Abstract
The proto-oncogene c-myc encodes a basic helix-loop-helix leucine zipper transcription factor (c-Myc). c-Myc plays a crucial role in cell growth and proliferation. Here, we examined how expression of c-Myc target genes and cell proliferation depend on variation of c-Myc protein levels. We show that proliferation rates, the number of cells in S-phase, and cell size increased in a dose-dependent manner in response to increasing c-Myc levels. Likewise, the mRNA levels of c-Myc responsive genes steadily increased with rising c-Myc levels. Strikingly, steady-state mRNA levels of c-Myc target genes did not saturate even at highest c-Myc concentrations. These characteristics predestine c-Myc levels as a cellular rheostat for the control and fine-tuning of cell proliferation and growth rates.
Keywords: gene activity, transcription factor, titration, transactivation
Introduction
The response of genes to the expression of a specific transcription factor (TF) is a complex issue and depends on many different parameters. These include the number, strength and location of binding sites of the respective TF in the target gene locus, its cooperation with other factors, the epigenetic stage of the target gene locus, and the activity of cellular signaling cascades, which can modify and modulate the activity of the TF.1,2 Thus, even if a gene locus has bona fide binding sites for a given TF and binding of the factor is traceable in chromatin immunoprecipitation (ChIP) experiments, the outcome of TF binding for gene activity is hardly predictable. An important question in this context is how much TF is required for the activation of a target gene and how do the activation curve characteristics of target genes mirror increasing levels of a TF? For example, does a 10-fold higher abundance of a TF lead to a 10-fold higher expression level of a target gene? The graph of an activation curve can provide important information about the mode of action of a TF. A linear relation ship between TF concentration and the activity of the target gene would point to a high on/off rate of the factor at the target gene or may reflect its rapid consumption at the target gene. In contrast, uninterrupted binding of the TF should lead to its saturation at a binding site and may result in a constant gene activity. To study these alternative models, we analyzed the expression characteristics of target genes of the short-lived TF c-Myc3 in a dose-dependent manner. c-Myc forms heterodimers with the long-lived Max protein and activates a large panel of target genes implicated in cell cycle and growth control.4-9 In addition to an E-box motif Myc-binding-site recognition in the human genome appears to be determined by the chromatin context.10,11 After binding, c-Myc regulates gene activity by affecting the recruitment and activity of chromatin remodeling and modifying factors.12
Here, we studied the effect of fine-tuned c-Myc protein levels on cell proliferation, and global gene expression in the human B-cell line P493–6. This cell line contains a construct that regulates c-myc from a tetracycline-dependent promoter. The system allows titration of c-Myc protein levels in a very sensitive manner by addition of different amounts of tetracycline (Tc) to the medium.7,9,13-15 We observed that cell cycle parameters changed in relation to the respective c-Myc protein levels in P493–6 cells. In addition, the steady increase of c-Myc protein levels was accompanied by a steady increase of mRNA levels of c-Myc target genes, suggesting that the expression of target genes is not saturated even at extremely high c-Myc levels in P493–6 cells. This finding has major implications on the mode of action of the transcription factor c-Myc.
