not classified as human subjects research and no Institutional Review Board approval was needed.
Results
Gene expression profiles of PC3 cells were analyzed 8 h after exposure to different doses of carbon ion (LET=33.7 keV/μm) and X-irradiation. Microarray data analysis followed by GO analysis allowed the identification of genes, as well as pathways, which were enriched among statistically significant genes. Finally our data were cross-referenced with observations from other studies based on clinical data (47,48).
Principal component analysis (PCA)
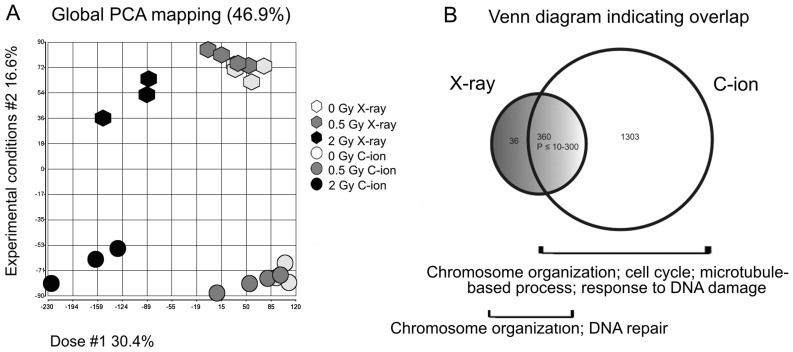
PCA representing the complete gene expression profile presented in two dimensions is shown in Fig. 1A. Data indicated similarity between samples, whereby shorter distance corresponded with greater similarity. Samples clustered together according to the dose (first component) and experimental conditions (second component), which described respectively 30.4% and 16.6% of the variance within the data when considering all genes. Differences in non-irradiated control samples with respect to experimental conditions were most likely due to transport of the cells to the irradiation facility in France. Low biological variance within control samples and samples irradiated with 0.5 Gy resulted in distinct clusters for these conditions. Samples irradiated with 2.0 Gy were more widely distributed over the plot indicating increased biological variation after irradiation with this dose. PCA also demonstrated a distinct shift in gene expression after 2.0 vs. 0.0 Gy for both carbon ion and X-irradiation, while only a small shift was observed after 0.5 Gy of either beam type.
Figure 1.
Radiation-induced changes in global gene expression. (A) 2D Principal component analysis (PCA) for PC3 cells after various doses of carbon ion irradiation or X-rays. PCA is based on global gene expression patterns for each irradiation condition. Analysis revealed distinct clustering in four groups of which the percent variability explained by the components dose and experimental conditions was 30.4 and 16.6%, respectively. (B) Venn diagrams showing overlap of significantly altered genes after 2.0 Gy irradiation of either beam quality. After 2.0 Gy carbon ion irradiation (white circle) and 2.0 Gy X-rays (grey circle) there are respectively 1,663 and 396 genes differentially expressed (DEX). There was a very significant overlap in gene expression changes between the conditions.
Differential gene expression after exposure to carbon ion irradiation and X-rays
Carbon ion irradiation induced very profound effects on gene expression levels. Therefore, very strict criteria (−2≥ FC ≥2; FDR ≤0.05) were used in our analysis for considering genes as being DEX. We found that 8 h after exposure to 2.0 Gy carbon ion irradiation the expression of 1,663 genes was changed at least two-fold (Table III). Of these, 69% were downregulated and 31% were upregulated. In contrast, after 2.0 Gy X-radiation only 396 genes were significantly changed. Similar to carbon ion irradiation, the majority of the DEX genes after X-ray exposure were downregulated (79%). Out of these 396 genes, 360 genes were also DEX after exposure to carbon ion irradiation which shows that there was a very significant (P≤10−300) overlap in the molecular response of PC3 cells to similar doses of high- and low-LET radiation (Fig. 1B). Under these strict statistical conditions, no genes were significantly changed after exposure to 0.5 Gy of either radiation type.
Table III.
Significantly expressed genes: FC ≤-2 or FC ≥2 and FDR ≤0.05.
| Radiation | Total no. of genes | Downregulated | Upregulated | No. of unidentified genes |
|---|---|---|---|---|
| 0.5 Gy C-ion | 0 | |||
| 2.0 Gy C-ion | 1663 | 1145 | 518 | 248 |
| 0.5 Gy X-ray | 0 | |||
| 2.0 Gy X-ray | 396 | 312 | 84 | 39 |
FDR, false discovery rate; FC, fold-change.
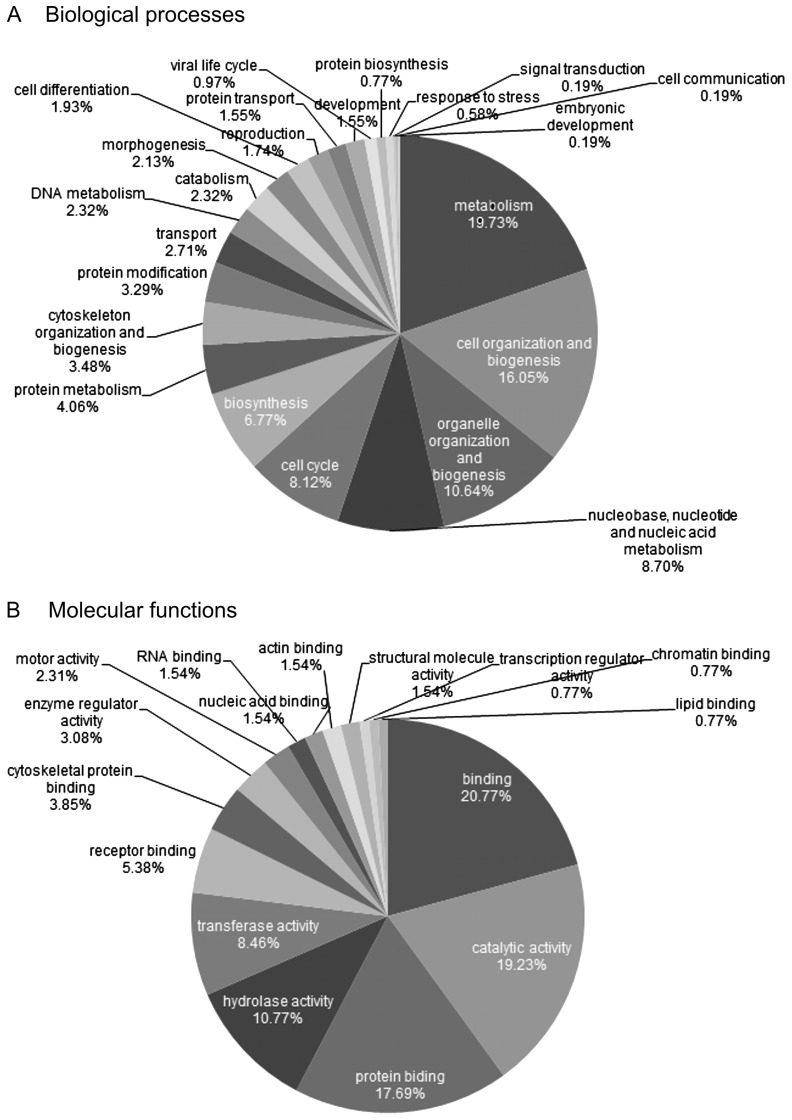
Functional enrichment of the DEX genes after 2.0 Gy carbon ion irradiation was examined using the ToppFun tool (data not shown). In addition, in order to get a better idea of the categories to which the gene sets belong to, the full gene set was slimmed with CateGOrizer. For the enriched biological processes CateGOrizer identified 21 classes (Fig. 2A), of which the majority (19.73%) was involved in cell metabolism. Next to this, a high percentage was involved in cell (16.05%) and organelle organization (10.64%). In addition, 8.12% of the gene sets were involved in cell cycle related processes. For the affected molecular functions 16 categories were identified (Fig. 2B). Many binding processes were affected (20.77% general binding; 17.69% protein binding; 5.38% receptor binding; 3.85% cytoskeletal protein binding; 1.54% RNA binding; 1.54% nucleic acid binding; 1.54% actin binding; 0.77% chromatin binding and 0.77% lipid binding). Also various enzymatic activities were influenced by radiation (19.23% catalytic; 10.77% hydrolase; 8.46% transferase activity and 3.08% enzyme regulator activity).
Figure 2.
Pie chart of the most important (A) biological processes and (B) molecular functions which are affected by 2.0 Gy carbon ion irradiation.
