Abstract
To clarify the role of potassium inwardly-rectifying-channel, subfamily-J, member 11 (KCNJ11) variation in susceptibility to type 2 diabetes (T2D), we performed a systematic meta-analysis to investigate the association between the KCNJ11 E23K polymorphism (rs5219) and the T2D in different genetic models. Databases including PubMed, Medline, EMBASE, and ISI Web of Science were searched to identify relevant studies. A total of 48 published studies involving 56,349 T2D cases, 81,800 controls, and 483 family trios were included in this meta-analysis. Overall, the E23K polymorphism was significantly associated with increased T2D risk with per-allele odds ratio (OR) of 1.12 (95% CI: 1.09–1.16; P<10−5). The summary OR for T2D was 1.09 (95% CI: 1.03–1.14; P<10−5), and 1.26 (95% CI: 1.17–1.35; P<10−5), for heterozygous and homozygous, respectively. Similar results were also detected under dominant and recessive genetic models. When stratified by ethnicity, significantly increased risks were found for the polymorphism in Caucasians and East Asians. However, no such associations were detected among Indian and other ethnic populations. Significant associations were also observed in the stratified analyses according to different mean BMI of cases and sample size. Although significant between study heterogeneity was identified, meta-regression analysis suggested that the BMI of controls significantly correlated with the magnitude of the genetic effect. The current meta-analysis demonstrated that a modest but statistically significant effect of the 23K allele of rs5219 polymorphism in susceptibility to T2D. But the contribution of its genetic variants to the epidemic of T2D in Indian and other ethnic populations appears to be relatively low.
Introduction
Type 2 diabetes (T2D) is a complex metabolic disease resulting from reduced insulin secretion and peripheral insulin resistance. By coupling cell metabolism with membrane potential, adenosine triphosphate-sensitive potassium channel (KATP) play a central role in regulation of insulin secretion in pancreatic-β cells. [1]. The KATP channel is a hetero-octamer of K+ inward rectifier Kir6.2 (KCNJ11) and regulatory sulfonylurea receptor SUR1 subunits (ABCC8) [2]. Mutations in both KCNJ11 and ABCC8 cause neonatal diabetes and congenital hyper-insulinemia in humans [3], [4]. In addition, KCNJ11 gene knock-out mice are characterized by defects in insulin secretion in response to either glucose or tolbutamide [5].
As a candidate gene for T2D in humans, a nonsynonymous E23K variant (rs5219) which results from a G → A transition in codon 23 in the NH2-terminal tail of Kir6.2 was identified [6]. With spectacular advance in genotyping method in recent years, larger-scale genetic association study concerning the relationship between the E23K polymorphism and T2D susceptibility has been conducted in various populations. However, inconsistent results have appeared in the literature. Such inconsistence may be due to chance, insufficient power of limited sample size, or bias in study design (e.g., inappropriate control selection). Alternatively, these disparate findings may reflect ethnic diversity (e.g., population stratification) or phenotypic heterogeneity. As a powerful tool for summarizing the results from different studies to estimate the major effect with enhanced precision, meta-analysis has generally been used in quantitative assessment of genetic variation and disease. Here we present the most comprehensive meta-analysis for the effects of E23K polymorphism of KCNJ11 on T2D risk.
Materials and Methods
Literature search strategy and inclusion criteria
To identify eligible literatures, we conducted a computer-based search of PubMed, Medline, EMBASE and ISI Web of Science databases without language restrictions. Studies published before the end of Mar. 2013 on T2D and the E23K polymorphism in the KCNJ11 gene were retrieved. Search keywords combinations were “potassium inwardly-rectifying-channel, subfamily-J, member 11”, “KCNJ11”, “Kir6.2”, “type 2 diabetes”, “type 2 diabetes mellitus”, “T2D”, “T2DM”, “non-insulin-dependent diabetes mellitus”, “NIDDM”, “polymorphism” or “variation”. The titles and abstracts were read to determine their relevance, and potentially relevant studies were retained for further evaluation. For retrieved articles, the full texts were carefully read to determine whether they meet the purpose of the present meta-analysis. Furthermore, the references of these studies were checked to identify other relevant publications.
Eligible studies should meet following criteria: (1) focusing on the association of the KCNJ11 E23K polymorphism with T2D risk (2) being case-control or cohort studies (3) diagnosis of T2D patient was confirmed pathologically and (4) providing sufficient data for calculation of odds ratio (OR) with its 95% confidence interval (95% CI) and P-value. The major reasons for exclusion were (1) case-only studies (2) overlapping data (3) review papers. If more than two studies reported the same sample, only the study providing more information or latest published was selected.
Data extraction
The following information was carefully extracted from all eligible publications: the first author, year of publication, country of origin, ethnicity of subjects, study design, sample size, sex distribution among cases and controls, mean age and body mass index (BMI) of cases and controls, source of control, Hardy–Weinberg equilibrium (HWE) status in controls, genotyping method, number of genotypes in cases and controls. Two authors independently assessed the articles for compliance with the inclusion criteria, and disagreement was followed by discussion until consensus was reached.
Statistical methods
The association of the KCNJ11 E23K polymorphism with T2D was evaluated by calculating a pooled OR and 95% CI for allele contrast (K vs. E allele), heterozygous (KE vs. EE) and homozygote (KK vs. EE). Then, we examined the association between the polymorphisms and T2D risk using dominant and recessive genetic models. The standard Q-statistic test was performed to evaluate whether the variation between studies was due to heterogeneity or due to chance [7]. ORs were calculated according to the method of DerSimonian and Laird, and 95% CI was constructed by Woolf's method [8], [9]. In addition, subgroup analysis was used to investigate potential sources of heterogeneity by stratified meta-analyses based on ethnic group, sample size (No. cases ≥1000 or <1000) and mean BMI of cases (<25, 25∼30, or >30). Ethnic group was defined as East Asians, Caucasians (e.g., people of European origin), Indians and others (e.g., African American, Jews, and Arabian). Subsequently, ethnicity, sample size, BMI, age and sex were analyzed as covariates in meta-regression to further investigate potential sources of heterogeneity. For family-based association studies, the transmission disequilibrium test (TDT) was used to analyze effect size of the polymorphism. In general, the OR was calculated from the ratio of transmitted alleles to non-transmitted alleles from heterozygous parents to affected offspring [10], [11]. Combined effect size from both case–control and family-based association studies were calculated according to the method described previously by Lohmueller et al [12]. The Z-test was used to determine the significance of overall OR.
