Abstract
Nascent untranslated transcripts in bacteria are prone to generating RNA-DNA hybrids (R-loops); Rho-dependent transcription termination acts to reduce their prevalence. Here we discuss the mechanisms of R-loop formation and growth inhibition in bacteria.
Keywords: R-loop, Rho, NusG, transcription termination, backtracking
Introduction and Historical Perspective
The term R-loop refers to a nucleic acid structure that is comprised of single-stranded RNA base-paired with one strand of duplex DNA leading to displacement of the second (complementary) DNA strand. This nomenclature derives from the term “D-loop,” which describes an intermediate structure that is formed during recombination and is comprised of two strands of DNA base-paired with one another and a third displaced DNA strand. R-loops are comprised of one strand of RNA and two of DNA.1 R-loops can occur either during replication, when DnaG (or an equivalent) primase synthesizes the RNA primer for Okazaki fragments,2 or from transcripts synthesized by RNA polymerase (RNAP). The focus of this article is limited to transcription-associated R-loops in bacterial cells.
Historically, a role for R-loops in bacterial physiology was first invoked by Tomizawa’s group in the 1980s, when they showed that, during replication of the ColE1 family of plasmids, the plasmid-encoded RNA-II transcript forms an R-loop, which is cleaved by the enzyme RNase HI (that targets RNA exclusively in RNA-DNA hybrids) to generate the 3′-OH end needed for initiation of DNA synthesis.3,4 Subsequently, Kogoma’s group provided evidence that E. coli cells deficient for either RNase HI or RecG (a helicase that is able to unwind R-loops) exhibit a phenomenon referred to as constitutive stable DNA replication (cSDR), in which transcription-generated R-loops mediate the establishment of replication forks for chromosomal DNA replication.1 Kogoma and colleagues also showed that mutants defective for both RNase HI and RecG are inviable, from which they inferred that R-loops are constantly being generated in E. coli and are lethal unless they are removed.1 Finally, since R-loops constrain DNA supercoils, their formation may be expected to be promoted by increased negative supercoiling of DNA; work from Drolet’s group5,6 has convincingly demonstrated that transcription-associated R-loop formation is a major contributor to sickness in topoisomerase I-deficient mutants of E. coli.
R-loop Mechanisms: The Questions
The studies mentioned above have presented a fairly simple picture of R-loops: they are formed by the heteroduplexing of RNA transcripts with their cognate DNA template strands, and are toxic when they occur in excess. However, the devil, as they say, is in the detail, and the following questions are of interest in understanding R-loop mechanisms (see Figure 1).
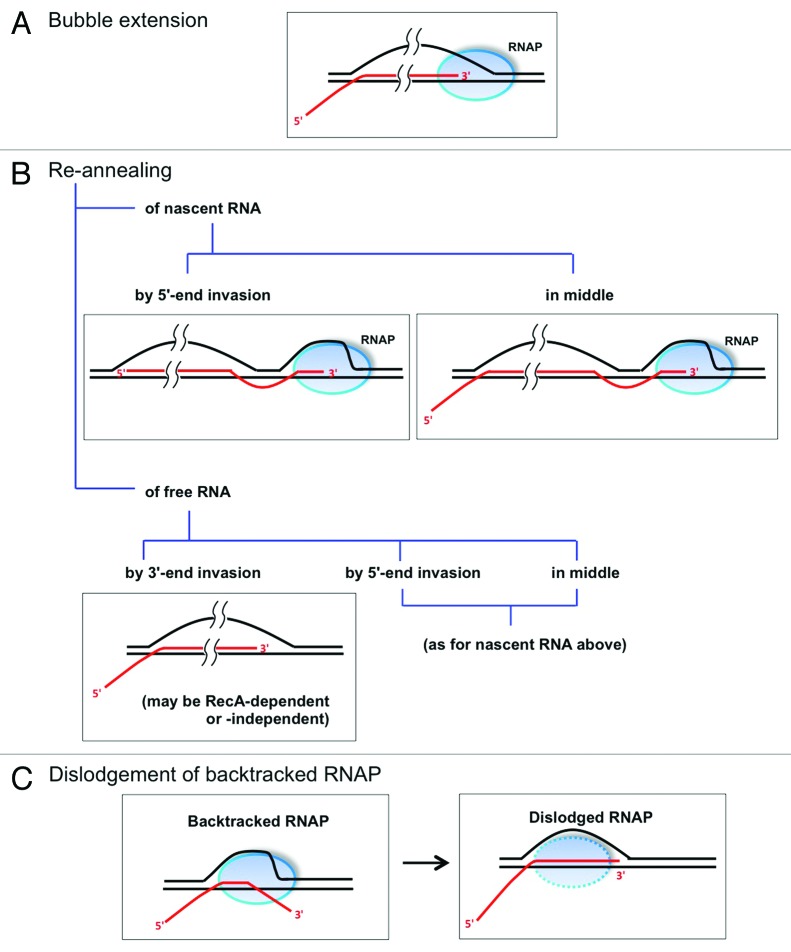