Functional enrichment analysis
For GSEA, 376, 845 and 202 gene sets, related to molecular functions (c5.mf.v3.1), biological processes (c5bp.v3.1) and cellular components (c5.cc.v3.1) respectively, were examined for enrichment among the DEX genes. In general, gene sets were mostly enriched (upregulated) in the control samples, corresponding with the vast amount of genes which are downregulated after irradiation. After 0.5 Gy carbon ion irradiation, 16 gene sets related to molecular functions were enriched in the control samples, while only one set was enriched in irradiated samples. After 2.0 Gy carbon ion radiation 57 sets were enriched in the controls. For 0.5 Gy X-irradiation no gene sets were found to be enriched whereas samples irradiated with 2.0 Gy X-rays had 93 enriched sets in control samples compared to three enriched gene sets in the irradiated samples. Gene sets involved in biological processes (data not shown) were only found to be enriched in the control samples. After 2.0 Gy X-irradiation 100 enriched gene sets were found. After carbon ion irradiation, 31 and 62 gene sets were found to be enriched after 0.5 and 2.0 Gy, respectively. Finally, 13 gene sets coding for cellular components (data not shown) were enriched in samples irradiated with 2.0 Gy carbon ion irradiation, whereas for X-rays 11 gene sets were enriched after irradiation with 2.0 Gy X-rays. Most of these gene sets related to mitochondrial structures. In the control samples 17 sets were enriched after 0.5 Gy carbon ion irradiation. For the X-irradiated samples four sets were enriched after 0.5 Gy, while 42 were enriched after 2.0 Gy.
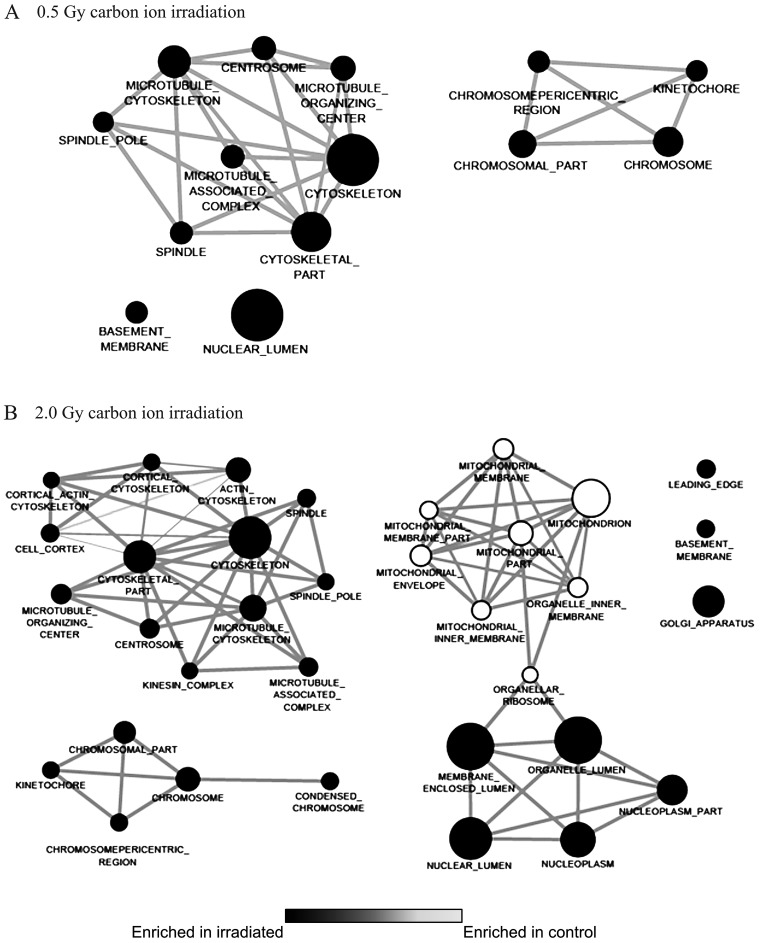
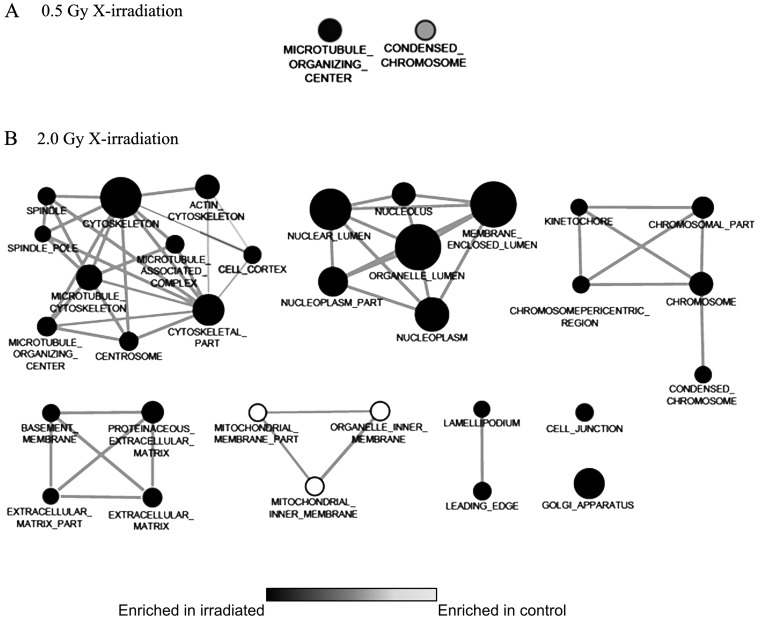
In order to get a more comprehensive view of the enriched gene sets in different conditions, enrichment maps for GO_cellular_components were created with cytoscape (Figs. 3 and 4). The figures show that after 0.5 Gy X-irradiation no related gene sets coding for cellular components were found (Fig. 4). After 0.5 Gy carbon ion irradiation two clusters were identified, which were associated with cytoskeleton and chromosomal components. After 2.0 Gy carbon ion or X-irradiation a cluster associated with chromosomal components were identified, while the cytoskeleton-associated cluster included more gene sets compared to 0.5 Gy irradiation, indicating a dose-dependent effect. Additionally, after 2.0 Gy of either beam quality two clusters associated with nucleus and mitochondrial components were found as well. Finally for the 2.0 Gy X-irradiation also a cluster associated with extracellular matrix were identified.
Figure 3.
Enrichment maps based on gene set enrichment analysis (GSEA) for GO cellular components after carbon ion irradiation (A, 0.5 Gy, and B, 2.0 Gy). White nodes represent gene sets enriched in irradiated samples (i.e. upregulated in irradiated samples), black nodes represent gene sets enriched in control samples (i.e. downregulated in irradiated samples). Node size correlates with the number of genes within each gene set. Edge width represents the overlap of member genes between gene sets. Enriched gene sets included have a P<0.001 and a false discovery rate (FDR) value P<0.05.
Figure 4.
Enrichment Maps based on gene set enrichment analysis (GSEA) for GO cellular components after X-irradiation (A, 0.5 Gy, and B, 2.0 Gy). White nodes represent gene sets enriched in irradiated samples (i.e. upregulated in irradiated samples), black nodes represent gene sets in control samples (i.e. downregulated in irradiated samples). Node size correlates with the number of genes within each gene set. Edge width represents the overlap of member genes between gene sets. Enriched gene sets included have a P<0.001 and a false discovery rate (FDR) value <0.05.
Genes involved in cell motility
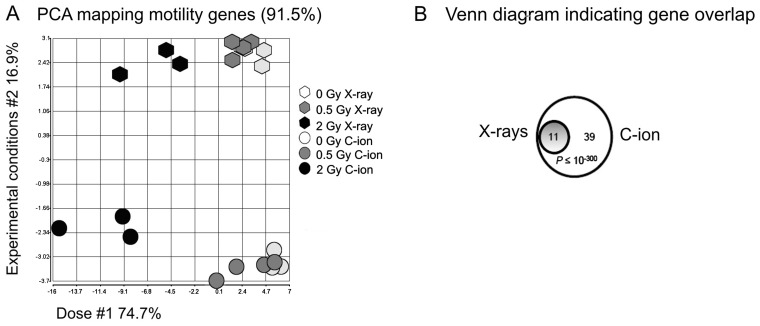
Given the lack of knowledge of potential long-term effects of carbon ion beams, we were interested in the radiation-induced response of genes related to cell migration. Therefore, we filtered the full gene set based on a list from the Gene Ontology website (GO0048870 - cell motility) containing 715 cell motility related genes. PCA revealed a similar spreading of the clusters (Fig. 5A) compared to the entire transcriptome (Fig. 1A). However, in this case a much larger proportion of the total variance could be explained by the dose (74.7%), indicating that the radiation dose had a very profound effect on the general expression of these genes.
Figure 5.
Radiation-induced changes in motility related gene expression. (A) Principal component analysis (PCA) of motility genes revealed distinct clusters similar to global PCA analysis. The percent variability is ∼91.5% explained by the components dose (74.7%) and experimental conditions (16.9%). (B) Venn diagram showing the overlap of motility genes differentially expressed after 2.0 Gy irradiation. All genes found to be differentially expressed by 2.0 Gy X-rays were also regulated by 2.0 Gy carbon ion irradiation.
