Abstract
Introduction
Circulating tumor cells (CTCs) that present mesenchymal phenotypes can escape standard methods of isolation, thus limiting possibilities for their characterization. Whereas mesenchymal CTCs are considered to be more malignant than epithelial CTCs, factors responsible for this aggressiveness have not been thoroughly defined. This study analyzed the molecular profile related to metastasis formation potential of CTC-enriched blood fractions obtained by marker unbiased isolation from breast cancer patients without (N−) and with lymph nodes metastases (N+).
Materials and Methods
Blood samples drawn from 117 patients with early-stage breast cancer were enriched for CTCs using density gradient centrifugation and negative selection with anti-CD45 covered magnetic particles. In the resulting CTC-enriched blood fractions, expression of CK19, MGB1, VIM, TWIST1, SNAIL, SLUG, HER2, CXCR4 and uPAR was analyzed with qPCR. Results were correlated with patients' clinicopathological data.
Results
CTCs (defined as expression of either CK19, MGB1 or HER2) were detected in 41% (20/49) of N− and 69% (34/49) of N+ patients (P = 0.004). CTC-enriched blood fractions of N+ patients were more frequently VIM (P = 0.02), SNAIL (P = 0.059) and uPAR-positive (P = 0.03). Positive VIM, CXCR4 and uPAR status correlated with >3 lymph nodes involved (P = 0.003, P = 0.01 and P = 0.045, respectively). In the multivariate logistic regression MGB1 and VIM-positivity were independently related to lymph node involvement with corresponding overall risk of 3.2 and 4.2. Moreover, mesenchymal CTC-enriched blood fractions (CK19−/VIM+ and MGB1+ or HER2+) had 4.88 and 7.85-times elevated expression of CXCR4 and uPAR, respectively, compared with epithelial CTC-enriched blood fractions (CK19+/VIM− and MGB1+ or HER2+).
Conclusions
Tumors of N+ patients have superior CTC-seeding and metastatic potential compared with N- patients. These differences can be attributed to VIM, uPAR and CXCR4 expression, which endow tumor cells with particularly malignant phenotypes.
Introduction
Presence of circulating tumor cells (CTCs) in blood of patients with epithelial cancer was first demonstrated by Ashworth in 1869 [1]. As techniques were developed to capture, enumerate and characterize circulating and disseminated tumor cells, significant progress was made in understanding metastatic processes [2], [3], [4], [5]. The number of CTCs detected in blood samples carry prognostic information in early [6], [7] and metastatic breast cancer [8], [9]. Also, CTCs detected via PCR-based methods (without the possibility of cell enumeration) have been associated with poor prognosis in a number of studies [10], [11], described in a recent meta-analysis [12].
As CTCs originate from the epithelium, use of epithelial markers (eg, cytokeratins, EpCAM) for their detection seems reasonable. Cytokeratin 19 (CK19) is a cytoskeletal protein of epithelial cells (both normal and cancerous) and is widely used for detection of CTCs [13], [14], [15] and DTCs [15], [16], [17], [18]. However, discovery of epithelial–mesenchymal transition (EMT) in cancer educes a reconsideration of CTCs as having exclusively epithelial phenotype [2], [19], [20], [21]. Moreover, the role of EMT in cancer implies that detection methods that rely solely on epithelial markers (or other markers downregulated during EMT) are likely to miss the most aggressive fraction of CTCs [2], [19], [22]. Thus, to increase sensitivity it is suggested to include additional mammary transcripts, like mammaglobin 1 (MGB1), which was shown to be a useful marker for detecting disseminated breast cancer cells in blood [23], [24], [25], bone marrow [26], [27] and lymph nodes [28], [29], [30]. Additionally to CK19 and MGB1, detection of HER2 transcripts, which is frequently overexpressed in breast cancers, strengthened prognostic value of the RT-qPCR based CTCs detecting assay [23].
Activation of EMT is linked to motility, stem cell characteristics, enhanced chemo- and radiotherapy resistance [19], [31], [32], [33]. The fraction of CTCs with a mesenchymal phenotype reportedly reaches almost 100% in the blood of some breast cancer patients [3]. Moreover, in some patients, disease progression during treatment was related to increased number of mesenchymal CTCs compared with their pre-treatment state [2]. The ability of tumor cells to metastasize can be modified by expression of various invasion and metastasis-related factors. Plasminogen activator, urokinase receptor (uPAR) constituting a part of uPA-PAI extracellular matrix degradation system might facilitating tumor cells invasion, migration and growth [34], [35]. uPAR was also shown to be amplified together with HER2 in breast cancer CTCs [36] and decreased expression of uPAR related to tumor cell dormancy [35]. Yet another protein, CXCR4 chemokine receptor, apart from being involved in metastases formation and migration of cancer cells to specific organs [37], [38] is functionally linked with HER2 signalling and malignant progression. CXCR4 expression is enhanced by HER2, which can together act in multiple steps of metastatic cascade [39].
Inherent increased malignancy of mesenchymal CTCs could also contribute to higher metastatic potential, which in early-stage breast cancer could be measured by lymph-node involvement. We have hypothesized that CTCs isolated from lymph node-negative (N−) and -positive (N+) patients could differ in expression of malignancy-associated genes. We therefore used a marker-unbiased CTC-enrichment method that enriches both epithelial and mesenchymal CTCs, in which we measured expression of mammary epithelial transcripts (CK19, MGB1), EMT-related factors (VIM, TWIST1, SNAIL, SLUG) and invasion- and metastasis- related genes (HER2, CXCR4, uPAR).
Methods
Patients
The study included 117 breast cancer patients, stages I–III, treated in the Medical University Hospital in Gdansk between April 2011 and May 2013. Tumor stage and node positivity were defined according to AJCC cancer staging manual version 7. Micrometastases were considered and there was one case in examined group identified, which we classified as N+ patient. There were no patients with isolated tumor cells in the N- group. Cancers were graded according to modified Bloom and Richardson system based on semi-quantitative method for assessing histological grade in breast tumours [40]. Median age of the patients was 61 years (28–89 years) ( Table 1 ). Inclusion criteria were primary operable breast cancer confirmed by histological examination, and signed consent form. Peripheral blood samples (5–10 mL) were drawn to EDTA-coated tubes before tumor excision and therapy initiation. The first few milliliters of blood were discarded to minimize possibility of keratinocyte contamination. Samples were stored at 4 °C until analysis, but no longer than 24 h. Blood samples from 17 healthy women (median age 36 years; range 20–73 years) and three women with benign breast disease (ductal carcinoma in situ – DCIS, median age 65 years; range 46–71) were similarly drawn and processed.
Table 1. Patients' characteristics (N = 117).
| Variable | Number of cases (%) | |
| Age – median (range) | 61 | (28–89) |
| T stage | ||
| T1 | 50 | (43) |
| T2 | 58 | (50) |
| T3 | 5 | (4) |
| T4 | 3 | (3) |
| Missing data | 1 | (1) |
| N stage | ||
| N- | 60 | (51) |
| N+ | 57 | (49) |
| Grade | ||
| G1 | 15 | (13) |
| G2 | 63 | (54) |
| G3 | 39 | (33) |
| HER2 status | ||
| Negative | 88 | (75) |
| Positive | 26 | (22) |
| Missing data | 3 | (3) |
| ER status | ||
| Negative | 23 | (20) |
| Positive | 94 | (80) |
| PR status | ||
| Negative | 29 | (25) |
| Positive | 88 | (75) |
| Histological type | ||
| Ductal | 88 | (75) |
| Lobular | 16 | (14) |
| Other | 12 | (10) |
| Missing data | 1 | (1) |
Median follow-up time was 1.5 years (0.2 to 2.2 years). Including the last follow-up data six deaths were observed, which is insufficient for performing survival analysis; however, follow-up data continue to be collected.