We calculated the sample size required for 80% power with the summary OR estimated from each ethnic populations, assuming an equal number of cases and controls, risk allele frequency (RAF) in controls estimated from different ethnicity. Furthermore, population attributable risk (PAR) was calculated to get a comprehensive view of the impact of the E23K variant on T2D at population level. PAR was calculated by the following formula: (OR-1)/OR * risk allele frequency [13].
Egger's test and funnel plots were used to assess small studies effects [14]. Sensitivity analysis was performed by excluding one study at a time to assess the stability of the results. The type I error rate was set at 0.05 for two-sided analysis. All of the calculations were performed using the STATA 10.0 (STATA Corporation, College Station, TX) and SAS (version 9.1; SAS Institute, Cary, NC).
Results
Characteristics of included studies
The literature search yielded 159 studies using keywords listed above. Figure S1 shows the literature search and selection process for eligible studies (Figure S1). Finally, a total of 48 studies including 56,349 cases, 81,800 controls and 483 family trios, were retrieved based on the search criteria for T2D susceptibility related to the KCNJ11 E23K polymorphism [15]–[62]. In addition, almost all studies indicated that the distribution of genotype frequencies among the control groups were consistent with HWE. The detailed characteristics of all the included studies of this meta-analysis were summarized in Table 1.
Table 1. Characteristics of the studies included in the meta-analysis.
| Study | Year | Ethnicity | Diagnostic criteria | Study design | No. of cases | No. of controls | MAF in cases/controls | Genotyping method | P HWE |
| Sakura [15] | 1996 | Caucasian | T2D patients | Population based-study | 133 | 82 | 0.30/0.30 | PCR-SSCP | 0.05 |
| Inoue [16] | 1997 | Caucasian | T2D patients | Population based-study | 291 | 164 | 0.34/0.34 | PCR-RFLP | >0.05 |
| Hani [17] | 1998 | Caucasian | T2D patients | Population based-study | 191 | 114 | 0.49/0.37 | PCR-SSCP | 0.95 |
| Altshuler [18] | 2000 | Caucasian | T2D per WHO criteria | Family based-study | 333 trios | / | / | SBE-FRET, SBE-FP | / |
| Yamada [19] | 2001 | East Asian | T2D per WHO criteria | Population based-study | 103 | 73 | 0.39/0.34 | PCR-SSCP | 0.20 |
| Gloyn [20] | 2001 | Caucasian | T2D patients | Population based-study | 360 | 307 | 0.40/0.36 | PCR-SSCP | 0.09 |
| Florez [21] | 2004 | Caucasian | T2D per WHO criteria | Population based-study | 1077 | 1077 | 0.47/0.61 | Flight mass spectroscopy | 0.71 |
| Barroso [22] | 2003 | Caucasian | T2D patients | Population based-study | 499 | 494 | 0.38/0.34 | FP-TDI | 0.82 |
| Gloyn [23] | 2003 | Caucasian | T2D per WHO criteria | Population based-study, Family based-study | 854, 150 trios | 1182 | 0.41/0.34 | PCR-RFLP | 0.53 |
| Hansen [24] | 2005 | Caucasian | T2D per WHO criteria | Population based-study | 1164 | 4733 | 0.40/0.36 | PCR-RFLP, LNA | 0.52 |
| van Dam [25] | 2005 | Caucasian | T2D per WHO criteria | Population based-study | 323 | 296 | 0.41/0.36 | PCR-RFLP | 0.56 |
| Yokoi [26] | 2006 | East Asian | T2D per WHO criteria | Population based-study | 1590 | 1244 | 0.38/0.37 | MassARRAY | 0.64 |
| Liu [27] | 2006 | East Asian | T2D per WHO criteria | Population based-study | 502 | 501 | 0.43/0.38 | Sequencing | >0.05 |
| Weedon [28] | 2006 | Caucasian | T2D per WHO criteria | Population based-study | 2332 | 3592 | 0.38/0.35 | TaqMan | >0.05 |
| Sale [29] | 2007 | Other | T2D patients | Population based-study | 572 | 587 | 0.06/0.07 | MassARRAY | 0.22 |
| Koo [30] | 2007 | East Asian | T2D per WHO criteria | Population based-study | 758 | 630 | 0.44/0.38 | TaqMan | 0.05 |
| Sakamoto [31] | 2007 | East Asian | T2D per WHO criteria | Population based-study | 906 | 889 | 0.39/0.34 | TaqMan | 0.72 |
| Saxena [32] | 2007 | Caucasian | T2D per WHO criteria | Population based-study | 5065 | 5785 | 0.49/0.47 | Affymetrix GeneChip, MassARRAY | >0.05 |
| Vaxillaire [33] | 2007 | Caucasian | T2D per ADA criteria | Population based-study | 287 | 2684 | 0.41/0.39 | TaqMan | 0.68 |
| Scott [34] | 2007 | Caucasian | T2D per WHO criteria | Population based-study | 2295 | 2363 | 0.49/0.46 | Illumina GeneChip, MassARRAY | 0.72 |
| Willer [35] | 2007 | Caucasian | T2D per WHO criteria | Population based-study | 1087 | 953 | 0.49/0.44 | MassARRAY | 0.32 |
| Qi [36] | 2007 | Caucasian | T2D patients | Population based-study | 682 | 1078 | 0.40/0.35 | TaqMan | 0.38 |
| Cejková [37] | 2007 | Caucasian | T2D per WHO criteria | Population based-study | 172 | 113 | 0.37/0.37 | PCR-RFLP | 0.26 |
| Doi [38] | 2007 | East Asian | T2D per WHO criteria | Population based-study | 550 | 2322 | 0.39/0.34 | TaqMan | 0.46 |
| Lyssenko [39] | 2008 | Caucasian | T2D per WHO criteria | Population based-study | 2201 | 16034 | 0.41/0.40 | TaqMan | >0.05 |