Figure 1. Mechanisms and models for R-loop generation during bacterial transcription. Three models of R-loop formation are schematically depicted, as explained in the text: (A) bubble extension, (B) re-annealing of nascent or free transcripts and, (C) following dislodgement of backtracked RNAP. DNA, RNA and RNAP are shown in black, red and blue, respectively. Direction of transcription elongation complex movement is from left to right in all panels.
1. How does an RNA transcript insert itself into duplex DNA to generate the R-loop? There are two dominant models that are called the “bubble extension” and “re-annealing” models, respectively. (Some authors7 have also referred to them synonymously as the “extended hybrid” and “thread back” models, respectively.) In the former (Fig. 1A), the RNA synthesized within the transcription elongation complex remains associated as a persistent RNA-DNA hybrid, even as the complex continues to translocate catalytically along DNA (that is, the RNA does not undergo the normal process8 of its separation from DNA, beyond the short stretch of 8 to 9 nucleotides at its 3′-end, to emerge through the exit channel of RNAP). Tomizawa’s group3 had suggested this mechanism for formation of the persistent RNA-DNA hybrid from the RNA-II transcript during ColE1 plasmid replication, but this was before the atomic structure of RNAP had been determined. In light of the structural information now available,8 it is difficult to envisage how the mechanism to separate RNA from DNA within the enzyme can be bypassed. In the second model (Fig. 1B), the R-loop is formed not as an extension of the transcription bubble but by the re-annealing of RNA with its cognate template DNA strand. More recently, a third model of R-loop formation has also been proposed,9,10 by which a longer RNA-DNA hybrid (about 25 bp) than that in a normal transcription bubble can be formed after dislodgement of a backtracked RNAP (Fig. 1C); this model is further discussed in a subsequent section below.
2. In a re-annealing model of R-loop formation, how is the RNA strand inserted into DNA? Three mechanistic possibilities may be envisaged (Fig. 1B): 3′-end invasion, 5′-end invasion, or insertion from the middle, of the RNA molecule into duplex DNA. Of these, the 3′-end invasion mechanism has been proposed to occur for cSDR, wherein the R-loop is believed to be formed much like the D-loop, with the 3′-end of RecA-bound nucleic acid (RNA or DNA) invading into duplex DNA;1 5′-end invasion has been suggested as the mechanism for R-loop formation in Rho-deficient strains, as further discussed below. The third possibility, insertion of the middle of an RNA molecule into DNA, has been schematically depicted5 as an apparent mechanism of R-loop generation in topoisomerase I-deficient E. coli, but it would require the action of a nicking-closing enzyme that can act on RNA-DNA hybrids (the existence of such an enzyme is currently unknown). On the other hand, an endonucleolytic cleavage within such RNA would enable R-loop formation by the 3′- or 5′-end invasion approaches.