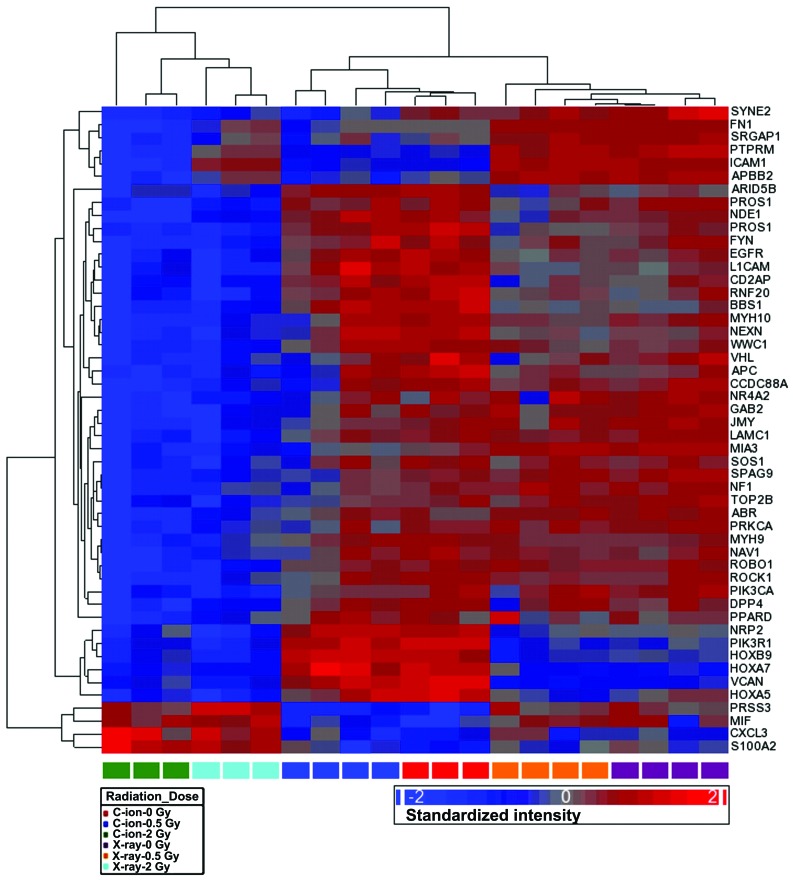
Out of these 746 genes, 50 genes were found to be DEX (−2≥ FC ≥2; FDR ≤0.05) after 2.0 Gy carbon ion radiation (46 downregulated and 4 upregulated), while only 15 genes were changed (all downregulated) after 2.0 Gy X-irradiation (Table IV). All of the latter were also significantly regulated by 2.0 Gy carbon ion irradiation (Fig. 5B). Unsupervised hierarchical clustering of the 50 motility genes that were DEX after 2.0 Gy carbon ion irradiation is shown in Fig. 6. A clear separation was visible between samples irradiated with 2.0 Gy (of either beam quality) and non-irradiated or low dose (0.5 Gy) irradiated samples. Within this last group two other distinct clusters were visible, separating X-ray and carbon ion samples.
Table IV.
Significantly regulated motility genes: FC ≤-2 or FC ≥2 and FDR ≤0.05.
| Radiation | Total no.of genes | Downregulated genes | Upregulated genes |
|---|---|---|---|
| 0.5 Gy C-ion | 0 | ||
| 2.0 Gy C-ion | 50 | 46 | 4 |
| ABR, APBB2, APC, ARID5B, BBS1, CCDC88A, CD2AP, DPP4, EGFR, FN1, FYN, GAB2, HOXA5, HOXA7, HOXB9, ICAM1, JMY, L1CAM, LAMC1, MIA3, MYH10, MYH9, NAV1, NDE1, NEXN, NF1, NR4A2, NRP2, PIK3CA, PIK3R1, PPARD, PRKCA, PROS1, PROS1, PTPRM, RNF20, ROBO1, ROCK1, SOS1, SPAG9, SRGAP1, SYNE2, TOP2B, VCAN, VHL, WWC1 | CXCL3, MIF, PRSS3, S100A2 | ||
| 0.5 Gy X-ray | 0 | ||
| 2.0 Gy X-ray | 15 | 15 | 0 |
| CCDC88A, FN1, GAB2, JMY, L1CAM, LAMC1, MIA3, MYH10, NEXN, PROS1, PROS1, ROBO1, ROCK1, SYNE2, TOP2B |
FDR, false discovery rate; FC, fold-change.
Figure 6.
Heat map of the hierarchical clustering for the 50 motility genes which were significantly regulated by 2.0 Gy carbon ion irradiation. First hierarchical separation distinguished samples irradiated with 2.0 Gy on the left of the heat map. Out of the remaining samples, two groups can be distinguished based on beam quality [carbon ions (middle) vs. X-ray (right)].
Six out of these 50 motility genes (NEXN, CCDC88A, FN1, MYH9, MYH10 and ROCK1) showed at least a four-fold decrease in gene expression after 2.0 Gy carbon ion radiation (Table V). Gene expression levels of these six genes were found to be two to four times more decreased after 2.0 Gy carbon ion radiation compared with 2.0 Gy X-rays. Gene expression changes after 0.5 Gy of both radiation types were not significantly altered, although a dose-dependent trend in expression levels can be seen for both radiation types.
Table V.
Microarray data of motility genes selected for RT-qPCR confirmation.
| Gene symbol | FC 0.5 Gy C-ion | FC 2.0 Gy C-iona | FC 0.5 Gy X-ray | FC 2.0 Gy X-raya |
|---|---|---|---|---|
| FN1 | −1.27 | −8.73 | −1.03 | −2.06 |
| CCDC88A | −2.27 | −13.87 | −1.32 | −4.21 |
| ROCK1 | −1.57 | −11.86 | −1.09 | −3.27 |
| MYH9 | −1.18 | −4.46 | −1.02 | −1.73 |
| NEXN | −1.49 | −8.36 | −1.16 | −2.49 |
| MYH10 | −1.36 | −4.81 | −1.15 | −2.21 |
Statistically significant: FDR P≤0.05. FDR, false discovery rate; FC, fold-change.
RT-qPCR analysis
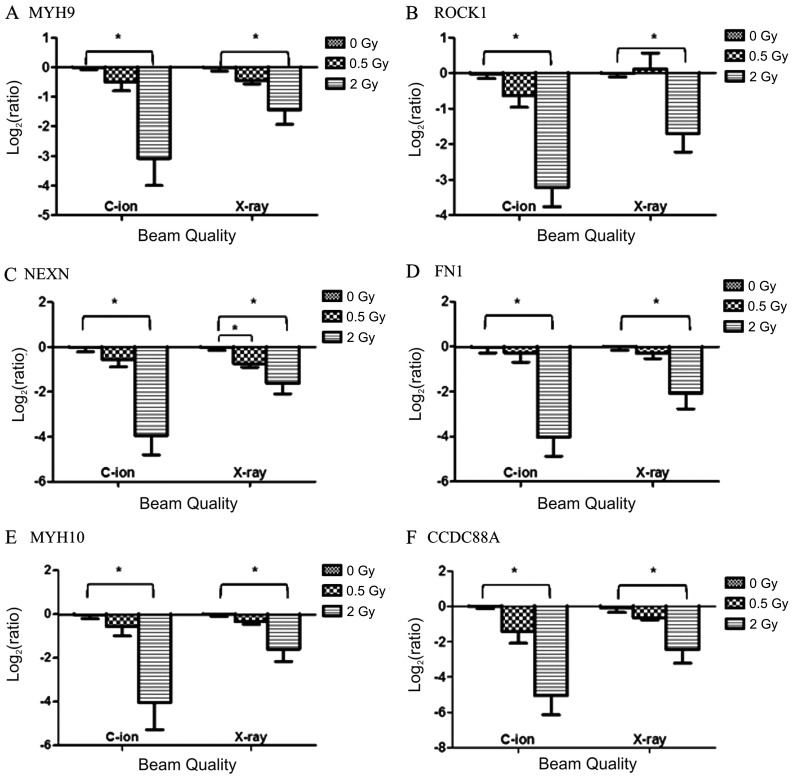
Expression levels of NEXN, CCDC88A, FN1, MYH9, MYH10 and ROCK1 were validated using quantitative real-time PCR. Average log2(R) is presented in Fig. 7. For all but one gene the expression levels tended to decrease after 0.5 Gy (although not significantly). After 2.0 Gy the expression of all six genes significantly decreased. This effect was more visible in carbon ion irradiated samples [log2(R) from −3 to −5] compared to X-irradiation [log2(R) from −1.5 to −2.5].
Figure 7.