| Alsmadi [40] | 2008 | Other | T2D per ADA criteria | Population based-study | 550 | 335 | 0.21/0.14 | TaqMan | 0.40 |
| Takeuchi [41] | 2008 | East Asian | T2D per WHO criteria | Population based-study | 7954 | 8809 | 0.38/0.35 | Illumina GeneChip, MassARRAY, TaqMan | 0.91 |
| Peng [42] | 2008 | East Asian | T2D per ADA criteria | Population based-study | 275 | 168 | 0.69/0.57 | PCR-RFLP | >0.05 |
| Bronstein [43] | 2008 | Other | T2D patients | Population based-study | 1131 | 1147 | 0.36/0.61 | KASPar | 0.58 |
| Sanghera [44] | 2008 | Indian | T2D per ADA criteria | Population based-study | 532 | 374 | 0.34/0.38 | TaqMan | 0.45 |
| Cauchi [45] | 2008 | Caucasian | T2D per WHO criteria | Population based-study | 2734 | 4234 | 0.37/0.37 | TaqMan | 0.69 |
| Ezzidi [46] | 2009 | Other | T2D per ADA criteria | Population based-study | 805 | 503 | 0.32/0.29 | TaqMan | 0.56 |
| Zhou [47] | 2009 | East Asian | T2D per WHO criteria | Population based-study | 1848 | 1910 | 0.41/0.39 | TaqMan | 0.39 |
| Chistiakov [48] | 2009 | Caucasian | T2D per WHO criteria | Population based-study | 129 | 117 | 0.50/0.39 | PCR-RFLP | >0.05 |
| Wang [49] | 2009 | East Asian | T2D per WHO criteria | Population based-study | 396 | 387 | 0.46/0.37 | SNapShot | 0.46 |
| Tabara [50] | 2009 | East Asian | T2D per ADA criteria | Population based-study | 484 | 397 | 0.41/0.37 | TaqMan | 0.30 |
| Thorsby [51] | 2009 | Caucasian | T2D patients | Population based-study | 750 | 1879 | 0.41/0.41 | PCR-RFLP | 0.18 |
| Hu [52] | 2009 | East Asian | T2D per WHO criteria | Population based-study | 1849 | 1785 | 0.42/0.39 | MassARRAY | >0.05 |
| Yamauchi [53] | 2010 | East Asian | T2D per WHO criteria | Population based-study | 4470 | 3071 | 0.38/0.37 | Illumina GeneChip | >0.05 |
| Neuman [54] | 2010 | Other | T2D patients | Population based-study | 573 | 843 | 0.37/0.36 | Pyrosequencing | 0.22 |
| Chauhan [55] | 2010 | Indian | T2D per WHO criteria | Population based-study | 2434 | 2403 | 0.39/0.32 | Golden Gate assay | 0.41 |
| Gupta [56] | 2010 | Indian | T2D per WHO criteria | Population based-study | 209 | 179 | 0.40/0.47 | Sequencing | 0.12 |
| Wen [57] | 2010 | East Asian | T2D per WHO criteria | Population based-study | 1165 | 1135 | 0.41/0.40 | MassARRAY | 0.10 |
| Rees [58] | 2011 | Indian | T2D per WHO criteria | Population based-study | 1663 | 1567 | 0.38/0.38 | TaqMan | 0.13 |
| Chavali [59] | 2011 | Indian | T2D per WHO criteria | Population based-study | 1017 | 1006 | 0.39/0.35 | Golden Gate assay | >0.05 |
| Cheung [60] | 2011 | Chinese | T2D per WHO criteria | Population based-study | 198 | 1185 | 0.33/0.33 | TaqMan | 0.41 |
| Gamboa-Meléndez [61] | 2012 | Other | T2D per ADA criteria | Population based-study | 1027 | 990 | 0.40/0.37 | KASPAR | >0.05 |
| Gonen [62] | 2012 | Other | T2D per ADA criteria | Population based-study | 162 | 79 | 0.34/0.30 | PCR-SSCP | NA |
WHO: World health organization, ADA: American diabetes association, MAF: minor allele frequency, LNA: locked nucleic acid assay, FP-TDI: fluorescence polarization template-directed incorporation. SBE-FRET: single-base extension with fluorescence resonance energy transfer; SBE-FP: single-base extension with fluorescence polarization.
Meta-analysis results
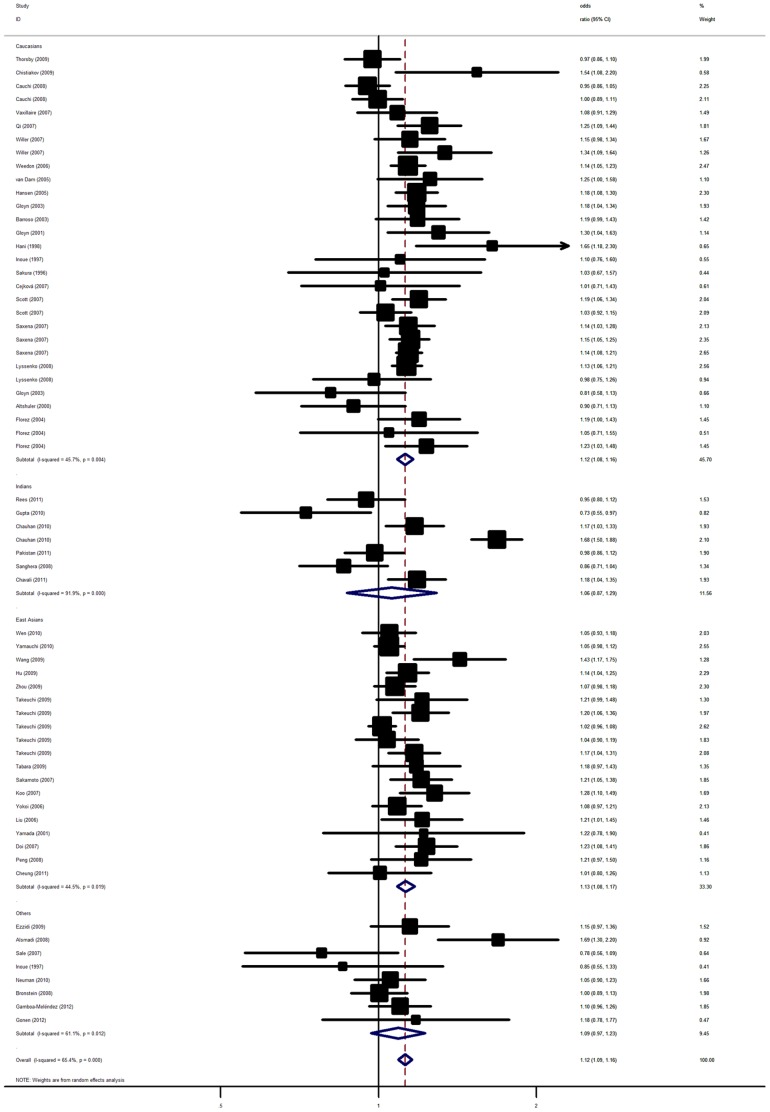
Overall, significant associations between KCNJ11 E23K polymorphism and T2D were detected when all the eligible studies were pooled into the meta-analysis (Table 2). The overall result showed that the 23K allele of rs5219 polymorphism was significantly associated with elevated T2D risk with per-allele OR of 1.12 (95% CI: 1.08–1.17, P<10−5; Figure 1). Significant increased T2D risks were also detected for heterozygous (OR = 1.09, 95% CI: 1.03–1.14, P<10−5) and homozygous (OR = 1.26, 95% CI: 1.17–1.35, P<10−5) when compared with wild type homozygous. Similar results still maintained using dominant and recessive genetic models (Table S1). When studies were stratified for ethnic populations, significant associations were also observed among East Asian and Caucasian populations with per-allele OR of 1.13 (95% CI: 1.08–1.17, P<10−5) and of 1.12 (95% CI: 1.08–1.16, P<10−5) respectively. Significantly increased risks were also found for heterozygous and homozygous (Table 2). However, no such association was detected in Indian and other ethnic populations in all genetic models. In the subgroup analysis by sample size, significant associations were also observed for both large and small studies in all genetic models. When stratified by mean BMI of cases, statistically significant results were also observed for T2D cases with different BMI (Table 2). For two family-based association studies including a total of 483 family trios, we failed to detect statistically significant evidence for the risk 23K allele over-transmission from heterozygous parents to their T2D offspring (pooled ORTDT = 0.87, 95% CI: 0.72–1.05; P = 0.14).
