Abstract
Non-canonical cytoplasmic activities and signal transduction of retinoic acid (RA) expand RA’s pleiotropic effects in coordinating the epigenome in embryonic stem cell (ESC). Examples include RA-bound cellular retinoic acid binding protein I, which activates ERK2. By engaging both cytosolic and nuclear mediators, RA can efficiently augment ESC’s epigenome.
Keywords: retinoic acid, epigenetic, embryonic stem cell, OCT4, CRABPI, ERK, TR2
Introduction
Maintaining healthy embryonic stem cells (ESCs) in culture requires delicate procedures to ensure their unlimited proliferation with pluripotency, a “stemness” feature unique to transiently existing cells in pre-implantation embryos. The “stemness” property of ESCs critically depends on tight control of the expression of a core group of four transcription factors, octamer-binding transcription factor 4 (OCT4, also named POU5F1), NANOG, SRY-box 2 (SOX2) and Kruppel-like factor 4 (KLF4).1-3 OCT4 protein expression, particularly, should be maintained at a constant, balanced level; over- or under-expression of OCT4 results in the loss of stemness.4-6 This is controlled by tight regulation of the Oct4 gene transcription program. To this end, extracellular factors and intracellular signal transduction pathways can modulate chromatin remodeling machinery and/or transcription complexes of many gene loci, including the Oct4 gene locus, and, therefore, are important in the epigenetic control of the ESC genome.7-9 Leukemia inhibitory factor (LIF) is the key signal to maintain pluripotency, while various growth factors/hormones, such as FGF/TGFβ/activin and retinoic acid (RA), induce cell differentiation.1-3 In particular, RA is most commonly used to induce ESC for various differentiation processes because it acts on RA receptors (RARs) that are widely expressed in almost all tissue/cell types to regulate almost all the known cellular processes, such as differentiation, proliferation and apoptosis.10-12 The canonical activity and signaling pathway of RA and RARs in these cellular processes is well known, and it is to provide hormone/transcription factor complexes to regulate specific gene expression.13 But, recently, certain non-canonical cytoplasmic activities and signal transduction mechanisms of RA have begun to be noticed.14-18 While the physiological implication of these interesting new observations remains debatable, their potential deserves attention, especially in our attempt to understand the increasingly complex picture about epigenetic regulation of the ESC genome.
The Non-Canonical Activity and Signal Transduction of RA
RA, either entering cells through a paracrine route or generated in situ, first binds two members of the cytosolic receptors named cellular RA binding proteins (CRABPs) I and II. Classical studies have proposed and demonstrated different functional roles for these two CRABPs. CRABPI binding to RA is involved in its intracellular metabolism and, thus, is likely to be responsible, at least partially, for maintaining intracellular RA concentrations; CRABPII binding to RA delivers RA to the nucleus for RA to access RARs.19-21 In this canonical signal transduction pathway, CRABPII and nuclear RARs gate the intra-nuclear signal of RA. Therefore, it has been thought that RA signaling plays out only in the nucleus, and the epigenetic control of RA-targeted gene expression relies primarily on the actions of RARs and CRABPII.
Recently, two lines of studies revealed certain non-canonical, cytoplasmic activities and mediators of RA, which may contribute to additional roles for RA in epigenetic regulation. The first has to do with an interesting behavior of RARs: In certain cell types such as neurons, RARs can also cytoplasmically localize to dendrites where they can modulate local translation.22,23 Second, multiple studies have revealed that RA, rapidly and non-canonically, activates extracellular signal-regulated kinase 2 (ERK2),14,16,22,24 a process that appears to involve CRABPI, but not RAR.14 This latter observation is of particular interest regarding epigenetic control of ESC differentiation, because ESC commitment and differentiation critically depends upon the MAPK pathway, where ERK is one of the most important upstream kinases that respond to a wide range of ESC growth/differentiation factors.24,25 While the precise molecular details underlying RA/CRABPI-elicited ERK activity remain to be discerned, this observation has generated insights into the plasticity of ESC cultures. In the following, I summarize a tentative signaling pathway that has been constructed based upon new data to illustrate how RA can be an epigenetic factor that exploits both nuclear (canonical) and cytosolic (non-canonical) mechanisms to affect ESC proliferation/differentiation programs.