3. Further, in a re-annealing model, does the RNA substrate exist as a nascent transcript (that is, still bound to RNAP) or as a free transcript in the cytoplasm? For a nascent transcript, the 3′-end invasion possibility for R-loop generation will not apply since the RNA 3′-end is held within the RNAP. Additionally, in accord with the twin-supercoiling domain concept of transcription elongation,11 it is the nascent transcript alone that will be associated with a region of negative supercoiling behind the moving RNAP, which could act to facilitate R-loop formation.
4. Is R-loop formation RecA-dependent or -independent? More generally, are there factors that directly modulate the efficiency of R-loop formation? As mentioned above, the proposal for cSDR is that it is RecA-dependent.1
5. Is the propensity for R-loop formation dictated by nucleotide sequence? The answer would appear to be in the affirmative, with G-rich segments in the RNA being conducive for both the nucleation and extension of the RNA-DNA hybrid.12
6. Finally, since transcription and translation are coupled to each other and occur in a single cytoplasmic compartment in bacteria, what is the effect (if any) of ribosomal engagement of the nascent and/or free transcripts on R-loop formation? Drolet and colleagues6 have provided evidence that the frequency of R-loops is increased in topoisomerase I-deficient mutants when translation is inhibited, suggesting that ribosome-free transcripts are more prone to generating R-loops.
R-loops and Rho-dependent Transcription Termination
It is in the context of these questions and models that we wish to discuss the findings of our recent paper13 on the role of Rho-dependent transcription termination (RDTT) in the avoidance of excessive R-loops in E. coli. RDTT is a process14,15 in which the Rho protein binds a nascent unstructured transcript that is not being simultaneously translated, and then acts to terminate transcription. Another protein that participates in RDTT is NusG, and both Rho and NusG are individually essential for viability in E. coli.
We had earlier proposed16,17 a unitary model in which: (i) nascent transcripts in bacteria are inherently prone to re-annealing with upstream DNA to generate growth-inhibitory R-loops; (ii) two general mechanisms that act to preclude such R-loop generation are transcription-translation coupling (for mRNAs of protein-coding genes) and secondary structure formation in RNA (e.g., in rRNA) and; (iii) when a nascent unstructured transcript fails to be engaged by ribosomes for translation, RDTT acts to abrogate synthesis of the transcript so that the substrate for R-loop formation itself is not available. This model predicts that R-loops would occur at increased frequency in mutants that are deficient for RDTT; several indirect lines of genetic evidence have been obtained by us earlier in support of this prediction.16,18
The results from our recent study13 have shown conclusively that the essential role of RDTT is the prevention of excessive genome-wide R-loops in E. coli. One of the assays for detection of R-looped regions exploits the sensitivity to sodium bisulfite of the displaced non-template DNA strand that results in C-to-T changes in nucleotide sequence of the latter.7,12 With the aid of this assay, we could show that R-loops occur genome-wide at a basal level in wild-type E. coli, and that the frequency of their occurrence is elevated in a mutant deficient for RDTT.13 Further, this study also established that excessive R-loops are indeed responsible for the lethality associated with complete absence of Rho or NusG in wild-type E. coli, since such lethality could be rescued by expression of UvsW, an R-loop helicase from phage T4.19
Thus, the model that RDTT acts as a back-up measure to prevent the default occurrence of R-loops from nascent untranslated transcripts is strongly supported by the new findings. This is important, not just because of the value of the model’s intrinsic message, but also because many of the questions regarding R-loop mechanisms can now be more definitively answered (at least with respect to those R-loops whose formation is modulated by RDTT). For example, the following mechanisms suggested to operate in R-loop formation are excluded in this model (since, in these cases, RDTT would not be able to contribute to reduced R-loop occurrence): bubble extension, 3′-end invasion, the involvement of free (as opposed to nascent) transcripts, and RecA-dependence. The last is excluded because our studies have indicated that R-loop formation in RDTT-defective mutants is equally feasible in both RecA-proficient and -deficient cells.13,16 Furthermore, clues to the nature of nucleotide sequence preferences for R-loop generation in vivo are likely to be available from the bisulfite-sensitive hotspots that have been identified in the genome-wide mapping studies.13