Hormone receptors status (ER and PgR) was evaluated using classical Allred scoring method with 3 being a cut-off point for positive result. Standard criteria for evaluating HER2 positivity were applied, being 3+ score in immunohistochemistry or positive result in fluorescence in situ hybridization (FISH), as previously described [41].
Ethics statement
The study was approved by the local Ethical Committee of the Medical University of Gdansk and the manuscript was prepared according to the REMARK criteria [42].
CTCs isolation/enrichment
For the immunofluorescence experiment, blood samples were first centrifuged at 200 g for 20 min at 20 °C to remove excessive platelets that hampered cell adhesion to polylysine slides after CTC-enrichment. The top serum layer, which contained platelets, was collected and discarded; the remaining fraction was then processed with the full blood sample intended for RNA isolation. Blood samples, or platelet-removed fractions (5 mL), were subjected to CTC-enrichment as described before [43] ( Figure 1 ). Briefly, phosphate buffered saline (PBS) diluted blood sample was transferred into a 15 ml tube containing two-layer density gradient (upper and lower gradient) and centrifuged to separate tumor cells containing fraction from erythrocytes and blood cells. The tumor cell containing fraction was collected and subjected to further depletion of CD45-positive cells with anti-CD45-covered magnetic particles (CD45 Dynabeads, Invitrogen). After depletion, the obtained cells pellets were used in the RNA isolation procedure, or if cells were to be visualized by immunofluorescence, pellets were suspended in 1 mL PBS buffer and spun down on 2–4 polylysine-coated glass slides in Rotofix 32A (Hettich).
Figure 1. Schematic representation of blood samples analysis process.
Collected blood sample was layered on density gradient. After centrifugation fraction containing tumor cells (marked by green box) was collected and subjected to negative selection. After CD45-depletetion, CTC-enriched blood sample was subjected to RNA isolation, reverse transcription and gene expression analysis with qPCR.
To evaluate cell recovery rates in the developed CTC-enrichment method, a spike-in experiment was carried out twice. On average, 6 and 11 MDA-MB-361 cells (5 and 11 in the first experiment; 6 and 10 cells in the second experiment) were spiked to peripheral blood samples from healthy women, which were then processed as the patients' samples. Four cytospins were prepared from each sample, slides were stained for CK19 and CD45 as in the immunofluorescence experiment and analyzed under Axiovert 200 (Zeiss) microscope.
RNA isolation and qRT-PCR
RNA was isolated with Trizol reagent (Invitrogen) according to manufacturer's instructions. Up to 10 μl of RNA was used in a reverse transcription reaction (Transcriptor FirstStrand Synthesis Kit, Roche) with random hexamers according to manufacturer's instructions. Presence of inhibitors in the isolated RNA was tested using exogenous RNA molecules (Solaris RNA Spike Control Kit, Thermo Scientific) according to manufacturer's instructions, adding 1 μl of 100× Solaris Spike solution per 1 μg of RNA. cDNA was diluted and 10 ng of cDNA (4 μL) was used in a single 20-μL qPCR reaction. Universal PCR Mastermix (with UNG AmpErase) and TaqMan Gene Expression Assays were used to measure expression of nine genes of interest: TWIST1 (Hs00361186_m1; UniGene Hs.644998), SNAIL (also known as SNAI1, Hs00195591_m1; UniGene Hs.48029), SLUG (also known as SNAI2, Hs00950344_m1; UniGene Hs.360174), CK19 (Hs01051611_gH; UniGene Hs.654568), MGB1 (Hs00935948_m1; UniGene Hs.46452), HER2 (Hs99999005_mH; UniGene Hs.446352), CXCR4 (Hs00237052_m1; UniGene Hs.593413), uPAR (Hs00182181_m1; UniGene Hs.466871), VIM (Hs00185584_m1; UniGene Hs.455493) and two reference genes GAPDH (Hs99999905_m1; UniGene Hs.544577) and YWHAZ (Hs03044281_g1; UniGene Hs.492407). Reference genes were chosen for their expression stability as assessed by geNorm according to the provided manual [43]. qPCR reactions were performed in duplicate on 96-well plates in a CFX96 thermal cycler (Bio-Rad). Cycling parameters were as follows: 10 min 95 °C, 45 cycles of 1 min at 60 °C followed by 30 s at 95 °C. For the TaqMan assays detecting genomic DNA (CK19 and YWHAZ), sample-specific controls (10 ng of untranscribed RNA) were included on every plate. Samples were assumed to be genomic DNA-free if differences between control and test samples were ≥5 CT. Gene expression was calculated using a modified ΔΔCt method that corrects for a run-to-run variation [44]. Therefore, every plate included an inter-run calibrator that allowed calculation of calibrator-normalized relative quantities using qBasePLUS software version 2.1 [44]. Gene expression levels were scaled to the minimal expression level of each gene in the patients' samples.
Immunofluorescence
As PCR-based techniques do not allow for cells visualization, immunofluorescence experiments were carried out for some patient samples to visualize CTCs isolated using the developed method (described in “CTCs isolation/enrichment”). Double-staining experiments were performed for the following proteins combinations: CK19 and CD45, CK19 and MGB1, CXCR4 and CD45, HER2 and CD45, SNAIL and CD45. Primary mouse monoclonal (CK19, CXCR4, SNAIL, HER2) or rabbit polyclonal (CD45, MGB1) antibodies were used (Santa Cruz Biotechnology; Abnova for anti-SNAIL antibody). Cytospins, prepared on polylysine-coated slides as described above, were fixed with cold methanol for 5 minutes and incubated with appropriate pairs of primary antibodies for 1 hour at room temperature. Pairs of mouse monoclonal antibodies and rabbit polyclonal antibodies were used. Slides were then stained for 1 h with corresponding secondary antibodies: Sheep Anti-Mouse DyLight 549 and Goat Anti-Rabbit DyLight 488 (JIR). Cell nuclei in all slides were stained with 4,6-diamidino-2-phenylindole (Sigma). Slides were evaluated using inverted fluorescent microscope Axiovert 200 (Zeiss) and analyzed with AxioVision software (Zeiss).
Statistical analysis
Statistical analyses were performed with STATISTICA software version 10. Categorical variables were analysed using contingency tables with χ2 statistics or Fisher's exact test where applicable. Spearman rank coefficient was used when continuous variables were correlated; Mann-Whitney test for analyzing continuous variables distribution in two groups. Logistic regression analysis was used to identify the gene predictors of lymph node involvement. Univariate predictors significant with a value of p≤0.10 were entered into a step-wise backwards multivariate model to identify those with independent prognostic information.
Unsupervised hierarchical clustering analysis was performed using GenePattern version 6 software (http://genepattern.broadinstitute.org/) [45]. Spearman's rank correlation was applied for columns and rows distance measure. Pairwise complete-linkage was used as a clustering method. Samples with missing gene expression values were removed from the analysis.
