Abstract
Self-assembled peptide/polypeptide nanofibers are appealing building blocks for creating complex three-dimensional structures. However, ordering assembled peptide/polypeptide nanofibers into three-dimensional structures on the microscale remains challenging and often requires the employment of top-down approaches. We report that silk-elastin-like protein polymers self-assemble into nanofibers in physiologically relevant conditions, the assembled nanofibers further form fiber clusters on the microscale, and the nanofiber clusters eventually coalesce into three-dimensional structures with distinct nanoscale and microscale features. It is believed that the interplay between fiber growth and molecular diffusion leads to the ordering of the assembled silk-elastin-like nanofibers at the microscale.
Self-assembled peptide/protein nanostructures may find important applications in various areas, including tissue engineering, biosensors, and drug delivery. As an example, self-assembled peptide nanofibers enable the cells to interact with the matrix in three-dimensional microenvironments, enhancing neurite outgrowth.1 However, it remains a daunting challenge to order self-assembled peptide nanofibers into three-dimensional structures on the microscale. It has been suggested to integrate top-down approaches and self-assembly in the fabrication of microscale-ordered nanostructures.2 Nevertheless, this strategy not only requires additional capital investment in fabrication facilities but also comprises the elegance of molecular self-assembly, including ease of processing and energy efficiency. Here, we report that, driven by the interplay of fiber nucleation/growth with molecular diffusion, silk-elastin-like protein (SELP) polymers self- assemble into three-dimensional structures with distinct nanoscale and microscale features.
Owing to relative ease of design, synthesis, and purification, most self-assembling peptides that have been extensively explored are small peptides, containing several to a few tens of amino acids.3 In contrast, living systems often utilize high molecular-weight proteins, such as actin and collagen, as building blocks to create nanofiber-based intracellular and extracellular matrices.4 Likely, large amino acid chains provide a rich source of structural/functional variability and complexity. Herein, two SELP polymers, SELP-47 K (MW: 69,814 theoretical calculation5 and 69 699 mass spectroscopy6) and SELP-815 K (MW: 65 374 theoretical calculation7), are used as a model system to study self-assembly of high molecular-weight proteins. The complete amino acid sequences of SELP-47K and SELP-815 K are MDPVVLQRRDWENPGVTQLNRLAAHPPFASDPMGAGSGAGAGS[(GVGVP)4GKGVP(GVGVP)3(GAGAGS)4]12(GVGVP)4GKGVP(GVGVP)3(GAGAGS)2GAGAMDPGRYQDLRSHHHHHH (one letter amino-acid abbreviation is used)5 and MDPVVLQRRDWENPGVTQLNRLAAHPPFASDPM[GAGS(GAGAGS)2(GVGVP)4GKGVP(GVGVP)11(GAGAGS)4GAGA]6MDPGRYQDLRSHHHHHH, respectively.6 In the design of SELPs, the elastin-derived GVGVP sequences are interspersed with the silk-derived GAGAGS sequences. The interspersion of the elastin unit prevents premature formation of large β-sheet crystals and renders SELP water soluble.6, 8 Compared to SELP-47 K, the larger silk-like block of each repeating unit of SELP-815 K improves its ability to form nanofibers. In this study, the self-assembly of SELP-47 K and SELP-815 K was analyzed in aqueous solution.