Table 2. Results of meta-analysis for KCNJ11 E23K polymorphism and T2D risk.
| Sub-group analysis | No. of studies | K allele | Heterozygous | Homozygous | |||||||||
| OR (95%CI) | P(Z) | P(Q)a | P(Q)b | OR (95%CI) | P(Z) | P(Q)a | P(Q)b | OR (95%CI) | P(Z) | P(Q)a | P(Q)b | ||
| Ethnicity | 0.10 | 0.06 | 0.01 | ||||||||||
| Caucasians | 22 | 1.12 (1.08–1.16) | <10−5 | 0.001 | 1.09 (1.06–1.13) | <10−5 | 0.13 | 1.33 (1.18–1.50) | <10−5 | 0.0008 | |||
| East Asians | 14 | 1.13 (1.08–1.17) | <10−5 | 0.02 | 1.13 (1.05–1.22) | 0.0009 | 0.02 | 1.30 (1.18–1.42) | <10−5 | 0.06 | |||
| Indians | 5 | 1.06 (0.87–1.29) | 0.56 | <10−5 | 1.00 (0.82–1.23) | 0.98 | 0.01 | 0.90 (0.69–1.18) | 0.45 | 0.02 | |||
| Others | 7 | 1.09 (0.97–1.23) | 0.15 | 0.01 | 0.98 (0.76–1.28) | 0.91 | <10−4 | 1.19 (1.00–1.43) | 0.05 | 0.31 | |||
| Sample size | 0.32 | 0.14 | 0.04 | ||||||||||
| Large | 22 | 1.12 (1.08–1.17) | <10−5 | <10−5 | 1.12 (1.10–1.15) | <10−4 | 0.12 | 1.17 (1.07–1.28) | 0.007 | 0.02 | |||
| Small | 26 | 1.13 (1.07–1.18) | <10−5 | <10−5 | 1.10 (1.02–1.19) | 0.009 | 0.002 | 1.32 (1.20–1.46) | <10−5 | 0.0007 | |||
| Mean BMI of cases | 0.37 | 0.20 | 0.03 | ||||||||||
| <25 | 12 | 1.15 (1.11–1.21) | <10−5 | 0.15 | 1.15 (1.08–1.22) | <10−5 | 0.21 | 1.40 (1.28–1.55) | <10−5 | 0.74 | |||
| 25∼30 | 25 | 1.12 (1.06–1.19) | <10−4 | <10−4 | 1.13 (1.06–1.21) | <10−4 | 0.004 | 1.18 (1.03–1.34) | 0.008 | <10−4 | |||
| >30 | 6 | 1.10 (1.03–1.18) | 0.008 | 0.07 | 1.11 (1.04–1.19) | 0.001 | 0.09 | 1.16 (1.01–1.33) | 0.02 | 0.10 | |||
| Total | 48 | 1.12 (1.09–1.16) | <10−5 | <10−5 | 1.09 (1.03–1.14) | <10−5 | 0.0001 | 1.26 (1.17–1.35) | <10−5 | <10−5 | |||
P(Z): Z test used to determine the significance of the overall OR.
P(Q)a: Cochran's chi-square Q statistic test used to assess the heterogeneity in subgroups.
P(Q)b: Cochran's chi-square Q statistic test used to assess the heterogeneity between subgroups.
Figure 1. Forest plot from the meta-analysis of T2D risk and KCNJ11 rs5219 polymorphism using random-effects model.
Significant heterogeneity was found among the 46 included studies (P<10−5). Hence, meta-regression was further conducted to investigate the source of heterogeneity. In meta-regression analysis, ethnicity (P = 0.79), sample size (P = 0.61), mean age (P = 0.36) of cases and controls (P = 0.61), gender distribution in cases (P = 0.96) and controls (P = 0.30) did not explain a large part of the heterogeneity among the individual study. By contrast, mean BMI (P = 0.03) explained about 11% of the heterogeneity.
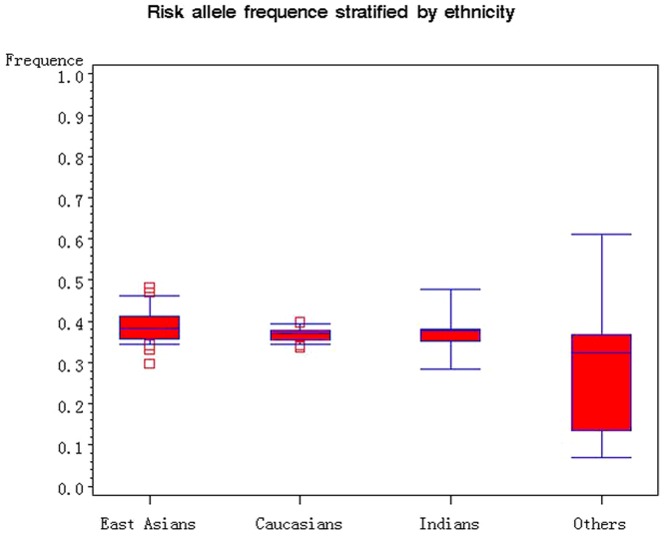
The 23K allele frequency of the rs5219 polymorphism varies in the control groups across different ethnic populations, ranging from 0.07 to 0.61 (Figure 2). In Caucasian controls, the K allele frequency was 0.40 (95% CI: 0.37–0.42), which was higher than that of East Asian controls (0.36; 95% CI: 0.34–0.38), Indian controls (0.34; 95% CI: 0.27–0.41). 2500 and 2200 case-control pairs will be required for 80% power to detect the risk allele among Caucasian and East Asian population respectively. The population attributable risk (PAR) of T2D related to E23K polymorphism was 4.6% overall, 4.4% for Caucasians and 4.5% for East Asians.