Given that Rho function is correlated with reduced R-loop occurrence in E. coli, is it possible that it does so, not by preventing their occurrence, but through its enzymatic activity as a 5′-3′ RNA-DNA helicase15 (that is, by unwinding R-loops which, under this scenario, could have arisen from nascent or free transcripts and by any of the three insertion mechanisms described above)? We think it unlikely because, if true, it would represent a role for Rho where it acts independently of NusG, since there is no evidence that NusG can directly stimulate Rho’s helicase activity; on the other hand, our results have shown that the increased occurrence of R-loops is manifest in both Rho- and NusG-deficient mutants.13,16,18
Continuing with the assumption that R-loops detected by the bisulfite-sensitivity assay (even in wild-type cells) are a proxy for the occurrence of nascent untranslated transcripts in E. coli, we may infer that widespread low-level transcription from both DNA strands exists across the entire genome, and that this may account for around 5% of all transcription within the cells.13 Such an inference is well supported by the recent findings of Peters et al.20 for pervasive antisense transcription in E. coli that is suppressed by RDTT. These observations are reminiscent also of those from the ENCODE project,21 which have demonstrated that > 70% of the human genome is represented in primary transcripts of different cells.
If excessive R-loops are lethal and RDTT acts to reduce their occurrence, why is Rho dispensable in some bacteria, for example, Bacillus subtilis and Staphylococcus aureus?15 One possibility is that in such bacteria, the enzymes for R-loop removal, such as RNase HI and RecG, are more active than they are in E. coli.
R-loops, RNAP Backtracking, and Transcription-Replication Conflict
In any discussion on R-loops, it is important to consider their inter-relationship with transcription-replication conflicts and with backtracked RNAP. Using E. coli mutants in which rRNA operon transcription was oriented head-on against replication fork direction, Michel and coworkers22 showed that R-loops are generated at the collision loci, which are toxic unless they are removed; the toxicity is associated with replication fork stalling and/or collapse. Several other studies9,13,23 have also concluded that R-loops cause problems for replication, although Aguilera and colleagues24 have suggested that R-loop toxicity is associated with transcription elongation impairment.
Evidence for the notion that RNAP backtracking is associated with R-loop formation comes from several different observations. First, the lethality of E. coli mutants lacking Rho or NusG is rescued by a mutation in RNAP that renders the enzyme resistant to backtracking.13 Second, the toxicity of R-loop-associated transcription-replication conflicts in strains with inverted rRNA operons is also alleviated by an apparently similar kind of RNAP mutation.22 Finally, Dutta et al.9 have shown that transcription that is not coupled to translation is associated with RNAP backtracking as well as replication-dependent generation of DNA double-strand ends, with R-loops being implicated in the process, since it is reversed upon overexpression of RNase HI.
It therefore appears that RNAP backtracking and R-loop formation are intimately correlated; this notion is supported also by the model that the leading ribosome on a nascent transcript is in physical contact (via NusG) with RNAP to promote transcription elongation and to prevent the latter from entering a backtracked conformation.25,26 Whether it is backtracking that promotes R-loop formation, or R-loops that promote backtracking, or both, remains to be determined. A related question will be whether transcription-replication conflicts promote R-loops, or vice versa. The findings of Boubakri et al.22 with the inverted rRNA operons do provide clear support for the former, whereas those of Dutta et al.9 do the same for the latter. A model that envisages promotion of R-loop formation by backtracking will explain our earlier finding18 that, in mutants defective for RDTT, R-loops apparently provide a pathway of mRNA turnover that is an alternative to that catalyzed by RNase E.