Relative gene expression changes of six selected motility genes 8 h after carbon ion or X-irradiation. Log2(ratio) of the expression of (A) MYH9, s(B) ROCK1, (C) NEXN, (D) FN1, (E) MYH10 and (F) CCDC88A is presented. Significantly altered gene expression compared to CTRL samples (*P≤0.05) based on one-tailed Mann-Whitney tests. RT-qPCR results confirm the downregulation observed by microarray analysis after radiation which was more pronounced after carbon ion radiation when compared to X-rays.
Cross-reference of our results with existing patient data
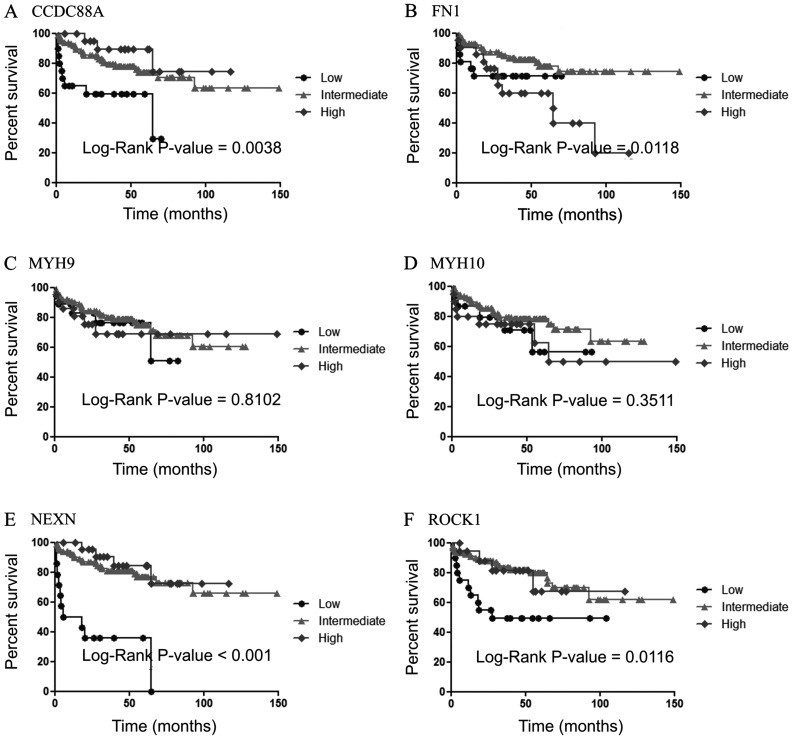
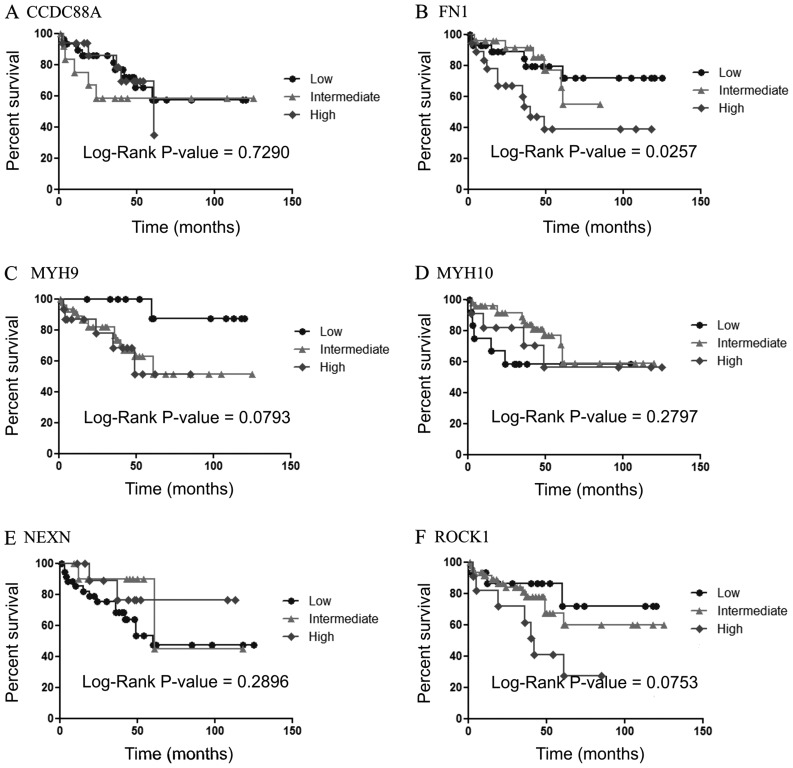
Because of the potential beneficial implications of a down-regulation of motility-related genes for the prognosis of prostate cancer patients, we verified the prognostic power of these genes. We used publicly accessible data sets from two prostate cancer studies (47,48) to generate Kaplan-Meier (recurrence-free) survival curves based on differential gene expression of our six motility genes (Figs. 8 and 9). Within the data set of Taylor et al (48) 150 samples were available for analysis for the six genes. Due to missing values in the data set of Gulzar et al (47) this data set was limited to 61 samples for CCDC88A; 74 for FN1; 60 for NEXN; 79 for MYH9; 77 for MYH10 and 75 for ROCK1. When samples were stratified according to FN1 expression both data sets correlated with high gene expression and poor prognosis of the patients (log-rank P=0.0118 and 0.0257, respectively). This was the only gene which was concordantly significant in both data sets. High and intermediate levels of CCDC88A correlated with a higher overall survival rate (log-rank P=0.0038) within the data set of Taylor et al, while analysis of the data set of Gulzar et al seemed to show an opposite trend however without statistical significance (log-rank P= 0.7290). High ROCK1 expression correlated with better survival rates (log-rank P=0.0116) among the patients of Taylor et al, whereas within the data set of Gulzar et al an opposite trend could be seen. Discrimination based on NEXN expression was highly significant within the data set of Taylor et al, however in this case low gene expression was associated with poor prognosis (log-rank P≤0.0001). This was not the case within the data set of Gulzar et al where no statistical discrimination could be made (log-rank P=0.2896). MYH9 expression did not show any difference in patient survival in the group of Taylor et al. However, within the data set of Gulzar et al data appeared to show a trend which correlated intermediate-high expression of MYH9 with a lower overall survival rate. Therefore, this analysis was performed again by dividing patient samples in two categories, low and intermediate-high. This resulted in a good discrimination between good (low MYH9 expression) and poor prognosis (log-rank low vs. intermediate P=0.0079; log-rank low vs. intermediate P=0.0073; data not shown). Finally, based on MYH10 gene expression patient samples could not discriminate between good and poor prognosis in either data set.
Figure 8.
Kaplan-Meier survival analysis of (A) CCDC88A, (B) FN1, (C) MYH9, (D) MYH10, (E) NEXN and (F) ROCK1 gene expression performed on the data set of Taylor et al (48). Tumor samples were divided into three groups based on whether the gene expression value was high (♦, dark grey); intermediate (▴, light grey); or low (•, black). Differences in survival were found to be significant for CCDC88A, FN1, NEXN and ROCK1 when log-rank P≤0.05.
Figure 9.
Kaplan-Meier survival analysis of (A) CCDC88A, (B) FN1, (C) MYH9, (D) MYH10, (E) NEXN and (F) ROCK1 gene expression performed on the data set of Gulzar et al (47). Tumor samples were divided into three groups based on whether the gene expression value was high (♦, dark grey); intermediate (▴, light grey); or low (•, black). Differences in survival were found to be significant for FN1 (log-rank P=0.0257).
Discussion
Radiation therapy plays an important role in the management of several types of cancer. The fate of an irradiated cancer cell is believed to be controlled by a network of signaling pathways that lead to different modes of cell death or survival. Although most of the cells will die due to the lethal dose, some cells will manage to survive after radiotherapeutic intervention, because they successfully utilize their repair mechanisms. Numerous studies have demonstrated enhanced aggressiveness of surviving cancer cells after conventional radiotherapy, accompanied with an upregulation of genes that favor cell migration, invasion and angiogenesis (21,22,26,49,50). It is well known that carbon ion beams have an increased biolo gical effectiveness compared to X-rays. Therefore, it would not be surprising that both types of radiation can induce different effects at the level of gene expression. So far, the observed differences in gene expression between different radiation qualities are not completely understood. In the present study, we compared the differences in the transcriptional response 8 h after carbon ion (LET=33.7 keV/μm) and X-irradiation of the human PC3 prostate adenocarcinoma cell line. To our knowledge, we are the first to describe global gene expression changes after exposure to different radiation qualities in prostate cancer cells.
