Abstract
The oriental fruit fly, Bactrocera dorsalis, is a destructive pest in tropical and subtropical areas. In this study, we performed transcriptome-wide analysis of the fat body of B. dorsalis and obtained more than 59 million sequencing reads, which were assembled into 27,787 unigenes with an average length of 591 bp. Among them, 17,442 (62.8%) unigenes matched known proteins in the NCBI database. The assembled sequences were further annotated with gene ontology, cluster of orthologous group terms, and Kyoto encyclopedia of genes and genomes. In depth analysis was performed to identify genes putatively involved in immunity, detoxification, and energy metabolism. Many new genes were identified including serpins, peptidoglycan recognition proteins and defensins, which were potentially linked to immune defense. Many detoxification genes were identified, including cytochrome P450s, glutathione S-transferases and ATP-binding cassette (ABC) transporters. Many new transcripts possibly involved in energy metabolism, including fatty acid desaturases, lipases, alpha amylases, and trehalose-6-phosphate synthases, were identified. Moreover, we randomly selected some genes to examine their expression patterns in different tissues by quantitative real-time PCR, which indicated that some genes exhibited fat body-specific expression in B. dorsalis. The identification of a numerous transcripts in the fat body of B. dorsalis laid the foundation for future studies on the functions of these genes.
Introduction
The oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae), is a highly destructive pest, and is distributed widely in tropical and subtropical areas around the world. It can feed and reproduce on more than 250 species of plants, causing enormous losses in many economically important agriculture products, such as fruits and vegetables [1]. Currently, insecticides are the most effective way to control this pest. However, resistance monitoring programs found that various populations of B. dorsalis have become resistant to different kinds of insecticides, resulting in their resurgence and environmental risks [2]. Therefore, more potent and powerful strategies are required to manage this pest. In recent years, RNA interference (RNAi)-based technology has shown great potential in pest control by silencing vital genes [3], [4]. For this method, abundant genomic resources are essential and necessary to identify suitable target genes for RNAi [5]. Unfortunately, molecular genetic information for B. dorsalis is relatively limited, and the entire genome is not yet available, although the analysis is in progress (Bactrobase: http://www.bactrobase.org/). However, transcriptome sequencing of B. dorsalis has been performed successfully, and has proved to be an effective method to gather genetic information from either the whole body or the midgut [6], [7]. These data have provided comprehensive gene expression information regarding detoxification, digestion, development and the RNAi mechanism. However, further tissue-specific transcriptome analyses are required to obtain a comprehensive view of the temporal and spatial changes in gene expression related to unique biological features.
The insect fat body is a dynamic tissue whose function is equivalent to the vertebrate liver. It is a major organ involved in various physiological and biological processes, including detoxification, developmental regulation and immunity [8], [9], [10]. Previously, because of technical limitations, molecular studies on the fat body initially focused on characterization of individual genes, particularly those involved in detoxification [11] or energy storage and release [10]. With the development of next generation sequencing (high-throughput deep sequencing) technology, characterization of the entire set of genes expressed in the insect fat body has become feasible. Such works have been conducted in the fruit fly (Drosophila melanogaster) [12], the tsetse fly (Glossina morsitans morsitans) [8], and the yellow fever mosquito (Aedes aegypti) [13], [14]. Despite the importance of this tissue, little is known about the molecular basis of physiological and immunological mechanisms in the fat body of B. dorsalis. In this study, we performed high-throughput Illumina Solexa sequencing to acquire a comprehensive view of the genes expressed in the fat body of B. dorsalis. We focused particularly on genes related to immune defense, detoxification and energy metabolism because of their importance for insect development. Furthermore, we investigated the gene expression patterns of several key genes in different tissues by quantitative real-time PCR. The results obtained from this study will provide an invaluable resource for future functional studies on fat bodies in B. dorsalis.
Results and Discussion
Illumina sequencing and reads assembly
Illumina sequencing was used to sequence a cDNA library from the fat body of B. dorsalis, and generated 59,158,922 reads with a length of 4,959,025,200 bp (Runs accession number: SRR1026844). To facilitate sequence assembly, the Trinity program assembled these raw reads into 46,444 contigs with an average length of 360 bp. Paired end-joining and gap-filling further assembled the contigs into 27,787 unigenes with an average length of 591 bp (Table 1). The analysis of the length distribution of the unigenes revealed that 4,336 unigenes (15.6%) were more than 1,000 bp, 5,039 unigenes (18.1%) were between 500 and 1,000 bp, and 18,412 unigenes (66.3%) ranged from 100 to 500 bp (Figure S1).
Table 1. Sequencing summary for the fat body transcriptome of Bactrocera dorsalis.
| Sequencing Summary | Fat body specific transcriptome |
| Total number of reads | 59,158,922 |
| Total base pairs (bp) | 4,959,025,200 |
| Average read length (bp) | 84 |
| Total number of contigs | 46,444 |
| Mean length of contigs (bp) | 360 |
| N50d of contigs | 628 |
| Total number of unigenes | 27,787 |
| Mean length of unigenes (bp) | 591 |
| N50d of unigenes | 945 |
| Sequences with E-value <10−5 | 17,442 (62.8%) |
For BLASTX annotation, the unigenes were searched against the non-redundant (nr) NCBI nucleotide database. A total of 17,442 unigenes (62.8%) returned positive BLAST results at a cut-off E-value of 10−5. However, the remaining 10,345 unigenes (37.2%) could not be matched to known genes in the NCBI database, either because of a lack of B. dorsalis genome information or their short nucleotide length.
Sequence homology distribution
The E-value distribution of the 17,442 annotated unigenes showed that 40.2% of the sequences had significant homology (E-value less than 1.0E−45) matching in the NCBI database, while 59.8% of the sequences had E-values that ranged from 1.0E−5 to 1.0E−45 (Figure S2A). The similarity distribution showed that 27.1% of the sequences had significant similarity higher than 80%, followed by 60.4% of the sequences with homology between 40 and 80%; only 12.5% of the sequences had similarity lower than 40% (Figure S2B). For species distribution, the majority of unigenes (72.0%) showed significant similarity with Drosophila. Of these, 13.0% of the unigene sequences had top matches (lowest E-value) with sequences from D. virilis, followed by D. willistoni (12.6%), D. melanogaster (11.6%), D. mojavensis (11.0%), and other Drosophila species (Figure S2C). This was in accord with our previously study of transcriptome profiling of the whole body and midgut of B. dorsalis, in which over 79% genes were most closely related to Drosophila [6], [7]. There were 249 unigene sequences (1.4%) that matched genes from B. dorsalis and the majority of these hits matched to cytochrome P450s and glutathione S-transferases (data not shown). In addition, 279 unigenes had a best hit to other insects from the Bactrocera, such as B. oleae (165 unigenes), B. cucurbitae (36), B. tryoni (32), B. papaya (19), B. correcta (18), and B. minax (9).
Functional classification and pathway analysis
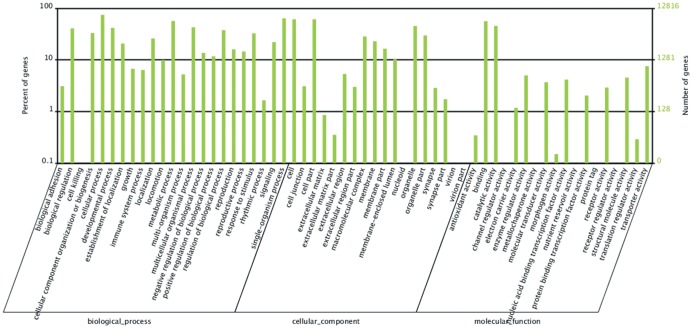
Gene ontology (GO) assignments were used to classify the functions of the predicted unigenes from fat body of B. dorsalis. According to the sequence similarity, 12,816 unigenes (46.0% of total) were categorized into 50 functional groups among the three main ontologies (Figure 1). In each of the three main categories (biological process, cellular component, and molecular function) of the GO classification, “Cellular process and Single-organism process”, “Cell or Cell part”, and “Binding and Catalytic activity” terms were the most abundant. In contrast, the “Cell killing” term under the Biological process category was the least abundant, and contained only three unigenes. The unigenes associated with the “virion” and “virion part” under the Cellular component had no more than three unigenes associated with each term. There were also a low percentage of genes from the categories of “metallochaperone activity”, “protein tag”, and “receptor regulator activity”.
