Abstract
A study on the occurrence of Aspergillus section Nigri species on grapes from four traditional grape-producing areas in Greece during the 2011/2012 vintage, and their capability to produce OTA was conducted. One hundred and twenty-eight black aspergilli isolates were characterised at the species level initially by the use of morphological criteria in accordance with appropriate keys, followed by molecular characterisation performed with Polymerase Chain Reaction–Restriction Fragment Length Polymorphism (PCR-RFLP) of the 5.8 ribosomal RNA gene Internal Transcribed Spacer region (5.8 rRNA ITS). Restriction enzyme digestion of the ITS amplicons using the HhaI, HinfI and RsaI, endonucleases distinguished eleven different patterns of restriction fragment length polymorphism (RFLP), four for each of the HhaI and RsaI digests and three for HinfI. From a total number of 128 individual isolates, 124 were classified into four Aspergillus species corresponding to A. carbonarius, A. tubingensis, A. japonicus and A. ibericus, and the remaining 4 were classified as members of the A. niger aggregate. A. carbonarius and A. tubingensis being the main representative species were equally counted, with higher geographical representation of the former in southern and the latter in northern regions, respectively. All isolates were tested for their ochratoxigenic potential by use of High Performance Liquid Chromatography (HPLC) and Enzyme Linked Immuno Sorbent Assay (ELISA), resulting in significant interspecies differences in OTA production.
Introduction
Ochratoxin A (OTA) is a naturally occurring mycotoxin, produced principally by a wide range of Aspergillus and Penicillium species [1] while various nephrotoxic, carcinogenic, immunotoxic, genotoxic and teratogenic effects [2], [3], [4], [5] as well as Balkan Endemic Nephropathy [6], [7] have been attributed to this mycotoxin.
OTA is receiving attention due to its high incidence in a wide range of food commodities such as cereal-based products, coffee, spices, nuts, olives and grape-derived products [8], [9], [10], [11], [12], [13]. RASFF Annual Report places OTA as the mycotoxin with most notifications for fruit and vegetables in 2012, while is being always second after aflatoxins in notifications on mycotoxins for food and feed for over the last decade [14]. Several publications report the high occurrence of OTA in wine and grape products from different European countries, with higher OTA levels reported for products originating from southern European regions and in particular in regions with a Mediterranean climate [15], [16], [17], [18], [19], [20]. Since 2005 the European Commission has imposed regulatory limits for OTA, and has established a 2 μg l−1 maximum level of OTA in wine and grape products [21], [22].
Several studies performed worldwide have shown that OTA is produced during infection of grapes in vineyards mainly by mycotoxigenic strains of black aspergilli (section Nigri), in particular Aspergillus carbonarius and species belonging to the Aspergillus niger aggregate such as Aspergillus niger and Aspergillus tubingensis, [18], [19], [20], [23], [24], [25]. A. carbonarius although less common than other black aspergilli, is considered to be the predominant species responsible for OTA contamination in grapes and wine, because of the ability of almost all of its strains to produce high levels of the toxin [26], [27]. Due to the variations in the toxin production potency of different aspergilli, it is of great importance to identify the Aspergillus species accurately in order to define potential toxicological risks at an early stage [28].
Black aspergilli are difficult to classify following only morphological criteria [29]. Though A. carbonarius can be easily recognized, closely related morphospecies like the biseriate species included in the A. niger aggregate, or uniseriate species such as Aspergillus aculeatus and Aspergillus japonicus, have been always difficult to distinguish. Among the molecular approaches used to decipher the Aspergillus taxonomy [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], Polymerase Chain Reaction - Restriction Fragment Length Polymorphism (PCR-RFLP) has been used successfully to identify Aspergillus species [33], [39]. The non-coding Internal Transcribed Spacer regions (ITS) due to their high degree of sequence polymorphism have been used widely for fungi molecular systematics at the species level [40], [41], [42]. Standard primer pairs have been used for selective amplification of these fungal sequences and have been proved useful for the identification of Aspergillus species [33], [39].
The objective of this study was to characterize (i) the biodiversity of black aspergilli isolates from vineyards of four traditional grape producing regions in Greece during the 2011/2012 vintage using molecular techniques and (ii) study their ochratoxigenic potential. To our knowledge this is the first study with the objective to monitor the population of black aspergilli by molecular means in Greece.
Materials and Methods
Ethics Statement
Part of this study was carried out in private lands. No specific permissions were required for these locations/activities that took place and all the land owners agreed to perform this research in their site. The field studies did not involve endangered or protected species.
Sampling and culturing conditions
Thirty-three vineyards, located at four traditional grape-producing regions of Greece, were chosen for sampling during the 2012 harvesting period (from August to October). The sampled vineyards were located in Iraklion prefecture of Crete (n = 8), Mesogeia province of Attica (n = 11), Corinthia and Arcadia prefectures of Peloponnese (n = 7), Thessaloniki, Imathia, Florina and Pella prefectures of Macedonia (n = 7). Selection of sampling areas was made with provision to represent the whole range of the typical climatic conditions of Greece, i.e. the Peloponnese from Southern Greece and Attica from Central Greece, having both a landlocked Mediterranean climate with vineyards of high and low altitudes, respectively, Crete from the Southern Aegean Sea with insular Mediterranean climate, and Macedonia from Northern Greece having a typical mountainous climate with vineyards of high altitude.
From every vineyard five plants were selected along two major diagonal transects and two bunches were collected from each plant, resulting in a total of 10 bunches per sampled vineyard. Bunches were kept separate in sterile plastic bags and stored in portable refrigerators during transfer to the laboratory where the analysis took place within 24 hours from sampling.
From each bunch five healthy berries were randomly selected and placed directly on the surface of Dichloran Rose Bengal Chloramphenicol (DRBC) medium (LabM, UK). The plates were incubated in the dark at 25°C for 7 days.
Mycoflora enumeration and black aspergilli isolation and identification
After incubation, incidence of infected with black aspergilli berries and distribution of most dominant grape mycoflora at genus level, were recorded. A representative number from the black aspergilli from every vineyard were isolated from DRBC and sub-cultured on Malt Extract Agar (MEA, LabM) and Czapek Dox Agar (CD, Oxoid) for further identification at species level. The initial identification of the different strains of Aspergillus section Nigri was performed using macroscopic and microscopic morphological criteria in accordance with appropriate keys [29], [43], [44]. The reference strains of A. carbonarius, A. niger, A. tubingensis, A. ochraceus and A. westerdijkiae were kindly provided by Prof. N. Magan from the Mycology Group of Cranfield University. The isolates were preserved at −80°C in the culture collection of the Department of Food Science and Human Nutrition of the Agricultural University of Athens, Greece.
DNA extraction
For DNA extraction all strains were grown in Yeast Extract Sucrose broth (YES; contained per litre 20 g yeast extract and 150 g sucrose) at 30°C for 2 days. Mycelia were collected, washed briefly with ethanol 96%, and dried using Whatman No. 1 filter paper. Approximately 200 mg of mycelia from each individual strain were frozen in liquid nitrogen and ground to a fine powder. DNA extractions were performed using the NucleoSpin Plant DNA kit (Macherey-Nagel, Germany) according to the manufacturer's instructions.
PCR reactions and Restriction Endonuclease DNA digestions
The 5.8 S-ITS region was amplified by PCR using universal primers ITS 1 and ITS 4 [45]. PCR reactions were performed in a 50 μL final volume, containing 1× standard reaction buffer (New England Biolabs, UK), 2.0 mM MgCl2, 300 μM dNTPs (each), 300 nM primers (each), 100 ng DNA template and 1.25 U of Taq DNA polymerase (New England Biolabs, UK). The PCR reactions were performed in a MJ Research PTC-200 thermal cycler (Bio-Rad Laboratories, USA), starting with an initial denaturation step at 95°C for 5 min, followed by 37 cycles consisting of 30 sec at 95°C, 30 sec at 52°C and 40 sec at 72°C, and a final extension step at 72°C for 10 min. PCR products were digested with the HhaI, HinfI and RsaI (New England Biolabs, UK) restriction endonucleases. Digestions were performed at 37°C for 3 h, in a 20 μL reaction volume containing 2 μL of 10X reaction buffer, 10 μL amplicon, 1.5 U restriction endonuclease, and 0.2 μL BSA (10 μg μL−1) for HhaI digestions. PCR amplicons and their restriction digestion fragments were separated by electrophoresis at 100 V, 1×TAE buffer, in 1% and 3% agarose gels, respectively. Agarose gels were subsequently stained in ethidium bromide solution (0.5 mg ml−1), and DNA bands were visualized under Ultra Violet (UV) light using a Gel Doc XR+ system (Bio-Rad Laboratories, USA). Molecular sizes were estimated by comparison with the DNA standard GeneRuler 50 bp and 100 bp DNA ladders (Thermo Scientific, USA).
Sequencing and Phylogenetic Analysis
PCR products were purified using a QIAquick PCR purification kit (Qiagen, USA). Sequencing was performed for both strands using primers ITS1 and ITS4 with the BigDye Terminator v3.1 cycle sequencing kit (Life Technologies, USA) in an ABI3730 xl Genetic Analyzer (Applied Biosystems, Life Technologies, USA) automatic DNA sequencer (Cemia, Greece).