Global radiation-induced gene expression changes
We found that 2.0 Gy carbon ion irradiation induced more pronounced changes in gene expression compared to similar doses of X-rays both in terms of number of genes and magnitude of changes. After carbon ion irradiation four times more genes were altered in PC3 cells. Our results demonstrated that almost 90% of the genes that were significantly altered after 2.0 Gy X-ray overlapped with those that were changed after 2.0 Gy carbon ions, indicating that similar molecular pathways were affected. Furthermore, 69% of the significantly altered genes were downregulated. Under our strict statistical criteria, no significant changes were induced after 0.5 Gy irradiation for both radiation qualities. Our results are in agreement with previous findings of Matsumoto et al (34), who examined gene expression in six different melanoma cell lines 1 h and 3 h after exposure to X-rays or carbon ion beams (average dose-LET=50 keV/μm). They found that, after exposure to 2 Gy of both radiation types, several hundreds of genes were differentially expressed at both time-points. Many of these genes showed a greater response to carbon ions compared to X-rays. Similar to our results they found that most of the altered genes were downregulated after radiation exposure. Higo et al (37) irradiated three oral squamous cell carcinoma cell lines with X-rays (2, 4 and 8 Gy) and carbon ions (1, 4 and 7 Gy; LET=78 keV/μm). They also found that carbon ion irradiation induced at least a two fold-change in 98 genes for all doses and all cell lines of which 85 genes were upregulated and 13 down-regulated. In agreement with our results, they found more genes altered by carbon ion radiation compared to X-rays (30 genes upregulated and 4 genes downregulated for all doses in all cell lines). However, in contrast with the study by Matsumoto et al (34) and our findings most of the genes found in the study of Higo et al (37) were upregulated.
All these studies observed a greater impact of carbon beam irradiation on changes in gene expression compared to conventional photon irradiation. However, there are differences in the number of genes that are upregulated or downregulated. Many factors can be responsible for this including the chosen time-point for sample collection. Furthermore, differences in cell type, as well as, physical aspects including irradiation conditions and LET of the beam may have an impact on radiation-induced genome changes.
One limitation of this study is that our results only compare equal doses. We are aware that, considering the higher RBE of carbon ion beams for a similar dose, carbon beams may induce more changes at the gene level compared to X-rays. However, because of limited beam time, only a small number of experiments could be performed and gene expression experiments were selected as first priority over an estimation of the RBE. We therefore analyzed radiation-induced changes in gene expression after equal doses. In view of time-dependent fluctuations (29) in gene expression induced after irradiation and the difficulty of correlating RBE to these time-dependent changes, comparing equal doses for gene expression profiling was considered as a suitable alternative. Indeed Matsumoto et al (34) determined RBE for their cell lines and beam qualities but still compared equal doses at different time-points. However, future experiments should include clonogenic survival assay of PC3 cells exposed to carbon ions and X-rays to determine the RBE. Additionally, experiments including more doses and more time-points would greatly contribute to the characterization of differences in carbon ion and X-irradiation.
Radiation-induced signaling pathways
Enrichment analysis of the DEX genes indicated that the cell cycle, cellular organization and metabolism are affected by 2.0 Gy carbon ion irradiation. Interestingly, although differential expression after 0.5 Gy was not found to be significant for single genes, GSEA did reveal significant changes.
Also other studies have indicated similar pathways being affected by high-LET radiation. Higo et al (37) performed Ingenuity Pathway Analysis which indicated significant alteration of several pathways, amongst which the cell cycle, cell movement and cell assembly and organization in carbon irradiated squamous cell carcinoma. Also Matsumoto et al (34) observed many changes in the cell cycle-related genes after carbon ion irradiation. In addition, they found that, in four out of six cell lines, upregulated genes were mostly p53 target genes. In our PC3 cell line, which has a mutated p53 status (51), we did not observe many significantly upregulated genes involved in the p53 pathway. Matsumoto et al (34) made similar observations with two of their melanoma cell lines, one of which being wild-type, and another having a mutated p53 status. Based on their findings, they suggested that downregulation of gene expression plays a key role in the extra effect of carbon ions compared with X-rays (i.e. an increased radiobiological effectiveness), whereas upregulation of genes relates to sensitivity to both beam qualities (i.e. inherent radiosensitivity of the cell to radiation).
Cell motility-related gene expression changes
An interesting phenomenon observed in in vitro studies is the effect of radiation on cell motility, migration and invasion. Previous studies on low-LET photon irradiation already reported an elevated migration potential of cancerous cells (21,22,26,49,50). So far, however, most studies comparing X-irradiation with particle beams indicated that particle radiation attenuates or even decreases the migration potential of most cancer cell lines (28–32). Therefore, in this study, we more specifically focused our gene expression analysis on genes involved in cell migration and motility. We observed a dose-dependent downregulation after carbon ion irradiation in several genes involved in these pathways, which was significant after 2.0 Gy radiation. Again, the number and amplitude of gene expression changes after X-irradiation of motility related genes was lower compared to carbon ion irradiation. For further confirmation by RT-qPCR, we focused on six genes of which the expression was at least four times decreased after 2.0 Gy of carbon ion irradiation (CCDC88A, ROCK1, NEXN, FN1, MYH10 and MYH9). These RT-qPCR results validated the observed changes as seen in the microarray analysis.
Fibronectin 1
Downregulation of FN1 gene expression was about four times stronger after 2.0 Gy carbon ion irradiation compared to 2.0 Gy X-rays, whereas irradiation with a dose of 0.5 Gy of either beam quality did not induce a significant change. FN1 codes for a glycoprotein and is involved in the integrin signaling pathway (52,53), thereby playing an important role in tissue organization, cell adhesion and cell migration. In addition, fibronectin has been reported to be involved in tumor metastasis (54–56). Influence of low-LET irradiation on fibronectin expression has been reported previously by Andarawewa et al (57). They found that human mammary epithelial cells exposed to a combination of 2 Gy X-ray and TGF-β underwent a morphological shift from an epithelial to a mesenchymal phenotype. This shift included a higher expression of fibronectin and led to increased cell motility and elevated invasion potential of epithelial cells in vitro. They suggested that irradiation of these preneoplastic cells with moderate doses prime them to undergo epithelial to mesenchymal transition (EMT), which could accelerate cancer progression. Additionally, Yang et al (58) irradiated a radio-sensitive and a radioresistant non-small cell lung carcinoma cell line with 2 Gy γ-irradiation and analyzed amongst others the expression of FN1. Their results indicated that 4 h after irradiation gene expression of FN1 was not changed in either cell lines. On the other hand, Hei et al (59) found that exposure of immortalized human lung and breast epithelial cells to 0.6 Gy high LET α-particles (LET=150 keV/μm) induced changes in fibronectin gene expression. Although, northern blot analysis could not confirm these results.
So far, different studies observed different effects of radiation on FN1 expression. Whether downregulation of FN1 in irradiated PC3 cells will result in changes in cellular behavior needs further investigation.
Actin-binding proteins
The other five motility genes (ROCK1, CCDC88A, NEXN, MYH9 and MYH10), which we selected for further investigation, code for actin-binding proteins. It is well known that remodeling of the actin-myosin skeleton by activation of the serine/threonine kinase Akt can lead to changes in migration, invasion and metastasis (60).
The ROCK1 gene codes for the Rho-associated, coiled coil containing protein kinase 1, which is a downstream effector of the Rho-pathway, that seems to play a role in tumor metastasis and invasion by influencing cell adhesion and migration capacity (61,62). Lin et al (62) transfected PC3 cells with mir-146a, a microRNA found to be downregulated in hormone-refractory prostate tumors, which targets ROCK1. Suppression of ROCK1 in these prostate cancer cells reduced cell proliferation, invasion capacity and their adhesion potential to bone marrow endothelial cells in vitro. Therefore, it was suggested that ROCK1 suppression could aid in reducing the metastatic potential of prostate cancer cells. Furthermore, the effect of radiation on ROCK1 expression has been previously studied. Zhai et al (63) irradiated three glioblastoma cell lines with different doses (ranging from 0 to 8 Gy) of photon radiation and observed a dose-dependent, enhanced invasive potential using in vitro assays. They hypothesized that radiation-induced activation of the PI-3K pathway affected Rho/ ROCK1 signaling. They found that ROCK inhibition significantly reduced radiation-induced cellular invasion, whereas inhibition of ROCK without radiation exposure had no effect on the invasion potential. In a study of Fujita et al (31), pancreatic cancer cell lines were irradiated with either carbon ions or X-rays. They observed that cancer cells were able to switch from a mesenchymal mode of motility to a protease-independent mechanism of invasion which was based on actomyosin contractility dependent on ROCK signaling. They found that although carbon ion radiation was capable of decreasing the metastatic potential of these cells, ROCK inhibition was needed to block invasiveness. In the present study, we found that radiation exposure decreased the expression of ROCK1 in a dose-dependent way in prostate cancer cells. This effect was more pronounced after 2.0 Gy carbon ion irradiation compared to 2.0 Gy X-rays, indicating that the magnitude of downregulation is radiation quality-dependent. Since previous results demonstrated that inhibition of ROCK attenuated cell migration capacity, our findings suggest that carbon ion irradiation could be more efficient in decreasing the migration potential of some cell lines.