Results
CTC-enriched blood samples characterization
For the average 6 and 11 MDA-MB-361 cells added to blood samples, the CTC-enrichment method showed average recovery rate of 54% and 72%, respectively. In every case, 2–4 cells were lost during sample processing. Double staining (staining of CD45 and another protein of interest) revealed CK19+/CD45−, HER2+/CD45−, CXCR4+/CD45− and SNAIL+/CD45− cells isolated from breast cancer patients (Figure S1). In CK19/MGB1 double staining, following phenotypes occurred: CK19+/MGB1+, CK19+/MGB1− and CK19−/MGB1+ (Figure S1).
Functional RNA was successively isolated from 84% (98/117) of patient samples and 65% (11/17) of controls. None of the control samples was positive for TWIST1, SNAIL, SLUG, HER2 or uPAR; 1 out of 11 showed CXCR4 expression (relative gene expression level 4.54); and 9 (82%; 9/11) showed VIM expression (median relative gene expression level 11.44, range 0–30.67). Expression of MGB1 and CK19 was analyzed in a qualitative manner. Patient samples were considered positive when measured relative gene expression level was higher than the highest measured relative expression level in the control samples. Table 2 presents the number of samples expressing analyzed genes according to this positivity criterion. Fifty-five percent (54/98) of the patient samples with invasive tumors were positive for either CK19, MGB1 or HER2. In total, 26% of the samples expressed at least one EMT marker (VIM, TWIST1, SNAIL or SLUG). No healthy control expressed CTCs markers, whereas in all three cases of DCIS expression of at least one CTC marker was detected ( Table 2 and Figure S2).
Table 2. Relative gene expression levels in CTC-enriched blood fractions from breast cancer patients.
| Gene | Number of samples* | % of positive samples | Average expression | SD | Median | Expression minimum | Expression maximum | ||
| Breast cancer patients | positive | negative | |||||||
| TWIST1 | 2 | 94 | 2 | 0.05 | 0.39 | 0 | 0 | 3.71 | |
| SNAIL | 8 | 88 | 8 | 0.32 | 1.64 | 0 | 0 | 14.38 | |
| SLUG | 0 | 92 | 0 | - | - | - | - | - | |
| VIM | 19 | 75 | 20 | 19.40 | 17.70 | 14.27 | 0 | 105.10 | |
| HER2 | 34 | 61 | 36 | 8.33 | 18.26 | 0 | 0 | 96.50 | |
| CXCR4 | 40 | 57 | 41 | 5.34 | 7.79 | 3.34 | 0 | 48.42 | |
| uPAR | 57 | 40 | 59 | 14.65 | 22.73 | 4.50 | 0 | 118.80 | |
| CK19 | 26 | 72 | 27 | 1056.88 | 1054 | 0 | 0 | 2585 | |
| MGB1 | 23 | 71 | 24 | 1714.53 | 1871 | 0 | 0 | 4424 | |
| DCIS | |||||||||
| TWIST1 | 0 | 3 | 0 | - | - | - | - | - | |
| SNAIL | 1 | 2 | 33 | 3.01 | 5.21 | 0 | 0 | 9.02 | |
| SLUG | 0 | 3 | 0 | - | - | - | - | - | |
| VIM | 0 | 3 | 0 | 20.18 | 7.17 | 22.28 | 12.2 | 26.07 | |
| HER2 | 2 | 1 | 67 | 7.66 | 8.76 | 5.78 | 0 | 17.21 | |
| CXCR4 | 1 | 2 | 33 | 7.07 | 9.06 | 3.93 | 0 | 17.28 | |
| uPAR | 2 | 1 | 67 | 9.79 | 8.48 | 14.49 | 0 | 14.88 | |
| CK19 | 1 | 2 | 33 | 17.81 | 30.85 | 0 | 0 | 53.43 | |
| MGB1 | 3 | 0 | 100 | 973.24 | 1567.26 | 17.93 | 17.93 | 2782 | |
| Healthy controls ** | |||||||||
| VIM | Not applicable | Not applicable | Not applicable | 13 | 9.40 | 11.44 | 0 | 30.70 | |
| CXCR4 | Not applicable | Not applicable | Not applicable | 0.40 | 1.40 | 4.54 | 0 | 4.54 | |
*Number of positive and negative samples for a particular gene, percentages of samples classified as marker-positive with the chosen cut-off levels as well as average expression with standard deviation (SD), minimal and maximal measured relative expression level.
**In the healthy control group only genes which expression was found in the samples are shown.
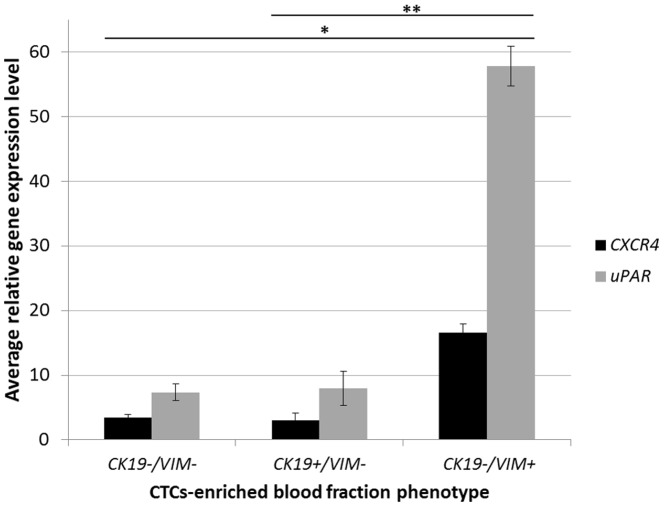
CTC-enriched samples expressing at least one of CTCs markers – MGB1 or HER2 were divided by CK19 and VIM gene expression status into four groups with positive or negative expression status. Phenotype frequencies were as follow: CK19+/VIM− 17% (16/94), CK19+/VIM+ 1% (1/94), CK19−/VIM+ 13% (12/94) and CK19−/VIM− 69% (65/94). CK19+/VIM+ sample was excluded from statistical analysis to avoid biased classification to either CK19+/VIM− or CK19−/VIM+ group. Expression of CXCR4 and uPAR was higher in VIM-positive fraction (CK19−/VIM+) than in the CK19−/VIM− CTC-enriched blood fraction ( Figure 2 ). In CK19−/VIM+ CTC-enriched blood fractions, average expression of CXCR4 and uPAR was 4.88 and 7.85-fold higher than in CK19−/VIM− fraction, respectively (P < 0.00001 for both).
Figure 2. Relative expression level of CXCR4 and uPAR in CTC-enriched blood fractions.
CTC-enriched blood fractions positive for either MGB1 or HER2 were divided according to VIM and CK19 expression status into three groups (CK19−/VIM−; CK19+/VIM−; CK19−/VIM+). Error bars depict standard error. * - statistically significant difference in CXCR4 and uPAR relative expression level (P<0.00001 for both) between CK19−/VIM− and CK19−/VIM+; ** - statistically significant difference in CXCR4 (P = 0.00002) and uPAR (P = 0.00004) relative expression level between CK19+/VIM− and CK19−/VIM+.