Alignment of the 5.8S-ITS region sequences was performed using the CLUSTALΩ multiple sequence alignment program available at ΕΒΙ (EMBL, UK). The following Aspergillus spp. 5.8S-ITS sequences: FJ450778, A. acidus; KF815051, A. aculeatinus; AJ279997, A. aculeatus; AM087614, A. awamori; AJ280010, A. brasiliensis; AF459734, A. carbonarius; FJ491684, A. coreanus; FJ629326, A. costaricaensis; AY656631, A. ellipticus; KC796400, A. eucalypticola; HE818074, A. fijiensis; AY373850, A. foetidus; AJ280013, A. heteromorphus; FJ629334, A. homomorphus; AY656624, A. ibericus; FJ629335, A. japonicus; FJ629336, A. lacticoffeatus; KC796401, A. neoniger; AJ223852, A. niger; FJ629352, A. piperis; EU159216, A. sclerotiicarbonarius; HM853552 A. saccharolyticus; FJ629353 A. sclerotioniger; FJ629367, A. tubingensis; AM745757, A. uvarumn; FJ629368, A. vadensis and FR733805, A. violaceofuscus, were included as outgroups for the phylogenetic analysis. Genetic distances were calculated using the Jukes-Cantor, parameter model and the phylogenetic inference was obtained by the Neighbour-Joining (NJ) method [46]. The NJ tree and the statistical confidence of a particular group of sequences in the tree, evaluated by bootstrap test (1000 pseudoreplicates), were performed using the computer program MEGA version 3.0 [47].
OTA extraction
All 128 black aspergilli isolates were centre inoculated to CYA medium (which contained per litre: K2HPO, 1 g; Czapek concentrate, 10 ml; trace metal solution, 1 ml; yeast extract, 5 g; sucrose, 30 g; agar, 15 g) [41] and incubated at 25°C for 7 days in order to assess their ochratoxigenic potential. Aspergillus section Nigri isolates was subjected to Ochratoxin A determination according to the Bragulat et al. protocol [48] with a slight modification in the OTA extraction step. Instead of removing 3 small agar plugs, the whole content of the Petri dish was removed and used for OTA extraction [49], [50]. Specifically, the content of the Petri dish (substrate and mycelium) was weighted in order to express OTA production per g of substrate and extraction took place with 100 ml of an 80/20 methanol/water solution of HPLC grade purity when prepared for HPLC analysis and a 50/50 solution when prepared for the ELISA method. The weighted substrates were blended with the solutions for 2 min and left for a total of 30 min before filtered, at first through a Whatman No 1 filter paper, and subsequently through Millex nylon membrane filter of 0.2 μm pore size (EMD Millipore Corp. Billerica, USA), and kept at −80°C until analysis. Additionally, known concentrations of OTA (50, 100 & 500 ppb) were spiked on CYA and recovery rates for both HPLC and ELISA methods were estimated, resulting in satisfactory recovery percentages of 96–99% and 84–95% for the former and latter method, respectively.
HPLC Analysis
Ochratoxin A analysis was performed using reverse-phase High Performance Liquid Chromatography with fluorometric detection (HPLC-FLD). This consisted of a JASCO AS-2055Plus autosampler, a JASCO LC-Net II/ADC system controller, a JASCO PU-980/LC-980-02 pump, and a JASCO FP-2020Plus fluorescence detector (JASCO Inc., Easton, USA). The samples were separated using a C18 analytical column (250×4.6 nm, 4 μm, Resteck Co., Pinnacle II, Bellefonte, USA) under isocratic conditions at a flow rate of 1 ml min−1 of the mobile phase (water/acetonitrile/acetic acid: 99/99/2). All chemicals used for HPLC analysis were HPLC grade (methanol and acetic acid: Sigma-Aldrich Co., Germany; water and acetonitrile: Carlo Erba Reactifs SDS, Val de Ruill, France). An excitation wavelength of 333 nm and an emission wavelength at 460 nm were used for UV detection. Standard solutions were made from stock ochratoxin A solution (10.06 μg ml−1 in acetonitrile; Biopure, Romer Labs Diagnostics GmbH, Tulln, Austria) in mobile phase and a recovery study took place by spiking known concentration solutions to substrate and following the same extraction procedure as for samples. Run time for samples was 14 min with OTA being detected at about 11 min. The limit of quantification was 2.0 ng OTA g−1 CYA (ppb), while the limit of detection was 1.0 ng OTA g−1 CYA.
ELISA
Ochratoxin A quantification was additionally performed with a competitive direct enzyme-linked immunosorbent assay (CD ELISA). Veratox ELISA quantitative test kit (Neogen Corp. Ltd, Lansing, USA) was applied to the samples, having a detection limit of 1 ppb (ng g−1) and range of quantification of 2–25 ppb. ELISA was strictly performed with accordance to the manufacturer's protocol and optical densities were determined at 650 nm absorbance with a spectrophotometer (Synergy HT, Biotek, USA). Quantification of OTA concentration was estimated with the aid of standard curves obtained from standard solutions provided within the test kit. When the determination of OTA by HPLC revealed concentrations greater than the upper limit of quantification by the ELISA method, serial dilutions of the extracts with the extraction solution took place in order to accomplish the ELISA method without extrapolation of its standard curves. Finally, non-ochratoxigenic isolates were omitted from screening with the ELISA method.
Results
Diversity of grapes' fungal population and Aspergillus section Nigri presence during the harvesting period
The genera of filamentous fungi most abundant on sampled grapes were, in descending order, Aspergillus (39%), Fusarium (15%), Alternaria (12%), Cladosporium (11%), Rhizopus (8%), Penicillium (7%) and Botrytis spp. (5)%. With respect to the different regions studied, Aspergillus was the most frequently isolated species in all regions (32%–55%) with the exception of Macedonia in Northern Greece where Alternaria was dominant (36%).
Among the potential OTA-producing fungi, only black aspergilli (Aspergillus section Nigri group) were isolated for further identification to species level. Initially, the 128 different black aspergilli species isolated from grapes were classified according to their morphological characteristics into three subgroups namely, Aspergillus niger aggregate, uniseriate species and A. carbonarius, and thereafter, a partial molecular identification took place.
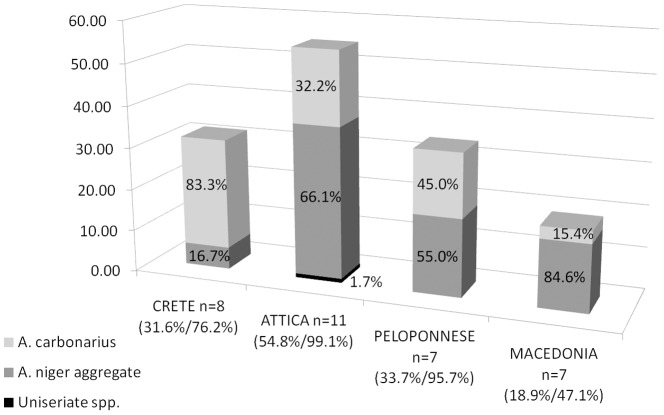
Incidence of black aspergilli isolation from the sampled vineyards is presented in Figure 1. Regarding the presence of Aspergillus section Nigri, Macedonia presented very low numbers, namely 47% incidence (i.e., % of contaminated out of total analysed grapes) with 18% distribution in the isolated mycoflora (i.e., % of group in total recorded fungi), while between the other regions occurrence of black aspergilli was similar and always the dominant genera. The highest incidence and distribution were observed in Attica (99% incidence/55% distribution), followed by the Peloponnese (96%/34%) and Crete (76%/32%).
Figure 1. Aspergillus section Nigri strains distribution.
A. section Nigri distribution in total mycoflora (bars) and different species distribution within the A. section Nigri group (percentages inside bars) for each sampled prefecture. The percentages underneath prefectures' names refer to distribution and incidence of A. section Nigri.
Regarding A. carbonarius, the highest distribution occurred for isolates from Crete, with 83% of black aspergilli identified, followed by the Peloponnese and Attica with 45% and 32%, respectively, while A. carbonarius isolation was most scarce in vineyards of Macedonia (15%). As regards uniseriate spp. a sole isolate was identified originating from Attica prefecture. According to the spore size that can discriminate between black uniseriate aspergilli, the isolate could be characterized as A. japonicus.