Figure 2. Frequencies of the 23K allele of KCNJ11 rs5219 polymorphism among controls stratified by ethnicity.
The “□” represent outlier.
Sensitivity analyses and publication bias
The results of sensitivity analysis confirmed the significant associations of the KCNJ11 E23K polymorphism with T2D risk, and no single study influenced the overall OR qualitatively (Figure S2). The Egger's test and funnel plots indicated no publication bias for the association of KCNJ11 E23K polymorphism and T2D (Figure S3; Egger test, P>0.05).
Discussion
Limited statistical power of relative small sample size is a common problem in genetic association for individual T2D studies. Therefore, sufficient sample power is necessary in deciphering genetic architecture of T2D, but it is sometimes very difficult for a single study to collect enough amounts of data to reach a reliable conclusion. By pooling of data from individual association studies, meta-analysis is an effective approach of increasing the sample size under investigation, thus enhancing the statistical power for the estimation of genetic effects.
Our results indicated that the rs5219 polymorphism of KCNJ11 is a risk factor for developing T2D. In the subgroup analyses by ethnicity, we found the rs5219 polymorphism was associated with T2D among East Asians and Caucasians, but not Indians or other ethnic populations. Of note, different ethnic populations were pooled in the other ethnic group and only a few studies were available in the subgroup, so the result must be interpreted with caution. There are several other possible reasons which may account for such differences. First, T2D is a complex disease and different genetic backgrounds may cause the discrepancy since the distributions of the risk-association alleles in KCNJ11 were different between various ethnicities. The K allele frequency of the rs5219 polymorphism was ∼36%, ∼40%, and ∼34%, among East Asians, Indians and Caucasians populations, respectively. Such result could also be due different linkage disequilibrium (LD) pattern of the polymorphism and nearby causal variant among different ethnic populations. Moreover, inter-individual difference like age, sex, dietary intake of nutrients, in addition to phenotype heterogeneity, such as years from onset and severity of the disease may also explain the discrepancy. Furthermore, study design and/or small sample size or some environmental factors may also affect the results. Therefore, more studies are needed to further validate the effect of the polymorphism on T2D risk among difference ethnic populations.
The KCNJ11 gene has attracted considerable attention as a promising candidate for T2D based on its position and its function as a key factor in the regulation of glucose-induced insulin secretion, since normoglycemic lysine carriers are shown to consistently display a defect in insulin secretion [21], [63], [64]. Functional studies suggested that the KK genotype might induce a critical inhibition of glucose-induced insulin release from pancreatic β-cells [65]. Furthermore, the KCNJ11 E23K variant was found to be associated with glucose intolerance and conversion from impaired glucose tolerance to T2D among Caucasians [66], [67]. Previous studies indicated that the E23K variant is functional by affecting in vitro properties of KATP channel via increasing the threshold of ATP concentration for insulin secretion [65], [68].
The distribution of the E23K variant in controls across various studies showed global variation (Figure 2), suggesting the possibility of population stratification. However, empirical evidence indicated that well-designed population-based association studies can keep the effects of population stratification to a minimum [69]. Almost all the population-based genetic association studies included in the present meta-analysis were well-designed by recruiting cases and controls from the same geographic region and ethnicity, which may help to reduce the effects of population stratification. Moreover, the effects of potential population stratification in any individual study may be in a random direction, so that one individual study with a small amount of stratification should have very limited effect on the overall results [70]. By combining results from TDT studies, which are robust to potential population stratification, we failed to detect an over-transmission of the 23K allele from heterozygous parents to their T2D offspring. Given the small number of studies and relative small sample size, the combined results from TDT studies should be interpreted with caution.
Power analysis revealed a hint in design for future association studies. So far, most association studies have included at the most several hundred subjects and results from these studies should be treated with caution as for limited statistic power to reach a reliable conclusion. According to our power analysis, powerful association studies on the E23K polymorphism and T2D risk may need several thousand individuals. Compared with candidate gene approach, genome wide association study (GWAS) with large sample size and unbiased to genomic structure is a powerful approach in susceptible gene identification for T2D. Recently, Tsai et al. [71] reported a two-stage genome-wide association (GWA) conducted in Chinese and identified KCNJ11 as a risk region in T2D susceptibility which was in line with the results the present meta-analysis.
In comparison with the previous published meta-analysis [63], [66], [72]–[74], the current study included more than ten times as many cases as the earlier meta-analysis. In addition, we assessed the effect of risk allele and T2D using various genetic models and reached consistent results. Furthermore, we systematically explored potential sources of heterogeneity across studies and the possibility of publication bias. In addition to combine those newly published data, the present study also statistically joined population-based and family-based genetic association studies into a single meta-analysis, which allowed us to enhance the power of the meta-analyses and also establish a comprehensive picture of the relationship between KCNJ11 and genetic susceptibility to T2D.
Heterogeneity among pooled studies is a frequently encountered issue in meta-analyses. Of note, our meta-analyses joined population bases association studies and family based association studies, which could enhance the heterogeneity. Hence, we performed subgroup analysis and meta-regression to identify potential source of between-study heterogeneity. However, they revealed that only BMI of controls could explain a small part of significant heterogeneity between studies. There may be a number of possible underlying reasons. Firstly, results from case–control studies may differ because of ethnic diversity (e.g., variation in allele frequencies) and geographic variation. Secondly, variations in methods of sample ascertainment and diagnosis may also contribute to such inconsistence. Thirdly, environmental factors, such as alcohol drinking, smoking behavior, obesity might also result in variability between studies.
To conclude, this may be the most comprehensive meta-analysis of KCNJ11 and T2D. Our results suggest a modest but statistically significant effect of the 23K allele of rs5219 in susceptibility to T2D, particularly in East Asians and Caucasians. More work will be required for future association studies, especially those which are properly powered, effectively control for confounding factors, employing family-based design. Moreover, gene–environment and gene–gene interactions should also be taking into consideration for future studies.
Supporting Information
(DOC)
Flow chart of literature search for studies examining KCNJ11 rs5219 polymorphism and risk of T2D.
(TIF)
Result of sensitivity analyses.
(TIF)
Begg's funnel plot of rs5219 polymorphism and T2D risk.