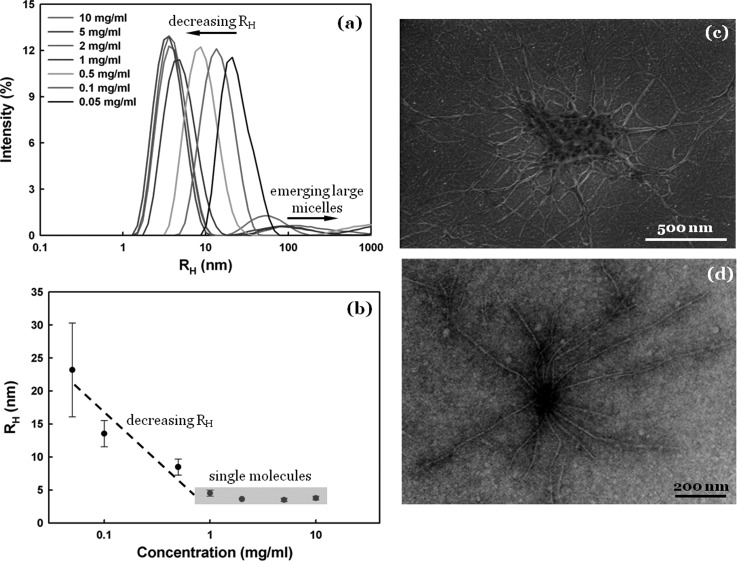
We first used dynamic light scattering (DLS) to examine the self-assembly of SELP-47 K in aqueous solutions at various concentrations and at room temperature. At a concentration of 0.05 mg/ml, a strong intensity of scattered light at a hydrodynamic radius (Rh) of 23 ± 8 nm suggests that the protein polymer forms small micelle-like nanoparticles [Fig. 1a]. As the concentration increases, the DLS analysis reveals two interesting phenomena. First, the strong scattering peak shifts to the low Rh range. As shown in Fig. 1b, SELP-47 K solutions at concentrations of 1∼10 mg/ml display a strong intensity of scattered light at Rh of 3.5 ∼ 4.5 nm, suggesting the presence of free chains in solution.9 Second, a weak and broad peak appears in the Rh range of 10 ∼ 1000 nm. It indicates the formation of micelle aggregates that are several tens of nanometers to a few hundred nanometers in size. It is likely that micelle aggregates, the formation of which by nature is stochastic, attract a considerable portion of SELP-47 K molecules in solution, reducing small micelles to eventually free chains. Indeed, SELP-47 K molecules in 1 ∼ 10 mg/ml solutions exist in the form of either free chains or highly heterogeneous micelle aggregates.
Figure 1.
Micelle formation and nanofiber nucleation. (a) DLS of SELP-47 K aqueous solutions at various concentrations. (b) Position of the strong DLS peak as a function of SELP-47 K concentration. (c) SELP-47 K nanofiber assembly in water at 37 °C may be nucleated at micelle aggregates. (d) The growth of SELP-47 K nanofibers in water at 4 °C may be initiated at a preexisting seed.
The formation of micelle aggregates is important to the initiation of SELP-47 K nanofibers. Although nanofiber assemblies can be initiated at preexisting seeds or micelle-converted nuclei,10 Liang et al. reported that a greater number of amyloid assemblies emerge from within larger micelle aggregates than from smaller nucleation sites.11 Xia et al. also suggested that the formation of micelles or micelle-like aggregates precedes the assembly of SELP fibers.12 Presumably, the high density of proteins in micelle aggregates is the key factor that is responsible for the conversion of soluble molecules to solid nanofibers. Consistent with those studies, scanning electron microscopy (SEM) analysis shows that many a nanofiber extends from SELP-47 K aggregates [Fig. 1c]. Transmission electron microscopy (TEM) analysis also suggests that a number of nanofibers may be initiated at a preexisting seed [Fig. 1d]. Previously, we reported that addition of precrystallized SELP-47 K greatly accelerates the gelation process of aqueous solutions.5 This is because precrystallized SELP-47 K as preexisting seeds can initiate nanofiber assemblies faster than micelle-converted nuclei. Still, the formation of numerous individual nanofibers was observed in SELP-47 K solutions.13 It is possible that those short nanofibers were initiated at micelle-converted nuclei or directly from free chains, which have a much higher number density in SELP-47 K solutions, in comparison to preexisting seeds and micelle aggregates.
Once polypeptide nanofibers are initiated, reaction and diffusion will dictate fiber growth.4 Reaction consists of binding of soluble molecules to preformed fibers and conversion of bound molecules to solid filaments. In many self-assembling peptides, molecular binding precedes conformational change.14 Binding of soluble SELP molecules to preformed nanofibers is mediated by electrostatic attraction. The head and tail tags of SELP contain both positive and negative charges, and the complementary charges renders the ends of SELP “sticky,” enabling the binding of soluble SELP molecules to preformed nanofibers. To support fiber growth, soluble molecules need to be transported to the vicinity of growing tips of nanofibers via diffusion. The growth rate of nanofibers may be limited by reaction or diffusion.4 Here, we propose that molecular diffusion may interplay with nanofiber nucleation and subsequently define the spatial arrangement of SELP nanofibers on the microscale.