Results
Titration of c-Myc levels
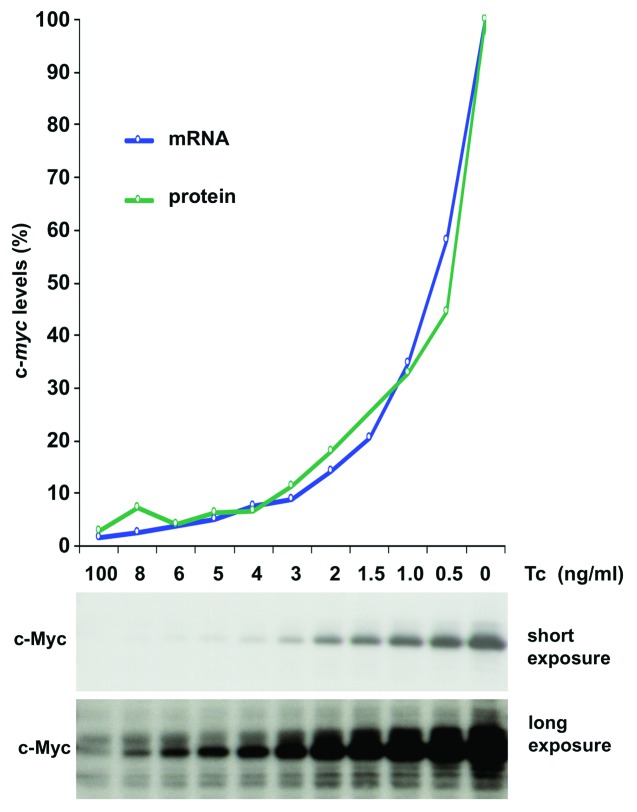
Growth and proliferation of P493–6 cells depend on the expression of a tetracycline (Tc) regulated c-myc gene.15 Various Tc concentrations, 0 – 100 ng/ml, were adjusted in the medium to titrate c-Myc protein levels in P493–6 cells. C-myc mRNA levels were analyzed by expression profiling and c-Myc protein levels by western blotting 48 h after adjustment of Tc concentrations (Fig. 1). Steady-state levels of c-Myc protein were reached approximately 4 h after adjustment to the respective Tc concentration (data not shown). Notably, c-Myc protein levels were > 100-fold higher in cells in the absence of Tc compared with levels in the presence of 100 ng/ml Tc.
Figure 1. Titration of c-Myc protein levels. (A) Analysis of c-Myc protein levels in P493–6 cells. P493–6 cells were treated for 48 h with the indicated concentrations of Tc to obtain total RNA and protein lysates. c-Myc levels were analyzed by expression profiling (blue line) and by fluorescent protein labeling (green line). The highest expression of c-Myc was set as 100% in the diagram. The panel below the graph contains a western blot showing the c-Myc levels in P493–6 cells with varying concentrations of Tc.
c-Myc levels accelerate cell proliferation and enlarge the population of cells in S-phase in a dose-dependent manner
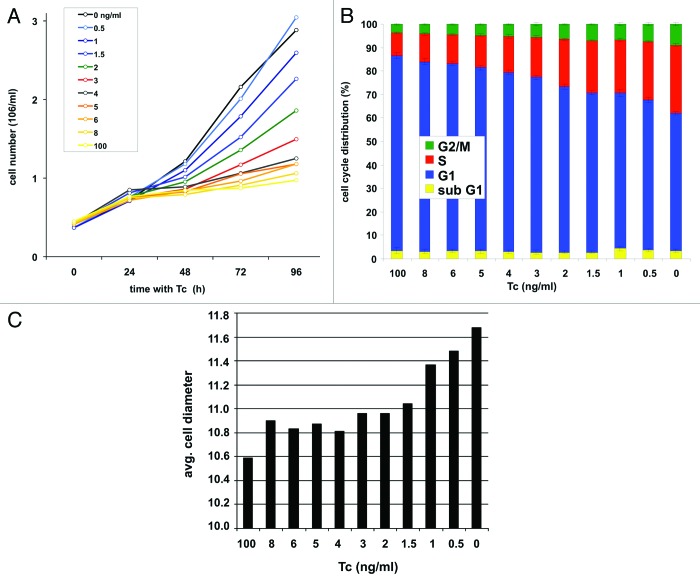
Treatment of cells with Tc (100 ng/ml) for 48 h shuts off expression of the recombinant c-myc and leads to cell cycle arrest of P493–6 cells.13,15 Arrested cells were washed, adjusted to new Tc concentrations (0h), and cell counts were determined in intervals of 24 h. The proliferation rates were similar for all Tc concentrations between 0 h and 24 h, but thereafter were strictly Tc-dependent (Fig. 2A). A little increase of c-Myc levels was accompanied by a respective little increase of proliferation rates at all Tc concentrations. Cells with the lowest c-Myc levels (corresponding to 100 ng/ml Tc) showed the lowest proliferation rate. Only the proliferation rate measured at the highest c-Myc concentration (0 ng/ml Tc) at day 4 showed a little drop probably owing to high density of cells in the culture medium. Taken together, the titration experiment demonstrates a direct correlation between c-Myc protein levels and cell proliferation rates over a large range of c-Myc concentrations.