Figure 1. Gene Ontology (GO) classification of unigenes in the fat body of Bactrocera dorsalis.
11,575 sequences were annotated within the GO database and summarized in three main categories: biological process, cellular component and molecular function. The left and right y-axes indicate the percentage and number of genes in each category, respectively.
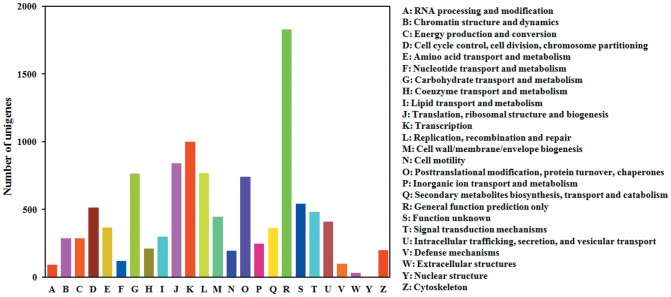
To further analyze the putative protein functions, the assembled unigenes were compared against Clusters of Orthologous Groups (COG). Out of 17,442 nr hits, 11,122 sequences were assigned a COG classification (Figure 2). Among the 25 COG categories, the cluster for “General function prediction” represented the largest group (1,828, 16.4%), followed by “Transcription” (998, 9.0%), “Translation, ribosomal structure and biogenesis” (839, 7.5%) and “Replication, recombination and repair” (768, 6.9%). In contrast, “Extracellular structures” and “Nuclear structure” represented relatively smaller clusters, containing only 30 and four unigenes, respectively.
Figure 2. Clusters of Orthologous Groups (COG) classification of unigenes in the fat body of Bactrocera dorsalis.
Among 17,442 nr hits, 5,185 sequences had a COG classification among the 25 categories.
Subsequently, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed on unigenes to identify the biological pathways that are active in the fat body of B. dorsalis. There were 11,857 unigenes assigned to 254 KEGG pathways (Table S1). The pathways with most representation among the unique sequences were “Metabolic pathways” (1611 members), “Pathways in cancer” (423 members), and “RNA transport” (400 members). These annotations are helpful to further investigate specific biological processes, functions and pathways occurring in the fat body of B. dorsalis.
Putative immunity-related genes
The insect fat body serves as a barrier between the internal and external environment, and protects the host against infection by microbes and foreign objects [15]. Recent studies found that a significant number of immunity-related genes were highly expressed in this tissue, and participated in triggering an immune response or preventing infection by microbial and protozoan pathogens [8], [16]. These genes are considered as the potential targets for pest control, and their specific sequence information is of great importance. In this study, fat body-specific transcriptome analysis revealed a surprising number of transcripts implicated in the immune response. A total of 71 unigenes encoding various immune-related enzymes were identified, which corresponded to 25 serine proteinase inhibitors or serpins, 17 immuno- and C-type lectins, 11 transferrins, six peptidoglycan recognition proteins (PGRPs), three lysozymes, three defensins, three gram-negative bacteria-binding proteins (GNBP), two β-1,3-glucan recognition proteins (β-GRP), and one defense protein Hdd11-like homolog.
The most striking group is the 25 unigenes corresponding to putative serine proteinase inhibitors or serpins. Among these sequences, 10 matched known B. dorsalis serpins, and other 15 unigenes were novel. Serpins are irreversible inhibitors of serine proteases that regulate various physiological and pathological reactions in humans and insects, especially in innate immune responses [17]. To date, multiple serpin genes have been identified in insect genomes [18]. Previous studies have shown that the serpin genes were highly enriched in the fat body, reflecting this organ's involvement in the immune response of insects [19]. Here, we analyzed the carboxyl-terminal inhibitor domain of seven complete sequences of serpins in the fat body of B. dorsalis. Phylogenetic analysis showed that the seven were classified into different clades (Figure 3). In serpin proteins, there is a highly conserved tertiary structure with a reactive center loop region near the C-terminus, which acts as the bait for the target protease [20]. Functional analyses of serpins in Manduca sexta found that there were also seven groups of serpin proteins: most of them are inhibitory and regulate proteases that have a role in cascades leading to activation of prophenoloxidase (proPO) and the cytokine spätzle [19]. In addition, Mamestra configurata serpin-1 homologs, serpins-1b and serpins-1c, are possibly involved in developmental processes related to the molt [21]. The multiple serpin family genes may have diverse functions. Therefore, further investigations are needed to determine the roles of these serpins in B. dorsalis.
Figure 3. Phylogenetic analysis of serpins from Bactrocera dorsalis (Bd), Drosophila melanogaster (Dm), Anopheles gambiae (Ag), Tribolium castaneum (Tc) and Glossina morsitans morsitans (Gmm).
The tree was created using the neighbor-joining method. Bootstrap values represent the percentage of 1000 replicates. Reference sequences were obtained from the NCBI database.
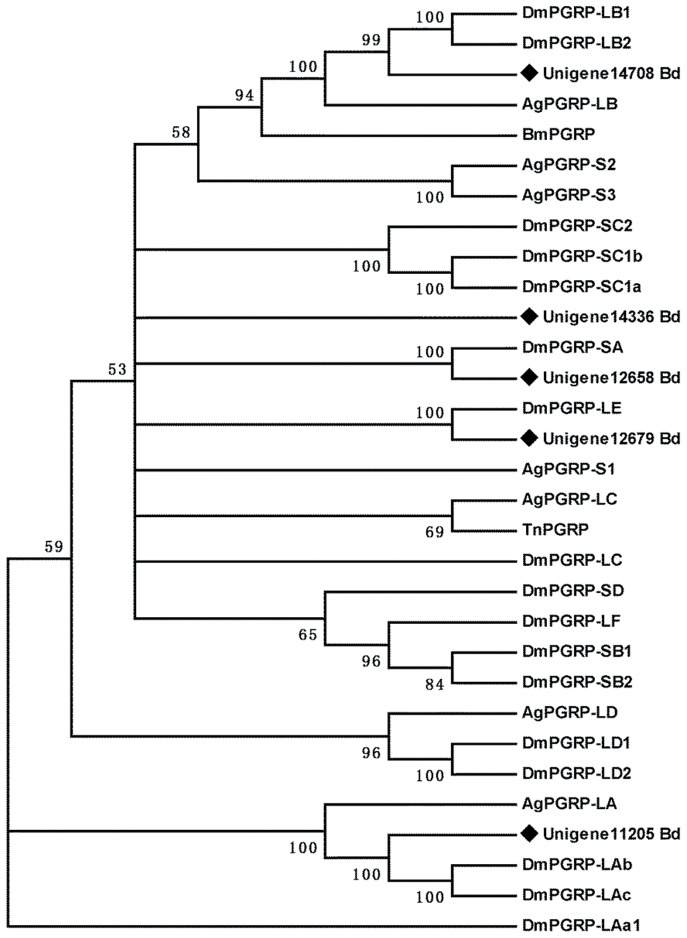
In the insect immune system, PGRPs are important pattern recognition molecules that recognize bacteria and their unique cell wall component, and are conserved from insects to mammals [22]. Based on structural analyses, insect PGRPs can be classified into two different types: short (PGRP-S) and long (PGRP-L) [23]. PGRP-Ss are small extracellular proteins, while PGRP-Ls are long intracellular or membrane-spanning proteins. Insect PGRPs have various functions that are unique to insects, including activation of the proPO cascade, Toll receptor and Imd pathways, as well as induction of phagocytosis [24]. In this study, six complete sequences encoding PGRPs with an average length of 1001 bp were identified from the fat body of B. dorsalis. Phylogenetic analysis together with genes from D. melanogaster and Anopheles gambiae showed that three could be assigned to the PGRP-L group and the other two were assigned to the PGRP-S group (Figure 4). The functional PGRP groups form discrete branches on phylogenetic tree of insect PGRPs: thus, the amino acid sequences of PGRPs correlate with the functions of PGRPs, permitting us to form an initially prediction of the functions of the PGRPs in B. dorsalis. In addition to PGRPs, GNBP and β-GRP are also important pattern recognition molecules. In Nilaparvata lugens, significantly higher expression levels of these two types of genes were observed in fat bodies compared with other tissues, which is consistent with their role in immunity [25]. Here we identified three GNBP genes and two beta-GRP genes in the fat body of B. dorsalis for the first time.