Molecular characterization of Aspergillus isolates by PCR-RFLP
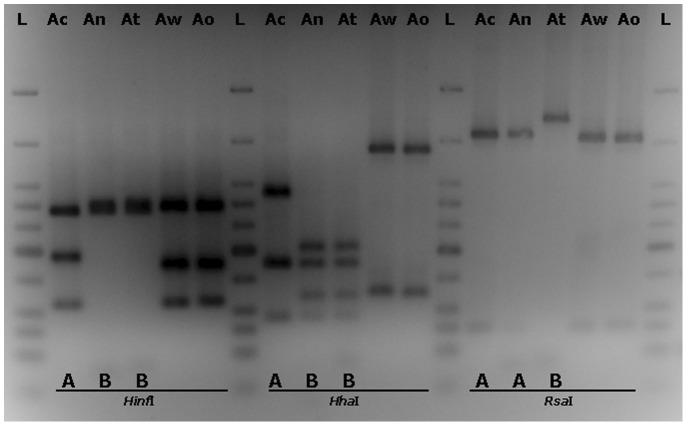
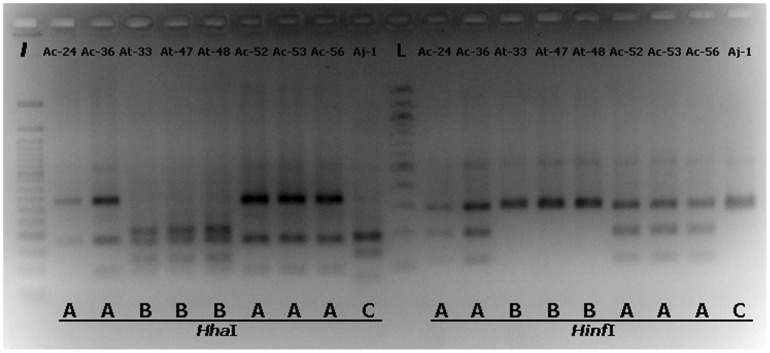
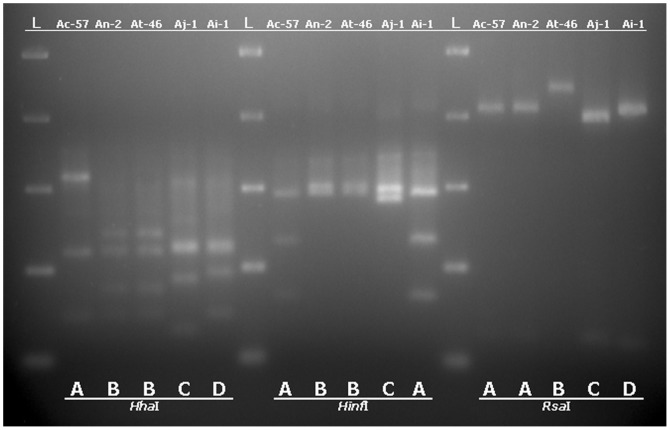
In order to monitor the representation of black aspergilli (A. aculeatus, A. awamori, A. brasiliensis, A. carbonarius, A. foetidus, A. ibericus, A. japonicus, A. niger, A. tubingensis and A. uvarumn) reported to be isolated from grapes [20], [25], [34], [37], [38] we focused on a partial molecular characterization of the 128 isolates at the species level by use of PCR-RFLP of the 5.8S-ITS region. PCR mediated amplification of the ribosomal region was performed using the ITS1 and ITS4 primers resulting to amplicon molecular sizes of approximately 600 bp for all samples. RFLP analysis was done using the HhaI, HinfI and RsaI restriction endonucleases in order to differentiate among species. The restriction patterns of reference strains as presented in Figure 2 were used for comparison with the individual isolates restriction digestion patterns. HhaI and HinfI enzyme digestion can differentiate A. carbonarius from A. niger/A. tubingensis since they both generate 2 different restriction patterns designated here as A (for A. carbonarius) and B (for A. niger and A. tubingensis). RsaI digestion can distinguish further A. niger from A. tubingensis generating different restriction patterns for these two species, pattern A (common for A. niger and A. carbonarius) and pattern B (for A. tubingensis). A total of 128 black Aspergillus isolates previously characterized by morphological criteria were analyzed by PCR-RFLP and their restriction patterns were compared with those obtained from the reference strains. Digestion with HinfI resulted to 61 A and 66 B pattern profiles and 1 for a new profile designated as C and closely resembles profile B. HhaI digestion resulted to 60 A, 66 B pattern profiles and 2 for new pattern profiles designated as C and D. These new profiles for both enzymatic digestions can be seen for samples Aj-1 and Ai-1 in Figures 3 and 4. Restriction digestion with HhaI can also differentiate uniseriate (A. japonicus, A. aculeatus, A. uvarum) from all biseriate black aspergilli species [33], [39]. Depending on the pattern which will result from the HinfI digestion, A. japonicus can be further distinguished from A. aculeatus [39]. According to restriction endonuclease site profile study (data not shown) of characterized black uniseriate aspergilli ITS sequences deposited in public sequence databases, HinfI digestion results in a double band visible pattern for either A. japonicus and A. uvarum and a three band visible pattern for A. aculeatus. The 66 strains which gave a B–B pattern combination after HinfI and HhaI digestions were subjected to digestion with the RsaI, resulting to 62 B and 4 A type restriction patterns. The strain with the C-C HinfI-HhaI, restriction digestion pattern when digested with RsaI, resulted to a new profile designated as C. This profile closely resembles RsaI profile A and can be seen in Figure 4. The strain with the A–D HinfI-HhaI, restriction digestion pattern, when digested with RsaI resulted to a pattern nearly indistinguishable from profile A (Figure 4). Though the molecular size of the largest DNA fragment generated (slightly above 500 bp) runs somehow faster that the corresponding band from profile A, the visible difference is not so obvious. This new profile was designated as D. According to the PCR-RFLP results for the 128 isolates, 60 (46.88%) were characterized as A. carbonarius, 62 (48.44%) as A. tubingensis, 4 (3.12%) as members of the A. niger aggregate, 1 (0.78%) – resembling the pattern C-C-C for HinfI, HhaI and RsaI digestions – could be characterized as A. japonicus/A. uvarum, and 1 (0.78%) with an A-D-D HinfI, HhaI and RsaI digestion pattern could not be clearly classified to any of the previously referred species. A detailed classification according to both phenotypical and molecular data can be seen in Table 1.
Figure 2. Ribosomal 5.8S-ITS region restriction digestion patterns of Aspergillus reference strains.
Restriction digestion patterns (designated as A and B) of ribosomal 5.8S-ITS DNA amplicons from Aspergillus reference strains, after digestion with the restriction endonucleases Hinf I, Hha I and Rsa I. Ac: Aspergillus carbonarius, An: Aspergillus niger, At: Aspergillus tubingensis, Aw: Aspergillus westerdijkiae, Ao: Aspergillus ochraceus, L: Low molecular weight DNA ladder (molecular sizes are 766, 500, 350, 300, 250, 200, 150, 100, 75, 50 and 25 bp respectively).
Figure 3. Ribosomal 5.8S-ITS region restriction digestion patterns of Aspergillus grape isolates.
Restriction digestion patterns (designated as A, B and C) of ribosomal 5.8S-ITS DNA amplicons from various Aspergilli grape isolates (presented as isolate designations), after digestion with the restriction endonucleases Hha I and Hinf I. 50 bp ( l ) and 100 bp (L) DNA ladders are also shown.
Figure 4. Restriction digestion patterns of sequenced ribosomal 5.8S-ITS region amplicons.
Restriction digestion patterns (designated as A, B, C and D) of five sequenced ribosomal 5.8S-ITS DNA amplicons from five different Aspergilli grape isolates (presented as isolate designations), after digestion with the restriction endonucleases Hha I, Hinf I and Rsa I. Each isolate is a representative of the five different Aspergillus species characterized in this study. L: DNA ladder.
Table 1. Identification, origin and OTA production of A. section Nigri isolates.
| Spp. - No | Species | Origin | OTA level (ppb) | |
| HPLC | ELISA | |||
| At-1 | A. tubingensis | Crete | <LOD | |
| At-2 | A. tubingensis | Crete | <LOD | |
| Ac-1 | A. carbonarius | Crete | 31.01 | 32.20 |
| Ac-2 | A. carbonarius | Crete | 27.79 | 43.16 |
| Ac-3 | A. carbonarius | Crete | 42.40 | 44.85 |
| Ac-4 | A. carbonarius | Crete | 1208.42 | 803.52 |
| Ac-5 | A. carbonarius | Crete | 814.63 | 665.20 |
| Ac-6 | A. carbonarius | Crete | 1208.79 | 906.39 |
| Ac-7 | A. carbonarius | Crete | 1040.77 | 550.92 |
| Ac-8 | A. carbonarius | Crete | 1290.46 | 590.72 |
| Ac-9 | A. carbonarius | Crete | 1270.25 | 585.80 |
| Ac-10 | A. carbonarius | Crete | 116.90 | 62.46 |
| Ac-11 | A. carbonarius | Crete | 6512.18 | 4828.77 |
| Ac-12 | A. carbonarius | Crete | 442.60 | 290.79 |
| Ac-13 | A. carbonarius | Crete | 1013.88 | 405.31 |
| Ac-14 | A. carbonarius | Crete | 1274.33 | 837.26 |
| Ac-15 | A. carbonarius | Crete | 706.68 | 415.62 |
| Ac-16 | A. carbonarius | Crete | 62.44 | 31.88 |
| Ac-17 | A. carbonarius | Crete | 68.88 | 13.42 |
| Ac-18 | A. carbonarius | Crete | 347.18 | 61.63 |
| Ac-19 | A. carbonarius | Crete | 229.36 | 79.67 |
| Ac-20 | A. carbonarius | Crete | 397.93 | 107.69 |
| Ac-21 | A. carbonarius | Crete | 556.58 | 140.77 |
| Ac-22 | A. carbonarius | Crete | 44.58 | 23.65 |
| Ac-23 | A. carbonarius | Crete | 355.52 | 279.08 |
| Ac-24 | A. carbonarius | Crete | 94.86 | 18.25 |
| Ac-25 | A. carbonarius | Crete | 760.72 | 299.86 |
| Ac-26 | A. carbonarius | Crete | 644.05 | 615.97 |
| At-3 | A. tubingensis | Crete | 227.37 | 314.39 |
| Ac-27 | A. carbonarius | Crete | <LOD | |
| At-4 | A. tubingensis | Crete | <LOD | |
| At-5 | A. tubingensis | Crete | <LOD | |
| At-6 | A. tubingensis | Crete | <LOD | |
| Ac-28 | A. carbonarius | Crete | 12782.62 | 5010.56 |
| Ac-29 | A. carbonarius | Crete | 34825.69 | 30456.77 |
| Ac-30 | A. carbonarius | Crete | 86.00 | 58.42 |
| At-7 | A. tubingensis | Attica | <LOD | |
| Ac-31 | A. carbonarius | Attica | 2263.18 | 742.60 |
| At-8 | A. tubingensis | Attica | <LOD | |