Figure 2. c-Myc regulates cell proliferation, cell cycle activity, and cell growth in P493–6 cells. (A) Growth curves of cells over a period of four days after adjustment of different Tc concentrations. (B) Cell cycle profile and (C) average cell size (μm) 48 h after adjustment of Tc concentrations.
Similarly, small differences in c-Myc levels caused a respective change in cell cycle distribution of P493–6 cells. An increase of c-Myc protein levels was followed by a steady increase in the percentage of cells in S-phase and G2/M, and a decline of cells in G1-phase (Fig. 2B). The same correlation was seen also for c-Myc levels and cell size (Fig. 2C). We conclude that growth-associated phenotypes as proliferation rates, cell cycle distribution, and cell size are regulated by c-Myc levels and can be fine-tuned by titration of c-Myc protein levels in P493–6 cells.
Expression levels of c-Myc-target genes do not saturate with increasing c-Myc levels
Next we studied the induction kinetics of c-Myc target genes in response to increasing c-Myc levels. Gene probes corresponding to the 3′ region of c-Myc target genes were sp.otted pair-wise on a microarray. We identified 446 genes with increasing expression in response to rising c-Myc levels. The c-Myc target genes included in this evaluation were induced at least 2-fold at the highest c-Myc concentration (Table S1). For details of the statistical analysis of the expression data see Materials and Methods.
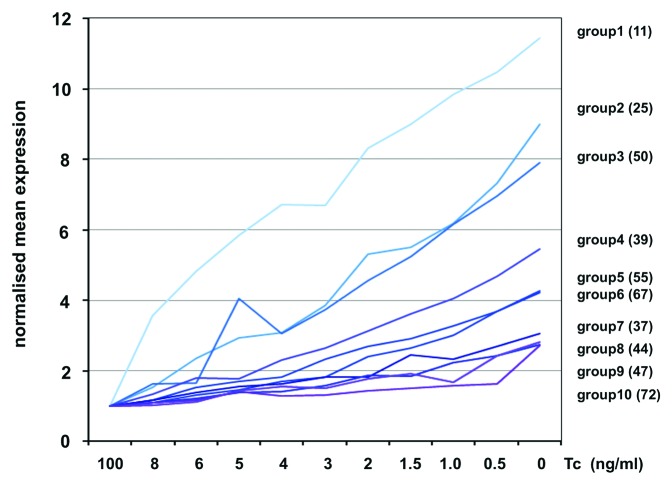
In P493–6 cells c-Myc protein reaches levels higher than observed in tumor cell lines with amplified c-myc (data not shown).16 To get a better survey of c-Myc regulated genes, we sorted induced genes in groups according to their response to c-Myc changes. The expression of c-Myc responsive genes at each c-Myc level was compared with expression levels in control cells (100 ng/ml Tc). By this approach, ten groups of c-Myc regulated genes were defined. Members of each group fulfilled the criterion that they were at least 2-fold induced at a specific c-Myc protein level. The average expression profile of the genes belonging to each group is shown (Fig. 3). For example, the mRNA levels of the 11 genes of group 1 are upregulated already 2-fold by little increases of c-Myc (8ng/ml Tc) and thereafter steadily increased with increasing c-Myc levels. In contrast, the 72 genes of group 10 show a significant induction only at the highest c-Myc concentration. Thus, the responsiveness of target genes to different c-Myc protein levels differed largely from highly to little responsive genes.
Figure 3. Expression of c-Myc target genes at different Tc concentrations. Average expression profiles of groups 1 - 10 were calculated from 447 induced genes. Genes in the different groups required a 2-fold or higher change in expression compared with control cells (100ng/ml Tc). The graphs of groups 1 - 10 show the normalized average expression profiles calculated from the genes that belong to each group (number of genes per group in brackets).