One of the functions of ROCK1 is the phosphorylation of the myosin light chains. Together with the myosin heavy chains (MYH), these proteins are responsible for the motor function of the actin-myosin complex, thereby driving various motility-based processes such as cytokinesis, cell rounding and cell migration (64). MYH9 and MYH10 genes encode for isoforms of non-muscle myosin II heavy chains (65,66). MYH9 was reported to be involved in the formation of lamellopodia at the leading edge of breast cancer cells (67). Furthermore, reducing the expression of MYH9 in an invasive form of MCF-7 breast cancer cells blocked the invasive potential of these cells (68). Also for CCDC88A and NEXN, the involvement of these genes in metastases has been described (60,69–71). CCDC88A, also known as GIV or Girdin, codes for coiled-coil domain containing 88A, and was found to be more highly expressed in samples of colorectal cancers with liver metastasis compared to cancers without metastasis (60). Garcia-Marcos et al (69) found the full length protein exclusively expressed in highly invasive colon, breast and pancreatic tumors, suggesting the protein to be a useful clinical marker for metastatic potential. Finally, NEXN was reported to be involved in cell motility and adhesion of HeLa cancer cells through the binding link from actin and the plasma membrane (71). To our knowledge no studies specifically focused on the influence of radiation on the expression of genes coding for the actin-binding proteins CCDC88A, NEXN, MYH9 and MYH10. However, Fushimi et al (36) irradiated oral squamous cell carcinoma cell lines with X-rays (2, 4 and 8 Gy), carbon ions or neon ions (1, 4 and 7 Gy; LET=78 keV/μm) and performed Ingenuity pathway analysis. They indicated integrin, actin cytoskeleton and PI3K/Akt signaling as affected pathways. The genes found in our study also play a role in these motility regulating pathways. Additionally, Akino et al (29) found that 12 h after irradiation of non-small-cell lung (NSLC) cancer cells with 5 Gy carbon ions the expression of the actin-binding protein anillin (ANLN) was downregulated, while irradiation with X-rays increased ANLN expression.
As we mentioned before differences in genes which are significantly expressed after irradiation can be due to use of other time-points taken for the sampling. Therefore, it could be informative to further investigate how the expression of actin-binding proteins in response to different radiation qualities changes during time.
In this study, we found that exposure of prostate cancer cells to different radiation qualities decreased the expression of these five genes coding for actin-binding proteins. Furthermore, enrichment analysis indicated that 1.54% of all significantly regulated genes are involved in actin-binding processes (Fig. 2), and further GSEA analysis indicated many gene sets involved in actin-regulation. In view of the importance of actin-binding and actin remodeling in migration processes, it will be important to investigate whether this downregulation may lead to changes in cellular behavior, thereby influencing the migration potential of the cells. Important to mention is that during radiotherapy most of the cancer cells will be eliminated because of the lethal dose. However, some cells can manage to survive, either because they receive sub-lethal doses and/or because they successfully induce their repair mechanisms. We are aware that in this study, only one time-point was analyzed (8 h) after irradiation with a single dose. Further studies could include more time-points, as well as fractionated irradiation to get a more complete picture of the differences in cell migration after low- and high-LET radiation.
Clinically relevant biomarkers in prostate cancer
In view of the involvement of these six genes in cancer cell motility we also investigated the importance of our selected genes in prostate cancer prognoses by performing a recurrence-free survival analysis on two independent sets of prostate cancer patients (47,48). FN1 was the only gene of which high expression was significantly correlated with higher recurrence in both data sets. This gene has been previously identified as a potential biomarker for radiation resistance after genomic analysis of several radioresistant head and neck squamous cell carcinomas (72). Futhermore, Hébrant et al (70) observed that not only FN1 but also CCDC88A and MYH9 were more highly expressed in more aggressive anaplastic thyroid carcinomas compared to papillary thyroid carcinomas. We observed an obvious drop in survival rates in both patient groups with high FN1 expression although there was a difference in the percentage of recurrence-free survival. However, this may be explained by the fact that we took publicly accessible data which were processed in a different manner by the original authors. We appointed high-intermediate-low gene expression based on different criteria in both sets. Adjusting the criteria slightly could equalize the difference in survival percentage. Performing the same analysis on an even larger study population will give more information on how recurrence-free survival may be correlated to FN1 gene expression.
Survival analysis was also in agreement for both data sets for MYH10 expression and NEXN expression. MYH10 expression could not be used to distinguish differences in recurrence between expression groups, while low NEXN expression correlated with poor survival rates. On the other hand CCDC88A, MYH9 and ROCK1 expression predicted a different patient outcome in the two study populations. Low MYH9 or ROCK1 expression tended to be correlated with better recurrence-free survival for the patients of Gulzar et al (47), while CCDC88A did not distinguish survival between low and high gene expression groups. However, for the patients of Taylor et al (48) low CCDC88A or low ROCK1 expression predicted a poor prognosis for the patients and MYH9 expression could not distinguish differences in survival based on low or high expression. We only found one other study investigating the effect of ROCK1 protein expression in osteosarcoma samples on patient survival (73). Liu et al correlated high ROCK1 presence with lower overall survival rates (73). Within our analysis both patient groups gave contrary results. Also other authors pointed out the difficulties of finding reliable prognostic markers within different sets of prostate cancer patients (47,48). We conclude that these five actin-binding genes may not be suitable biomarkers for prostate cancer prognosis, however, our finding that they are downregulated after radiation exposure in PC3 cells may lead to changes in cell behavior after irradiation.
Although we focused on these five genes in the context of cell migration, it should be noted that actin remodeling is also a very important process during cell cycle progression and mitosis as well. Therefore, it is possible that the observed changes are part of, or a side effect of, alterations in cell cycle due to radiation exposure rather than an actual change in migration potential of the cell. The present study should thus be extended with in vitro and in vivo assays to verify whether the observed changes are permanent, thereby leading to a change in cellular phenotype or returning to baseline without major consequences for the cell. These assays could bring more insight into the clinical radiation effects induced in patients.
The main goal of this study was to analyze the effects of different types of radiation (carbon ions vs. X-rays) on the gene expression of prostate cancer cells. It is well known that changes in gene expression play a key role in cancer progression (18,19).
Dysregulation of signaling pathways in surviving cells after radiotherapy, such as those associated with cell migration and motility, can determine the fate of the tumor and consequently the fate of the patient. Since most prostate cancer-related deaths are due to metastases (9–11) it is of pivotal importance to understand how therapeutic intervention can influence metastatic development of irradiated cancer cells. Furthermore, cases of radiation-induced metastasis in prostate cancer patients have been reported (74–77). Since clinical trials, using optimized settings for carbon ion treatment, have shown high survival rates and good local control (8,12–15), carbon ion irradiation has been approved as a valid treatment for prostate cancers (8). However, in view of this new treatment option it is of high importance to further investigate changes in cancer cells that survived the treatment in the context of potential long-term effects.
We found that, under the conditions investigated, carbon ion irradiation induced more pronounced changes in PC3 cells in terms of number of genes and magnitude of changes compared to X-rays. We more specifically focused on genes involved in cell motility and found that these genes were generally downregulated after irradiation, an effect which was more pronounced after carbon ion irradiation compared to X-rays. This irradiation-induced downregulation may suggest a suppressed migration potential of PC3 cells, especially after carbon ion irradiation. Further research is needed to investigate whether PC3 cells show a decrease in migration potential after exposure to different radiation qualities. In this regard, functional assays on cell motility and invasion, such as the scratch healing assay and Boyden chamber assay can help in determining whether the observed changes also influence the cellular behavior after irradiation. Understanding how different radiation qualities affect the migration potential of prostate cancer cells is important for improving the clinical outcome of cancer radiation therapy.
Acknowledgments
This study is partly supported by the Federal Public Service in the context of the feasibility study ‘Application of hadron-therapy in Belgium’, which is part of action 30 of the Belgian cancer plan. We would like to thank the iPAC committee of GANIL for the beam time granted (P911-H), the staff of the LARIA for allowing us access to and use of their facility and the team of CIMAP for performing the dosimitry. We also thank Bart Marlein and Ludo Melis for their continued assistance during the X-irradiations at SCK•CEN. We are grateful to John Gueulette, Stefaan Vynckier, Pierre Scalliet, Emiliano d’Agostino and Filip Vanhavere for their guidance in dosimetry and to Maïté Hanot and Claire Rodriguez-Lafrasse (Laboratoire de Radiobiologie Cellulaire et Moleculaire, Lyon, France) for their strategic advice in planning the carbon ion irradiation experiments at GANIL.