To explain their observations of R-loop mediated and replication-dependent toxicity of backtracked RNAP, Nudler's group9,10 has proposed a model (Fig. 1C) in which (i) an RNA-DNA hybrid (up to 25 bp long) is formed (following dislodgement of a backtracked RNAP by a co-directional replication fork) through restoration of the register between the 3′-end of the nascent transcript and DNA and; (ii) re-initiation of replication from the RNA 3′-end results in a residual nick in the leading strand that is converted into a double strand end upon passage of a subsequent replication fork through this region. This model represents the third possible mechanism of R-loop generation (in addition to the bubble extension and re-annealing mechanisms described above). However, this model will not account for R-loops several hundreds of bp long that have been detected in both bacteria and eukaryotes,4,7,12 as is also evident from our genome-wide bisulfite sensitivity studies.13 We therefore suggest that the R-loops associated with RNAP backtracking are generated by 5′-end invasion and re-annealing of nascent transcripts.
Beneficial Effects of R-loops?
Given that a basal level of genome-wide R-loops exists even in wild-type E. coli, the possibility should be considered whether, at this level of occurrence, they confer a beneficial effect to the cells.27 Thus, they may assist in the mechanisms of DNA damage repair, for example by serving to bridge double-strand breaks in the DNA or by priming replication restart through cSDR. It is interesting to note in this context that R-loop “annealases,” that is, enzymes that promote the invasion of RNA into a DNA duplex, appear to exist in E. coli.28
R-loops in Eukaryotic Transcription
Finally, the structure of RNAP as well as the processes of transcription initiation and elongation are quite well conserved across bacteria and eukaryotes.8,29 It is not unusual, therefore, that pathological R-loops occur also in eukaryotic cells, arising from nascent transcripts that fail to be engaged by proteins in co-transcriptional processes, such as splicing, polyadenylation or export from the nucleus into the cytoplasm.30 Physiologically, R-loops have been shown in cells of the immune system to participate in class switch recombination and somatic hypermutation7,12 and, in this case too, evidence for a re-annealing mechanism of R-loop formation has been obtained.7
Concluding Remarks
In summary, transcription-associated R-loops are a feature of all living cells. In E. coli, they apparently arise by 5′-end invasion and re-annealing of nascent untranslated transcripts across the entire genome, and the frequency of their occurrence is kept in check by RDTT. Although R-loops do serve some physiological functions, such as during plasmid replication and cSDR in bacteria and for immune cell maturation in eukaryotes, for the most part they are pathological and give rise to transcription-replication conflicts and genome instability. We believe that the stage is now set for a more comprehensive understanding of how R-loops are generated and of the consequences of their occurrence, in both bacteria and eukaryotic cells.
Acknowledgments
Work in the authors’ laboratory is supported by a Centre of Excellence in Microbial Biology research grant of the Department of Biotechnology, and J.G. is recipient of the J. C. Bose Fellowship of the Department of Science and Technology, Government of India.