Discussion
c-Myc levels can be titrated in a fine-tuned manner in P493–6 cells. We see that even slight increases in c-Myc levels lead to measurable effects on cell proliferation rates, cell cycle distribution, and expression of target genes. The expression of c-Myc target genes steadily increased in response to increasing c-Myc levels even at the highest concentration of c-Myc, which by far exceeds c-Myc levels in normal cells. This finding was unexpected since we anticipated a saturation of target gene activity for very high c-Myc concentrations. Thus, c-Myc levels behave as a rheostat for the regulation of target gene activity and thereby for regulation of cell proliferation rates. We note that we currently do not know whether all cells of a population respond equally to changes in tetracycline levels, or if variations in c-Myc levels at the single cell could influence the response of c-Myc target genes.
Interestingly, both the response of target genes and of cell proliferation increased further even at extreme high levels of c-Myc. How can this failure of saturation of the c-Myc response be explained? The mode of action of TFs in gene regulation is still far from being understood. While questions as binding specificity to DNA, interaction with co-activators, co-repressor or other factors have successfully been addressed, little is known about the dynamic and affinity of TF-binding to gene loci in living cells and how binding of TFs correlates with gene activity.1 An open question is, whether the residence time of a TF at a gene locus can be used as a measure for the activity of the factor and for the activity of the target gene? In the past, the presence of low and high affinity binding sites has been reported for c-Myc in promoter regions,16 but these sites have not been correlated yet with the activity of the target gene. Examination of the gene list with high affinity c-Myc binding sites published by the Amati laboratory16 revealed the presence of genes with high affinity binding sites in all gene groups (1–10) described in Figure 3. However, the biological relevance of high affinity sites is still elusive. It could well be that low affinity sites with a high turn-over of c-Myc binding represent much better c-Myc responsive elements than high affinity c-Myc binding sites do.
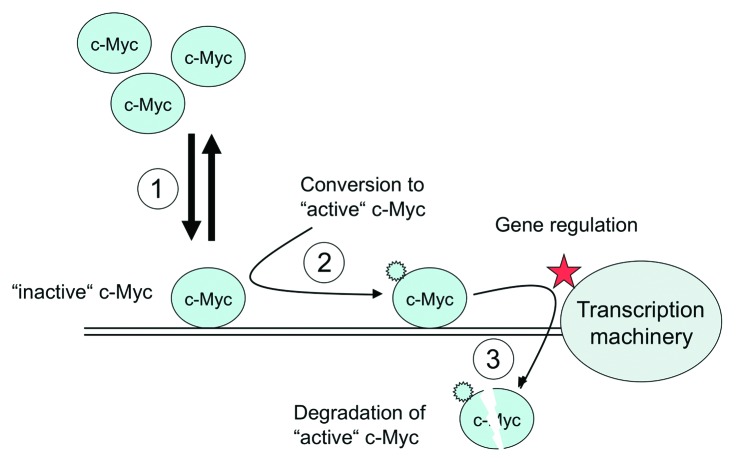
It‘s also unclear whether binding of c-Myc to a bona fide binding site is sufficient for gene activation, or whether c-Myc has to undergo additional modifications/alterations at a binding site to gain its gene regulatory activity. c-Myc is highly modified by phosphorylation, acetylation, and ubiquitinylation.17,18 If specific modifications are required, c-Myc binding sites can in principal accommodate “inactive” and “active” forms of c-Myc (Fig. 4). In this scenario c-Myc may bind as an “inactive” precursor and must be converted into an “active” form, before it facilitates gene regulation. The transition of c-Myc from the “inactive” into the “active” state could be regulated in a target site-specific manner. However, currently little is known about the specificity of such a process and how efficiently it could occur.
Figure 4. Model for c-Myc function. High levels of c-Myc cannot saturate functional c-Myc binding sites (1). After binding c-Myc can be converted into its “active” form (2). The active form of c-Myc is unstable and degraded or removed/inactivated24,25 by other mechanisms (3).
c-Myc is a very short-lived molecule with a half-life of 15 – 30 min in all phases of the cell cycle except mitosis.3 This instability, which c-Myc shares with other transcription factors, has been implicated in its gene regulatory function. The removal of “active” c-Myc from its binding site may be accomplished by ubiquitinylation-coupled degradation. Since ubiquitinylation of c-Myc’s transactivation domain has also been reported to be a possible mechanism for its conversion into the “active” form,19-21 activation and degradation of c-Myc may be coupled processes occurring as consecutive steps at the target site (see model in Figure 4). Alternatively, the activation signal may be removed from the “active” form and c-Myc returns to the “inactive” pool. If c-Myc requires an activation mechanism at its target site, this could be an additional rate-limiting step in gene activation. In this case, c-Myc binds to the target site with high or low affinity, but gene activation does not occur as long as c-Myc is not converted into its “active” form. Alternatively, c-Myc is directly converted into its “active” form after binding, and degraded or released thereafter.