RA-CRABPI-Activated ERK Modulates Specific Transcription Factors for Epigenetic Regulation of Oct4 in ESC
Among the four core transcription factors important for ESC’s pluripotency, OCT4 is known as the master regulator. Studies of both ESC and induced pluripotent stem cell (iPSC) have established the requirement for OCT4 levels to be kept in a narrow window in order to maintain ESC pluripotency: either over- or under-expression of OCT4 can disturb normal propagation of ESC cultures.4,5 Chromatin studies show that certain gene loci, including the Oct4 gene locus, exist in a “bivalent” state in normally proliferating ESC cultures—although they are constitutively activated (transcribed), they can be rapidly repressed in response to certain signal inputs that trigger the loss of pluripotency.26,27
The key question is: how are bivalent loci guarded in proliferating ESCs? In other words, how do ESCs maintain the strictly required expression level of OCT4? Traditional molecular biology studies have attributed this control to the presence of positive and negative regulatory elements localized in the Oct4 gene promoter and enhancer, which provide binding sites for activators like testis receptor 2 (TR2), liver receptor homolog-1 (LRH-1) and steroidogenic factor -1 (SF-1) and for repressors like sumoylated TR2, RAR, germ cell nuclear factor (GCNF) and chicken ovalbumin upstream promoter transcription factors (COUP-TFs).16,18,28 A critical issue, then, is understanding how ESCs coordinate the dynamic and behavior of these activators and repressors in order to maintain the intricate balance between activation and repression of a specific gene locus like Oct4. Of particular importance is how the ESC epigenome is controlled to specify differentiation. This knowledge requires understanding the whole spectrum of epigenetic mechanisms that a cell can exploit to execute a specific and precise differentiation program. We have studied the orphan nuclear receptor TR2, as well as its intriguing response to RA, to illuminate the delicate balance of epigenetic mechanisms that can modulate the OCT4 level in ESC.
TR2 is most highly expressed in pre-implantation embryos.29 It regulates the Oct4 gene through binding to hormone response elements in its promoter, acting as an activator or a repressor depending upon the culture condition. In a normal ESC culture, TR2 is an activator and a proliferating factor; but in RA-treated ESC culture, TR2 becomes a repressor and a differentiation factor.18 Interestingly, TR2 does not bind to RA, making it intriguing how it can rapidly sense the RA signal to become a repressor. Also, sumoylation appears critical to TR2’s biological activity, because unsumoylated TR2 recruits coactivators such as p300/CBP associated factor (PCAF), whereas sumoylated TR2 preferentially recruits co-repressors, including receptor interacting protein 140 (RIP140).16,17 Relevant to the rapid effect of RA on TR2 is the finding that RA rapidly activates ERK2, which phosphorylates threonine-210 of TR2 to stimulate its subsequent sumoylation.18 This occurs within minutes of exposure to RA, before the RAR-mediated canonical activity of RA can be detected, and cannot be reversed by RAR antagonists. Thus, in healthy proliferating ESC cultures, TR2 is maintained at its dephosphorylated and unsumoylated state, which provides an activator function to help maintain constitutively active transcription of the Oct4 gene. However, when the culture is exposed to a differentiating agent like RA, at least two events are initiated. First, RA exerts a rapid, non-canonical activation of crucial signaling molecules such as ERK2, which rapidly reverses certain key transcription factors’ property (like TR2’s) to augment crucial stemness loci such as the Oct4 locus, thus reducing ESC proliferation potential and disturbing stemness. Importantly, this occurs even before the ESC genome exhibits any sign of RA-induced differentiation. Second, RA enters the nuclei to elicit its canonical activity—binding to RARs to act on specific gene targets such as those for differentiation.