References
- 1.Karger CP, Jäkel O. Current status and new developments in ion therapy. Strahlenther Onkol. 2007;183:295–300. doi: 10.1007/s00066-007-1645-x. [DOI] [PubMed] [Google Scholar]
- 2.Krämer M, Weyrather WK, Scholz M. The increased biological effectiveness of heavy charged particles: from radiobiology to treatment planning. Technol Cancer Res Treat. 2003;2:427–436. doi: 10.1177/153303460300200507. [DOI] [PubMed] [Google Scholar]
- 3.Iwadate Y, Mizoe J, Osaka Y, Yamaura A, Tsujii H. High linear energy transfer carbon radiation effectively kills cultured glioma cells with either mutant or wild-type p53. Int J Radiat Oncol Biol Phys. 2001;50:803–808. doi: 10.1016/s0360-3016(01)01514-0. [DOI] [PubMed] [Google Scholar]
- 4.Wada S, Kobayashi Y, Funayama T, Natsuhori M, Ito N, Yamamoto K. Detection of DNA damage in individual cells induced by heavy-ion irradiation with an non-denaturing comet assay. J Radiat Res. 2002;43(Suppl):S153–S156. doi: 10.1269/jrr.43.s153. [DOI] [PubMed] [Google Scholar]
- 5.Hamada N. Recent insights into the biological action of heavy-ion radiation. J Radiat Res. 2009;50:1–9. doi: 10.1269/jrr.08070. [DOI] [PubMed] [Google Scholar]
- 6.Rossi CJ, Jr, Slater JD, Reyes-Molyneux N, et al. Particle beam radiation therapy in prostate cancer: is there an advantage? Semin Radiat Oncol. 1998;8:115–123. doi: 10.1016/s1053-4296(98)80007-6. [DOI] [PubMed] [Google Scholar]
- 7.Orecchia R, Fossati P, Rossi S. The National Center for Oncological Hadron Therapy: status of the project and future clinical use of the facility. Tumori. 2009;95:169–176. doi: 10.1177/030089160909500207. [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa H, Tsuji H, Kamada T, et al. Carbon-ion radiation therapy for prostate cancer. Int J Urol. 2012;19:296–305. doi: 10.1111/j.1442-2042.2012.02961.x. [DOI] [PubMed] [Google Scholar]
- 9.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 10.Fidler IJ, Kim SJ, Langley RR. The role of the organ micro-environment in the biology and therapy of cancer metastasis. J Cell Biochem. 2007;101:927–936. doi: 10.1002/jcb.21148. [DOI] [PubMed] [Google Scholar]
- 11.Hagen RM, Ladomery MR. Role of splice variants in the metastatic progression of prostate cancer. Biochem Soc Trans. 2012;40:870–874. doi: 10.1042/BST20120026. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa H, Tsuji H, Kamada T, et al. Risk factors of late rectal bleeding after carbon ion therapy for prostate cancer. Int J Radiat Oncol Biol Phys. 2006;66:1084–1091. doi: 10.1016/j.ijrobp.2006.06.056. [DOI] [PubMed] [Google Scholar]
- 13.Wakatsuki M, Tsuji H, Ishikawa H, et al. Quality of life in men treated with carbon ion therapy for prostate cancer. Int J Radiat Oncol Biol Phys. 2008;72:1010–1015. doi: 10.1016/j.ijrobp.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 14.Akakura K, Tsujii H, Morita S, et al. Phase I/II clinical trials of carbon ion therapy for prostate cancer. Prostate. 2004;58:252–258. doi: 10.1002/pros.10328. [DOI] [PubMed] [Google Scholar]
- 15.Tsuji H, Yanagi T, Ishikawa H, et al. Hypofractionated radiotherapy with carbon ion beams for prostate cancer. Int J Radiat Oncol Biol Phys. 2005;63:1153–1160. doi: 10.1016/j.ijrobp.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 16.Vicini FA, Kestin LL, Martinez AA. The importance of adequate follow-up in defining treatment success after external beam irradiation for prostate cancer. Int J Radiat Oncol Biol Phys. 1999;45:553–561. doi: 10.1016/s0360-3016(99)00235-7. [DOI] [PubMed] [Google Scholar]
- 17.Zietman AL, Bae K, Slater JD, et al. Randomized trial comparing conventional-dose with high-dose conformal radiation therapy in early-stage adenocarcinoma of the prostate: long-term results from proton radiation oncology group/american college of radiology 95-09. J Clin Oncol. 2010;28:1106–1111. doi: 10.1200/JCO.2009.25.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 20.Msaouel P, Pissimissis N, Halapas A, Koutsilieris M. Mechanisms of bone metastasis in prostate cancer: clinical implications. Best Pract Res Clin Endocrinol Metab. 2008;22:341–355. doi: 10.1016/j.beem.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Wild-Bode C, Weller M, Rimner A, Dichgans J, Wick W. Sublethal irradiation promotes migration and invasiveness of glioma cells: implications for radiotherapy of human glioblastoma. Cancer Res. 2001;61:2744–2750. [PubMed] [Google Scholar]
- 22.Zhou YC, Liu JY, Li J, et al. Ionizing radiation promotes migration and invasion of cancer cells through transforming growth factor-beta-mediated epithelial-mesenchymal transition. Int J Radiat Oncol Biol Phys. 2011;81:1530–1537. doi: 10.1016/j.ijrobp.2011.06.1956. [DOI] [PubMed] [Google Scholar]
- 23.Yao H, Zeng ZZ, Fay KS, et al. Role of α(5)β(1) integrin up-regulation in radiation-induced invasion by human pancreatic cancer cells. Transl Oncol. 2011;4:282–292. doi: 10.1593/tlo.11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordes N, Blaese MA, Meineke V, Van Beuningen D. Ionizing radiation induces up-regulation of functional beta1-integrin in human lung tumour cell lines in vitro. Int J Radiat Biol. 2002;78:347–357. doi: 10.1080/09553000110117340. [DOI] [PubMed] [Google Scholar]
- 25.De Bacco F, Luraghi P, Medico E, et al. Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. J Natl Cancer Inst. 2011;103:645–661. doi: 10.1093/jnci/djr093. [DOI] [PubMed] [Google Scholar]
- 26.Fujita M, Otsuka Y, Yamada S, Iwakawa M, Imai T. X-ray irradiation and Rho-kinase inhibitor additively induce invasiveness of the cells of the pancreatic cancer line, MIAPaCa-2, which exhibits mesenchymal and amoeboid motility. Cancer Sci. 2011;102:792–798. doi: 10.1111/j.1349-7006.2011.01852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mikheeva SA, Mikheev AM, Petit A, et al. TWIST1 promotes invasion through mesenchymal change in human glioblastoma. Mol Cancer. 2010;9:194. doi: 10.1186/1476-4598-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goetze K, Scholz M, Taucher-Scholz G, Mueller-Klieser W. The impact of conventional and heavy ion irradiation on tumor cell migration in vitro. Int J Radiat Biol. 2007;83:889–896. doi: 10.1080/09553000701753826. [DOI] [PubMed] [Google Scholar]
- 29.Akino Y, Teshima T, Kihara A, et al. Carbon-ion beam irradiation effectively suppresses migration and invasion of human non-small-cell lung cancer cells. Int J Radiat Oncol Biol Phys. 2009;75:475–481. doi: 10.1016/j.ijrobp.2008.12.090. [DOI] [PubMed] [Google Scholar]
- 30.Ogata T, Teshima T, Kagawa K, et al. Particle irradiation suppresses metastatic potential of cancer cells. Cancer Res. 2005;65:113–120. [PubMed] [Google Scholar]
- 31.Fujita M, Otsuka Y, Imadome K, Endo S, Yamada S, Imai T. Carbon-ion radiation enhances migration ability and invasiveness of the pancreatic cancer cell, PANC-1, in vitro. Cancer Sci. 2012;103:677–683. doi: 10.1111/j.1349-7006.2011.02190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goetze K, Scholz M, Taucher-Scholz G, Mueller-Klieser W. Tumor cell migration is not influenced by p21 in colon carcinoma cell lines after irradiation with X-ray or (12)C heavy ions. Radiat Environ Biophys. 2010;49:427–435. doi: 10.1007/s00411-010-0297-x. [DOI] [PubMed] [Google Scholar]
- 33.Sharungbam GD, Schwager C, Chiblak S, et al. Identification of stable endogenous control genes for transcriptional profiling of photon, proton and carbon-ion irradiated cells. Radiat Oncol. 2012;7:70. doi: 10.1186/1748-717X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsumoto Y, Iwakawa M, Furusawa Y, et al. Gene expression analysis in human malignant melanoma cell lines exposed to carbon beams. Int J Radiat Biol. 2008;84:299–314. doi: 10.1080/09553000801953334. [DOI] [PubMed] [Google Scholar]
- 35.Girdhani S, Lamont C, Hahnfeldt P, Abdollahi A, Hlatky L. Proton irradiation suppresses angiogenic genes and impairs cell invasion and tumor growth. Rad Res. 2012;178:33–45. doi: 10.1667/rr2724.1. [DOI] [PubMed] [Google Scholar]