Correlations between gene expression of examined markers
SNAIL expression was found in 8% (8/96) of the samples; its expression correlated with positive status of VIM (P = 0.03), HER2 (P = 0.0002), CXCR4 (P = 0.006) and uPAR (P = 0.02) ( Table 3 ). CTC-enriched HER2-positive blood samples were more frequently VIM-positive (38% vs. 12%, P = 0.004), SNAIL-positive (24% vs. 0%, P = 0.0002), CXCR4-positive (53% vs. 33%, P = 0.05) and uPAR-positive (100% vs. 34%, P<0.00001) than HER2-negative samples ( Table 3 ). Moreover, 82% (32/39) of CTC-enriched CXCR4-positive samples were also uPAR-positive (P = 0.0001; Table 3 ). Spearman rank correlations between relative expression of the tested genes showing similar gene dependence are presented in Table S1.
Table 3. Correlations between genes expression status in CTC-enriched blood fractions.
| TWIST1 | SNAIL | CK19 | MGB1 | VIM | HER2 | CXCR4 | ||||||||||||||||
| Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | ||
| SNAIL | Neg. | 87 | 1 | 0.16 | ||||||||||||||||||
| Pos. | 7 | 1 | ||||||||||||||||||||
| Total | 94 | 2 | ||||||||||||||||||||
| CK19 | Neg. | 70 | 2 | 1 | 65 | 7 | 0.67 | |||||||||||||||
| Pos. | 24 | 0 | 23 | 1 | ||||||||||||||||||
| Total | 94 | 2 | 88 | 8 | ||||||||||||||||||
| MGB1 | Neg. | 70 | 1 | 0.43 | 65 | 6 | 1 | 61 | 10 | 0.00004 | ||||||||||||
| Pos. | 22 | 1 | 22 | 1 | 10 | 13 | ||||||||||||||||
| Total | 92 | 2 | 87 | 7 | 71 | 23 | ||||||||||||||||
| VIM | Neg. | 72 | 1 | 0.37 | 70 | 3 | 0.03 | 52 | 23 | 0.076 | 55 | 18 | 1 | |||||||||
| Pos. | 18 | 1 | 14 | 4 | 17 | 2 | 15 | 4 | ||||||||||||||
| Total | 90 | 2 | 85 | 7 | 69 | 25 | 70 | 22 | ||||||||||||||
| HER2 | Neg. | 61 | 0 | 0.13 | 61 | 0 | 0.0002 | 48 | 13 | 0.24 | 49 | 12 | 0.21 | 53 | 7 | 0.004 | ||||||
| Pos. | 32 | 2 | 26 | 8 | 23 | 11 | 22 | 10 | 20 | 12 | ||||||||||||
| Total | 93 | 2 | 87 | 8 | 71 | 24 | 71 | 22 | 73 | 19 | ||||||||||||
| CXCR4 | Neg. | 56 | 1 | 1 | 56 | 1 | 0.006 | 40 | 17 | 0.42 | 42 | 15 | 0.45 | 53 | 3 | 0.00001 | 41 | 16 | 0.05 | |||
| Pos. | 37 | 1 | 31 | 7 | 31 | 9 | 29 | 7 | 22 | 16 | 20 | 18 | ||||||||||
| Total | 93 | 2 | 87 | 8 | 71 | 26 | 71 | 22 | 75 | 19 | 61 | 34 | ||||||||||
| uPAR | Neg. | 40 | 0 | 0.51 | 40 | 0 | 0.02 | 33 | 7 | 0.12 | 34 | 6 | 0.07 | 34 | 5 | 0.13 | 40 | 0 | <0.00001 | 33 | 7 | 0.0001 |
| Pos. | 53 | 2 | 47 | 8 | 39 | 18 | 37 | 17 | 41 | 14 | 21 | 33 | 24 | 32 | ||||||||
| Total | 93 | 2 | 87 | 8 | 72 | 25 | 71 | 23 | 75 | 19 | 61 | 33 | 57 | 39 | ||||||||
Number of cases classified as positive (Pos.) or negative (Neg.) for a particular marker are shown. Statistically significant P values are given in bold.
Correlation with clinicopathological data
In CTC-enriched blood fractions of N+ patients, at least one of the markers – CK19, MGB1 or HER2 – was more frequently (69%, 34/49) detected than in N- patients (41%, 20/49; P = 0.004, Table 4 ). Concerning phenotype of CTC-enriched blood fractions, there was no difference between lymph node involvement in patients with epithelial and mesenchymal CTC-enriched blood fraction phenotype (P = 0.69), however mesenchymal phenotype was more frequently found in patients with more than 3 lymph nodes involved −50% in comparison to patients with epithelial CTC-enriched blood fraction phenotype −7% (P = 0.008, Table 5 ). Moreover, in N+ patients, CTC-enriched blood fractions were more often MGB1, SNAIL, VIM and uPAR-positive (P = 0.03, P = 0.059, P = 0.02 and P = 0.03, respectively) ( Table 5 ). Positive status of CXCR4 was associated with higher T stage (P = 0.01). There was no correlation between HER2 status of primary tumors (PT) and HER2-status of CTC-enriched blood fractions (P = 0.87) ( Table 5 ).
Table 4. Results of univariate and multivariate logistic regression analysis in relation to lymph node involvement.
| Gene expression status | Number of cases (%) | Univariate analysis | Multivariate analysis | |||
| N- | N+ | OR (95% CI) | P | OR (95% CI) | P | |
| CK19 | ||||||
| Negative | 39 (80) | 33 (67) | 1.9 (0.75–4.8) | 0.17 | ||
| Positive | 10 (20) | 16 (33) | ||||
| MGB1 | ||||||
| Negative | 40 (85) | 31 (66) | 2.9 (1.06–8.2) | 0.03 | 3.2 (1.1–9.2) | 0.029 |
| Positive | 7 (15) | 16 (34) | ||||
| HER2 | ||||||
| Negative | 32 (67) | 29 (62) | 1.24 (0.5–2.9) | 0.6 | ||
| Positive | 16 (33) | 18 (38) | ||||
| VIM | ||||||
| Negative | 43 (90) | 32 (70) | 3.76 (1.2–11.7) | 0.02 | 4.2 (1.3–13.5) | 0.01 |
| Positive | 5 (10) | 14 (30) | ||||
| TWIST1 | ||||||
| Negative | 47 (98) | 47 (98) | 1 (0.6–17) | 1 | ||
| Positive | 1 (2) | 1 (2) | ||||
| SNAIL | ||||||
| Negative | 47 (98) | 41 (85) | 8.02 (0.92–69.9) | 0.06 | NS | |
| Positive | 1 (2) | 7 (15) | ||||
| CXCR4 | ||||||
| Negative | 32 (65) | 25 (52) | 1.73 (0.76–3.9) | 0.19 | ||
| Positive | 17 (35) | 23 (48) | ||||
| uPAR | ||||||
| Negative | 25 (52) | 15 (31) | 2.46 (1.06–5.7) | 0.03 | NS | |
| Positive | 23 (48) | 34 (69) | ||||
| CK19/MGB1/HER2 | ||||||
| Negative | 29 (59) | 15 (31) | 3.29 (1.41–7.64) | 0.005 | Not applicable | |
| Positive | 20 (41) | 34 (69) | ||||
N- lymph node involvement absent, N+ lymph node involvement present. OR-overall risk; CI – confidence interval, NS-not statistically significant.