| Ac-32 | A. carbonarius | Attica | 31.68 | 17.79 |
| At-9 | A. tubingensis | Attica | <LOD | |
| At-10 | A. tubingensis | Attica | <LOD | |
| At-11 | A. tubingensis | Attica | <LOD | |
| Ac-33 | A. carbonarius | Attica | 18752.53 | 7221.62 |
| Ac-34 | A. carbonarius | Attica | 748.87 | 216.94 |
| At-12 | A. tubingensis | Attica | <LOD | |
| At-13 | A. tubingensis | Attica | <LOD | |
| At-14 | A. tubingensis | Attica | <LOD | |
| Ac-35 | A. carbonarius | Attica | 553.06 | 341.50 |
| At-15 | A. tubingensis | Attica | <LOD | |
| Ac-36 | A. carbonarius | Attica | 121.32 | 116.19 |
| An-1 | A. niger aggr. | Attica | <LOD | |
| Ac-37 | A. carbonarius | Attica | 54.13 | 69.34 |
| Ac-38 | A. carbonarius | Attica | 57.92 | 43.92 |
| At-16 | A. tubingensis | Attica | <LOD | |
| At-17 | A. tubingensis | Attica | <LOD | |
| At-18 | A. tubingensis | Attica | <LOD | |
| At-19 | A. tubingensis | Attica | <LOD | |
| Ac-39 | A. carbonarius | Attica | 618.33 | 393.06 |
| At-20 | A. tubingensis | Attica | <LOD | |
| At-21 | A. tubingensis | Attica | <LOD | |
| Ac-40 | A. carbonarius | Attica | 182.46 | 97.34 |
| Ac-41 | A. carbonarius | Attica | 194.28 | 107.03 |
| Ac-42 | A. carbonarius | Attica | 17.64 | 7.85 |
| At-22 | A. tubingensis | Attica | <LOD | |
| At-23 | A. tubingensis | Attica | <LOD | |
| At-24 | A. tubingensis | Attica | <LOD | |
| Ac-43 | A. carbonarius | Attica | 4.71 | 7.73 |
| Ac-43 | A. carbonarius | Attica | 2.07 | 2.03 |
| At-25 | A. tubingensis | Attica | <LOD | |
| Ac-45 | A. carbonarius | Attica | 671.44 | 496.88 |
| Ai-1 | A. ibericus | Attica | <LOD | |
| At-26 | A. tubingensis | Attica | <LOD | |
| At-27 | A. tubingensis | Attica | <LOD | |
| Ac-46 | A. carbonarius | Attica | 4.26 | 3.83 |
| An-2 | A. niger aggr. | Attica | <LOD | |
| At-28 | A. tubingensis | Attica | <LOD | |
| At-29 | A. tubingensis | Attica | <LOD | |
| At-30 | A. tubingensis | Attica | <LOD | |
| Ac-47 | A. carbonarius | Attica | 3924.93 | 4041.81 |
| At-31 | A. tubingensis | Attica | <LOD | |
| Ac-48 | A. carbonarius | Attica | 2725.74 | 2998.55 |
| At-32 | A. tubingensis | Attica | <LOD | |
| At-33 | A. tubingensis | Attica | <LOD | |
| Ac-49 | A. carbonarius | Attica | 23.24 | 9.87 |
| At-34 | A. tubingensis | Attica | <LOD | |
| At-35 | A. tubingensis | Attica | <LOD | |
| At-36 | A. tubingensis | Attica | <LOD | |
| At-37 | A. tubingensis | Attica | <LOD | |
| At-38 | A. tubingensis | Attica | <LOD | |
| At-39 | A. tubingensis | Attica | <LOD | |
| At-40 | A. tubingensis | Attica | <LOD | |
| At-41 | A. tubingensis | Attica | <LOD | |
| At-42 | A. tubingensis | Attica | <LOD | |
| Aj-1 | A. japonicus | Attica | <LOD | |
| At-43 | A. tubingensis | Peloponnese | <LOD | |
| At-44 | A. tubingensis | Peloponnese | <LOD | |
| At-45 | A. tubingensis | Peloponnese | <LOD | |
| At-46 | A. tubingensis | Peloponnese | <LOD | |
| At-47 | A. tubingensis | Peloponnese | <LOD | |
| At-48 | A. tubingensis | Peloponnese | <LOD | |
| Ac-50 | A. carbonarius | Peloponnese | 90.34 | 59.40 |
| Ac-51 | A. carbonarius | Peloponnese | 141.51 | 82.34 |
| Ac-52 | A. carbonarius | Peloponnese | 82.00 | 56.81 |
| Ac-53 | A. carbonarius | Peloponnese | 65.44 | 55.56 |
| An-3 | A. niger aggr. | Peloponnese | <LOD | |
| Ac-54 | A. carbonarius | Peloponnese | 684.39 | 675.38 |
| Ac-55 | A. carbonarius | Peloponnese | 741.88 | 662.32 |
| Ac-56 | A. carbonarius | Peloponnese | 247.80 | 152.97 |
| At-49 | A. tubingensis | Peloponnese | <LOD | |
| At-50 | A. tubingensis | Peloponnese | <LOD | |
| Ac-57 | A. carbonarius | Peloponnese | 745.14 | 686.79 |
| At-51 | A. tubingensis | Peloponnese | <LOD | |
| Ac-58 | A. carbonarius | Peloponnese | 167.52 | 116.37 |
| At-52 | A. tubingensis | Peloponnese | <LOD | |
| At-53 | A. tubingensis | Macedonia | <LOD | |
| At-54 | A. tubingensis | Macedonia | <LOD | |
| Ac-59 | A. carbonarius | Macedonia | 1496.38 | 605.75 |
| At-55 | A. tubingensis | Macedonia | <LOD | |
| At-56 | A. tubingensis | Macedonia | <LOD | |
| An-4 | A. niger aggr. | Macedonia | <LOD | |
| At-57 | A. tubingensis | Macedonia | <LOD | |
| At-58 | A. tubingensis | Macedonia | <LOD | |
| At-59 | A. tubingensis | Macedonia | <LOD | |
| At-60 | A. tubingensis | Macedonia | <LOD | |
| At-61 | A. tubingensis | Macedonia | <LOD | |
| Ac-60 | A. carbonarius | Macedonia | 3654.06 | 2273.10 |
| At-62 | A. tubingensis | Macedonia | <LOD | |
Species identification and origin of A. section Nigri spp. isolated from the present study and their ochratoxigenic potential assayed by HPLC and ELISA methods.
Limit of Quantification (LOQ) 2 ng OTA g−1 CYA and Limit of Detection (LOD) 1 ng OTA g−1 CYA.
Sequencing and Phylogeny study
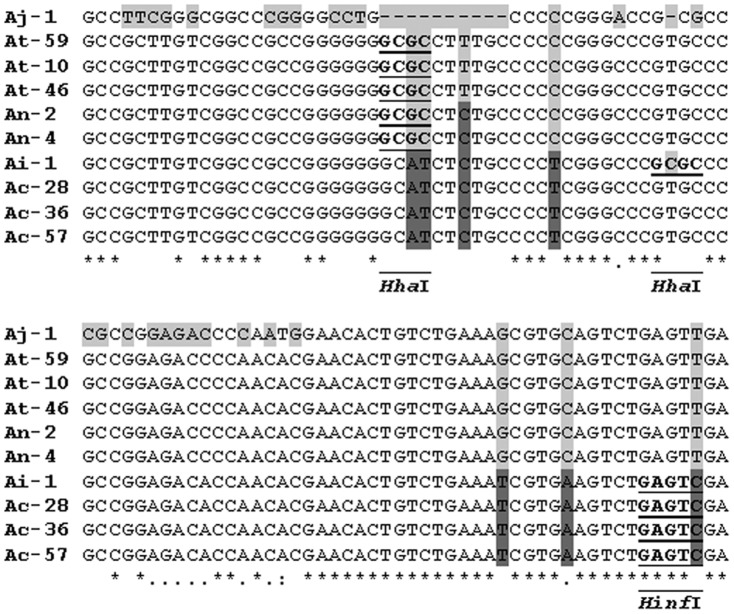
Representative isolates (spp. Ac-28, At-10, Ac-36, Ai-1, An-2, Aj-1, At-46, Ac-57, An-4 and At-59) for each species as resulted from PCR-RFLP analysis were subjected to sequencing for results verification. A part from the resulted sequences alignment can be seen in Figure 5. The DNA sequence restriction pattern analysis was in accordance with these of reference strains for all isolates sequenced except isolate Ai-1. Isolate Ai-1 had a HinfI restriction site pattern common to A. carbonarius, however, the presence of an extra HhaI restriction site (nucleotides 143 to 146) generated a HhaI restriction site profile similar to this of A. japonicus. A presentation of the DNA fragments generated according to the HinfI, HhaI and RsaI restriction sites, in all sequenced isolates and representative reference isolates of black aspergilli species isolated from grapes can be seen in Table 2. The fragment sizes that are generated resemble the sizes of the fragments visualized after gel electrophoresis of the amplicon digests.
Figure 5. Alignment of the ribosomal 5.8S-ITS amplicon sequences of representative Aspergilli isolates.
A part of the amplicon sequences (nucleotides 101 to 200) alignment is presented. Restriction sites are presented as bold-underlined, and variable nucleotides are highlighted. The extra HhaI restriction site present in the Ai-1 sequence is also shown.
Table 2. 5.8S-ITS RFLP DNA fragment sizes profiles.
| Isolate | Rest. Endonuclease | ||
| HinfI | HhaI | RsaI | |
| Ac-28, Ac-36, Ac-57 (A. carbonarius) | 288, 191, 110, 8 | 330, 178, 89 | 521, 76 |
| At-10, At-46, At-59 (A. tubingensis) | 302, 289, 8 | 207, 178, 124, 90 | 599 |
| An-2, An-4 (A. niger aggregate) | 302, 289, 8 | 207, 178, 124, 90 | 523, 76 |
| Ai-1 (A. ibericus) | 289, 192, 110, 8 | 186, 178, 145, 90 | 523, 76 |
| Aj-1 (A. japonicus) | 293, 274, 8 | 185, 178, 137, 75 | 497, 78 |
| A. carbonarius AF459734 | 288, 191, 110, 8 | 330, 178, 89 | 521, 76 |
| A. ibericus AY656624 | 289, 192, 110, 8 | 186, 178, 145, 90 | 523, 76 |
| A. brasiliensis AJ280010 | 290, 192, 110, 8 | 207, 178, 124, 91 | 524, 76 |
| A. niger AJ223852 | 301, 290, 8 | 207, 178, 123, 91 | 524, 75 |
| A. tubingensis FJ629367 | 302, 289, 8 | 207, 178, 124, 90 | 599 |
| A. awamori AM087614 | 302, 289, 8 | 207, 178, 124, 90 | 523, 76 |
| A. foetidus AY373850 | 302, 286, 8 | 207, 178, 124, 87 | 520, 76 |
| A. aculeatus AJ279997 | 273, 183, 110, 8 | 185, 177, 137, 75 | 496, 78 |
| A. japonicus FJ629335 | 293, 274, 8 | 185, 178, 137, 75 | 497, 78 |
| A. uvarum AM745757 | 293, 273, 8 | 185, 177, 137, 75 | 496, 78 |
Individual isolates and reference strain isolates from grapes are clustered according to their RFLP profile deduced after sequencing of the 5.8S-ITS amplicon. Fragment sizes generated respect to the restriction endonuclease site position in the sequence, are given in bp.