As illustrated in Fig. 2a, different nucleation sites possess different abilities to initiate fiber assemblies. In particular, larger micelle aggregates initiate the growth of more nanofibers than smaller nucleation sites such as micelle-converted nuclei. Highly heterogeneous fiber assemblies initiated at nucleation sites of different sizes can alter the diffusion patterns of soluble SELP and the spatial distribution of nanofiber assemblies. To support the growth of a single nanofiber at an average rate (the number of monomers per time), soluble SELP will be added to the fiber at a monomer rate ; if a nucleation site initiates the simultaneous growth of N nanofibers at an average rate , soluble SELP will be added to the fibers at a higher monomer rate . Soluble SELP can be added to growing nanofiber from a local population or from surrounding areas via diffusion. When the local population of soluble SELP is being depleted, the growth of a single nanofiber will create a concentration gradient and induce an influx of soluble SELP toward the tip of the growing fiber. Following a detailed mathematical model,13 the influx is
| (1) |
Here, r is the distance from the fiber tip. When a larger nucleation site initiates the simultaneous growth of N nanofibers, a stronger concentration gradient will be created and the influx of SELP will be increased by N-fold
| (2) |
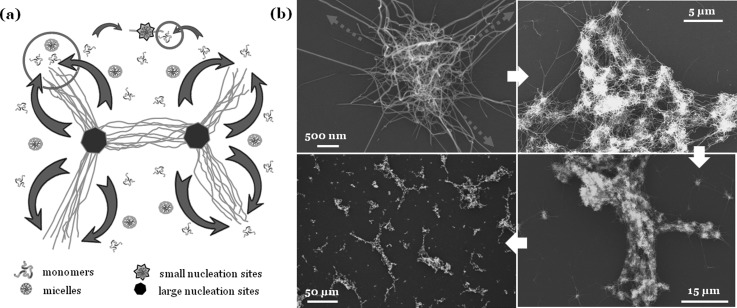
Figure 2.
(a) A model is proposed for diffusion-regulated nanofiber assembly. An isolated growing nanofiber or a small fiber cluster initiated at a small nucleation site functions as a moving sink (represented by a small circle), attracting a small number of soluble SELP-47 K, and a large fiber cluster initiated at a large nucleation site functions as a powerful vacuum (represented by a large circle), attracting a large number of soluble SELP-47 K. The diffusion-regulated assembly of nanofibers and the subsequent formation of large fiber structures were observed in SELP-815 K (b) and SELP-47 K (c).
Figuratively, isolated growing nanofibers and smaller clusters of growing nanofibers function as a smaller moving sink, attracting a smaller number of soluble SELP molecules; larger clusters of growing nanofibers function as a more powerful vacuum, removing a greater number of soluble molecules from the solution. Consequently, water-rich microdomains will be generated and left behind, leading to formation of micrometer-sized voids.
The proposed diffusion-regulated formation of microscale nanofiber assemblies was observed in SELP-47K13 and SELP-815 K [Fig. 2b] aqueous solutions. As illustrated in Fig. 2b, numerous nanofibers are formed from and randomly oriented within micelle aggregates that are about 2 μm in size, after 24 h of incubation at 37 °C; in the outlet of an aggregate, however, most fibers are aligned and extended in a few directions. This observation is consistent with the proposed model of nanofiber growth. As the density of SELP molecules outside micelle aggregates drops greatly, a fiber cluster can gain comparative advantages at attracting soluble SELP if nanofibers grow preferentially in a few directions than uniformly in all directions. Notably, nanofibers emerging from two clusters that are several micrometers apart can grow toward each other, linking the two clusters together. Further, many clusters are linked together, forming nanofiber micro-assemblies that are more than 100 μm in length and 10 ∼ 20 μm in width. In comparison to individual fiber clusters, the microscale nanofiber assemblies possess a greater ability to attract soluble SELP. As soluble SELP molecules flow toward the nanofiber micro-assemblies, isolated nanofibers and small fiber clusters may also move toward the fiber micro-assemblies, join them, and enhance their ability to attract soluble SELP. Consequently, the growth of remaining isolated fiber clusters or nanofibers would be greatly suppressed. Removal of soluble molecules, isolated nanofibers, and small fiber clusters from the solution creates SELP-free microdomains, a microscale feature of the microscale nanofiber assemblies. Many microscale nanofiber assemblies are branched, suggesting that interconnected macroscopic nanofiber structures may be formed in SELP aqueous solutions at higher concentrations.