Figure 4. Phylogenetic analysis of peptidoglycan recognition proteins from Bactrocera dorsalis (Bd), Drosophila melanogaster (Dm), Anopheles gambiae (Ag), Bombyx mori (Bm) and Trichoplusia ni (Tn).
The tree was created using the neighbor-joining method. Bootstrap values represent the percentage of 1000 replicates. Reference sequences were obtained from the NCBI database.
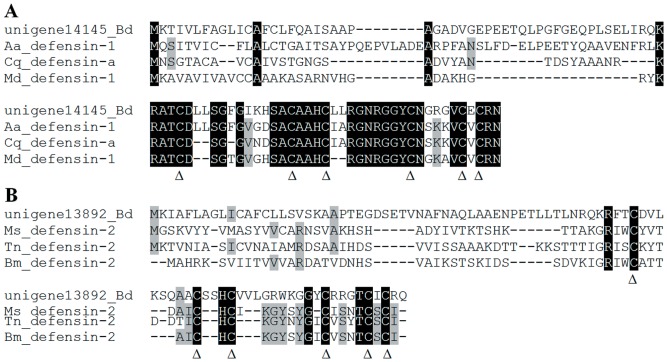
Defensins, a class of small and diverse cysteine-rich proteins, have broad-spectrum antimicrobial activity against bacteria, fungi and enveloped viruses [26]. Here, we newly identified three defensin-like protein genes in the fat body of B. dorsalis. We compared the predicted amino acid sequences of these three new defensin-like proteins, termed defensin-like-1, -2 and -3, with other insect defensins. Two of them represented full length of open reading frames (ORFs), whereas the defensin-like-3 lacked a coding sequence at the 5′ end. Furthermore, multiple protein alignments showed that the two complete defensin-like proteins have six highly conserved cysteine residues (Figure 5). Such defensin-like proteins were also identified in the midgut-specific transcriptomes of M. sexta and Plutella xylostella [27], [28].
Figure 5. Amino acid alignment of two new defensin-like proteins in Bactrocera dorsalis.
The ClustalW multiple alignment program aligned the amino acid sequences. Identical residues are shown in white with black background. The defensin family motif and six highly conserved residues are indicated with a triangle. Aedes aegypti (Aa), Bactrocera dorsalis (Bd), Bombyx mori (Bm), Culex quinquefasciatus (Cq), Musca domestica (Md), Manduca sexta (Ms), Trichoplusia ni (Tn).
To determine whether these genes are related to the immune response, we randomly selected six genes to characterize their expression patterns in different tissues. Interestingly, all the selected genes demonstrated significantly higher expression levels in the fat body compared with other tissues (Figure 6). The six genes showed fat body-specific expression, which implied that the fat body is the primary site of immunity-related genes expression. These results will provide comprehensive information to enable the determination of the molecular mechanism underlying the immune response in B. dorsalis.
Figure 6. Tissue specificity of immunity-related genes expression in Bactrocera dorsalis.
Expression levels in head (HD), thorax (TH), midgut (MG), Malpighian tubules (MT) and fat body (FB) were detected by qPCR. The relative expression was calculated based on the value of the lowest expression, which was ascribed an arbitrary value of 1. Different letters above the bars indicate significant differences based on Duncan's new multiple range tests (DMRT, P<0.05).
Detoxification-related genes
In the last decade, B. dorsalis has become a serious problem in fruit production, and has developed resistance to various insecticides [2]. The insect fat body, classically viewed as a tissue primarily involved in xenobiotic metabolism, harbors several detoxification enzymes [9], [11]. In this study, we obtained abundant transcripts in the fat body of B. dorsalis, especially the unigenes encoding cytochrome P450, glutathione S-transferases and ATP-binding cassette transporters, which were mainly involved in detoxification of xenobiotic [29], [30].
Cytochrome P450 (P450s)
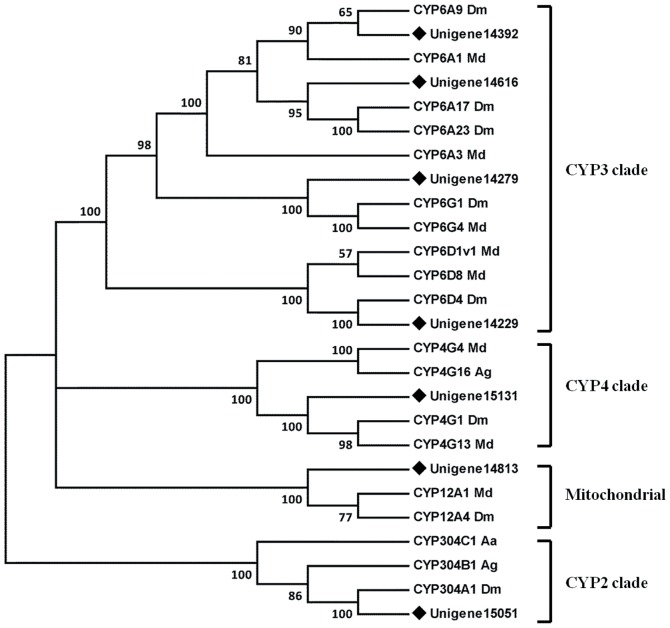
P450s compose a superfamily that is involved mainly in the metabolism of a variety of endogenous and exogenous compounds [31]. In this study, 37 distinct sequences with an average length of 1089 bp encoding P450s were identified. Based on the closest BLAST hit in the NCBI nr database, these unigenes were divided into four clades and 14 families, including four families of CYP18, CYP 304, CYP305 and CYP307 in the CYP2 clade; three families of CYP6, CYP9, CYP309 and CYP317 in the CYP3 clade, two families of CYP4 and CYP311 in the CYP4 clade; and four families of CYP12, CYP301, CYP302 and CYP314 in the mitochondrial CYP clade (Table 2). Interestingly, 17 of these unigenes predicted novel P450 genes in the fat body of B. dorsalis. Among these, seven unigenes appeared to be full length, and were further subjected to phylogenetic analysis with other known insect P450s. The results showed that four unigenes belonged to the CYP3 clade, and one each to the CYP4, CYP2 and mitochondrial CYP clades (Figure 7).