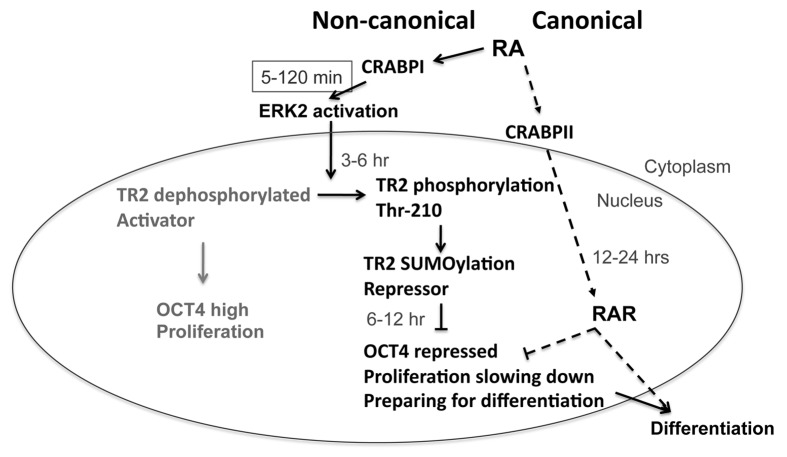
Thus, for ESCs exposed to a differentiation agent like RA, there appears to be intricate signal integration to alter the dynamics of intracellular signaling pathways and coordinate both the cytosolic and the nuclear environments. In the cytosol, RA acts on its cytosolic receptor CRABPI to rapidly jump-start the ERK pathway to change those factors critical to the maintenance of stemness (such as TR2’s regulation of Oct4), disrupting ESC pluripotency and proliferation. Subsequently or simultaneously, RA enters the nucleus to engage its nuclear signal mediators, the RARs, which repress the Oct4 gene by binding to its promoter’s RA response elements and alter the expression of other genes, leading to differentiation. By engaging both the cytosolic and nuclear mediators of RA, the ESC can very rapidly and efficiently convert its transcription program from proliferation to differentiation. Figure 1 illustrates the integration of the non-canonical (cytosolic) and canonical (nuclear) RA signaling pathway to epigenetically alter the ESC genome.
Figure 1. Non-canonical (solid arrows) and canonical (dashed arrows) activities/signaling pathways of RA to augment the ESC epigenome. RA enters ESC, first encountering cytosolic receptors CRABPI or CRABPII. The canonical pathway takes the nuclear route via CRABPII, which delivers RA to nuclear RARs and elicits changes in RA targeted gene expression, mostly triggering ESC differentiation detectable in 12–24 h. The non-canonical pathway takes the cytosolic route via CRABPI, which rapidly (5–120 min) activates cytosolic ERK that can further act on numerous substrates including, for instance, the transcription activator for Oct4, TR2, that then becomes a repressor of Oct4, thereby lowering OCT4 level, slowing down ESC proliferation and disturbing its pluripotency. This tips the balance of the ESC genome and prepares ESC for specific differentiation programs that will be elicited by the nuclear activity of RA via RARs.
Are There Additional Cytosolic RA Signaling Pathways That May Also Contribute to Epigenetic Regulation in ESC?
Using gene knockout/knockdown strategies, we established the functional role for CRABPI in mediating RA-activation of ERK without involving RARs.14 In examining its subsequent action on TR2 and then the Oct4 gene, we delineated a specific pathway relevant to the coordination of the cytosol and the genome of ESC, which ultimately plays out in its epigenetic regulation and is important to ESC biology (Fig. 1). It is unclear, however, whether holo-CRABPI activates only ERK, or may also modulate the activities of other signaling molecules. Additionally, more studies are needed to uncover other potential targets of RA-activated ERK. Further, classical studies have proposed a canonical function of CRABPI in regulating RA concentration, presumably by interaction with RA metabolizing machineries.21,30,31 But this has not been experimentally demonstrated. Finally, it is also unclear whether and how ERK activation by CRABPI/RA may integrate with any of the RA metabolic pathways. Regardless of the spectrum of activity that holo-CRABPI may exhibit, an important lesson learned from studying these RA signal effectors is our past lack of recognition for cytosolic players that not merely control RA concentration but may also modulate the nuclear events triggered by RA. Recognizing this gap in our knowledge is crucial to fully understand the widely documented pleiotropism of RA’s effects. To understand the extreme sensitivity of ESC to extracellular conditions, including to the status of RA, it will behoove us to identify the cytosolic games that are afoot in contributing to the mystifying plasticity of the ESC genome. Filling this gap in our knowledge may be the Holy Grail in our attempt to understand the full spectrum of epigenetic factors in ESC biology.