(TIF)
Results of meta-analysis for KCNJ11 E23K polymorphism and T2D risk using dominant and recessive genetic models.
(DOCX)
Funding Statement
This work was supported by the research foundation of Shanghai Municipal Health Bureau (No. 20124276). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bell GI, Polonsky KS (2001) Diabetes mellitus and genetically programmed defects in beta-cell function. Nature 414: 788–791. [DOI] [PubMed] [Google Scholar]
- 2. Aguilar-Bryan L, Bryan J (1999) Molecular biology of adenosine triphosphate-sensitive potassium channels. Endocr Rev 20: 101–135. [DOI] [PubMed] [Google Scholar]
- 3. Smith AJ, Taneja TK, Mankouri J, Sivaprasadao A (2007) Molecular cell biology of KATP channels: implications for neonatal diabetes. Expert Rev Mol Med 9: 1–17. [DOI] [PubMed] [Google Scholar]
- 4. Nichols CG, Koster JC, Remedi MS (2007) b-cell hyperexcitability: from hyperinsulinism to diabetes. Diabetes Obes Metab 9: 81–88. [DOI] [PubMed] [Google Scholar]
- 5. Miki T, Nagashima K, Tashiro F, Kotake K, Yoshitomi H, et al. (1998) Defective insulin secretion and enhanced insulin action in KATP channel-deficient mice. Proc Natl Acad Sci USA 95: 10402–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sakura H, Wat N, Horton V, Millns H, Turner RC, et al. (1996) Sequence variations in the human Kir6.2 gene, a subunit of the beta-cell ATP-sensitive K-channel: no association with NIDDM in white Caucasian subjects or evidence of abnormal function when expressed in vitro. Diabetologia 39: 1233–1236. [DOI] [PubMed] [Google Scholar]
- 7. Colditz GA, Burdick E, Mosteller F (1995) Heterogeneity in meta-analysis of data from epidemiologic studies: a commentary. Am J Epidemiol 142: 371–382. [DOI] [PubMed] [Google Scholar]
- 8. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 9. Woolf B (1955) On estimating the relation between blood group and disease. Ann Hum Genet 19: 251–253. [DOI] [PubMed] [Google Scholar]
- 10. Spielman RS, McGinnis RE, Ewens WJ (1993) Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). Am J Hum Genet 52: 506–516. [PMC free article] [PubMed] [Google Scholar]
- 11. Song Y, Niu T, Manson JE, Kwiatkowski DJ, Liu S (2004) Are variants in the CAPN10 gene related to risk of type 2 diabetes? A quantitative assessment of population and family-based association studies. Am J Hum Genet 74: 208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN (2003) Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat Genet 33: 177–182. [DOI] [PubMed] [Google Scholar]
- 13. Cugino D, Gianfagna F, Santimone I, de Gaetano G, Donati MB, et al. (2012) Type 2 diabetes and polymorphisms on chromosome 9p21: A meta-analysis. Nutr Metab Cardiovasc Dis 22: 619–625. [DOI] [PubMed] [Google Scholar]
- 14. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. 80 Ardlie KG, Lunetta KL, Seielstad M (2002) Testing for population subdivision and association in four case–control studies. Am J Hum Genet 71: 304–311. [Google Scholar]
- 15. Sakura H, Wat N, Horton V, Millns H, Turner RC, et al. (1996) Sequence variations in the human Kir6.2 gene, a subunit of the beta-cell ATP-sensitive K-channel: no association with NIDDM in while Caucasian subjects or evidence of abnormal function when expressed in vitro. Diabetologia 39: 1233–1236. [DOI] [PubMed] [Google Scholar]
- 16. Inoue H, Ferrer J, Warren-Perry M, Zhang Y, Millns H, et al. (1997) Sequence variants in the pancreatic islet beta-cell inwardly rectifying K+ channel Kir6.2 (Bir) gene: identification and lack of role in Caucasian patients with NIDDM. Diabetes 46: 502–507. [DOI] [PubMed] [Google Scholar]
- 17. Hani EH, Boutin P, Durand E, Inoue H, Permutt MA, et al. (1998) Missense mutations in the pancreatic islet beta cell inwardly rectifying K+ channel gene (KIR6.2/BIR): a meta-analysis suggests a role in the polygenic basis of Type II diabetes mellitus in Caucasians. Diabetologia 41: 1511–1515. [DOI] [PubMed] [Google Scholar]
- 18. Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, et al. (2000) The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26: 76–80. [DOI] [PubMed] [Google Scholar]
- 19. Yamada Y, Kuroe A, Li Q, Someya Y, Kubota A, et al. (2001) Genomic variation in pancreatic ion channel genes in Japanese type 2 diabetic patients. Diabetes Metab Res. Rev 17: 213–216. [DOI] [PubMed] [Google Scholar]
- 20. Gloyn AL, Hashim Y, Ashcroft SJ, Ashfield R, Wiltshire S, et al. (2001) Association studies of variants in promoter and coding regions of beta-cell ATP-sensitive K-channel genes SUR1 and Kir6.2 with Type 2 diabetes mellitus (UKPDS 53). Diabet Med 18: 206–212. [DOI] [PubMed] [Google Scholar]
- 21. Florez JC, Burtt N, de Bakker PI, Almgren P, Tuomi T, et al. (2004) Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes 53: 1360–1368. [DOI] [PubMed] [Google Scholar]
- 22. Barroso I, Luan J, Middelberg RP, Harding AH, Franks PW, et al. (2003) Candidate gene association study in type 2 diabetes indicates a role for genes involved in beta-cell function as well as insulin action. PLoS Biol 1: E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gloyn AL, Weedon MN, Owen KR, Turner MJ, Knight BA, et al. (2003) Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes 52: 568–572. [DOI] [PubMed] [Google Scholar]
- 24. Hansen SK, Nielsen EM, Ek J, Andersen G, Glümer C, et al. (2005) Analysis of separate and combined effects of common variation in KCNJ11 and PPARG on risk of type 2 diabetes. J Clin Endocrinol Metab 90: 3629–3637. [DOI] [PubMed] [Google Scholar]
- 25. van Dam RM, Hoebee B, Seidell JC, Schaap MM, de Bruin TW, et al. (2005) Common variants in the ATP-sensitive K+ channel genes KCNJ11 (Kir6.2) and ABCC8 (SUR1) in relation to glucose intolerance: population-based studies and meta-analyses. Diabet Med 22: 590–598. [DOI] [PubMed] [Google Scholar]