Table 5. Correlation between CTC-enriched blood fractions gene expression status and clinicopathological parameters.
| TWIST1 | SNAIL | CK19 | MGB1 | VIM | CXCR4 | uPAR | HER2 | CTC-EBF phenotype* | |||||||||||||||||||
| Clinical variable | Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | Neg. | Pos. | P | CK19-/VIM+ ** | CK19+/VIM- ** | P |
| T | |||||||||||||||||||||||||||
| T1 | 41 | 1 | 1 | 39 | 3 | 1 | 29 | 14 | 0.25 | 35 | 5 | 0.02 | 38 | 3 | 0.009 | 31 | 12 | 0.01 | 21 | 21 | 0.10 | 29 | 13 | 0.34 | 1 | 7 | 0.04 |
| T2-4 | 52 | 1 | 48 | 5 | 42 | 12 | 35 | 18 | 37 | 15 | 25 | 28 | 18 | 36 | 31 | 21 | 11 | 9 | |||||||||
| N | |||||||||||||||||||||||||||
| N- | 47 | 1 | 1 | 47 | 1 | 0.059 | 39 | 10 | 0.17 | 40 | 7 | 0.03 | 43 | 5 | 0.02 | 32 | 17 | 0.19 | 25 | 23 | 0.03 | 32 | 16 | 0.61 | 3 | 6 | 0.69 |
| N+ | 47 | 1 | 37 | 7 | 33 | 16 | 31 | 16 | 32 | 14 | 25 | 23 | 15 | 34 | 29 | 18 | 9 | 10 | |||||||||
| Number of lymph nodes involved | |||||||||||||||||||||||||||
| ≤3 | 78 | 2 | 1 | 74 | 6 | 0.62 | 59 | 23 | 0.55 | 60 | 18 | 0.53 | 67 | 11 | 0.003 | 52 | 29 | 0.01 | 37 | 44 | 0.045 | 52 | 27 | 0.46 | 6 | 15 | 0.008 |
| >3 | 16 | 0 | 14 | 2 | 13 | 3 | 11 | 5 | 8 | 8 | 5 | 11 | 3 | 13 | 9 | 7 | 6 | 1 | |||||||||
| Grade | |||||||||||||||||||||||||||
| G1-2 | 65 | 1 | 0.53 | 62 | 4 | 0.25 | 49 | 18 | 0.91 | 48 | 16 | 0.86 | 52 | 11 | 0.34 | 40 | 26 | 0.59 | 30 | 36 | 0.22 | 41 | 24 | 0.73 | 7 | 13 | 0.23 |
| G3 | 29 | 1 | 26 | 4 | 23 | 8 | 23 | 7 | 23 | 8 | 17 | 14 | 10 | 21 | 20 | 10 | 5 | 3 | |||||||||
| HR status | |||||||||||||||||||||||||||
| Negative | 14 | 0 | 1 | 13 | 1 | 1 | 10 | 5 | 0.53 | 11 | 3 | 1 | 13 | 2 | 0.73 | 8 | 7 | 0.64 | 8 | 7 | 0.30 | 12 | 2 | 0.07 | 1 | 1 | 1 |
| Positive | 80 | 2 | 75 | 7 | 62 | 21 | 60 | 20 | 62 | 17 | 49 | 33 | 32 | 50 | 49 | 32 | 11 | 15 | |||||||||
| HER2 status | |||||||||||||||||||||||||||
| Negative | 70 | 2 | 1 | 66 | 6 | 1 | 52 | 21 | 0.58 | 53 | 17 | 0.96 | 56 | 14 | 1 | 43 | 30 | 0.72 | 30 | 42 | 0.66 | 46 | 26 | 0.87 | 9 | 14 | 0.62 |
| Positive | 21 | 0 | 19 | 2 | 17 | 5 | 16 | 5 | 18 | 4 | 12 | 10 | 8 | 14 | 13 | 8 | 3 | 2 | |||||||||
Number of cases classified as positive (Pos.) or negative (Neg.) for a particular marker in each group are shown. Statistically significant P values are given in bold.
*CTC-enriched blood fraction phenotype – only MBG1+ and/or HER2+ fractions were considered
** CK19−/VIM+ - mesenchymal phenotype; CK19+/VIM− - epithelial phenotype.
Multivariate analysis revealed that independent predictors of lymph nodes involvement were positive status of MGB1 – OR 3.2 (95% CI 1.1–9.2, P = 0.029) and VIM – OR 4.2 (95% CI 1.3–13.5, P = 0.01) ( Table 4 ).
In hierarchical clustering, the study population was divided into two main groups, which differed in expression of VIM, CXCR4, uPAR, HER2 ( Figure 3 ). Patients in the cluster with elevated expression of these genes showed more frequent lymph node involvement (58%) than patients from the cluster with lower expression (35%; P = 0.03).
Figure 3. Unsupervised hierarchical clustering of relative gene expression values in CTC-enriched blood fraction.

Samples containing missing values were excluded from the analysis. On the left-hand side cluster of CTC-enriched blood samples with elevated VIM (P = 0.068), HER2, CXCR4 and uPAR (P<0.00001, for all three) expression in which lymph node involvement was more frequently observed (P = 0.04).
Discussion
Dissemination of cancer cells is an early event and disseminated tumor cells can be found in bone marrow of patients with carcinoma in situ [46]. Nevertheless, in metastatic breast cancer more CTCs are seen than in early breast cancer patients [13], [16], [47]. No clear association is apparent between lymph node involvement and CTC detection rate. Some studies show similar CTCs detection rate in N− and N+ breast cancer patients [23], [48], which would indicate similar tumor seeding potential, but dissimilar colonization potential for disseminated cells. However, preliminary results from a large SUCCESS trial revealed a correlation between CTC presence and lymph node involvement [49], implying different seeding, and possibly colonization, potentials in N− and N+ patients. To explore differences in seeding and colonization potential we analyzed expression of mammary epithelial transcripts (CK19, MGB1), EMT-related factors (TWIST1, SNAIL, SLUG, VIM) and invasion and metastasis-related genes (HER2, CXCR4, uPAR) in CTC-enriched blood fractions from N- and N+ breast cancer patients. Considering the unresolved discussion concerning markers for CTC detection and characterization [50] and drawbacks of CTC-positive selection, we decided to apply a negative selection method for CTC-enrichment. Similar to prior research identifying CTCs with PCR-based methods, we used a “classical” definition of CTCs presence, meaning expression of either CK19, MGB1 or HER2 [23]. However, growing body of evidence shows that mesenchymal CTCs are not a rare event in cancers, what made us apply an extended definition of CTCs phenotypes – epithelial or mesenchymal – which includes VIM as an additional marker related to mesenchymal state. As a result we consider CTCs-enriched blood fractions being CK19+/VIM− and MGB1+ or HER2+ as containing CTCs in epithelial state, and CK19−/VIM+ and MGB1+ or HER2+ fractions as carrying mesenchymal CTCs. Even though the phrase describing MGB1 as “mammary epithelial transcript” implies it is present in mammary epithelium, it does not mean that it is expressed only by cells in the epithelial state. Although MGB1 function is not known, structurally related proteins are a group of secretory proteins binding steroid ligands that might present anti inflammatory activity [51], [52]; thus implying related function in human breast tissue, not restricted to epithelial state.