Table 2. Different CYPs P450 clades, families and GSTs classes in the fat body transcriptome of Bactrocera dorsalis.
| Enzymes | Occurrence | Family members with corresponding number |
| CYPs P450 | ||
| CYP2 clade (4 family) | ||
| CYP18 | 2 | CYP18A1 (2) |
| CYP304 | 3 | CYP304A1 (3) |
| CYP305 | 1 | CYP305A1 (1) |
| CYP307 | 3 | CYP307A1 (3) |
| CYP3 clade (3 family) | ||
| CYP6 | 30 | CYP6A2 (1), CYP6A9 (6), CYP6A13 (4), CYP6A14 (4), CYP6A21 (1), CYP6A22 (1), CYP6D4 (1), CYP6D5 (1), CYP6G1 (5), CYP6U1 (3), CYP6V1 (3) |
| CYP9 | 6 | CYP9B2 (3), CYP9F2 (1), CYP9H1 (2) |
| CYP309 | 4 | CYP309A1 (4) |
| CYP317 | 3 | CYP317A1 (3) |
| CYP4 clade (2 family) | ||
| CYP4 | 20 | CYP4AC1 (3), CYP4AE1 (1), CYP4D1 (2), CYP4D2 (1), CYP4E1 (2), CYP4E2 (1), CYP4G1 (5), CYP4P1 (3), CYP4S3 (2) |
| CYP311 | 2 | CYP311A1 (2) |
| Mitochondrial CYP clade (4 family) | ||
| CYP12 | 15 | CYP12A4 (7), CYP12B1 (2), CYP12B2 (4), CYP12C1 (2) |
| CYP301 | 1 | CYP301A1 (1) |
| CYP302 | 4 | CYP302A1 (4) |
| CYP314 | 1 | CYP314A1 (1) |
| GSTs classes | ||
| Delta | 9 | Delta (3), Delta1 (3), Delta2 (1), Delta5 (1), Delta6 (1) |
| Epsilon | 5 | Epsilon2 (1), Epsilon4 (1), Epsilon5 (2), Epsilon9 (1) |
| Omega | 3 | Omega1 (3) |
| Sigma | 1 | Sigma1 (1) |
| Theta | 3 | Theta1 (1), Theta3 (1), Theta5 (1) |
| Microsomal | 3 | Microsomal (2), Microsomal-like (1) |
| Zeta | 3 | Zeta1 (1), Zeta2 (2) |
Figure 7. Phylogenetic analysis of cytochrome P450s from Bactrocera dorsalis (Bd), Drosophila melanogaster (Dm), Musca domestica (Md), Anopheles gambiae (Ag) and Aedes aegypti (Aa).
The tree was created using the neighbor-joining method. Bootstrap values represent the percentage of 1000 replicates. Reference sequences were obtained from the NCBI database.
In the present study, more than half of the P450s belonged to the CYP3 and CYP4 clades, which is in accordance with data from other insects [25], [28]. CYP3 and CYP4 in other insects are also the most abundant P450 clades, and their function is related to xenobiotic metabolism [31]. For example, overexpression of CYP 6G1 in D. melanogaster resulted in the increased metabolic detoxification of insecticides [32]. In addition, our previous study of P450s in B. dorsalis identified nine genes that are mainly expressed in the fat body, and were upregulated in response to insecticide exposure [33]. This suggested that P450s detoxification probably takes place mainly in the fat body of B. dorsalis. Therefore, we believe that the new P450 genes identified here may contribute to insecticide resistance. Further investigations are required to determine the roles of these novel P450 genes in B. dorsalis.
Glutathione S-transferases (GSTs)
GSTs are a group of multifunctional enzymes that play important roles in the detoxification of hydrophobic toxic compounds as well as mediating the oxidative stress response [34]. After removing redundant sequences, 18 different sequences with an average length of 762 bp encoding specific GSTs were identified in the fat body of B. dorsalis (Table 2). These GSTs were assigned to the delta, epsilon, sigma, theta, zeta and microsomal classes. Among them, 10 correspond to known B. dorsalis GSTs, and the other eight sequences were newly reported. Specifically, six of the new GSTs appeared to be complete, and these sequences were further subjected to phylogenetic analysis with other known insect GSTs. The results showed that two unigenes belonged to the delta class, and one unigene was assigned to each of the epsilon, theta, sigma and microsomal GST classes (Figure 8).
Figure 8. Phylogenetic analysis of the glutathione S-transferases from Bactrocera dorsalis (Bd), Drosophila melanogaster (Dm), Musca domestica (Md), Aedes aegypti (Aa), Locusta migratoria (Lm) and Glossina morsitans morsitans (Gmm).
The tree was created using the neighbor-joining method. Bootstrap values represent the percentage of 1000 replicates. Reference sequences were obtained from the NCBI database.
Half of GSTs identified in the fat body of B. dorsalis belong to the delta and epsilon classes, members of which are insect-specific and are implicated mainly in insecticide resistance and detoxification [35]. Earlier studies reported that these two classes of GSTs were responsible for organophosphate, DDT, and pyrethroid resistance [34], [36]. Interestingly, we identified complete sequences of two delta and one epsilon; these genes appear to be important in the detoxification of various insecticides. Notably, compared with our previous transcriptomes of B. dorsalis, we found a new sigma gene in the fat body. Evidence suggests that insect sigma GSTs are not only involved in detoxification of exogenous compounds and oxidative stress resistance, but also probably play a role in muscle structure [37], [38]. Hence, the sigma GST in B. dorsalis is likely to have multiple functions. It is generally presumed that GST detoxification occurs mainly in the insect fat body [34]. We identified several GSTs in the fat body of B. dorsalis; however, the detailed functions of these genes remain to be elucidated.
ATP-binding cassette (ABC) transporters
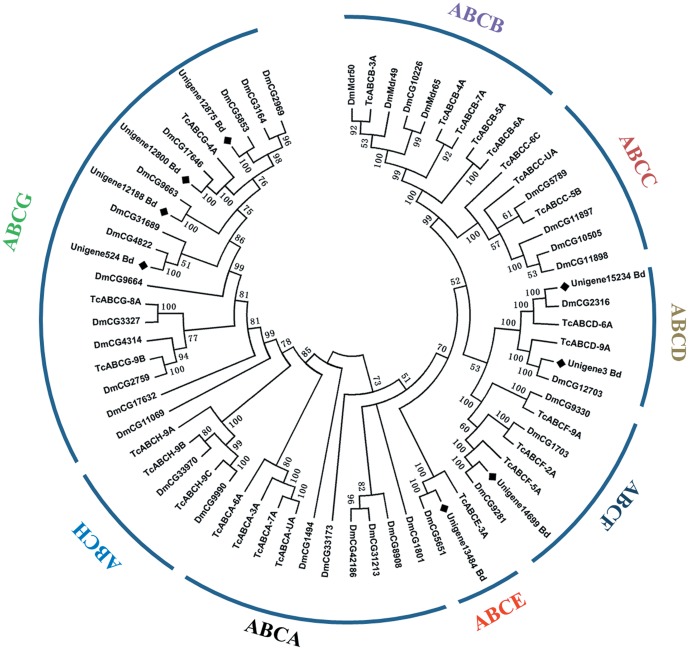
In insects, ABC transporters are not only responsible for transporting molecules, but also are involved in the biochemical defense against toxicants [39]. From the fat body transcriptome, 29 different sequences with an average length of 1467 bp encoding ABC transporters were identified; and no allelic variants were found. These sequences were classified into seven subfamilies (four belong to A subfamily, four belong to B subfamily, one belongs to C subfamily, two belong to D subfamily, one belongs to E subfamily, three belong to F subfamily, and four belong to G subfamily) (Table S2). Among these, eight sequences had a complete ORF and were subjected to phylogenetic analysis with D. melanogaster ABC transporters. We found that two belong to the D subfamily, one each to the E and F subfamilies, and four to the G subfamily (Figure 9).
Figure 9. Phylogenetic analysis of ABC transporters from Bactrocera dorsalis (Bd), Drosophila melanogaster (Dm) and Tribolium castaneum (Tc).
The tree was created using the neighbor-joining method. Bootstrap values represent the percentage of 1000 replicates. Reference sequences were obtained from the NCBI database.