- 26. Yokoi N, Kanamori M, Horikawa Y, Takeda J, Sanke T, et al. (2006) Association studies of variants in the genes involved in pancreatic beta-cell function in type 2 diabetes in Japanese subjects. Diabetes 55: 2379–2386. [DOI] [PubMed] [Google Scholar]
- 27. Liu Z, Zhang YW, Feng QP, Li YF, Wu GD, et al. (2006) Association analysis of 30 type 2 diabetes candidate genes in Chinese Han population. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 28: 124–128. [PubMed] [Google Scholar]
- 28. Weedon MN, McCarthy MI, Hitman G, Walker M, Groves CJ, et al. (2006) Combining information from common type 2 diabetes risk polymorphisms improves disease prediction. PLoS Med 3: e374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sale MM, Smith SG, Mychaleckyj JC, Keene KL, Langefeld CD, et al. (2007) Variants of the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in an African-American population enriched for nephropathy. Diabetes 56: 2638–2642. [DOI] [PubMed] [Google Scholar]
- 30. Koo BK, Cho YM, Park BL, Cheong HS, Shin HD, et al. (2007) Polymorphisms of KCNJ11 (Kir6.2 gene) are associated with Type 2 diabetes and hypertension in the Korean population. Diabet Med 24: 178–186. [DOI] [PubMed] [Google Scholar]
- 31. Sakamoto Y, Inoue H, Keshavarz P, Miyawaki K, Yamaguchi Y, et al. (2007) SNPs in the KCNJ11-ABCC8 gene locus are associated with type 2 diabetes and blood pressure levels in the Japanese population. J Hum Genet 52: 781–793. [DOI] [PubMed] [Google Scholar]
- 32. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336. [DOI] [PubMed] [Google Scholar]
- 33. Vaxillaire M, Veslot J, Dina C, Proença C, Cauchi S, et al. (2008) Impact of common type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes 57: 244–254. [DOI] [PubMed] [Google Scholar]
- 34. Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, et al. (2007) A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Willer CJ, Bonnycastle LL, Conneely KN, Duren WL, Jackson AU, et al. (2007) Screening of 134 single nucleotide polymorphisms (SNPs) previously associated with type 2 diabetes replicates association with 12 SNPs in nine genes. Diabetes 56: 256–264. [DOI] [PubMed] [Google Scholar]
- 36. Qi L, van Dam RM, Asselbergs FW, Hu FB (2007) Gene-gene interactions between HNF4A and KCNJ11 in predicting Type 2 diabetes in women. Diabet Med 24: 1187–1191. [DOI] [PubMed] [Google Scholar]
- 37. Cejková P, Novota P, Cerná M, Kolostová K, Nováková D, et al. (2007) KCNJ11 E23K polymorphism and diabetes mellitus with adult onset in Czech patients. Folia Biol (Praha). 53: 173–175. [PubMed] [Google Scholar]
- 38. Doi Y, Kubo M, Ninomiya T, Yonemoto K, Iwase M, et al. (2007) Impact of Kir6.2 E23K polymorphism on the development of type 2 diabetes in a general Japanese population: The Hisayama Study. Diabetes 56: 2829–2833. [DOI] [PubMed] [Google Scholar]
- 39. Lyssenko V, Jonsson A, Almgren P, Pulizzi N, Isomaa B, et al. (2008) Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med 359: 2220–2232. [DOI] [PubMed] [Google Scholar]
- 40. Alsmadi O, Al-Rubeaan K, Wakil SM, Imtiaz F, Mohamed G, et al. (2008) Genetic study of Saudi diabetes (GSSD): significant association of the KCNJ11 E23K polymorphism with type 2 diabetes. Diabetes Metab Res Rev 24: 137–140. [DOI] [PubMed] [Google Scholar]
- 41. Takeuchi F, Serizawa M, Yamamoto K, Fujisawa T, Nakashima E, et al. (2009) Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes 58: 1690–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Peng SB, Su Y, Han DL, Zhang ZR (2008) Association analyses of KCNJ11 gene polymorphisms with type 2 diabetes mellitus. Yueyang Zhi Ye Ji Shu Xue Yuan Xue Bao 23: 77–80. [Google Scholar]
- 43. Bronstein M, Pisanté A, Yakir B, Darvasi A (2008) Type 2 diabetes susceptibility loci in the Ashkenazi Jewish population. Hum Genet 124: 101–104. [DOI] [PubMed] [Google Scholar]
- 44. Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, et al. (2008) Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant risk. BMC Med Genet 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cauchi S, Nead KT, Choquet H, Horber F, Potoczna N, et al. (2008) The genetic susceptibility to type 2 diabetes may be modulated by obesity status: implications for association studies. BMC Med Genet 9: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ezzidi I, Mtiraoui N, Cauchi S, Vaillant E, Dechaume A, et al. (2009) Contribution of type 2 diabetes associated loci in the Arabic population from Tunisia: a case-control study. BMC Med Genet 10: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhou D, Zhang D, Liu Y, Zhao T, Chen Z, et al. (2009) The E23K variation in the KCNJ11 gene is associated with type 2 diabetes in Chinese and East Asian population. J Hum Genet 54: 433–435. [DOI] [PubMed] [Google Scholar]
- 48. Chistiakov DA, Potapov VA, Khodirev DC, Shamkhalova MS, Shestakova MV, et al. (2009) Genetic variations in the pancreatic ATP-sensitive potassium channel, beta-cell dysfunction, and susceptibility to type 2 diabetes. Acta Diabetol 46: 43–49. [DOI] [PubMed] [Google Scholar]
- 49. Wang F, Han XY, Ren Q, Zhang XY, Han LC, et al. (2009) Effect of genetic variants in KCNJ11, ABCC8, PPARG and HNF4A loci on the susceptibility of type 2 diabetes in Chinese Han population. Chin Med J (Engl) 122: 2477–2482. [PubMed] [Google Scholar]
- 50. Tabara Y, Osawa H, Kawamoto R, Onuma H, Shimizu I, et al. (2009) Replication study of candidate genes associated with type 2 diabetes based on genome-wide screening. Diabetes 58: 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thorsby PM, Midthjell K, Gjerlaugsen N, Holmen J, Hanssen KF, et al. (2009) Comparison of genetic risk in three candidate genes (TCF7L2, PPARG, KCNJ11) with traditional risk factors for type 2 diabetes in a population-based study–the HUNT study. Scand J Clin Lab Invest 69: 282–287. [DOI] [PubMed] [Google Scholar]