In our study, we found expression of either CK19, MGB1 or HER2 in 55% of the patients, which agrees with previous studies of early breast cancer patients [53], [54], [55]. Unlike in healthy controls, in DCIS cases expression of at least one CTC marker was detected, what supports results of the early cancer dissemination model [46]. We have observed significantly different CTCs detection rate (defined as expression of either CK19, MGB1 or HER2) in N− and N+ patients (41% vs. 69%, respectively, P = 0.005; Table 4 ), which supports the notion that tumors from N+ patients have superior seeding potential compared with N− patients. We would not have drawn the same conclusion if CK19 was the only CTC marker in our study, as in that case there would be no significant difference in CTC marker detection rate between N− and N+ patients (20% vs. 33%, P = 0.17; Table 4 ). Also, the study of Pecot indicates that cytokeratin is not an ultimate marker for CTC identification as cytokeratin-negative cancer cells can be found both in circulation and within PTs [56]. Multimarker approach for CTCs detection is therefore a necessity [14], [57]. Our immunofluorescence results also showed CK19−/MGB1+ cells in CTC-enriched blood fractions, which was also seen in prior research [58]. Analysis of expression of EMT markers, and of invasion- and metastasis-related genes revealed that CTC-enriched blood fractions of N+ patients are more frequently VIM-positive and uPAR-positive (and show a trend toward being SNAIL-positive); and patients with more than three involved lymph nodes are often VIM, CXCR4 and uPAR-positive. Increased metastatic abilities of tumor cells in N+ patients could be therefore attributed to expression of CXCR4, uPAR and VIM (and possibly SNAIL). CXCR4 is a receptor for CXCL12 chemokine, which is secreted by the common sites of breast cancer metastasis, including lymph nodes [3]. Thus, CXCR4 expression on CTCs could mediate homing of tumor cells to lymph nodes. Also, the role of uPAR in extracellular matrix degradation complex, migration and invasion could explain its metastasis-promoting function [34]. Amplification and increased expression of uPAR was observed before in CTCs [36], PTs, and disseminating tumor cells in bone marrow and in lymph nodes of breast cancer patients [59]. Co-expression of uPAR and HER2 shown by Meng et al. [36] also supports our observations in HER2-positive CTC-enriched blood fractions, which were all uPAR-positive.
Although induction and maintenance of EMT are thought to require TWIST1, SNAIL and SLUG, their expression was not found in CTCs expressing mesenchymal markers [2], [60], nor were they highly expressed in our CTC-enriched blood fractions; only 8% and 2% of samples were positive for either SNAIL or TWIST1, respectively and SLUG was not detected in any sample. Similar rates were detected by Mego et al., who analyzed CTC-enriched blood fractions depleted of EpCAM-positive CTCs [22]. Despite the small number of SNAIL-positive samples, SNAIL expression correlated with positive status of VIM, CXCR4, uPAR and HER2. We also saw positive VIM status in 20% of the CTC-enriched blood fractions, which correlated with SNAIL, CXCR4 and HER2 expression. This result supports data showing increased malignancy of CTCs with mesenchymal phenotype, possibly resulting from EMT [20], [33], [61]. No significant increase in expression of CXCR4 and uPAR in CK19+/VIM− CTC-enriched blood fraction would speak for the passive model of dissemination, in which tumor cells are physically translocated into the vasculature (or the neovasculature formed around tumor cells) [62]. In that case, cells would not need a migratory phenotype, typically associated with EMT, and could remain in their epithelial phenotype, which according to several theories would strengthen their metastatic ability because of increased adhesive properties. We presume that CTCs with mesenchymal phenotype (lacking epithelial adhesion molecules) acquire additional malignant properties (expression of CXCR4 and uPAR) that allow successful lymph node colonization. It would be interesting to check whether expression of CXCR4 and uPAR are related to any specific metastatic pattern other than the lymph node metastasis seen in our study.
Limitations of our study include relatively small sample size and short follow-up period, which hamper survival analysis. Moreover, the associations between variables are deduced from their inter-correlation, which does not necessarily inform about causal relationship. Therefore, our results should be seen as hypothesis-generating discoveries, and would obtain additional strength when supported by research aiming at deciphering molecular mechanisms behind them.
Conclusions
In summary, our results show increased CTCs detection rate as well as more frequent VIM, SNAIL, and uPAR–positive rates of CTC-enriched blood fraction in patients with lymph nodes metastases. This indicates differences in both seeding potential and increased ability of the tumor cells of N+ patients to reach and divide in lymph nodes. We have also shown that VIM-positive (CK19−/VIM+) CTC-enriched blood fractions, unlike CK19+/VIM− fractions, show elevated CXCR4 and uPAR levels, which might contribute to more aggressive tumor characteristics.
Supporting Information
Exemplary photos of immunostained cells isolated from CTC-enriched blood fractions of breast cancer patients.
(PDF)
Relative genes expression levels in breast cancer patients and controls. Expression of VIM, HER2, CXCR4, uPAR, TWIST1, SNAIL, CK19 and MGB1 in CTCs-enriched blood fractions of breast cancer patients with invasive carcinoma (BC), patients with ductal carcinoma in situ (DCIS) and healthy controls (HC). As no sample expressed SLUG, results for this gene are not presented.
(PDF)
Spearman's rank correlations coefficients of relative gene expression levels in CTC-enriched blood fractions.