BLAST analysis of all sequences indicated that 3 clones had high similarity to A. carbonarius, 3 to A. tubingensis, 2 to A. niger/A. awamori, 1 to A. japonicus/A. aculeatus and 1 to A. ibericus. The results support the characterization of the 10 isolates deduced from the RFLP analysis and suggest that the isolate with the A-D HinfI-HhaI RFLP pattern can be classified as A. ibericus.
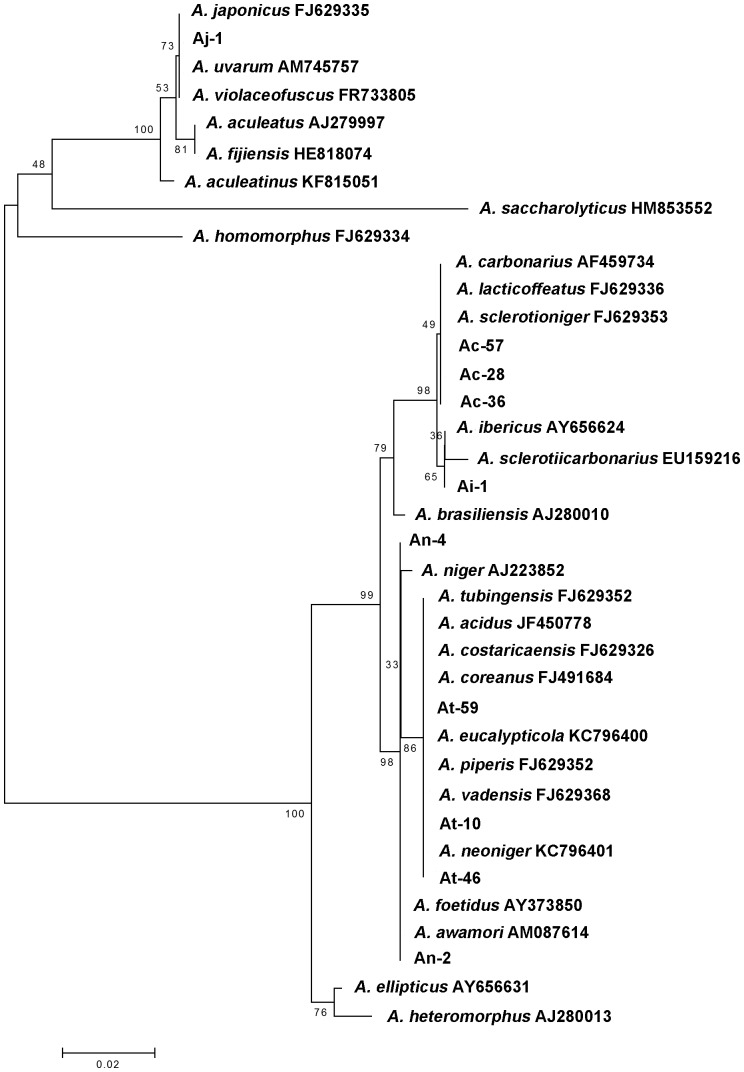
The phylogenetic relationship between the isolates of Aspergillus species is illustrated in an N-J method cluster analysis (Figure 6). The ten sequences were aligned together with reference strains sequences retrieved from NCBI resulting to the formation of five clades. Isolate Aj-1 was aligned with uniseriate A. japonicus, A. uvarum and A. aculeatus forming together with A. aculeatinus, A. fijiensis and A. violaceofuscus a clade separated from all other biseriate Aspergilli sequences with a 100% bootstrap value. Two other major clades were formed. The first consisted by A. carbonarius/A. ibericus isolates (Ac-28, Ac-36, Ac-57 and Ai-1) together with A. carbonarius, A. ibericus and A. brasiliensis outgroup reference sequences, each one of the last three species forming a separate subgroup inside the clade. A. carbonarius sequences were aligned together and were separated from the A. ibericus sequences with a 98% bootstrap value. A. brasiliensis formed a separate branch in this clade with a 79% bootstrap value. The second major clade was formed by A. tubingensis/A. niger aggregate isolates (At-10, At-46, At-59 and An-2, An-4 respectively) and A. niger aggregate outgroup reference sequences. A. tubingensis sequences were clustered together with other black aspergilli (not reported to be found in grapes), forming a subgroup separated from all other A. niger aggregate (A. niger, A. awamori and A. foetidus) species of the clade, with a 86% bootstrap value. Two other minor clades were also formed, one by A. saccharolyticus together with A. homomorphus and the other by A. ellipticus together with A. heteromorphus.
Figure 6. Neighbour-Joining phylogenetic tree based on divergences of ribosomal 5.8S-ITS sequences.
The alignment of ribosomal 5.8S-ITS sequences from 10 isolates and 10 reference strains of Aspergillus spp. was performed using the CLUSTALΩ program. Nucleotide divergences were estimated according to the Jukes–Cantor model. Node numbers represent the frequency (proportion) with which a cluster appears in 1000 bootstrap runs.
Ochratoxigenic potential of Aspergillus section Nigri species
A total of 128 isolates from grapes belonging to Aspergillus section Nigri, were tested in vitro for their potential to produce ochratoxin A. For the estimation of their ochratoxigenic potential a Competitive Direct ELISA procedure was also applied apart from the official HPLC method. Results showed that of the 60 A. carbonarius isolates 59 (98.33%) produced OTA from all geographic regions assayed. In contrast, among the A. niger aggregate, only 1 out of 62 isolates (1.61%), identified as A. tubingensis, had the potential to produce OTA. Finally, neither A. ibericus nor A. japonicus isolates were capable for OTA production.
Referring to ranges of OTA production, within the A. carbonarius species 1 isolate (1.67%) produced less than 1 ng OTA g−1 CYA, 19 isolates (31.67%) produced between 1 and 100 ng OTA g−1 CYA, 24 (40%) between 100 and 1000 ng OTA g−1 CYA, 13 (21.67%) between 1000 and 10000 ng OTA g−1 CYA and 3 isolates (5%) had an extremely high OTA potential with quantities greater than 10000 ng OTA g−1 CYA (Table 3). As regards the single A. tubingensis OTA producing isolate it had a potential of 227 ng OTA g−1 CYA.
Table 3. OTA ranges and distribution.
| A. carbonarius | A. tubingensis | A. niger aggregate | A. ibericus | A. japonicus | A. section Nigri | A. carbonarius OTA ranges (ppb or ng g−1 CYA) | |||||
| Prefecture | n (OTA producers) | <1 | 1–100 | 100–1000 | 1000–10000 | >10000 | |||||
| Crete | 30 (29) | 6 (1) | 0 | 0 | 0 | 36 (30) | 1 | 8 | 11 | 8 | 2 |
| Peloponnese | 9 (9) | 10 (0) | 1 (0) | 0 | 0 | 20 (9) | 0 | 3 | 6 | 0 | 0 |
| Attica | 19 (19) | 36 (0) | 2 (0) | 1 (0) | 1 (0) | 59 (19) | 0 | 8 | 7 | 3 | 1 |
| Macedonia | 2 (2) | 10 (0) | 1 (0) | 0 | 0 | 13 (2) | 0 | 0 | 0 | 2 | 0 |
| Total n (% ΟΤΑ producers) | 60 (98.33%) | 62 (1.61%) | 4 (0%) | 1 (0%) | 1 (0%) | 128 (46.88%) | 1 | 19 | 24 | 13 | 3 |
OTA-producers and their distribution within the identified species of A. section Nigri group, and ranges of OTA potential for A. carbonarius isolates per prefecture.
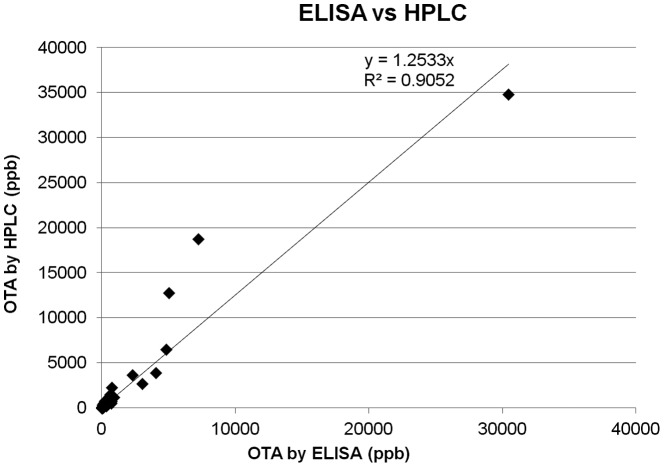
Regarding the performance of the ELISA test kit quantification, results were linearly correlated with those of the HPLC method with an r2 = 0.905 (Figure 7), but with an underestimation of the produced OTA at concentration levels above 5000 ppb. Results of both methods are presented in Table 1, together with the characterisation of the isolates after both phenotypic and molecular means.
Figure 7. Correlation of HPLC and ELISA values for OTA determination.
Linear correlation and R2 for the two methods assayed for OTA quantification (ELISA & HPLC).