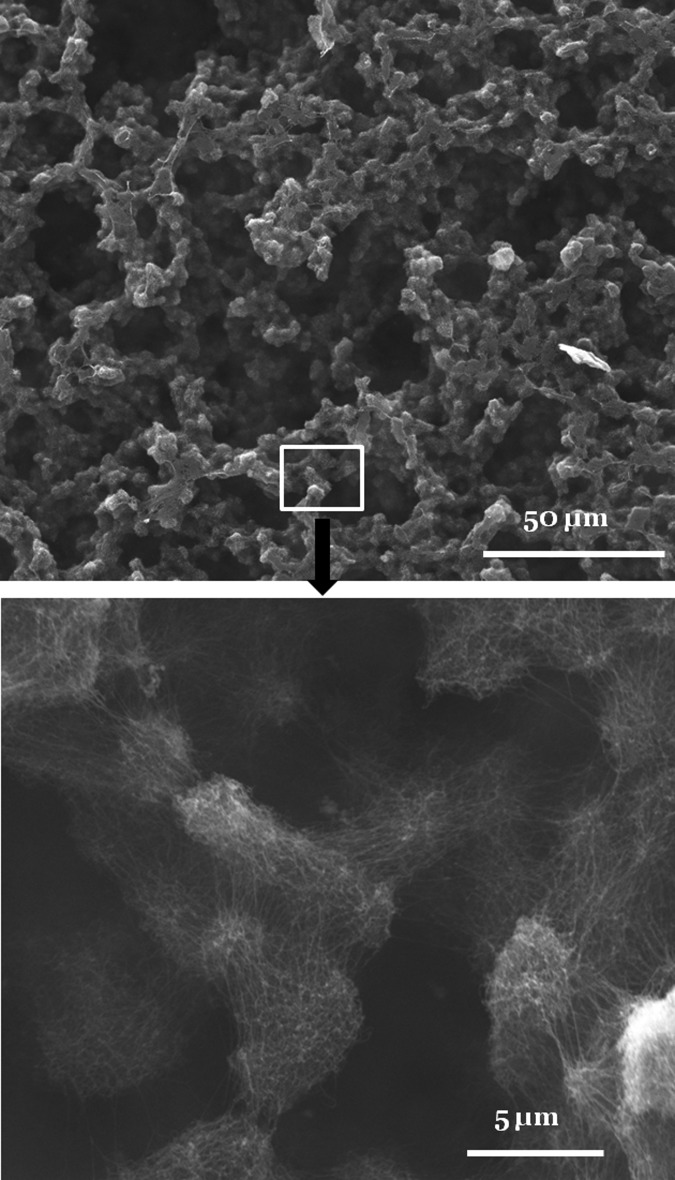
To examine the ability of SELPs to form even larger three-dimensional (3D) nanofiber structures, concentrated solutions were prepared. At a concentration of 5 mg/ml, SELP-47 K aqueous solution formed weak nanofiber hydrogels. At a concentration of 10 mg/ml (i.e., 1% w/w), a typical concentration for preparing peptide nanofiber scaffolds for tissue regeneration,1, 15 mechanically robust SELP-47 K nanofiber scaffolds with distinct nanoscale and microscale features were self-assembled [Fig. 3]. On the microscale, the nanofiber scaffold consists of two continuous phases: a water-rich phase that appears as interconnected voids of 20 ∼ 40 μm in size on dry samples, and a fiber-rich phase that is comprised of interconnected fiber clusters of 2 ∼ 10 μm in size. On the nanoscale, SELP-47 K nanofibers are randomly oriented and densely packed in the fiber clusters, and inter-fiber spacing varies from several to a few tens of nanometers. Formation of 3D nanofiber scaffolds was also observed in a 15 mg/ml SELP-815 K aqueous solution.13 Such 3D nanofiber scaffolds with distinct nanoscale and microscale features are very appealing for applications in tissue or cell engineering, as the polypeptide nanofibers enable the cells to interact with the matrix in 3D microenvironments and the micrometer-sized voids facilitate cell infiltration into the scaffolds. Further, the SELP nanofiber scaffolds are assembled in physiologically relevant conditions, including the use of water as a solvent and the incubation of polypeptide solutions at 37 °C. This makes the self-assembling process compatible with simultaneous cell seeding.