(PDF)
Funding Statement
This work was supported by the National Centre for Research and Development LIDER/13/117/L-1/09/NCBiR/2010. In addition, this work was supported by the system project “InnoDoktorant–Scholarships for PhD students, IIIrd edition”. The project is co-financed by the European Union in the frame of the European Social Fund. Publication cost supported by the European Commission from the FP7 project MOBI4Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ashworth T (1869) A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust Med J 14: 146–147. [Google Scholar]
- 2. Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, et al. (2013) Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339: 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stoecklein NH, Klein CA (2010) Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. International Journal of Cancer 126: 589–598. [DOI] [PubMed] [Google Scholar]
- 4. Banys M, Muller V, Melcher C, Aktas B, Kasimir-Bauer S, et al. (2013) Circulating tumor cells in breast cancer. Clinica Chimica Acta 423: 39–45. [DOI] [PubMed] [Google Scholar]
- 5. Chaffer CL, Weinberg RA (2011) A Perspective on Cancer Cell Metastasis. Science 331: 1559–1564. [DOI] [PubMed] [Google Scholar]
- 6. Lucci A, Hall CS, Lodhi AK, Bhattacharyya A, Anderson AE, et al. (2012) Circulating tumour cells in non-metastatic breast cancer: a prospective study. The Lancet Oncology 13: 688–695. [DOI] [PubMed] [Google Scholar]
- 7. Bidard FC, Mathiot C, Delaloge S, Brain E, Giachetti S, et al. (2010) Single circulating tumor cell detection and overall survival in nonmetastatic breast cancer. Annals of Oncology 21: 729–733. [DOI] [PubMed] [Google Scholar]
- 8. Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, et al. (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. New England Journal of Medicine 351: 781–791. [DOI] [PubMed] [Google Scholar]
- 9.Giuliano M, Giordano A, Jackson S, Hess KR, De Giorgi U, et al. (2011) Circulating tumor cells as prognostic and predictive markers in metastatic breast cancer patients receiving first-line systemic treatment. Breast Cancer Research 13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xenidis N, Ignatiadis M, Apostolaki S, Perraki M, Kalbakis K, et al. (2009) Cytokeratin-19 mRNA-Positive Circulating Tumor Cells After Adjuvant Chemotherapy in Patients With Early Breast Cancer. Journal of Clinical Oncology 27: 2177–2184. [DOI] [PubMed] [Google Scholar]
- 11. Stathopoulou A, Vlachonikolis I, Mavroudis D, Perraki M, Kouroussis C, et al. (2002) Molecular detection of cytokeratin-19-positive cells in the peripheral blood of patients with operable breast cancer: evaluation of their prognostic significance. Journal of Clinical Oncology 20: 3404–3412. [DOI] [PubMed] [Google Scholar]
- 12. Zhao S, Liu Y, Zhang Q, Li H, Zhang M, et al. (2011) The prognostic role of circulating tumor cells (CTCs) detected by RT-PCR in breast cancer: a meta-analysis of published literature. Breast Cancer Research and Treatment 130: 809–816. [DOI] [PubMed] [Google Scholar]
- 13. Strati A, Kasimir-Bauer S, Markou A, Parisi C, Lianidou ES (2013) Comparison of three molecular assays for the detection and molecular characterization of circulating tumor cells in breast cancer. Breast Cancer Research 15: R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Andergassen U, Kolbl AC, Hutter S, Friese K, Jeschke U (2013) Detection of Circulating Tumour Cells from Blood of Breast Cancer Patients via RT-qPCR. Cancers 5: 1212–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daskalaki A, Agelaki S, Perraki M, Apostolaki S, Xenidis N, et al. (2009) Detection of cytokeratin-19 mRNA-positive cells in the peripheral blood and bone marrow of patients with operable breast cancer. Br J Cancer 101: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pierga JY, Bonneton C, Vincent-Salomon A, de Cremoux P, Nos C, et al. (2004) Clinical significance of immunocytochemical detection of tumor cells using digital microscopy in peripheral blood and bone marrow of breast cancer patients. Clinical Cancer Research 10: 1392–1400. [DOI] [PubMed] [Google Scholar]
- 17. Tamaki Y, Akiyama F, Iwase T, Kaneko T, Tsuda H, et al. (2009) Molecular detection of lymph node metastases in breast cancer patients: results of a multicenter trial using the one-step nucleic acid amplification assay. Clinical Cancer Research 15: 2879–2884. [DOI] [PubMed] [Google Scholar]
- 18. Inokuchi M, Ninomiya I, Tsugawa K, Terada I, Miwa K (2003) Quantitative evaluation of metastases in axillary lymph nodes of breast cancer. Br J Cancer 89: 1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Konigsberg R, Obermayr E, Bises G, Pfeiler G, Gneist M, et al. (2011) Detection of EpCAM positive and negative circulating tumor cells in metastatic breast cancer patients. Acta oncologica 50: 700–710. [DOI] [PubMed] [Google Scholar]
- 20. Raimondi C, Gradilone A, Naso G, Vincenzi B, Petracca A, et al. (2011) Epithelial-mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Research and Treatment 130: 449–455. [DOI] [PubMed] [Google Scholar]
- 21. Bonnomet A, Brysse A, Tachsidis A, Waltham M, Thompson EW, et al. (2010) Epithelial-to-mesenchymal transitions and circulating tumor cells. J Mammary Gland Biol Neoplasia 15: 261–273. [DOI] [PubMed] [Google Scholar]
- 22. Mego M, Mani SA, Lee BN, Li C, Evans KW, et al. (2012) Expression of epithelial-mesenchymal transition-inducing transcription factors in primary breast cancer: The effect of neoadjuvant therapy. International Journal of Cancer 130: 808–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ignatiadis M, Kallergi G, Ntoulia M, Perraki M, Apostolaki S, et al. (2008) Prognostic value of the molecular detection of circulating tumor cells using a multimarker reverse transcription-PCR assay for cytokeratin 19, mammaglobin A, and HER2 in early breast cancer. Clinical Cancer Research 14: 2593–2600. [DOI] [PubMed] [Google Scholar]
- 24. Markou A, Strati A, Malamos N, Georgoulias V, Lianidou ES (2011) Molecular characterization of circulating tumor cells in breast cancer by a liquid bead array hybridization assay. Clin Chem 57: 421–430. [DOI] [PubMed] [Google Scholar]
- 25. Reinholz MM, Kitzmann KA, Tenner K, Hillman D, Dueck AC, et al. Cytokeratin-19 and mammaglobin gene expression in circulating tumor cells from metastatic breast cancer patients enrolled in North Central Cancer Treatment Group trials, N0234/336/436/437. Clinical Cancer Research 17: 7183–7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Corradini P, Voena C, Astolfi M, Delloro S, Pilotti S, et al. (2001) Maspin and mammaglobin genes are specific markers for RT-PCR detection of minimal residual disease in patients with breast cancer. Annals of Oncology 12: 1693–1698. [DOI] [PubMed] [Google Scholar]
- 27. Bossolasco P, Ricci C, Farina G, Soligo D, Pedretti D, et al. (2002) Detection of micrometastatic cells in breast cancer by RT-pCR for the mammaglobin gene. Cancer Detect Prev 26: 60–63. [DOI] [PubMed] [Google Scholar]
- 28. Marchetti A, Buttitta F, Bertacca G, Zavaglia K, Bevilacqua G, et al. (2001) mRNA markers of breast cancer nodal metastases: comparison between mammaglobin and carcinoembryonic antigen in 248 patients. J Pathol 195: 186–190. [DOI] [PubMed] [Google Scholar]
- 29. Leygue E, Snell L, Dotzlaw H, Hole K, Troup S, et al. (1999) Mammaglobin, a potential marker of breast cancer nodal metastasis. J Pathol 189: 28–33. [DOI] [PubMed] [Google Scholar]
- 30. Kataoka A, Mori M, Sadanaga N, Ueo H, Tsuji K, et al. (2000) RT-PCR detection of breast cancer cells in sentinel lymph modes. Int J Oncol 16: 1147–1152. [DOI] [PubMed] [Google Scholar]
- 31. Vazquez-Martin A, Oliveras-Ferraros C, Cufi S, Del Barco S, Martin-Castillo B, et al. (2010) Metformin regulates breast cancer stem cell ontogeny by transcriptional regulation of the epithelial-mesenchymal transition (EMT) status. Cell cycle 9: 3807–3814. [PubMed] [Google Scholar]
- 32. Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, et al. (2007) Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Research 67: 1979–1987. [DOI] [PubMed] [Google Scholar]
- 33. Theys J, Jutten B, Habets R, Paesmans K, Groot AJ, et al. (2011) E-Cadherin loss associated with EMT promotes radioresistance in human tumor cells. Radiotherapy and oncology 99: 392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andreasen PA, Kjoller L, Christensen L, Duffy MJ (1997) The urokinase-type plasminogen activator system in cancer metastasis: a review. International Journal of Cancer 72: 1–22. [DOI] [PubMed] [Google Scholar]
- 35. Aguirre Ghiso JA, Kovalski K, Ossowski L (1999) Tumor dormancy induced by downregulation of urokinase receptor in human carcinoma involves integrin and MAPK signaling. J Cell Biol 147: 89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meng S, Tripathy D, Shete S, Ashfaq R, Saboorian H, et al. (2006) uPAR and HER-2 gene status in individual breast cancer cells from blood and tissues. Proceedings of the National Academy of Sciences of the United States of America 103: 17361–17365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, et al. (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3: 537–549. [DOI] [PubMed] [Google Scholar]
- 38. Muller A, Homey B, Soto H, Ge N, Catron D, et al. (2001) Involvement of chemokine receptors in breast cancer metastasis. Nature 410: 50–56. [DOI] [PubMed] [Google Scholar]
- 39. Li YM, Pan Y, Wei Y, Cheng X, Zhou BP, et al. (2004) Upregulation of CXCR4 is essential for HER2-mediated tumor metastasis. Cancer Cell 6: 459–469. [DOI] [PubMed] [Google Scholar]
- 40.Lakhani SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ (2012) WHO Classification of Tumours of the Breast: International Agency for Research on Cancer.