Discussion
Incidence of isolation and distribution of Aspergillus section Nigri group among the fungal genera were in accordance with previous reports from Greece [20], [51] and other Mediterranean countries [52], [53], [54]. A very high incidence was observed in our samples since 23 out of 33 vineyards sampled presented 100% presence for black aspergilli, although some studies have reported lower indices for vineyards from some European and South American countries [19], [55], [56], [57].
Regarding the molecular characterisation of the isolated black aspergilli, a PCR-RFLP method was applied in order to differentiate the Aspergillus strains. RFLP analysis of the rDNA 5.8S ITS products was performed to identify different isolates of black Aspergillus species from grapes compared to reference strains. A total of 128 black Aspergilli isolate-ITS amplicons were digested using the HhaI, HinfI and RsaI restriction digestion endonucleases resulting in 4 different RFLP patterns for HhaI and RsaI respectively and 3 patterns for the HinfI enzyme digests. In previous studies [33], [39], [58], [59], [60] according to authors' descriptions these restriction endonucleases have been used successfully to differentiate the A. carbonarius, A. niger, A. tubingensis, A. japonicus and A. aculeatus species. It was shown that HinfI enzyme digestion can differentiate A. carbonarius from A. niger/A. tubingensis, and so does HhaI. Furthermore, HhaI can differentiate the uniseriate A. japonicus, A. aculeatus and A. uvarum species from all three previously reported biseriate aspergilli species. HinfI can distinguish further A. japonicus/A. uvarum from A. aculeatus and RsaI can distinguish A. tubingensis from all other species belonging to the A. niger aggregate [33], [39], [58], [59], [60].
According to the digestion pattern combinations resulted from the PCR-RFLP analysis of the 128 Aspergilli isolates and reference species, 60 were characterized as A. carbonarius, 62 as A. tubingensis and 4 as species belonging to the A. niger aggregate other than A. tubingensis. These results are in accordance with results presented by other authors, supporting the use of the HhaI, HinfI and RsaI restriction endonucleases for accurate characterization of A. carbonarius, A. tubingensis and, A. niger species [33], [39], [59], [60]. Two other digestion pattern combinations have resulted using the HhaI and HinfI enzymes. One combination resumed a pattern presented by Spadaro et al. [39], suggesting that the isolate possibly belongs to the A. japonicus/A. uvarum species. The other digest pattern combination suggested a characterization of the isolate as A. carbonarius according to HinfI digestion; however, this could not be supported from the HhaI digestion profile which closely resembled this of A. japonicus.
Sequencing of representative isolate DNAs supported the RFLP data since the restriction site profile of each isolate generated DNA fragments with molecular sizes that corresponded to the bands found after electrophoresis of the amplicon digests. An extra HhaI site is present in the sequence of isolate Ai-1 generating a restriction digestion profile different from the rest that are observed. A nucleotide substitution, C instead of T that is present in nearly all other sequences generates this extra HhaI site. The results presented in Table 2 show that the DNA fragments generated according to the HinfI restriction sites are nearly equal in size and close to the DNA fragment sizes that generate pattern B for (A. tubingensis and A. niger). Though the pattern generated for A. japonicus is not identical to pattern B, it is very similar and cannot be distinguished clearly from it in a gel electrophoresis due to similar sizes and the techniques' limited resolution. A similar conclusion for unisereriate/biseriate species may be also deduced when evaluating by gel electrophoresis a profile generated after RsaI digestion.
Sequence alignment with reference strain sequences retrieved from NCBI, together with BLAST analysis verified the RFLP results and showed that 3 out of the ten individual isolates were assigned to A. carbonarius, 3 to A. tubingensis, 2 showed high homology to members of the A. niger aggregate (A. niger/A. awamori) and 1 showed high homology to A. japonicus/A. uvarum species. Isolate Ai-1 was assigned to the species A. ibericus after BLAST analysis, showing 100% homology when aligned with A. ibericus AY656624.1, AY656623.1 and AY656622.1 strains. A. ibericus was first reported by Serra et al. [61] as a new Aspergillus species isolated from grapes and to our knowledge this is the first report that this species is isolated in Greece. Furthermore, as shown from a comparison of the A. ibericus 5.8S-ITS DNA sequences available at NCBI (data not shown), the fact that an extra HhaI site is present in all of these sequences makes it useful for characterization of A. ibericus by PCR-RFLP using this restriction endonuclease. The phylogenetic analysis performed using reference species' sequences as outgroups supports the sequence data and is in accordance with data presented by other authors using the ITS region for phylogenetic analysis of black aspergilli [37].
According to various authors [20], [34], [37], [38], [58], [62], [63,] black aspergilli that have been isolated from grapes fall into the biseriate A. carbonarius, A. tubingensis, A. niger, A. awamori, A. foetidus, A. brasiliensis and A. ibericus, and the uniseriate A. japonicus, A. uvarum and A. aculeatus species. Characterization of these isolates by molecular means has been based on different approximations using RFLP [25], [31], [33], [39], [64], AFLP [30], [32], [36], [65], or multilocus analysis [62], [66]. RFLP and AFLP have been used successfully for characterization of the main ochratoxigenic aspergilli isolated from grapes [25], [30], [31], [32], [33], [36], [39], [64], [65]. Multilocus analysis based on calmodulin, beta-tubulin and ITS sequences has proved a useful tool for extended characterization and comparative study of isolates from a great variety of sources such as grapes, coffee, soil, surfaces, or unknown origin [37], [62], [66]. In this work we have followed a similar approximation using ITS PCR-RFLP in order to characterize black aspergilli isolates from grapes. We have been able to characterize the isolates belonging to the A. carbonarius, A. tubingensis and A. ibericus species, since the RFLP patterns (Table 2) can clearly differentiate these species from all other black aspergilli species isolated from grapes.
The Aj-1 isolate could not be characterized only by molecular means since there is no difference between the RFLP patterns and ITS region DNA sequence for either species. According to the morphological observations the Aj-1 isolate was characterized as A. japonicus since its conidial size (4.3 μm) falls in the range of A. japonicus (4–5 μm) and not in the range of A. uvarum (3–4 μm) [34]. PCR-RFLP when used in combination with other means such as use of morphological criteria is a reliable approximation for black aspergilli characterization, where, for example, differentiation of aspergilli as biseriate/uniseriate can initially short the strains to two major groups. Isolates An-1 to An-4 have been characterized as A. niger aggregate members due to the limitation of the ITS PCR-RFLP to differentiate between A. niger, A. awamori and A. foetidus species, thus resulting to a partial molecular characterization of these four isolates.
With regard to the field study, Greece is a country with extended coast, although, areas of grape planting are characterized by different climatic characteristics. Vineyards localisation can vary from very low altitude, near seaside, either arid or wet, to very high altitudes and wet climate regions. Referring to the present study, influence of the geographic localization of the vineyards on the incidence of black aspergilli in grapes was significant. The vineyards selected for this study are representative of these different climatic profiles existing in the country. Most studies that correlated the geo-climatic conditions with the presence and distribution of black aspergilli reported a trend of higher isolation incidence in regions with higher mean temperatures, but more wet areas [19], [53], [66], [67]. These reports are partly in accordance with the present study, since the geographical localisation of sampled vineyards influenced black aspergilli presence, but with the higher incidences found in arid areas. In accordance with our results are the findings of Serra et al. [54], [56] for Spain where higher incidences for black aspergilli were reported from hot and dry regions. In addition, most of the ochratoxigenic isolates came from regions with arid climate, Crete and Attica prefecture, but this could be also correlated with the low altitude of these vineyards in contrast with the Peloponnese and Macedonia where altitudes of grape cultivation are higher. Moreover, the incidence of black aspergilli and the A. carbonarius isolation were much lower in Macedonia where temperatures are lower and the climate is wet compared with the other regions. In contrast, Chiotta et al. [65] have reported higher incidences of A. carbonarius from vineyards of higher altitudes. In addition, Perrone et al. [20] and Visconti et al. [24] denoted the strong correlation of relative humidity and rainfall with A. carbonarius presence and OTA accumulation in grapes. Finally, many works denote that, apart from the climate of a wider region, very important role have also the different practices used in grape cultivation and the specific microclimate of the vineyard [68], [69], [70].
A. niger aggregate and A. carbonarius were equally distributed in the present work, while only one uniseriate species was isolated and identified as A. japonicus. Similar distributions of A. carbonarius within the A. section Nigri group has been also reported by Bau et al. [69] for grapes from the Mediterranean Spanish coast and Tjamos et al. [51] for Rhodes island in Greece. These results are in accordance with our findings, since Crete, which has an insular Mediterranean climate, similar to those of Rhode and the Mediterranean Spanish coast, presented the higher incidence of A. carbonarius among black aspergilli. In contrast, El Khoury et al. [71] for Lebanon and Chiotta et al. [65], [66] for Argentina have reported relatively lower participation of A. carbonarius within black aspergilli, while Battilani et al. [19] reported very high occurrence (20%) of uniseriate species in Italian vineyards during grape ripening.
Several studies from countries all over the world during the last years have identified A. section Nigri group as the responsible fungi for OTA contamination of grapes and derived products. Specifically, A. carbonarius is considered as the main OTA contamination source, and to a lesser extend species of A. niger aggregate, mainly A. niger and A. tubingensis. This conclusion derives from the higher ochratoxigenic ability of A. carbonarius in contrast to A. niger aggregate isolates that, even when they are more abundant within the A. section Nigri group, they are either totally incapable of toxin production or their toxigenic ability is scarce and weak [19], [53], [72]. In our study, all A. carbonarius isolates, with one exception, were capable for OTA production with most of them having a high ochratoxigenic potential, while from A. niger aggregate only one isolate identified as Aspergillus tubingensis was capable to produce OTA in vitro (Table 3). Similar results have been reported for other Mediterranean countries [19], [54], [56], [69], [73], although there are reports of higher ochratoxigenic incidences among A. niger aggregate and uniseriate spp. [39]. Ranges of ochratoxigenic potential for A. carbonarius isolates varied from 0 to 34 ppm (μg OTA g−1 CYA), with most of them producing between 1 and 1000 ppb (Table 3). Our results are also in agreement with studies contacted in Greece [51], [68], although Greek isolates with higher ochratoxigenic potential have been reported [31]. Particularly, Tjamos et al. [51], [68] in their studies have also shown that most A. carbonarius isolates were strong OTA producers with capabilities greater than 25 ppb, while only 6% of the A. niger aggregate presented high OTA potential.