The characteristic to activate target genes over a wide range of concentrations enables c-Myc to fine tune cellular growth and proliferation rates and predestines c-Myc as an excellent master gene for proliferation and growth control. Consistently, the levels of c-Myc are tightly regulated by a large number of growth factors and intracellular gene regulatory feedback mechanisms,22,23 and little changes in c-Myc directly attenuate or enhance target gene activity and thereby cell proliferation. Two recent studies demonstrated that c-Myc acts a universal amplifier of expressed genes.24,25 Here we confirm this result and show that this function applies in addition to cell growth and proliferation.
Materials and Methods
Cell culture and cell count
P493–6 cells were grown as described earlier.13,15 c-Myc protein levels were titrated by incubation in medium with increasing Tc concentrations. Cell number and viability were determined by trypan blue staining using a cell counter (Cedex, Innovatis).
Immunoblotting
Lysates were prepared by treating 5x106 viable cells with 100 µl lysis buffer [100 mM HEPES, 150 mM NaCl, 10 mM NaF, 0,5 M EDTA, 10% glycerin, 1% Triton-X-100 and protease inhibitor (Roche)]. Ten µl of lysate was analyzed by western blotting. The polyclonal antibodies N-262 for c-Myc were obtained from Santa Cruz Biotechnology (sc-764). For quantification of western blot signals secondary alexafluor680-labeled antibodies and Odyssey-detection facilities were used from LI-COR Biosciences.
Cell cycle analysis
Cells were treated with Tc in 96 well plates and stained (end concentrations 0.1% citrate, 0.3% Nonidet P40, 0.5 mg/ml RNaseA, 50µg/ml propidiumiodide). The DNA content was directly analyzed with flow cytometry (FACSArray, Beckton Dickinson) for 6 individual samples per treatment.
Production of filter arrays with human gene probes
The array contains probes for genes, which have been described as c-Myc induced target genes previously (www.myccancergene.org) or as genes being involved in tumorigenesis, cell cycle control or apoptosis. Probes are synthetic oligonucleotides, 60 bp long, and complementary to the 3′ region of different human mRNAs. Each probe was spotted twice on each filter array. In addition, ten E. coli sequences and ten random sequences were included as negative controls. For induced genes see Table S1 in Supplementary data.
Labeling of total RNA and hybridization of arrays
Labeling, hybridization, scanning and data processing procedures were performed as described elsewhere.26 In brief, a standard RT-reaction was performed using 4µg of total RNA in the presence of 50µCi [α-33P]-dCTP (3000 Ci/mmol; Perkin Elmer Life Sciences, MA, USA) and an oligo-dT(20) primer. The hybridization temperature was 40°C.
Analysis of expression data
To ensure the highest data reliability we produced 18 data points in total per gene and the average was calculated from all 18 data points. Total RNA from three independent experiments was prepared from cells corresponding to each c-Myc level. The RNA samples were labeled individually and each sample was hybridized with three filters; each filter contained all gene probes twice. For evaluation of the results we differentiated between significance of expression, which determines whether a signal on the filter is above background, and significance of differential expression, which indicates whether expression of a gene is different in two samples. The expression data of two different probes were analyzed by means of a two-tailed t-test for independent samples (p ≤ 0.05).
c-Myc-responsive genes were determined by pair-wise comparisons of expression values at each c-Myc level with the expression values of the reference sample 'c-MycOFF' (100 ng/ml Tc). Genes that were defined c-Myc responsive had to fulfil the following criteria: (i) significance of differential expression at least for the comparison of c-Myc-OFF vs. c-Myc-ON (without Tc) (ii) the change in expression between c-Myc-OFF and c-Myc-ON had to be at least 2-fold (iii) significance of expression in the c-Myc-ON samples. The expression data are listed in Table S1. We used Spotfire Decision Site 8.2 software (Spotfire Inc.) for visualization of expression profiles and hierarchical clustering.