Acknowledgments
This work was supported by grants DK60521, DK54733, the Distinguished McKnight University Professorship, and the Dean’s Commitment. I thank Dr. FH Burton for discussion and editing.
Glossary
Abbreviations:
- RA
retinoic acid
- ESC
embryonic stem cell
- iPSC
induced pluripotent stem cell
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/25395
References
- 1.Christophersen NS, Helin K. Epigenetic control of embryonic stem cell fate. J Exp Med. 2010;207:2287–95. doi: 10.1084/jem.20101438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–71. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez CV, Garcia-Lavandeira M, Garcia-Rendueles MER, Diaz-Rodriguez E, Garcia-Rendueles AR, Perez-Romero S, et al. Defining stem cell types: understanding the therapeutic potential of ESCs, ASCs, and iPS cells. J Mol Endocrinol. 2012;49:R89–111. doi: 10.1530/JME-12-0072. [DOI] [PubMed] [Google Scholar]
- 4.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–77. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–75. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Mah N, Prigione A, Wolfrum K, Andrade-Navarro MA, Adjaye J. A transcriptional roadmap to the induction of pluripotency in somatic cells. Stem Cell Rev. 2010;6:282–96. doi: 10.1007/s12015-010-9137-2. [DOI] [PubMed] [Google Scholar]
- 7.Dvorak P, Dvorakova D, Hampl A. Fibroblast growth factor signaling in embryonic and cancer stem cells. FEBS Lett. 2006;580:2869–74. doi: 10.1016/j.febslet.2006.01.095. [DOI] [PubMed] [Google Scholar]
- 8.Pucéat M. TGFbeta in the differentiation of embryonic stem cells. Cardiovasc Res. 2007;74:256–61. doi: 10.1016/j.cardiores.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Pera MF, Tam PP. Extrinsic regulation of pluripotent stem cells. Nature. 2010;465:713–20. doi: 10.1038/nature09228. [DOI] [PubMed] [Google Scholar]
- 10.Samarut E, Rochette-Egly C. Nuclear retinoic acid receptors: conductors of the retinoic acid symphony during development. Mol Cell Endocrinol. 2012;348:348–60. doi: 10.1016/j.mce.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 11.Zile MH. Function of vitamin A in vertebrate embryonic development. J Nutr. 2001;131:705–8. doi: 10.1093/jn/131.3.705. [DOI] [PubMed] [Google Scholar]
- 12.Rhinn M, Dollé P. Retinoic acid signalling during development. Development. 2012;139:843–58. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 13.Wei L-N. Retinoid receptors and their coregulators. Annu Rev Pharmacol Toxicol. 2003;43:47–72. doi: 10.1146/annurev.pharmtox.43.100901.140301. [DOI] [PubMed] [Google Scholar]
- 14.Persaud SD, Lin YW, Wu CY, Kagechika H, Wei L-N. Cellular retinoic acid binding protein I mediates rapid non-canonical activation of ERK1/2 by all-trans retinoic acid. Cell Signal. 2013;25:19–25. doi: 10.1016/j.cellsig.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chuang Y-S, Huang W-H, Park SW, Persaud SD, Hung C-H, Ho P-C, et al. Promyelocytic leukemia protein in retinoic acid-induced chromatin remodeling of Oct4 gene promoter. Stem Cells. 2011;29:660–9. doi: 10.1002/stem.