- 36.Fushimi K, Uzawa K, Ishigami T, et al. Susceptible genes and molecular pathways related to heavy ion irradiation in oral squamous cell carcinoma cells. Radiother Oncol. 2008;89:237–244. doi: 10.1016/j.radonc.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 37.Higo M, Uzawa K, Kawata T, et al. Enhancement of SPHK1 in vitro by carbon ion irradiation in oral squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2006;65:867–875. doi: 10.1016/j.ijrobp.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 38.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3) Invest Urol. 1979;17:16–23. [PubMed] [Google Scholar]
- 39.Ziegler JF, Ziegler MD, Biersack JP. SRIM - The stopping and range of ions in matter. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms. 2010;268:1818–1823. [Google Scholar]
- 40.El-Saghire H, Thierens H, Monsieurs P, Michaux A, Vandevoorde C, Baatout S. Gene set enrichment analysis highlights different gene expression profiles in whole blood samples X-irradiated with low and high doses. Int J Radiat Biol. 2013;89:628–638. doi: 10.3109/09553002.2013.782448. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 42.Hu Z, Bao J, Reecy JM. CateGOrizer: A web-based program to batch analyze Gene Ontology Classification Categories. Online J Bioinformatics. 2008;9:108–112. [Google Scholar]
- 43.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 44.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gulzar ZG, McKenney JK, Brooks JD. Increased expression of NuSAP in recurrent prostate cancer is mediated by E2F1. Oncogene. 2013;32:70–77. doi: 10.1038/onc.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor BS, Schultz N, Hieronymus H, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sofia Vala I, Martins LR, Imaizumi N, et al. Low doses of ionizing radiation promote tumor growth and metastasis by enhancing angiogenesis. PLoS One. 2010;5:e11222. doi: 10.1371/journal.pone.0011222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pickhard AC, Margraf J, Knopf A, et al. Inhibition of radiation induced migration of human head and neck squamous cell carcinoma cells by blocking of EGF receptor pathways. BMC Cancer. 2011;11:388. doi: 10.1186/1471-2407-11-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isaacs WB, Carter BS, Ewing CM. Wild-type p53 suppresses growth of human prostate cancer cells containing mutant p53 alleles. Cancer Res. 1991;51:4716–4720. [PubMed] [Google Scholar]
- 52.Leiss M, Beckmann K, Giros A, Costell M, Fässler R. The role of integrin binding sites in fibronectin matrix assembly in vivo. Curr Opin Cell Biol. 2008;20:502–507. doi: 10.1016/j.ceb.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 53.Oppenheimer SB. Cellular basis of cancer metastasis: A review of fundamentals and new advances. Acta Histochem. 2006;108:327–334. doi: 10.1016/j.acthis.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 54.Ding J, Li D, Wang X, Wang C, Wu T. Fibronectin promotes invasiveness and focal adhesion kinase tyrosine phosphorylation of human colon cancer cell. Hepatogastroenterology. 2008;55:2072–2076. [PubMed] [Google Scholar]
- 55.Qian P, Zuo Z, Wu Z, et al. Pivotal role of reduced let-7g expression in breast cancer invasion and metastasis. Cancer Res. 2011;71:6463–6474. doi: 10.1158/0008-5472.CAN-11-1322. [DOI] [PubMed] [Google Scholar]
- 56.Nam JM, Onodera Y, Bissell MJ, Park CC. Breast cancer cells in three-dimensional culture display an enhanced radio-response after coordinate targeting of integrin alpha5beta1 and fibronectin. Cancer Res. 2010;70:5238–5248. doi: 10.1158/0008-5472.CAN-09-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andarawewa KL, Erickson AC, Chou WS, et al. Ionizing radiation predisposes nonmalignant human mammary epithelial cells to undergo transforming growth factor beta induced epithelial to mesenchymal transition. Cancer Res. 2007;67:8662–8670. doi: 10.1158/0008-5472.CAN-07-1294. [DOI] [PubMed] [Google Scholar]
- 58.Yang HJ, Kim N, Seong KM, Youn H, Youn B. Investigation of radiation-induced transcriptome profile of radioresistant non-small cell lung cancer A549 cells using RNA-seq. PLoS One. 2013;8:e59319. doi: 10.1371/journal.pone.0059319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hei TK, Zhao YL, Roy D, Piao CQ, Calaf G, Hall EJ. Molecular alterations in tumorigenic human bronchial and breast epithelial cells induced by high LET radiation. Adv Space Res. 2001;27:411–419. doi: 10.1016/s0273-1177(01)00009-6. [DOI] [PubMed] [Google Scholar]
- 60.Liu C, Xue H, Lu Y, Chi B. Stem cell gene Girdin: a potential early liver metastasis predictor of colorectal cancer. Mol Biol Rep. 2012;39:8717–8722. doi: 10.1007/s11033-012-1731-8. [DOI] [PubMed] [Google Scholar]
- 61.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28:65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 62.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 2008;14:417–424. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhai GG, Malhotra R, Delaney M, et al. Radiation enhances the invasive potential of primary glioblastoma cells via activation of the Rho signaling pathway. J Neurooncol. 2006;76:227–237. doi: 10.1007/s11060-005-6499-4. [DOI] [PubMed] [Google Scholar]
- 64.Sandquist JC, Swenson KI, Demali KA, Burridge K, Means AR. Rho kinase differentially regulates phosphorylation of nonmuscle myosin II isoforms A and B during cell rounding and migration. J Biol Chem. 2006;281:35873–35883. doi: 10.1074/jbc.M605343200. [DOI] [PubMed] [Google Scholar]
- 65.Cai Y, Rossier O, Gauthier NC, et al. Cytoskeletal coherence requires myosin-IIA contractility. J Cell Sci. 2010;123:413–423. doi: 10.1242/jcs.058297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 67.Morimura S, Suzuki K, Takahashi K. Nonmuscle myosin IIA is required for lamellipodia formation through binding to WAVE2 and phosphatidylinositol 3,4,5-triphosphate. Biochem Biophys Res Commun. 2011;404:834–840. doi: 10.1016/j.bbrc.2010.12.069. [DOI] [PubMed] [Google Scholar]
- 68.Derycke L, Stove C, Vercoutter-Edouart AS, et al. The role of non-muscle myosin IIA in aggregation and invasion of human MCF-7 breast cancer cells. Int J Dev Biol. 2011;55:835–840. doi: 10.1387/ijdb.113336ld. [DOI] [PubMed] [Google Scholar]
- 69.Garcia-Marcos M, Jung BH, Ear J, Cabrera B, Carethers JM, Ghosh P. Expression of GIV/Girdin, a metastasis-related protein, predicts patient survival in colon cancer. FASEB J. 2011;25:590–599. doi: 10.1096/fj.10-167304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hébrant A, Dom G, Dewaele M, et al. mRNA expression in papillary and anaplastic thyroid carcinoma: molecular anatomy of a killing switch. PLoS One. 2012;7:e37807. doi: 10.1371/journal.pone.0037807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang W, Zhang W, Han Y, et al. NELIN, a new F-actin associated protein, stimulates HeLa cell migration and adhesion. Biochem Biophys Res Commun. 2005;330:1127–1131. doi: 10.1016/j.bbrc.2005.03.082. [DOI] [PubMed] [Google Scholar]
- 72.Amundson SA, Smilenov LB. Integration of biological knowledge and gene expression data for biomarker selection: FN1 as a potential predictor of radiation resistance in head and neck cancer. Cancer Biol Ther. 2010;10:1252–1255. doi: 10.4161/cbt.10.12.13731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu X, Choy E, Hornicek FJ, et al. ROCK1 as a potential therapeutic target in osteosarcoma. J Orthop Res. 2011;29:1259–1266. doi: 10.1002/jor.21403. [DOI] [PubMed] [Google Scholar]
- 74.Tseng TY, Sevilla DW, Moul JW, Maloney KE. Prostatic carcinosarcoma 15 years after combined external beam radiation and brachytherapy for prostatic adenocarcinoma: a case report. Prostate Cancer Prostatic Dis. 2006;9:195–197. doi: 10.1038/sj.pcan.4500870. [DOI] [PubMed] [Google Scholar]
- 75.Mazzucchelli R, Lopez-Beltran A, Cheng L, Scarpelli M, Kirkali Z, Montironi R. Rare and unusual histological variants of prostatic carcinoma: clinical significance. BJU Int. 2008;102:1369–1374. doi: 10.1111/j.1464-410X.2008.08074.x. [DOI] [PubMed] [Google Scholar]
- 76.Hansel DE, Epstein JI. Sarcomatoid carcinoma of the prostate: a study of 42 cases. Am J Surg Pathol. 2006;30:1316–1321. doi: 10.1097/01.pas.0000209838.92842.bf. [DOI] [PubMed] [Google Scholar]
- 77.Huan Y, Idrees M, Gribetz ME, Unger PD. Sarcomatoid carcinoma after radiation treatment of prostatic adenocarcinoma. Ann Diagn Pathol. 2008;12:142–145. doi: 10.1016/j.anndiagpath.2006.08.008. [DOI] [PubMed] [Google Scholar]