Supplementary Material
Acknowledgments
We thank David Levens for discussion, Chi V. Dang for communicating data, and Markus Kietzmann and Christoph Schaab for software, data collection and analysis. This work was supported by SFB/TR 5 and SFB684.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/transcription/article/25907
References
- 1.Hawkins RD, Ren B. Genome-wide location analysis: insights on transcriptional regulation. Hum Mol Genet. 2006;15(Spec No 1):R1–7. doi: 10.1093/hmg/ddl043. [DOI] [PubMed] [Google Scholar]
- 2.Zeitlinger J, Simon I, Harbison CT, Hannett NM, Volkert TL, Fink GR, Young RA. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/S0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 3.Hann SR, Thompson CB, Eisenman RN. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. Nature. 1985;314:366–9. doi: 10.1038/314366a0. [DOI] [PubMed] [Google Scholar]
- 4.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–45. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 5.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–99. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 6.Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/S0065-230X(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 7.Schlosser I, Hölzel M, Hoffmann R, Burtscher H, Kohlhuber F, Schuhmacher M, Chapman R, Weidle UH, Eick D. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene. 2005;24:520–4. doi: 10.1038/sj.onc.1208198. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt EV. The role of c-myc in cellular growth control. Oncogene. 1999;18:2988–96. doi: 10.1038/sj.onc.1202751. [DOI] [PubMed] [Google Scholar]
- 9.Schuhmacher M, Kohlhuber F, Hölzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall’ Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–70. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci U S A. 2003;100:8164–9. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–34. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pajic A, Spitkovsky D, Christoph B, Kempkes B, Schuhmacher M, Staege MS, Brielmeier M, Ellwart J, Kohlhuber F, Bornkamm GW, et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int J Cancer. 2000;87:787–93. doi: 10.1002/1097-0215(20000915)87:6<787::AID-IJC4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 14.Schlosser I, Hölzel M, Mürnseer M, Burtscher H, Weidle UH, Eick D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003;31:6148–56. doi: 10.1093/nar/gkg794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuhmacher M, Staege MS, Pajic A, Polack A, Weidle UH, Bornkamm GW, Eick D, Kohlhuber F. Control of cell growth by c-Myc in the absence of cell division. Curr Biol. 1999;9:1255–8. doi: 10.1016/S0960-9822(99)80507-7. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–29. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eilers M, Eisenman RN. Myc’s broad reach. Genes Dev. 2008;22:2755–66. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vervoorts J, Lüscher-Firzlaff JM, Lüscher B. The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem. 2006;281:34725–9. doi: 10.1074/jbc.R600017200. [DOI] [PubMed] [Google Scholar]
- 19.Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, et al. The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell. 2005;123:409–21. doi: 10.1016/j.cell.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 20.Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell. 2003;11:1177–88. doi: 10.1016/S1097-2765(03)00173-4. [DOI] [PubMed] [Google Scholar]
- 21.Popov N, Schülein C, Jaenicke LA, Eilers M. Ubiquitylation of the amino terminus of Myc by SCF(β-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat Cell Biol. 2010;12:973–81. doi: 10.1038/ncb2104. [DOI] [PubMed] [Google Scholar]
- 22.Chung HJ, Levens D. c-myc expression: keep the noise down! Mol Cells. 2005;20:157–66. [PubMed] [Google Scholar]
- 23.Kelly K, Siebenlist U. The role of c-myc in the proliferation of normal and neoplastic cells. J Clin Immunol. 1985;5:65–77. doi: 10.1007/BF00915003. [DOI] [PubMed] [Google Scholar]
- 24.Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machl AW, Schaab C, Ivanov I. Improving DNA array data quality by minimising ‘neighbourhood’ effects. Nucleic Acids Res. 2002;30:e127. doi: 10.1093/nar/gnf127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.