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta P, Ho PC, Huq MM, Ha SG, Park SW, Khan AA, et al. Retinoic acid-stimulated sequential phosphorylation, PML recruitment, and SUMOylation of nuclear receptor TR2 to suppress Oct4 expression. Proc Natl Acad Sci U S A. 2008;105:11424–9. doi: 10.1073/pnas.0710561105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta P, Park SW, Farooqui M, Wei L-N. Orphan nuclear receptor TR2, a mediator of preadipocyte proliferation, is differentially regulated by RA through exchange of coactivator PCAF with corepressor RIP140 on a platform molecule GRIP1. Nucleic Acids Res. 2007;35:2269–82. doi: 10.1093/nar/gkl1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park SW, Hu X, Gupta P, Lin YP, Ha SG, Wei L-N. SUMOylation of Tr2 orphan receptor involves Pml and fine-tunes Oct4 expression in stem cells. Nat Struct Mol Biol. 2007;14:68–75. doi: 10.1038/nsmb1185. [DOI] [PubMed] [Google Scholar]
- 19.Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem J. 2000;348:481–95. doi: 10.1042/0264-6021:3480481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta. 2012;1821:152–67. doi: 10.1016/j.bbalip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei L-N, Chang L, Hu X. Studies of the type I cellular retinoic acid-binding protein mutants and their biological activities. Mol Cell Biochem. 1999;200:69–76. doi: 10.1023/A:1006906415388. [DOI] [PubMed] [Google Scholar]
- 22.Chen N, Napoli JL. All-trans-retinoic acid stimulates translation and induces spine formation in hippocampal neurons through a membrane-associated RARalpha. FASEB J. 2008;22:236–45. doi: 10.1096/fj.07-8739com. [DOI] [PubMed] [Google Scholar]
- 23.Poon MM, Chen L. Retinoic acid-gated sequence-specific translational control by RARalpha. Proc Natl Acad Sci U S A. 2008;105:20303–8. doi: 10.1073/pnas.0807740105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gotoh N. Control of stemness by fibroblast growth factor signaling in stem cells and cancer stem cells. Curr Stem Cell Res Ther. 2009;4:9–15. doi: 10.2174/157488809787169048. [DOI] [PubMed] [Google Scholar]
- 25.Binétruy B, Heasley L, Bost F, Caron L, Aouadi M. Concise review: regulation of embryonic stem cell lineage commitment by mitogen-activated protein kinases. Stem Cells. 2007;25:1090–5. doi: 10.1634/stemcells.2006-0612. [DOI] [PubMed] [Google Scholar]
- 26.Golob JL, Paige SL, Muskheli V, Pabon L, Murry CE. Chromatin remodeling during mouse and human embryonic stem cell differentiation. Dev Dyn. 2008;237:1389–98. doi: 10.1002/dvdy.21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Armstrong L. Epigenetic control of embryonic stem cell differentiation. Stem Cell Rev. 2012;8:67–77. doi: 10.1007/s12015-011-9300-4. [DOI] [PubMed] [Google Scholar]
- 28.Pesce M, Schöler HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–8. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- 29.Lee C-H, Chang L, Wei L-N. Molecular cloning and characterization of a mouse nuclear orphan receptor expressed in embryos and testes. Mol Reprod Dev. 1996;44:305–14. doi: 10.1002/(SICI)1098-2795(199607)44:3<305::AID-MRD4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 30.Fiorella PD, Napoli JL. Expression of cellular retinoic acid binding protein (CRABP) in Escherichia coli. Characterization and evidence that holo-CRABP is a substrate in retinoic acid metabolism. J Biol Chem. 1991;266:16572–9. [PubMed] [Google Scholar]
- 31.Boylan JF, Gudas LJ. Overexpression of the cellular retinoic acid binding protein-I (CRABP-I) results in a reduction in differentiation-specific gene expression in F9 teratocarcinoma cells. J Cell Biol. 1991;112:965–79. doi: 10.1083/jcb.112.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]