Conclusions
From our results there is a clear correlation of A. section Nigri group with the arid climate of Crete and Attica, whereas for colder and wet areas its presence is scarcer. Moreover, higher incidence of ochratoxigenic isolates and OTA production was recorded for the A. carbonarius isolates, confirming in this way that A. carbonarius is the main species responsible for OTA contamination in Greek grapes. Regarding PCR-RFLP our data indicate that A. ibericus can be differentiated from other aspergilli using the HhaI restriction endonuclease.
Acknowledgments
The authors are also grateful to Prof. N. Magan from Cranfield University for providing fungal reference strains and also to Assistant Prof. D. Tsitsigiannis from the Agricultural University of Athens for advisory support.
Funding Statement
This work has been supported by the project ‘Design and development of innovative tools for the detection of ochratoxigenic fungi in wine and table grapes – FungalPrognosis 242’ that is co-financed by the European Union (European Social Fund – ESF) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) - Research Funding Program: ARISTEIA-I, (www.fungalprognosis.gr). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. El Khoury A (2010) Atoui (2010) Ochratoxin A: general overview and actual molecular status. Toxins (Basel) (4): 461–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fuchs R, Peraica M (2005) Ochratoxin A in human kidney diseases. Food Addit Contam 22 (S1): 53–57. [DOI] [PubMed] [Google Scholar]
- 3. Pfohl-Leszkowicz A, Manderville RA (2007) Ochratoxin A: An overview on toxicity and carcinogenicity in animals and humans. Mol Nutr Food Res 51(1): 61–99. [DOI] [PubMed] [Google Scholar]
- 4. Reddy L, Bhoola K (2010) Ochratoxins-food contaminants: impact on human health. Toxins (Basel) (4): 771–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsuda Y, Wakai T, Kubota M, Osawa M, Sanpei A, et al. (2013) Mycotoxins are conventional and novel risk biomarkers for hepatocellular carcinoma. World J Gastroenterol 19(17): 2587–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abouzied MM, Horvath AD, Podlesny PM, Regina NP, Metodiev VD, et al. (2002) Ochratoxin A concentrations in food and feed from a region with Balkan Endemic Nephropathy. Food Addit Contam 19(8): 755–764. [DOI] [PubMed] [Google Scholar]
- 7. Stefanović V, Polenaković M (2009) Fifty years of research in Balkan endemic nephropathy: where are we now? Nephron Clin Pract 112(2): 51–56. [DOI] [PubMed] [Google Scholar]
- 8. Leong SL, Hien LT, An TV, Trang NT, Hocking AD, et al. (2007) Ochratoxin A-producing Aspergilli in Vietnamese green coffee beans. Lett Appl Microbiol 45(3): 301–106. [DOI] [PubMed] [Google Scholar]
- 9. Duarte SC, Pena A, Lino CM (2010) A review on ochratoxin A occurrence and effects of processing of cereal and cereal derived food products. Food Microbiol 27(2): 187–198. [DOI] [PubMed] [Google Scholar]
- 10. Palencia ER, Hinton DM, Bacon CW (2010) The black Aspergillus species of maize and peanuts and their potential for mycotoxin production. Toxins (Basel) 2(4): 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Velmourougane K, Bhat R, Gopinandhan TN (2010) Coffee berry borer (Hypothenemus hampei) - a vector for toxigenic molds and ochratoxin A contamination in coffee beans. Foodborne Pathog Dis 7(10): 1279–1284. [DOI] [PubMed] [Google Scholar]
- 12. Tittlemier SA, Varga E, Scott PM, Krska R (2011) Sampling of cereals and cereal-based foods for the determination of ochratoxin A: an overview. Food Addit Contam Part A 28(6): 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ponsone ML, Chiotta ML, Palazzini JM, Combina M, Chulze S (2012) Control of ochratoxin A production in grapes. Toxins (Basel) 4(5): 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.European Commission (EC) (2012) Rapid Alert System for Food and Feed (RASFF) Annual Report 2012. Directorate-General for Health and Consumers, Available: http://ec.europa.eu/food/food/rapidalert/docs/rasff_annual_report_2012_en.pdf. Accessed 1 October 2013.
- 15. Otteneder H, Majerus P (2001) Occurrence of ochratoxin A (OTA) in wines: influence of the type of wine and its geographical origin. Food Addit Contam 17(9): 793–798. [DOI] [PubMed] [Google Scholar]
- 16. Pietri A, Bertuzzi T, Pallaroni L, Piva G (2001) Occurrence of ochratoxin A in Italian wines. Food Addit Contam 18(7): 647–654. [DOI] [PubMed] [Google Scholar]
- 17. Lopez de Cerain A, González-Peñas E, Jiménez AM, Bello J (2002) Contribution to the study of ochratoxin A in Spanish wines. Food Addit Contam 19(11): 1058–1064. [DOI] [PubMed] [Google Scholar]
- 18. Battilani P, Pietri A, Bertuzzi T, Languasco L, Giorni P, et al. (2003) Occurrence of ochratoxin A-producing fungi in grapes grown in Italy. J Food Prot 66(4): 633–636. [DOI] [PubMed] [Google Scholar]
- 19. Battilani P, Barbano C, Marin S, Sanchis V, Kozakiewicz Z, et al. (2006) Mapping of Aspergillus Section Nigri in Southern Europe and Israel based on geostatistical analysis. Int J Food Microbiol 111 (S1): S72–82. [DOI] [PubMed] [Google Scholar]
- 20. Perrone G, De Girolamo A, Sarigiannis Y, Haidukowski ME, Visconti A (2013) Occurrence of ochratoxin A, fumonisin B2 and black aspergilli in raisins from Western Greece regions in relation to environmental and geographical factors. Food Addit Contam Part A 30(7): 1339–1347. [DOI] [PubMed] [Google Scholar]
- 21. European Commission (EC) (2005) Commission regulation (EC) no 123/2005 of 26 January 2005 amending regulation (EC) no 466/2001 as regards ochratoxin A. Off J Eur Union. L25: 3–5. [Google Scholar]
- 22. European Commission (EC) (2006) Commission regulation (EC) No 1881/2006 of 19 December 2006 setting maximum levels for certain contaminants in foodstuffs. Off J Eur Union L365: 6–24. [Google Scholar]
- 23. Leong SL, Hocking AD, Varelis P, Giannikopoulos G, Scott ES (2006) Fate of ochratoxin A during vinification of Semillon and Shiraz grapes. J Agric Food Chem 54(17): 6460–6464. [DOI] [PubMed] [Google Scholar]
- 24. Visconti A, Perrone G, Cozzi G, Solfrizzo M (2008) Managing ochratoxin A risk in the grape-wine food chain. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 25(2): 193–202. [DOI] [PubMed] [Google Scholar]
- 25. Martínez-Culebras PV, Crespo-Sempere A, Sánchez-Hervás M, Elizaquivel P, Aznar R, et al. (2009) Molecular characterization of the black Aspergillus isolates responsible for ochratoxin A contamination in grapes and wine in relation to taxonomy of Aspergillus section Nigri. Int J Food Microbiol 132(1): 33–41. [DOI] [PubMed] [Google Scholar]
- 26. Mateo R, Medina A, Mateo EM, Mateo F, Jiménez M (2007) An overview of ochratoxin A in beer and wine. Int J Food Microbiol 119(1–2): 79–83. [DOI] [PubMed] [Google Scholar]
- 27. Astoreca AL, Magnoli CE, Dalcero AM (2010) Ecophysiology of Aspergillus section nigri species potential ochratoxin a producers. Toxins (Basel) 2(11): 2593–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Magan N (2006) Mycotoxin contamination of food in Europe: early detection and prevention strategies. Mycopathologia 162(3): 245–253. [DOI] [PubMed] [Google Scholar]
- 29. Abarca ML, Accensi F, Cano J, Cabañes FJ (2004) Taxonomy and significance of black aspergilli. Antonie Van Leeuwenhoek 86(1): 33–49. [DOI] [PubMed] [Google Scholar]
- 30. Schmidt H, Niessen L, Vogel RF (2005) Molecular diagnosis of ochratoxinogenic Aspergillus species. Mycotoxin Res 21(1): 61–64. [DOI] [PubMed] [Google Scholar]
- 31. Bau M, Castellá G, Bragulat MR, Cabañes FJ (2005) DNA-based characterization of ochratoxin-A-producing and non-producing Aspergillus carbonarius strains from grapes. Res Microbiol 2005 156(3): 375–381. [DOI] [PubMed] [Google Scholar]
- 32. Perrone G, Susca A, Epifani F, Mulè G (2006) AFLP characterization of Southern Europe population of Aspergillus Section Nigri from grapes. Int J Food Microbiol 111 (S1): S22–27. [DOI] [PubMed] [Google Scholar]
- 33. Martínez-Culebras PV, Ramón D (2007) An ITS-RFLP method to identify black Aspergillus isolates responsible for OTA contamination in grapes and wine. Int J Food Microbiol 113(2): 147–153. [DOI] [PubMed] [Google Scholar]
- 34. Perrone G, Susca A, Cozzi G, Ehrlich K, Varga J, et al. (2007) Biodiversity of Aspergillus species in some important agricultural products. Stud Mycol. 59: 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oliveri C, Torta L, Catara V (2008) A polyphasic approach to the identification of ochratoxin A-producing black Aspergillus isolates from vineyards in Sicily. Int J Food Microbiol 127(1–2): 147–154. [DOI] [PubMed] [Google Scholar]
- 36. Esteban A, Leong SL, Hocking AD, Abarca ML, Cabañes FJ, et al. (2008) Utility of microsatellite markers and amplified fragment length polymorphism in the study of potentially ochratoxigenic black aspergilli. Curr Microbiol 57(4): 348–355. [DOI] [PubMed] [Google Scholar]
- 37. Varga J, Frisvad JC, Kocsubé S, Brankovics B, Tóth B, et al. (2011) New and revisited species in Aspergillus section Nigri. Stud Mycol. Jun 30 69(1): 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Perrone G, Stea G, Epifani F, Varga J, Frisvad JC, et al. (2011) Aspergillus niger contains the cryptic phylogenetic species A. awamori. Fungal Biol. Nov;115(11): 1138–50. [DOI] [PubMed] [Google Scholar]
- 39. Spadaro D, Patharajan S, Lorè A, Garibaldi A, Gullino ML (2012) Ochratoxigenic black species of Aspergilli in grape fruits of northern Italy identified by an improved PCR-RFLP procedure. Toxins (Basel) 4(2): 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cary JW, Ehrlich KC (2006) Aflatoxigenicity in Aspergillus: molecular genetics, phylogenetic relationships and evolutionary implications. Mycopathologia 162(3): 167–177. [DOI] [PubMed] [Google Scholar]
- 41. Borman AM, Linton CJ, Miles SJ, Johnson EM (2008) Molecular identification of pathogenic fungi. J Antimicrob Chemother 61 (S1): 7–12. [DOI] [PubMed] [Google Scholar]
- 42. Levin RE (2012) PCR detection of aflatoxin producing fungi and its limitations. Int J Food Microbiol 156(1): 1–6. [DOI] [PubMed] [Google Scholar]
- 43.Samson RA, Hoekstra ES, Frisvad JC (2000) Introduction to Food and Airborne Fungi. Netherlands, Utrecht: Centraalbureau voor Schimmelcultures. p.389.