Figure 3.
SEM images of SELP-47 K nanofiber structures assembled in 10 mg/ml aqueous solutions. Self-assembled SELP-47 K nanofibers form micrometer-sized clusters, which further assemble into macroscopically continuous structures with defined microscale order.
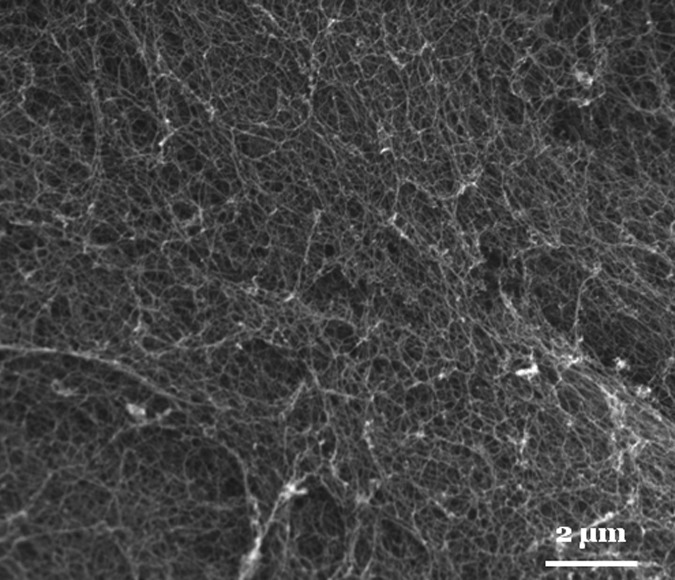
The key to the self-assembly of SELP nanofiber scaffolds with both nanoscale and microscale features is the interplay of fiber nucleation with molecular diffusion. To further test this diffusion-regulated assembly of SELP nanofiber scaffolds, we increased the concentration of SELP-47 K solutions and examined the self-assembly of concentrated solutions. At a concentration of 20 mg/ml, nanofiber clusters increase in size but micrometer-sized voids decrease in size.13 At a concentration of 40 mg/ml, SELP-47 K solutions self-assemble into relatively uniform nanofiber matrices with sub-micrometer-sized voids [Fig. 4], which are similar to other self-assembling peptide nanofiber structures.1, 15 Presumably, sufficient micelle aggregates exist in the SELP-47 K solution at this concentration, initiating the simultaneous growth of nanofibers uniformly; there are enough soluble molecules in the solution to support the growth of nanofibers in all directions. As a result, there is no driving force for the alignment of nanofibers in certain directions to enhance comparative advantages at attracting soluble SELP; soluble molecules are added to growing nanofibers locally; no flux of SELP occurs over a distance of several micrometers; and thus no micrometer-sized voids are generated.
Figure 4.
SEM images of uniform SELP-47 K nanofiber structures assembled in 40 mg/ml aqueous solutions.
To summarize, the interplay of nanofiber nucleation and molecular diffusion leads to the preferential growth of large fiber clusters that are likely initiated within micelle aggregates in SELP aqueous solutions. Consequently, three-dimensional nanofiber scaffolds with distinct nanoscale and microscale features are self-assembled at appropriate SELP concentrations (e.g., 1% ∼ 2% w/w). Studies to date have revealed how fiber nucleation, molecular diffusion and binding, and conformation conversion affect the self-assembly of protein or peptide nanofibers.4 Little is known about how the interplays of these events may define the spatial organization of nanofibers. Previously, it was reported that the silk-like sequences of SELPs can be spontaneously transformed into β-sheet crystals, critical to nanofiber self-assembly.16 Further, mechanical forces greatly improve the conversion of bound SELP molecules into nanofibers,17 and physical treatments such as autoclaving also facilitate conformational conversion in SELP nanofibers.18 In this study, we demonstrated the way the diffusion-regulated self-assembly defines the microarchitectures of SELP nanofiber scaffolds. Notably, the SELP nanofiber scaffolds with distinct nanoscale and microscale architectures may find important applications in cell and tissue engineering.
Acknowledgments
This work was supported by the US National Institutes of Health (EB009160, EB009801) and National Science Foundation (CMMI0856215).