Analyses of fully sequenced insect genomes have identified 68 ABC transporters in Tribolium castaneum, 56 in D. melanogaster, 52 in A. gambiae, 51 in Bombyx mori, and 43 in Apis mellifera (Table S2). Additionally, 18 ABC transporters were identified in the B. oleae transcriptome [40]. The numbers of ABC transporters were compared among seven different insect species; the 68 genes in T. castaneum form the largest ABC transporter data set, but the number of ABC transporters identified in this study is greater than that in B. oleae. The number of ABC transporters in the fat body of B. dorsalis is within the range of ABC transporters in other insect species (18-69) (Table S2), additional ABC transporters may await discovery because of their absence from the current transcriptomic database. In D. melanogaster, the ABC transporter was regulated by heavy metals via the metal-responsive transcription factor 1 and are associated with biochemical detoxification of zinc and copper [41]. Recently, three Lepidopteran ABC transporter subfamilies, including B, C, and G, were reported to confer resistance to xenobiotics, such as insecticides [30]. Therefore, over half of the ABC transporters identified in this study are believed to be involved in detoxification, which represent promising avenues for further investigation.
We randomly selected nine detoxification-related genes to determine their expression specificity. Four genes showed higher expression levels in the fat body than in other tissues, including three ABC transporters, D member 1 (unigene 15234), E member 1 (unigene 13484), G member 1 (unigene 12800), and CYP12A4 (unigene 14813), and displayed tissue-specific expression patterns (Figure 10). In contrast, the remaining three GSTs and two P450s showed low expression levels in the fat body. The exclusive expression of several genes in the fat body indicates that these detoxification-related genes probably play vital roles in the metabolism of xenobiotics, and thus permit substantial molecular studies of the mechanism of detoxification in B. dorsalis.
Figure 10. Tissue specificity of detoxification-related genes expression in Bactrocera dorsalis.
The qPCR analysis was performed as described in Figure 6.
Genes putatively involved in energy metabolism
The insect fat body, equivalent to vertebrate adipose tissue and liver, is a major organ involved in energy metabolism [10], [42]. During insect growth and development, insects store the energy reserves in the fat body in response to energy demands. It is generally accepted that energy is crucial for the activity and movement of organisms, and disruption of energy homeostasis affects natural development and vital movement, leading to eventual death of organisms. Therefore, identification of genes related to energy metabolism is significant and would help develop new methods to control pest insects via interference with the energy system. In this study, many unigenes were identified that share high similarity with genes related to energy metabolism in other insects, including fatty acid desaturase, lipase, alpha-amylase, trehalose-6-phosphate synthase, ATP synthase, glycerol-3-phosphate dehydrogenase, aldose 1-epimerase, beta-glucosidase, ribosomal protein S7 and aminopeptidase (Table 3).
Table 3. Details of sequences involved in energy metabolism.
| Unigene ID | Length | Homology genes in Nr/Gene ID | Homology species | E-value | |
| Fatty acid desaturase | Unigene14007 | 2136 | fatty acid desaturase/AAB17283 | Drosophila melanogaster | 0 |
| Unigene14769 | 446 | fatty acid desaturase 2/BAB21537 | Drosophila melanogaster | 3.00E−72 | |
| Lipase | Unigene4378 | 1296 | monoacylglycerol lipase ABHD12-like isoform 1/XP_001604091 | Nasonia vitripennis | 1.00E−60 |
| Unigene13291 | 784 | lipase 4, isoform A/NP_609418 | Drosophila melanogaster | 2.00E−82 | |
| Unigene14677 | 1540 | brummer lipase/ATGL-like protein, partial/ | Glossina morsitans morsitans | 0 | |
| Unigene13974 | 1382 | lipase 3-like/XP_003695770 | Apis florea | 2.00E−62 | |
| Alpha-amylase | Unigene12619 | 1059 | alpha-amylase 3/EFN65423 | Camponotus floridanus | 2.00E−50 |
| Unigene14089 | 1814 | alpha-amylase/XP_001649784 | Aedes aegypti | 1E-174 | |
| Trehalose-6-phosphate synthase | Unigene15155 | 2402 | trehalose-6-phosphate synthase/NP_608827 | Drosophila melanogaster | 0 |
| ATP synthase | Unigene9638 | 671 | V-ATPase G subunit/AEV43310 | Bactrocera dorsalis | 9.00E−38 |
| Unigene13304 | 1909 | vacuolar ATP synthase subunit S1/ABF18127 | Aedes aegypti | 4.00E−33 | |
| Unigene429 | 2252 | vacuolar ATP synthase subunit E/NP_001040451 | Bombyx mori | 1.00E−92 | |
| Unigene14983 | 2480 | ATP synthase alpha subunit vacuolar/XP_001849275 | Culex quinquefasciatus | 0 | |
| Unigene12971 | 2456 | ATP synthase/EHJ65007 | Danaus plexippus | 0 | |
| Unigene9659 | 765 | ATP synthase delta chain, mitochondrial/EAT46274 | Aedes aegypti | 3.00E−48 | |
| Unigene14297 | 600 | ATP synthase F0 subunit 6/YP_006883645 | Bactrocera tryoni | 2.00E−55 | |
| Unigene455 | 784 | H+ transporting ATP synthase O subunit isoform 1/NP_001040526 | Bombyx mori | 1.00E−62 | |
| Glyceraldehyde-3-phosphate dehydrogenase | Unigene11633 | 981 | glyceraldehyde-3-phosphate dehydrogenase/ADO24527 | Bactrocera dorsalis | 6.00E−157 |
| Aldose 1-epimerase | Unigene3995 | 327 | aldose 1-epimerase/XP_001843677 | Culex quinquefasciatus | 2.00E−40 |
| Beta-glucosidase | Unigene14223 | 1153 | beta-glucosidase/XP_001659855 | Coptotermes formosanus | 2.00E−84 |
| Ribosomal protein S7 | Unigene12342 | 555 | ribosomal protein S7, isoform A/NP_651782 | Drosophila melanogaster | 1.00E−87 |
| Unigene12887 | 772 | mitochondrial ribosomal protein S7, isoform A/NP_523537 | Drosophila melanogaster | 6.00E−71 | |
| Aminopeptidase N | Unigene6316 | 630 | Aminopeptidase N/EFN68544 | Camponotus floridanus | 2.00E−41 |
| Aminopeptidase P | Unigene15293 | 1954 | aminopeptidase P, isoform A/NP_477409 | Drosophila melanogaster | 0 |
The fatty acid desaturase gene, which encodes a predicted protein mainly expressed in the fat body, plays a pivotal role in mitochondrial energy metabolism as well as controlling body size [43]. In D. melanogaster, mutation in this gene can induce a sustained change in energy homeostasis [44]. Here we identified one B. dorsalis fatty acid desaturase (unigene14007, 2,136 bp), which is similar to the same functional gene in D. melanogaster (AAB17283).
Lipids represent the major component of the fat body and are the main source of metabolic fuel [45]. Lipid metabolism is required to meet the energy demand during non-feeding periods. Brummer lipase, or insect adipose triglyceride lipase (ATGL), belongs to the calcium-independent phospholipase A2 (iPAL2) family, and plays an important role in energy metabolism [46]. In D. melanogaster, loss of ATGL caused accumulation of triglyceride, leading to fat flies, whereas its overexpression produced lean flies [47]. In the fat body of B. dorsalis, we found unigene 14677 (1,540 bp) that was a full-length sequence encoding an ATGL, which showed similarity with a specific gene of G. morsitans morsitans, whose function was not annotated.
Alpha-amylase is a common enzyme that catalyzes hydrolysis of α-(1,4) glycosidic linkages in starch and related compounds to serve as an energy source [48]. This enzyme can encounter varying proportions of starch constituents in the diet to provide the energy required for all cellular functions [49]. In the present study we identified two sequences encoding alpha-amylase in the fat body of B. dorsalis, for the first time.