- 44.Pitt JI, Hocking AD (2009). Fungi and Food Spoilage. London - New York: Springer p.519.
- 45.White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ editors. PCR Protocols: a Guide to Methods and Applications. New York: Academic Press.pp. 315–322.
- 46. Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4): 406–425. [DOI] [PubMed] [Google Scholar]
- 47. Kumar S, Tamura K, Nei M (2004) “MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment”. Brief Bioinformatics 5(2): 150–163. [DOI] [PubMed] [Google Scholar]
- 48. Bragulat MR, Abarca ML, Cabanes FJ (2001) An easy screening method for fungi producing ochratoxin A in pure culture. Int J Food Microbiol 71: 139–144. [DOI] [PubMed] [Google Scholar]
- 49. Valero A, Farré JR, Sanchis V, Ramos AJ, Marín S (2006) Kinetics and spatial distribution of OTA in Aspergillus carbonarius cultures. Food Microbiol 23: 753–756. [DOI] [PubMed] [Google Scholar]
- 50. Kapetanakou AE, Panagou EZ, Gialitaki M, Drosinos EH (2009) Evaluating the combined effect of water activity, pH and temperature on ochratoxin A production by Aspergillus ochraceus and Aspergillus carbonarius on culture medium and Corinth raisins. Food Control 20: 725–732. [Google Scholar]
- 51. Tjamos SE, Antoniou PP, Tjamos EC (2006) Aspergillus spp., distribution, population composition and ochratoxin A production in wine producing vineyards in Greece. Int J Food Microbiol 111(S1): S61–S66. [DOI] [PubMed] [Google Scholar]
- 52. Bellí N, Bau M, Marín S, Abarca ML, Ramos AJ, et al. (2006) Mycobiota and ochratoxin A producing fungi from Spanish wine grapes. Int J Food Microbiol 111(S1): S40–S45. [DOI] [PubMed] [Google Scholar]
- 53. Lasram S, Oueslati S, Mliki A, Ghorbel A, Silar P, et al. (2012) Ochratoxin A and ochratoxigenic black Aspergillus species in Tunisian grapes cultivated in different geographic areas. Food Control 25: 75–80. [Google Scholar]
- 54. Serra R, Braga A, Venâncio A (2005) Mycotoxin-producing and other fungi isolated from grapes for wine production, with particular emphasis on ochratoxin A. Res Microbiol 156: 515–521. [DOI] [PubMed] [Google Scholar]
- 55. Magnoli C, Violante M, Combina M, Palacio G, Dalcero AM (2003) Mycoflora and ochratoxin-producing strains of Aspergillus section Nigri in wine grapes in Argentina. Lett Appl Microbiol 37: 179–184. [DOI] [PubMed] [Google Scholar]
- 56. Serra R, Abrunhosa L, Kozakiewicz Z, Venâncio A (2003) Black Aspergillus species as ochratoxin A producers in Portuguese wine grapes. Int J Food Microbiol 88: 63–68. [DOI] [PubMed] [Google Scholar]
- 57. Díaz GA, Torres R, Vega M, Latorre BA (2009) Ochratoxigenic Aspergillus species on grapes from Chilean vineyards and Aspergillus threshold levels on grapes. Int J Food Microbiol 133: 195–199. [DOI] [PubMed] [Google Scholar]
- 58. Accensi F, Abarca ML, Cano J, Figuera L, Cabañes FJ (2001) Distribution of ochratoxin A producing strains in the A. niger aggregate. Antonie van Leeuwenhoek 79: 365–370. [DOI] [PubMed] [Google Scholar]
- 59. Medina A, Mateo R, López-Ocaña L, Valle-Algarra FM, Jiménez M (2005) Study of Spanish grape mycobiota and ochratoxin A production by Isolates of Aspergillus tubingensis and other members of Aspergillus section Nigri. Appl Environ Microbiol. Aug;71(8): 4696–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Apriyanto DN, Francis MCSS, Baharuddin S, Endang SR (2013) Ochratoxigenic Black Aspergilli Isolated from Dried Agricultural Products in Yogyakarta, Indonesia. J Food Sci Engineer 3, 472–480. [Google Scholar]
- 61. Serra R, Cabañes FJ, Perrone G, Castellá G, Venâncio A, et al. (2006) Aspergillus ibericus: a new species of section Nigri isolated from grapes. Mycologia 98(2): 295–306. [DOI] [PubMed] [Google Scholar]
- 62. Susca A, Perrone G, Cozzi G, Stea G, Logrieco AF, et al. (2013) Multilocus sequence analysis of Aspergillus Sect. Nigri in dried vine fruits of worldwide origin. Int J Food Microbiol. Jul 15;165(2): 163–8. [DOI] [PubMed] [Google Scholar]
- 63. Varga J, Kocsubé S, Suri K, Szigeti G, Szekeres A, et al. (2010) Fumonisin contamination and fumonisin producing black Aspergilli in dried vine fruits of different origin. Int J Food Microbiol. Oct 15;143(3): 143–9. [DOI] [PubMed] [Google Scholar]
- 64. Bau M, Castellá G, Bragulat MR, Cabañes FJ (2006) RFLP characterization of Aspergillus niger aggregate species from grapes from Europe and Israel. Int J Food Microbiol. 1;111, 18–21. [DOI] [PubMed] [Google Scholar]
- 65. Chiotta ML, Ponsone ML, Sosa DM, Combina M, Chulze SN (2013) Biodiversity of Aspergillus section Nigri populations in Argentinean vineyards and ochratoxin A contamination. Int J Food Microbiol 149: 171–176. [DOI] [PubMed] [Google Scholar]
- 66. Chiotta ML, Susca A, Stea G, Mulè G, Perrone G, et al. (2011) Phylogenetic characterization and ochratoxin A – Fumonisin profile of black Aspergillus isolated from grapes in Argentina. Int J Food Microbiol 149: 171–176. [DOI] [PubMed] [Google Scholar]
- 67. Chiotta ML, Ponsone ML, Combina M, Torres AM, Chulze SN (2009) Aspergillus section Nigri species isolated from different wine-grape growing regions in Argentina. Int J Food Microbiol 136: 137–141. [DOI] [PubMed] [Google Scholar]
- 68. Tjamos SE, Antoniou PP, Kazantzidou A, Antonopoulos DF, Papageorgiou I, et al. (2004) Aspergillus niger and Aspergillus carbonarius in Corinth Raisin and Wine-Producing Vineyards in Greece: Population Composition, Ochratoxin A Production and Chemical Control. J Phytopathology 152: 250–255. [Google Scholar]
- 69. Bau M, Bragulat MR, Abarca ML, Minguez S, Cabañes FJ (2005) Ochratoxigenic species from Spanish wine grapes. Int J Food Microbiol 98: 125–130. [DOI] [PubMed] [Google Scholar]
- 70. Labrinea EP, Natskoulis PI, Spiropoulos AE, Naresh M, Tassou CC (2011) A survey of ochratoxin A occurrence in Greek wines, Food Addit Contam Part B. 4(1): 61–66. [DOI] [PubMed] [Google Scholar]
- 71. El Khoury A, Rizk T, Lteif R, Azouri H, Delia M-L, et al. (2008) Fungal contamination and Aflatoxin B1 and Ochratoxin A in Lebanese wine-grapes and musts. Food Chem Toxicol 46: 2244–2250. [DOI] [PubMed] [Google Scholar]
- 72. Palumbo JD, O'Keeffe TL, Vasquez SJ, Mahoney NE (2011) Isolation and identification of ochratoxin A-producing Aspergillus section Nigri strains from California raisins. Lett Appl Microbiol 52: 330–336. [DOI] [PubMed] [Google Scholar]
- 73. Lasram S, Bellí N, Chebil S, Nahla Z, Ahmed M, et al. (2007) Occurrence of ochratoxigenic fungi and ochratoxin A in grapes from a Tunisian vineyard. Int J Food Microbiol 114: 376–379. [DOI] [PubMed] [Google Scholar]