References
- Holmes T. C., de Lacalle S., Su X., Liu G. S., Rich A., and Zhang S. G., Proc. Natl. Acad. Sci. U.S.A. 97(12), 6728 (2000). 10.1073/pnas.97.12.6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. H., Tejeda-Montes E., Poch M., and Mata A., Chem. Soc. Rev. 40(9), 4563 (2011). 10.1039/c1cs15064b [DOI] [PubMed] [Google Scholar]
- Dong H., Paramonov S. E., and Hartgerink J. D., J. Am. Chem. Soc. 130(41), 13691 (2008); 10.1021/ja8037323 [DOI] [PubMed] [Google Scholar]; Papapostolou D., Smith A. M., Atkins E. D. T., Oliver S. J., Ryadnov M. G., Serpell L. C., and Woolfson D. N., Proc. Natl. Acad. Sci. U.S.A. 104(26), 10853 (2007); 10.1073/pnas.0700801104 [DOI] [PMC free article] [PubMed] [Google Scholar]; Smith A. M., Banwell E. F., Edwards W. R., Pandya M. J., and Woolfson D. N., Adv. Funct. Mater. 16(8), 1022 (2006); 10.1002/adfm.v16:8 [DOI] [Google Scholar]; von Maltzahn G., Vauthey S., Santoso S., and Zhang S. U., Langmuir 19(10), 4332 (2003). 10.1021/la026526+ [DOI] [Google Scholar]
- Howard J., Mechanics of Motor Proteins and the Cytoskeleton (Sinauer Associates, Publishers, Sunderland, Mass., 2001), p. xvi. [Google Scholar]
- Cappello J., Crissman J. W., Crissman M., Ferrari F. A., Textor G., Wallis O., Whitledge J. R., Zhou X., Burman D., Aukerman L., and Stedronsky E. R., J. Controlled Release 53(1-3), 105 (1998). 10.1016/S0168-3659(97)00243-5 [DOI] [PubMed] [Google Scholar]
- Teng W. B., Cappello J., and Wu X. Y., Biomacromolecules 10(11), 3028 (2009). 10.1021/bm900651g [DOI] [PubMed] [Google Scholar]
- Dandu R., Von Cresce A., Briber R., Dowell P., Cappello J., and Ghandehari H., Polymer 50(2), 366 (2009). 10.1016/j.polymer.2008.11.047 [DOI] [Google Scholar]
- Qiu W. G., Huang Y. D., Teng W. B., Cohn C. M., Cappello J., and Wu X. Y., Biomacromolecules 11(12), 3219 (2010). 10.1021/bm100469w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Thevenot J., Ibarboure E., Lecommandoux S., and Chaikof E. L., Angew. Chem. Int. Ed. 49(25), 4257 (2010). 10.1002/anie.201001356 [DOI] [PubMed] [Google Scholar]
- Lomakin A., Teplow D. B., Kirschner D. A., and Benedek G. B., Proc. Natl. Acad. Sci. U.S.A. 94(15), 7942 (1997). 10.1073/pnas.94.15.7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y., Lynn D. G., and Berland K. M., J. Am. Chem. Soc. 132(18), 6306 (2010). 10.1021/ja910964c [DOI] [PubMed] [Google Scholar]
- Xia X. X., Xu Q. B., Hu X., Qin G. K., and Kaplan D. L., Biomacromolecules 12(11), 3844 (2011). 10.1021/bm201165h [DOI] [PMC free article] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.4863077 for additional data on nanofiber self-assembly and a detailed mathematical model of soluble SELP diffusion.
- Scheibel T., Bloom J., and Lindquist S. L., Proc. Natl. Acad. Sci. U.S.A. 101(8), 2287 (2004). 10.1073/pnas.0308754101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. G., Nat. Biotechnol. 21(10), 1171 (2003). 10.1038/nbt874 [DOI] [PubMed] [Google Scholar]
- Hwang W., Kim B. H., Dandu R., Cappello J., Ghandehari H., and Seog J., Langmuir 25(21), 12682 (2009). 10.1021/la9015993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Peng X. F., Hijji K., Cappello J., Ghandehari H., Solares S. D., and Seog J., J. Am. Chem. Soc. 133(6), 1745 (2011). 10.1021/ja110191f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W. G., Cappello J., and Wu X. Y., Appl. Phys. Lett. 98(26), 263702 (2011). 10.1063/1.3604786 [DOI] [PMC free article] [PubMed] [Google Scholar]