- 52. Hu C, Zhang R, Wang C, Wang J, Ma X, et al. (2009) PPARG, KCNJ11, CDKAL1, CDKN2A-CDKN2B, IDE-KIF11-HHEX, IGF2BP2 and SLC30A8 are associated with type 2 diabetes in a Chinese population. PLoS One 4: e7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamauchi T, Hara K, Maeda S (2010) A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet 42: 864–868. [DOI] [PubMed] [Google Scholar]
- 54. Neuman RJ, Wasson J, Atzmon G, Wainstein J, Yerushalmi Y, et al. (2010) Gene-gene interactions lead to higher risk for development of type 2 diabetes in an Ashkenazi Jewish population. PLoS One 5: e9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chauhan G, Spurgeon CJ, Tabassum R, Bhaskar S, Kulkarni SR, et al. (2010) Impact of common variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2 and CDKAL1 on the risk of type 2 diabetes in 5164 Indians. Diabetes 59: 2068–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gupta V, Khadgawat R, Ng HK, Kumar S, Aggarwal A, et al. (2010) A validation study of type 2 diabetes-related variants of the TCF7L2, HHEX, KCNJ11, and ADIPOQ genes in one endogamous ethnic group of north India. Ann Hum Genet 74: 361–368. [DOI] [PubMed] [Google Scholar]
- 57. Wen J, Rönn T, Olsson A, Yang Z, Lu B, et al. (2010) Investigation of type 2 diabetes risk alleles support CDKN2A/B, CDKAL1, and TCF7L2 as susceptibility genes in a Han Chinese cohort. PLoS One 5: e9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rees SD, Hydrie MZ, Shera AS, Kumar S, O'Hare JP, et al. (2011) Replication of 13 genome-wide association (GWA)-validated risk variants for type 2 diabetes in Pakistani populations. Diabetologia 54: 1368–1374. [DOI] [PubMed] [Google Scholar]
- 59. Chavali S, Mahajan A, Tabassum R, Dwivedi OP, Chauhan G, et al. (2011) Association of variants in genes involved in pancreatic β-cell development and function with type 2 diabetes in North Indians. J Hum Genet 56: 695–700. [DOI] [PubMed] [Google Scholar]
- 60. Cheung CY, Tso AW, Cheung BM, Xu A, Fong CH, et al. (2011) The KCNJ11 E23K polymorphism and progression of glycaemia in Southern Chinese: a long-term prospective study. PLoS One 6: e28598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gamboa-Meléndez MA, Huerta-Chagoya A, Moreno-Macías H, Vázquez-Cárdenas P, Ordóñez-Sánchez ML, et al. (2012) Contribution of common genetic variation to the risk of type 2 diabetes in the Mexican Mestizo population. Diabetes 61: 3314–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gonen MS, Arikoglu H, Erkoc Kaya D, Ozdemir H, Ipekci SH, et al. (2012) Effects of single nucleotide polymorphisms in K(ATP) channel genes on type 2 diabetes in a Turkish population. Arch Med Res 43: 317–23. [DOI] [PubMed] [Google Scholar]
- 63. Nielsen EM, Hansen L, Carstensen B, Echwald SM, Drivsholm T, et al. (2003) The E23K variant of Kir6.2 associates with impaired post-OGTT serum insulin response and increased risk of type 2 diabetes. Diabetes 52: 573–577. [DOI] [PubMed] [Google Scholar]
- 64. Lyssenko V, Almgren P, Anevski D, Orho-Melander M, Sjogren M, et al. (2005) The Botnia Study Group: genetic prediction of future type 2 diabetes. PLoS Med 2: e345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schwanstecher C, Meyer U, Schwanstecher M (2002) KIR6.2 polymorphism predisposes to type 2 diabetes by inducing over-activity of pancreatic b-cell ATP-sensitive K channels. Diabetes 51: 875–879. [DOI] [PubMed] [Google Scholar]
- 66. van Dam RM, Hoebee B, Seidell JC, Schaap MM, de Bruin TWA, et al. (2005) Common variants in the ATP-sensitive K+ channel genes KCNJ11 (Kir6.2) and ABCC8 (SUR1) in relation to glucose intolerance: population-based studies and meta-analyses. Diabet Med 22:590–598. Li YY (2013) The KCNJ11 E23K gene polymorphism and type 2 diabetes mellitus in the Chinese Han population: a meta-analysis of 6,109 subjects. Mol Biol Rep 40: 141–6. [DOI] [PubMed] [Google Scholar]
- 67. Laukkanen O, Pihlajamäki J, Lindström J, Eriksson J, Valle TT, et al. (2004) Polymorphisms of the SUR1 (ABCC8) and Kir6.2 (KCNJ11) Genes predict the conversion from impaired glucose tolerance to type 2 diabetes. The Finnish Diabetes Prevention Study. J Clin Endocrinol Metab 89: 6286–6290. [DOI] [PubMed] [Google Scholar]
- 68. Riedel MJ, Light PE (2005) Saturated and cis/trans unsaturated acyl CoA esters differentially regulate wild-type and polymorphic b-cell ATP-sensitive K+ channels. Diabetes 54: 2070–2079. [DOI] [PubMed] [Google Scholar]
- 69. Ardlie KG, Lunetta KL, Seielstad M (2002) Testing for population subdivision and association in four case–control studies. Am J Hum Genet 71: 304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Song Y, Niu T, Manson JE, Kwiatkowski DJ, Liu S (2004) Are variants in the CAPN10 gene related to risk of type 2 diabetes? A quantitative assessment of population and family-based association studies. Am J Hum Genet 74: 208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu CH, et al. (2010) A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet 6: e1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang L, Zhou X, Luo Y, Sun X, Tang Y, et al. (2012) Association between KCNJ11 gene polymorphisms and risk of type 2 diabetes mellitus in East Asian populations: a meta-analysis in 42,573 individuals. Mol Biol Rep 39: 645–59. [DOI] [PubMed] [Google Scholar]
- 73. Li YY (2013) The KCNJ11 E23K gene polymorphism and type 2 diabetes mellitus in the Chinese Han population: a meta-analysis of 6,109 subjects. Mol Biol Rep 40: 141–6. [DOI] [PubMed] [Google Scholar]
- 74. Qin LJ, Lv Y, Huang QY (2013) Meta-analysis of association of common variants in the KCNJ11-ABCC8 region with type 2 diabetes. Genet Mol Res 12: 2990–3002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Flow chart of literature search for studies examining KCNJ11 rs5219 polymorphism and risk of T2D.
(TIF)
Result of sensitivity analyses.
(TIF)
Begg's funnel plot of rs5219 polymorphism and T2D risk.
(TIF)
Results of meta-analysis for KCNJ11 E23K polymorphism and T2D risk using dominant and recessive genetic models.
(DOCX)