- 41. Zaczek AJ, Markiewicz A, Seroczynska B, Skokowski J, Jaskiewicz J, et al. (2012) Prognostic significance of TOP2A gene dosage in HER-2-negative breast cancer. The oncologist 17: 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, et al. (2006) REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Research and Treatment 100: 229–235. [DOI] [PubMed] [Google Scholar]
- 43. Markiewicz A, Ksiazkiewicz M, Seroczynska B, Skokowski J, Szade J, et al. (2013) Heterogeneity of mesenchymal markers expression-molecular profiles of cancer cells disseminated by lymphatic and hematogenous routes in breast cancer. Cancers (Basel) 5: 1485–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8: R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, et al. (2006) GenePattern 2.0. Nature Genetics 38: 500–501. [DOI] [PubMed] [Google Scholar]
- 46. Husemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, et al. (2008) Systemic spread is an early step in breast cancer. Cancer Cell 13: 58–68. [DOI] [PubMed] [Google Scholar]
- 47. Molloy TJ, Devriese LA, Helgason HH, Bosma AJ, Hauptmann M, et al. (2011) A multimarker QPCR-based platform for the detection of circulating tumour cells in patients with early-stage breast cancer. British Journal of Cancer 104: 1913–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wulfing P, Borchard J, Buerger H, Heidl S, Zanker KS, et al. (2006) HER2-positive circulating tumor cells indicate poor clinical outcome in stage I to III breast cancer patients. Clinical Cancer Research 12: 1715–1720. [DOI] [PubMed] [Google Scholar]
- 49.Rack BK, Schindlbeck C, Andergassen U, Schneeweiss A, Zwingers T, et al.. (2010) Use of circulating tumor cells (CTC) in peripheral blood of breast cancer patients before and after adjuvant chemotherapy to predict risk for relapse: The SUCCESS trial. Journal of Clinical Oncology.
- 50. Lasa A, Garcia A, Alonso C, Millet P, Cornet M, et al. (2013) Molecular detection of peripheral blood breast cancer mRNA transcripts as a surrogate biomarker for circulating tumor cells. PLoS One 8: e74079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kundu GC, Mantile G, Miele L, Cordella-Miele E, Mukherjee AB (1996) Recombinant human uteroglobin suppresses cellular invasiveness via a novel class of high-affinity cell surface binding site. Proceedings of the National Academy of Sciences of the United States of America 93: 2915–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Heyns W, Bossyns D (1983) A comparative study of estramustine and pregnenolone binding to prostatic binding protein: evidence for subunit cooperativity. J Steroid Biochem 19: 1689–1694. [DOI] [PubMed] [Google Scholar]
- 53. Benoy IH, Elst H, Philips M, Wuyts H, Van Dam P, et al. (2006) Real-time RT-PCR detection of disseminated tumour cells in bone marrow has superior prognostic significance in comparison with circulating tumour cells in patients with breast cancer. British Journal of Cancer 94: 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Raynor MP, Stephenson SA, Pittman KB, Walsh DC, Henderson MA, et al. (2009) Identification of circulating tumour cells in early stage breast cancer patients using multi marker immunobead RT-PCR. Journal of hematology & oncology 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen Y, Zou TN, Wu ZP, Zhou YC, Gu YL, et al. (2010) Detection of cytokeratin 19, human mammaglobin, and carcinoembryonic antigen-positive circulating tumor cells by three-marker reverse transcription-PCR assay and its relation to clinical outcome in early breast cancer. Int J Biol Markers 25: 59–68. [DOI] [PubMed] [Google Scholar]
- 56. Pecot CV, Bischoff FZ, Mayer JA, Wong KL, Pham T, et al. (2011) A Novel Platform for Detection of CK+ and CK− CTCs. Cancer Discovery 1: 580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Andergassen U, Hofmann S, Kolbl AC, Schindlbeck C, Neugebauer J, et al. (2013) Detection of Tumor Cell-Specific mRNA in the Peripheral Blood of Patients with Breast Cancer—Evaluation of Several Markers with Real-Time Reverse Transcription-PCR. Int J Mol Sci 14: 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhao S, Yang H, Zhang M, Zhang D, Liu Y, et al. (2013) Circulating tumor cells (CTCs) detected by triple-marker EpCAM, CK19, and hMAM RT-PCR and their relation to clinical outcome in metastatic breast cancer patients. Cell biochemistry and biophysics 65: 263–273. [DOI] [PubMed] [Google Scholar]
- 59. Hemsen A, Riethdorf L, Brunner N, Berger J, Ebel S, et al. (2003) Comparative evaluation of urokinase-type plasminogen activator receptor expression in primary breast carcinomas and on metastatic tumor cells. International Journal of Cancer 107: 903–909. [DOI] [PubMed] [Google Scholar]
- 60. Bonnomet A, Syne L, Brysse A, Feyereisen E, Thompson EW, et al. (2012) A dynamic in vivo model of epithelial-to-mesenchymal transitions in circulating tumor cells and metastases of breast cancer. Oncogene 31: 3741–3753. [DOI] [PubMed] [Google Scholar]
- 61. Giordano A, Gao H, Anfossi S, Cohen E, Mego M, et al. (2012) Epithelial-mesenchymal transition and stem cell markers in patients with HER2-positive metastatic breast cancer. Molecular cancer therapeutics 11: 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Alpaugh ML, Tomlinson JS, Kasraeian S, Barsky SH (2002) Cooperative role of E-cadherin and sialyl-Lewis X/A-deficient MUC1 in the passive dissemination of tumor emboli in inflammatory breast carcinoma. Oncogene 21: 3631–3643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Exemplary photos of immunostained cells isolated from CTC-enriched blood fractions of breast cancer patients.
(PDF)
Relative genes expression levels in breast cancer patients and controls. Expression of VIM, HER2, CXCR4, uPAR, TWIST1, SNAIL, CK19 and MGB1 in CTCs-enriched blood fractions of breast cancer patients with invasive carcinoma (BC), patients with ductal carcinoma in situ (DCIS) and healthy controls (HC). As no sample expressed SLUG, results for this gene are not presented.
(PDF)
Spearman's rank correlations coefficients of relative gene expression levels in CTC-enriched blood fractions.
(PDF)