In insects, the blood sugar is trehalose instead of glucose; therefore, the synthesis and utilization of trehalose is unique to insect energy metabolism compared with other animals [50]. Trehalose-6-phosphate synthase (TPS) is the crucial enzyme for the biosynthesis of trehalose, and catalyzes the formation of trehalose-6-phosphate from uridine diphosphate glucose and glucose-6-phosphate [51]. Apart from the synthesizing trehalose in insects, TPS was also reported to be an important enzyme for insect development. For example, mutations of the TPS gene in D. malanogaster lead to early larval lethality and affected normal insect development [52]. Furthermore, the development of N. lugens and Spodotera exigua larvae treated with dsRNA of TPS was disturbed, resulting in lethality, suggesting that a potential way to control the pests is to disrupt TPS expression using RNAi [53], [54]. In the fat body of B. dorsalis, we found a 2,402 bp sequence (unigene 15155) encoding TPS, which showed similarity the same functional gene of D. melanogaster (NP_608827). Taken together, a better understanding of the proteins related to energy metabolism will help to develop new methods to control this insect pest.
In this study, we randomly selected six genes similar to fatty acid desaturase, lipase, and ATP synthase to analyze their expression differences among different tissues, and to detect whether these genes are involved in energy metabolism in B. dorsalis. The qPCR analysis showed significantly higher expression levels of two lipase genes, lipase 3 (unigene 13974) and lipase 4 (unigene 13291), and fatty acid desaturase (unigene 14007) in the fat body; much lower levels were observed in the head (Figure 11). Additionally, two ATP synthase genes were both mainly expressed in Malpighian tubules, while brummer lipase (unigene 14677) was mainly expressed in the midgut. Our results may reflect the functional diversification of these genes, thus providing a valuable resource for further studies on energy metabolism in B. dorsalis.
Figure 11. Tissue specificity of energy metabolism-related genes expression in Bactrocera dorsalis.
The qPCR analysis was performed as described in Figure 6.
Conclusions
In this study, we used Illumina sequencing technology to investigate the fat body transcriptome of B. dorsalis. We assembled the raw reads into 27,787 unigenes with an average length of 591 bp. Many genes that are potentially relevant to immunity, detoxification and energy metabolism were identified within the annotated unigenes. Specific information for these unigenes identified in this study is shown in Table S4. Several genes showed fat body-specific expression, implying that these genes might be potential targets for B. dorsalis management. Meanwhile, the fat body-specific transcriptome analysis generated a large number of genes newly identified in B. dorsalis, which will facilitate further functional study. These results provided basic information to enhance our understanding of physiological and immunological mechanism that occur in the B. dorsalis fat body, and laid the foundation for future functional genomics studies.
Materials and Methods
Ethics Statement
No specific permits were required for the insects collected in this study. No specific permissions were required for these locations/activities which the insect specimens were collected. We confirm that these locations are not privately-owned or protected in any way and the species collections did not involve endangered or protected species.
Insect samples
The stock colony of B. dorsalis was originally collected from Fujian province, China. The insects were reared under laboratory conditions at 27±1°C, 70±5% relative humidity, and a 14:10 h light: dark photoperiod, using an artificial diet, as described previously [55].
Collection of fat bodies
To obtain complete gene expression information, fat bodies of B. dorsalis were collected from different developmental stages, including the third-instar larvae, pupae and adults. The insects were anesthetized on ice for 30 min and dissected under a stereomicroscope (Olympus SZX12, Tokyo, Japan). The samples were isolated on ice, placed in a 2.0 mL DEPC-treated centrifuge tube containing RNA store reagent (TIANGEN, Beijing, China), placed immediately in liquid nitrogen and frozen at −80°C.
RNA isolation for transcriptome analysis
An RNeasy plus Micro Kit (QIAGEN, Hilden, Germany) extracted the total RNA from each sample, according to manufacturer's instructions, and gDNA Eliminator spin columns removed the genomic DNA. Measuring the absorbance at 260 nm using a NanoVue UV-Vis spectrophotometer (GE Healthcare Bio-Science, Sweden) quantified the RNA. The absorbance ratio of OD260/280 and OD260/230 was used to assess the purity of the RNA and 1% agarose gel electrophoresis confirmed the integrity of the RNA. A mixture of RNA from all these developmental stages at an equal ratio was used to construct the whole-set transcriptome.
Illumina sequencing analysis
The cDNA library of B. dorsalis fat body was constructed using an mRNA-Seq assay for paired-end transcriptome sequencing. The Beijing Genomics Institute (BGI, Shenzhen, China) performed the sequencing. Briefly, poly (A) mRNA was purified from 20 μg of total RNA using Oligo (dT) magnetic beads and then fragmented into short sequences in the presence of fragmentation buffer at 94°C for 5 min. These short fragments were used as templates for first-strand cDNA synthesis using random hexamer-primers. Subsequently, second-strand cDNA synthesis was performed using buffer, dNTPs, RNaseH, and DNA polymerase Ι. A QiaQuick PCR extraction kit (Qiagen) purified the short fragments, which were then washed with EB buffer for end reparation and single nucleotide adenine addition. After the end-repair and ligation of adaptors, PCR amplified the cDNA products to create a fat body specific sequencing library. The cDNA library was sequenced on an Illumina HiSeq 2000 instrument for 4 gigabase in-depth in a single run.
De novo assembly
Transcriptome de novo assembly was carried out using the short reads assembling program Trinity [56]. Firstly, reads with a certain length of overlap were combined to form longer fragments without Ns, forming contigs. Then, using paired-end reads, contigs from the same transcript, as well as the distances between these contigs were detected. Finally, Trinity connected these contigs to produce consensus sequences that could not be extended at either end, and were defined as unigenes. After clustering, the obtained unigenes were divided into two classes: clusters and singletons. The distinct sequences were compared to protein databases including nr, Swiss-Prot, KEGG and COG using the BLASTx algorithm with a significant cut-off E-value of <10−5. If the alignment results of different databases conflicted with each other, we followed the priority order of Nr, Swiss-Prot, KEGG, and COG when determining the unigene sequence direction. Blast2GO software with default parameters performed annotation with gene ontology terms (GO, http://www.geneontology.org). Blastall software against Cluster of Orthologous Groups database and Kyoto Encyclopedia of Genes and Genomes database performed the COG and KEGG pathway annotations, respectively.
Identification and analysis of interesting genes
Interesting sequences were identified from the BLAST results generated against the nr database with a cut-off E-value of <10−5. ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) determined the complete coding region and protein BLAST results were generated. MEGA 5.04 [57] was used to analyze the phylogenetic relationships between Serpins, PGRPs, P450, GSTs, and ABC transporter genes and those from other insects to infer their classification. The neighbor-joining method (with pairwise deletion) was used to create the phylogenetic trees. A bootstrap test was conducted (1000 replications) to calculate the percentage of replicate trees in which sequences clustered together. ClustalW software generated the multiple alignments of the various protein sequences [58].
Quantitative real-time PCR (qPCR) analysis
Different tissues, including the head, thorax, midgut, Malpighian tubules and fat body, were dissected from day-7 adults to investigate tissue-specific gene expression patterns. The TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from each tissue sample, which was then treated with DNase (TaKaRa, Dalian, China) for DNA digestion. RNAs (1 μg) were reverse transcribed into first-strand cDNAs using PrimeScript RT reagent Kit (TaKaRa, Dalian, China). The qPCR reactions were performed in a 20 μL total reaction volume including 10 μL GoTaq qPCR Master Mix (Promega, Madison, WI, USA), 1 μL of template cDNA, 7 μL ddH2O, and 1 μL of each primer (0.2 mM). The reaction was carried out on an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) under the following thermal program: 1 cycle of 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. At the end of each qPCR experiment, a melting curve was generated to rule out the possibility of primer-dimer formation. The sequences of the specific primer sets are listed in Table S3. The α-Tubulin (GU269902) gene from B. dorsalis was used as an internal control. The relative expression levels were calculated using 2−ΔΔCt method [59]. Results were expressed as the mean ± SE of three biological replications. Data were analyzed statistically using one-way analysis of variance (ANOVA), and the means were separated by Duncan's multiple range tests for significance (P<0.05) using SPSS 16.0 for windows (SPSS Inc., Chicago, IL, USA).