Glossary
Abbreviations:
- bp
base-pairs
- RDTT
Rho-dependent transcription termination
- RNAP
RNA polymerase
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/25101
References
- 1.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–38. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg A, Baker T. DNA replication. W.H.Freeman, New York 1992. [Google Scholar]
- 3.Itoh T, Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci U S A. 1980;77:2450–4. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selzer G, Tomizawa JI. Specific cleavage of the p15A primer precursor by ribonuclease H at the origin of DNA replication. Proc Natl Acad Sci U S A. 1982;79:7082–6. doi: 10.1073/pnas.79.23.7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massé E, Drolet M. R-loop-dependent hypernegative supercoiling in Escherichia coli topA mutants preferentially occurs at low temperatures and correlates with growth inhibition. J Mol Biol. 1999;294:321–32. doi: 10.1006/jmbi.1999.3264. [DOI] [PubMed] [Google Scholar]
- 6.Massé E, Drolet M. Escherichia coli DNA topoisomerase I inhibits R-loop formation by relaxing transcription-induced negative supercoiling. J Biol Chem. 1999;274:16659–64. doi: 10.1074/jbc.274.23.16659. [DOI] [PubMed] [Google Scholar]
- 7.Roy D, Yu K, Lieber MR. Mechanism of R-loop formation at immunoglobulin class switch sequences. Mol Cell Biol. 2008;28:50–60. doi: 10.1128/MCB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geszvain K, Landick R. The structure of bacterial RNA polymerase. In: Higgins NP, ed. The Bacterial Chromosome. Washington DC: ASM Press 2005:283-96. [Google Scholar]
- 9.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–43. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–45. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roy D, Lieber MR. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol Cell Biol. 2009;29:3124–33. doi: 10.1128/MCB.00139-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leela JK, Syeda AH, Anupama K, Gowrishankar J. Rho-dependent transcription termination is essential to prevent excessive genome-wide R-loops in Escherichia coli. Proc Natl Acad Sci U S A. 2013;110:258–63. doi: 10.1073/pnas.1213123110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peters JM, Vangeloff AD, Landick R. Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol. 2011;412:793–813. doi: 10.1016/j.jmb.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rabhi M, Rahmouni AR, Boudvillain M. Transcription termination factor Rho: a ring shaped RNA helicase from bacteria. In: Jankowsky E, ed. RNA Helicases. Cambridge UK, RSC publishing 2010: 243-71. [Google Scholar]
- 16.Harinarayanan R, Gowrishankar J. Host factor titration by chromosomal R-loops as a mechanism for runaway plasmid replication in transcription termination-defective mutants of Escherichia coli. J Mol Biol. 2003;332:31–46. doi: 10.1016/S0022-2836(03)00753-8. [DOI] [PubMed] [Google Scholar]
- 17.Gowrishankar J, Harinarayanan R. Why is transcription coupled to translation in bacteria? Mol Microbiol. 2004;54:598–603. doi: 10.1111/j.1365-2958.2004.04289.x. [DOI] [PubMed] [Google Scholar]
- 18.Anupama K, Leela JK, Gowrishankar J. Two pathways for RNase E action in Escherichia coli in vivo and bypass of its essentiality in mutants defective for Rho-dependent transcription termination. Mol Microbiol. 2011;82:1330–48. doi: 10.1111/j.1365-2958.2011.07895.x. [DOI] [PubMed] [Google Scholar]
- 19.Dudas KC, Kreuzer KN. UvsW protein regulates bacteriophage T4 origin-dependent replication by unwinding R-loops. Mol Cell Biol. 2001;21:2706–15. doi: 10.1128/MCB.21.8.2706-2715.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–33. doi: 10.1101/gad.196741.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boubakri H, de Septenville AL, Viguera E, Michel B. The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo. EMBO J. 2010;29:145–57. doi: 10.1038/emboj.2009.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, et al. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev. 2011;25:2041–56. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tous C, Aguilera A. Impairment of transcription elongation by R-loops in vitro. Biochem Biophys Res Commun. 2007;360:428–32. doi: 10.1016/j.bbrc.2007.06.098. [DOI] [PubMed] [Google Scholar]
- 25.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–8. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, et al. A NusE:NusG complex links transcription and translation. Science. 2010;328:501–4. doi: 10.1126/science.1184953. [DOI] [PubMed] [Google Scholar]
- 27.Camps M, Loeb LA. Critical role of R-loops in processing replication blocks. Front Biosci. 2005;10:689–98. doi: 10.2741/1564. [DOI] [PubMed] [Google Scholar]
- 28.Ivancic-Bace I, Radovcic M, Bockor L, Howard JL, Bolt EL. Cas3 stimulates runaway replication of a ColE1 plasmid in Escherichia coli and antagonises RNase HI. RNA Biol. 2013;10:1–9. doi: 10.4161/rna.23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebright RH. RNA polymerase: structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J Mol Biol. 2000;304:687–98. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 30.Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]