Supporting Information
Size distribution of unigenes in the fat body of Bactrocera dorsalis . The sizes of 27,787 unigenes that had a BLAST annotation in the NCBI database were calculated.
(TIF)
Homology analysis of unigenes in the fat body of Bactrocera dorsalis . All distinct gene sequences that had BLAST annotations against the nr database with a cut-off E-value of 1.0E−5 were analyzed. The sequences were summarized based on the (A) E-value distribution, (B) similarity distribution, and (C) species distribution, respectively.
(TIF)
KEGG annotation of unigenes.
(XLSX)
Comparatively analysis of ABC transporters in seven insect species.
(XLSX)
Primers used in real-time qPCR for tissue specific expression.
(XLSX)
Sequence information of all unigenes identified in this study.
(XLSX)
Acknowledgments
We thank Beijing Genomics Institute at Shenzhen (BGI Shenzhen) for help in sequencing and Yi Tian for keeping the laboratory colony of B. dorsalis.
Funding Statement
This research was funded in part by the Natural Science Foundation of Chongqing (CSTC, 2013jjB0176), the Program for Changjiang Scholars and Innovative Research Teams in Universities (IRT0976), the earmarked fund for Modern Agro-industry (Citrus) Technology Research System, and the Fundamental Research Funds for the Central Universities (XDJK2013A017) of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stephens AEA, Kriticos DJ, Leriche A (2007) The current and future potential geographical distribution of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Bull Entomol Res 97: 369–378. [DOI] [PubMed] [Google Scholar]
- 2. Jin T, Zeng L, Lin YY, Lu YY, Liang GW (2011) Insecticide resistance of the oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae), in mainland China. Pest Manag Sci 67: 370–376. [DOI] [PubMed] [Google Scholar]
- 3. Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, et al. (2007) Control of coleopteran insect pests through RNA interference. Nat Biotech 25: 1322–1326. [DOI] [PubMed] [Google Scholar]
- 4. Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, et al. (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotech 25: 1307–1313. [DOI] [PubMed] [Google Scholar]
- 5. Li H, Jiang WH, Zhang Z, Xing YR, Li F (2013) Transcriptome analysis and screening for potential target genes for RNAi-mediated pest control of the beet armyworm, Spodoptera exigua . PLoS One 8: e65931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shen GM, Dou W, Niu JZ, Jiang HB, Yang WJ, et al. (2011) Transcriptome analysis of the oriental fruit fly (Bactrocera dorsalis). PLoS One 6: e29127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shen GM, Dou W, Huang Y, Jiang XZ, Smagghe G, et al. (2013) In silico cloning and annotation of genes involved in the digestion, detoxification and RNA interference mechanism in the midgut of Bactrocera dorsalis [Hendel (Diptera: Tephritidae)]. Insect Mol Biol 22: 354–365. [DOI] [PubMed] [Google Scholar]
- 8. Attardo GM, Strickler-Dinglasan P, Perkin SAH, Caler E, Bonaldo MF, et al. (2006) Analysis of fat body transcriptome from the adult tsetse fly, Glossina morsitans morsitans . Insect Mol Biol 15: 411–424. [DOI] [PubMed] [Google Scholar]
- 9. Yang JL, McCart C, Woods DJ, Terhzaz S, Greenwood KG, et al. (2007) A Drosophila system approach to xenobiotic metabolism. Physiol Genomics 30: 223–231. [DOI] [PubMed] [Google Scholar]
- 10. Arrese EL, Soulages JL (2010) Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol 55: 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Snyder MJ, Stevens JL, Andersen JF, Feyereisen R (1995) Expression of cytochrome P450 genes of the CYP4 family in midgut and fat body of the tobacco hornworm, Manduca sexta . Arch Biochem Biophys 321: 13–20. [DOI] [PubMed] [Google Scholar]
- 12. Jiang Z, Wu XL, Michal JJ, McNamara JP (2005) Pattern profiling and mapping of the fat body transcriptome in Drosophila melanogaster . Obes Res 13: 1898–1904. [DOI] [PubMed] [Google Scholar]
- 13. Feitosa FM, Calvo E, Merino EF, Durham AM, James AA, et al. (2006) A transcriptome analysis of the Aedes aegypti vitellogenic fat body. J Insect Sci 6: 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Price DP, Nagarajan V, Churbanov A, Houde P, Milligan B, et al. (2011) The fat body transcriptomes of the yellow fever mosquito Aedes aegypti, pre- and post- blood meal. PLoS One 6: e22573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dunn PE (1986) Biochemical aspects of insect immunology. Annu Rev Entomol 31: 321–339. [Google Scholar]
- 16. Guz N, Kilincer N, Aksoy1 S (2012) Molecular characterization of Ephestia kuehniella (Lepidoptera: Pyralidae) transferrin and its response to parasitoid Venturia canescens (Hymenoptera: Ichneumonidae Gravenhorst). Insect Mol Biol 21: 139–147. [DOI] [PubMed] [Google Scholar]
- 17. Janciauskiene S (2001) Conformational properties of serine proteinase inhibitors (serpins) confer multiple pathophysiological roles. Biochim Biophys Acta 1535: 221–235. [DOI] [PubMed] [Google Scholar]
- 18. Zou Z, Picheng Z, Weng H, Mita K, Jiang H (2009) A comparative analysis of serpin genes in the silkworm genome. Genomics 93: 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suwanchaichinda C, Ochieng R, Zhuang SF, Kanost MR (2013) Manduca sexta serpin-7: a putative regulator of hemolymph prophenoloxidase activation. Insect Biochem Mol Biol 43: 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gettins PGW (2002) Serpin structure, mechanism, and function. Chem Rev 102: 4751–4803. [DOI] [PubMed] [Google Scholar]
- 21. Chamankhah M, Braun L, Visal-Shah S, O'Grady M, Baldwin D, et al. (2003) Mamestra configurata serpin-1 homologues: cloning, localization and developmental regulation. Insect Biochem Mol Biol 33: 355–369. [DOI] [PubMed] [Google Scholar]
- 22. Kang D, Liu G, Lundstrom A, Gelius E, Steiner H (1998) A peptidoglycan recognition protein in innate immunity conserved from insects to mammals. Proc Natl Acad Sci USA 95: 10078–10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Werner T, Liu G, Kang D, Ekengren S, Steiner H, Hultmark D (2000) A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster . Proc Natl Acad Sci USA 97: 13772–13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dziarski R (2004) Peptidoglycan recognition proteins (PGRPs). Mol Immun 40: 877–886. [DOI] [PubMed] [Google Scholar]
- 25. Bao YY, Wang Y, Wu WJ, Zhao D, Xue J, et al. (2012) De novo intestine-specific transcriptome of the brown planthopper Nilaparvata lugens revealed potential functions in digestion, detoxification and immune response. Genomics 99: 256–264. [DOI] [PubMed] [Google Scholar]
- 26. Pasupuleti M, Schmidtchen A, Malmsten M (2012) Antimicrobial peptides: key components of the innate immune system. Crit Rev Biotechnol 32: 143–171. [DOI] [PubMed] [Google Scholar]
- 27. Pauchet Y, Wilkinson P, Vogel H, Nelson DR, Reynolds SE, et al. (2010) Pyrosequencing the Manduca sexta larval midgut transcriptome: messages for digestion, detoxification and defence. Insect Mol Biol 19: 61–75. [DOI] [PubMed] [Google Scholar]
- 28. Xie W, Lei YY, Fu W, Yang ZX, Zhu X, et al. (2012) Tissue-specific transcriptome profiling of Plutella xylostella third instar larval midgut. Int J Biol Sci 8: 1142–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, et al. (2002) Evolution of supergene families associated with insecticide resistance. Science 298: 179–181. [DOI] [PubMed] [Google Scholar]
- 30. Liu SM, Zhou S, Tian L, Guo EE, Luan YX, et al. (2011) Genome-side identification and characterization of ATP-binding cassette transporter in the silkworm, Bombyx mori . BMC Genomics 12: 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feyereisen R (2011) Insect CYP Genes and P450 Enzymes. Insect Mol Biol 8: 236–316. [Google Scholar]
- 32. JouBen N, Heckel DG, Haas M, Schuphan I, Schmidt B (2008) Metabolism of imidacloprid and DDT by P450 CYP6G1 expressed in cell cultures of Nicotiana tabacum suggests detoxification of these insecticides in Cyp6g1-overexpressing strains of Drosophila melanogaster leading to resistance. Pest Manag Sci 64: 65–73. [DOI] [PubMed] [Google Scholar]
- 33. Huang Y, Shen GM, Jiang HB, Jiang XZ, Dou W, et al. (2013) Multiple P450 genes: Identification, tissue-specific expression and their responses to insecticide treatments in the oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidea). Pestic Biochem Physiol 106: 1–7. [Google Scholar]
- 34. Enayati AA, Ranson H, Hemingway J (2005) Insect glutathione transferases and insecticide resistance. Insect Mol Biol 14: 3–8. [DOI] [PubMed] [Google Scholar]
- 35. Hemingway J, Ranson H (2000) Insecticide resistance in insect vectors of human disease. Annu Rev Entomol 45: 371–391. [DOI] [PubMed] [Google Scholar]
- 36. Lumjuan N, McCarroll L, Prapanthadara LA, Hemingway J, Ranson H (2005) Elevated activity of an Epsilon class glutathione transferase confers DDT resistance in the dengue vector, Aedes aegypti . Insect Biochem Mol Biol 35: 861–871. [DOI] [PubMed] [Google Scholar]
- 37. Singh SP, Coronella JA, Benes H, Cochrane BJ, Zimniak P (2001) Catalytic function of Drosophila melanogaster glutathione S-transferase DmGSTS1-1 (GST-2) in conjugation of lipid peroxidation end products. Eur J Biochem 268: 2912–2923. [DOI] [PubMed] [Google Scholar]
- 38. Huang YF, Xu ZB, Lin XY, Feng QL, Zheng SC (2011) Structure and expression of glutathione S-transferase genes from the midgut of the Common cutworm, Spodoptera litura (Noctuidae) and their response to xenobiotic compounds and bacteria. J Insect Physiol 57: 1033–1044. [DOI] [PubMed] [Google Scholar]
- 39. Dassa E, Bouige P (2001) The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol 152: 211–229. [DOI] [PubMed] [Google Scholar]
- 40. Pavlidi N, Dermauw W, Rombauts S, Chrisargiris A, Van Leeuwen T, et al. (2013) Analysis of the Olive Fruit Fly Bactrocera oleae Transcriptome and Phylogenetic Classification of the Major Detoxification Gene Families. PLoS One 8: e66533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yepiskoposyan H, Egli D, Dergestad T, Selvaraj A, Treiber C, et al. (2006) Transcriptome response to heavy metal stress in Drosophila reveals a new zinc transporter that confers resistance to zinc. Nucleic Acids Res 34: 4866–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Y, Liu H, Liu S, Wang S, Jiang R, et al. (2009) Hormonal and nutritional regulation of insect fat body development and function. Arch Insect Biochem Physiol 71: 16–30. [DOI] [PubMed] [Google Scholar]
- 43. Proudlock JW, Haslam JM, Linnane AW (1971) Biogenesis of mitochondria 19 the effects of unsaturated fatty acid depletion on the lipid composition and energy metabolism of a fatty acid desaturase mutant of Saccharomyces cerevisiae . J Bioenerg 2: 327–349. [DOI] [PubMed] [Google Scholar]
- 44. Wicker-Thomas C, Henriet C, Dallerac R (1997) Partial characterization of a fatty acid desaturase gene in Drosophila melanogaster . Insect Biochem Mol Biol 27: 963–972. [DOI] [PubMed] [Google Scholar]
- 45. Gilby AR (1965) Lipids and their metabolism in insects. Annu Rev Entomol 10: 141–160. [Google Scholar]
- 46. Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, et al. (2004) Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem 279: 48968–48975. [DOI] [PubMed] [Google Scholar]
- 47. Grönke S, Mildner A, Fellert S, Tennagels N, Petry S, et al. (2005) Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila . Cell Metab 1: 323–330. [DOI] [PubMed] [Google Scholar]
- 48. Ikuo M, Shigetaka Y, Kazuhiko I, Sachio M, Sakuzo F, et al. (1994) Roles of the aromatic residues conserved in the active center of Saccharomycopsis alpha-amylase for transglycosylation and hydrolysis activity. Biochemistry 33: 451–458. [DOI] [PubMed] [Google Scholar]
- 49. Yu SM, Kuo YH, Sheu G, Sheu YJ, Liu LF (1991) Metabolic derepression of alpha-amylase gene expression in suspension-cultured cells of rice. J Biol Chem 266: 21131–21137. [PubMed] [Google Scholar]
- 50. Friedman S (1978) Trehalose regulation, one aspect of metabolic homeostasis. Annu Rev Entomol 23: 389–407. [Google Scholar]
- 51. Cabib E, Leloir L (1958) The biosynthesis of trehalose-phosphate. J Biol Chem 231: 259–275. [PubMed] [Google Scholar]
- 52. Chen Q, Ma E, Behar KL, Xu T, Haddad GG (2002) Role of trehalose phosphate synthase in anoxia tolerance and development in Drosophila melanogaster . J Biol Chem 277: 3274–3279. [DOI] [PubMed] [Google Scholar]
- 53. Tang B, Chen J, Yao Q, Pan ZQ, Xu WH, et al. (2010) Characterization of a trehalose-6-phosphate synthase gene from Spodoptera exigua and its function identification through RNA interference. J insect Physiol 56: 813–821. [DOI] [PubMed] [Google Scholar]
- 54. Chen J, Zhang D, Yao Q, Zhang J, Dong X, et al. (2010) Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens . Insect Mol Biol 19: 777–786. [DOI] [PubMed] [Google Scholar]
- 55. Cong L, Yang WJ, Shen GM, Dou W, Wang JJ (2012) Molecular characterization of the cDNA encoding ecdysone receptor isoform B1 and its expression in the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Fla Entomol 95: 650–658. [Google Scholar]
- 56. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li KB (2003) ClustalW-WPI: ClustalW analysis using distributed and parallel computing. Bioinformatics 19: 1585. [DOI] [PubMed] [Google Scholar]
- 59. Livak KJ. Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Size distribution of unigenes in the fat body of Bactrocera dorsalis . The sizes of 27,787 unigenes that had a BLAST annotation in the NCBI database were calculated.
(TIF)
Homology analysis of unigenes in the fat body of Bactrocera dorsalis . All distinct gene sequences that had BLAST annotations against the nr database with a cut-off E-value of 1.0E−5 were analyzed. The sequences were summarized based on the (A) E-value distribution, (B) similarity distribution, and (C) species distribution, respectively.
(TIF)
KEGG annotation of unigenes.
(XLSX)
Comparatively analysis of ABC transporters in seven insect species.
(XLSX)
Primers used in real-time qPCR for tissue specific expression.
(XLSX)
Sequence information of all unigenes identified in this study.
(XLSX)