Abstract
Background
The association between cytotoxic T-lymphocyte antigen 4 (CTLA-4) gene -1722T/C polymorphism (rs733618) and cancer has been widely assessed, and a definitive conclusion remains elusive. We first performed a hospital based case-control study to measure this association of esophageal cancer with CTLA-4 -1722T/C polymorphism in Han Chinese population, and then carried out a meta-analysis to obtain a comprehensive evaluation for this issue.
Methodology/Principal Findings
This case-control study involved 629 esophageal squamous cell carcinoma (ESCC) cases and 686 age and gender well matched cancer-free controls. PCR-LDR (polymerase chain reaction-ligase detection reactions) method was used to identify genotypes. Meta-analysis was conducted by STATA (v12.0) software. This case-control study showed no significant difference in the genotype and allele distributions of CTLA-4 -1722T/C polymorphism between esophageal cancer cases and control subjects, in accord with the findings of the further meta-analysis in all genetic models. Evidence of large heterogeneity was observed among all eligible studies in the recessive model. Further subgroup analyses by ethnicity, cancer type and system, detected null associations in this meta-analysis.
Conclusion
This case-control study and the further meta-analysis, failed to identify the association between CTLA-4 -1722T/C polymorphism and cancer risk.
Introduction
It is estimated that about 12.7 million multiple cancer cases and 7.6 million cancer deaths have occurred in 2008 worldwide, with more than half of the cases and about two-thirds of the deaths in the developing countries [1]. The evidence is mounting that cancer is a complex disease results from interactions between multiple genetic backgrounds and environmental factors [2], [3]. Of late, a number of studies demonstrate that genetic variants of the genes that regulate the activation and proliferation of T lymphocytes and nature killer (NK) cells may influence cancer risk [4], [5]. In the last decade, single nucleotide polymorphisms (SNPs) have been extensively investigated, and many studies have examined the hypothesis that genetic variants of the immune genes may be relevant to the risk of a variety of cancers [6], [7].
Cytotoxic T-lymphocyte antigen 4 (CTLA-4), also named CD152, is a member of the immunoglobulin superfamily. CTLA-4 is expressed mainly on activated T cells, acts as a vital restraining regulator of T-cell proliferation and activation, and induces Fas-independent apoptosis of activated T cells to further inhibit immune function of T-cell [6], [8]. Blocking CTLA-4 function and enhancing T cell activation, several different types of malignant neoplasms in tumor-transplanted mice were inhibited or cured, and owned long-lasting antitumor immunity [9]. It suggests that CTLA-4 plays an important role in carcinogenesis. CTLA-4 gene is located on chromosome 2q33, and is composed of four exons that encode several functional domains of the CTLA-4 protein and possess several vital SNPs, such as the +49A/G (rs231775), -318C/T (rs5742909), CT60G/A (rs3087243), -1661A/G (rs4553808), and -1722T/C (rs733618) SNPs, etc [6], [10].
A meta-analysis showed that CTLA-4 +49A/G polymorphism may be a risk factor for cancer, whereas -318C/T and +6230G/A (CT60) polymorphisms were lack of association with cancer [4]. Of late, Geng and colleagues reported a meta-analysis with a negative result on the association between CTLA-4 -1722T/C polymorphism and cancer risk [11]. Linkage disequilibrium (LD) plot of CTLA-4 (involving rs733618, rs4553808, rs5742909, rs231775 and rs3087243) was generated using Haploview 4.2 program and the results suggest that −1661A/G (rs4553808) and −318C/T (rs5742909) are in high LD; the others are in low LD [11]. The CTLA-4 -1722T/C polymorphism has not been investigated in esophageal cancer. To further investigate this potential relationship, we decided to evaluate the association of CTLA-4 -1722T/C polymorphism with esophageal cancer risk in a hospital based case-control study, and then performed a comprehensive meta-analysis to derive a more precise result.
Materials and Methods
Subjects
This hospital-based case–control study included 629 sporadic esophageal squamous cell carcinoma (ESCC) cases and 686 cancer-free subjects consecutively recruiting from the Affiliated People's Hospital of Jiangsu University and Affiliated Hospital of Jiangsu University (Zhenjiang City, Jiangsu Province, China), between October 2008 and December 2010. All recruited subjects were local residents of Han Chinese population, and all ESCC subjects were diagnosed by surgical resection and pathologic examination. The ESCC subjects who had a history of personal malignant tumor or autoimmune disorder, or had undergone radiotherapy or chemotherapy were excluded. Ethnicity, gender and average age (±5 years) of the controls were well matched to esophageal cancer cases. The control individuals were selected from the two hospitals for cure of fracture. At recruitment, this hospital based case-control study was approved by the Ethics Committee of Jiangsu University (Zhenjiang, China). Information of all subjects was collected from a structured questionnaire which was administered by two experienced research doctors. The information of demographic data (e.g. age, gender) and related risk factors (such as, tobacco use and alcohol consumption) is listed in Table 1 . Each subject signed the written informed consent and donated 2-ml sample of peripheral blood.
Table 1. Distribution of selected demographic variables and risk factors in ESCC cases and controls.
| Variable | Cases (n = 629) | Controls (n = 686) | P a | |||
| n | % | n | % | |||
| Age (years) mean ± SD | 62.85 (±8.13) | 62.58 (±7.89) | 0.541 | |||
| Age (years) | 0.155 | |||||
| <63 | 310 | 49.28 | 365 | 53.21 | ||
| ≥63 | 319 | 50.72 | 321 | 46.79 | ||
| Sex | 0.185 | |||||
| Male | 444 | 70.59 | 461 | 67.20 | ||
| Female | 185 | 29.41 | 225 | 32.80 | ||
| Tobacco use | <0.001 | |||||
| Never | 355 | 56.44 | 499 | 72.74 | ||
| Ever | 274 | 43.56 | 187 | 27.26 | ||
| Alcohol use | <0.001 | |||||
| Never | 428 | 68.04 | 526 | 76.68 | ||
| Ever | 201 | 31.96 | 160 | 23.32 | ||
Two-sided χ 2 test and student t test; Bold values are statistically significant (P<0.05).
DNA extraction, SNP selection, and genotyping
Blood samples were collected with ethylenediamine tetra-acetic acid (EDTA) anticoagulant vacutainer tubes (BD Franklin Lakes NJ, USA). Genomic DNA was extracted from lymphocytes using the QIAamp DNA Blood Mini Kit (Qiagen, Berlin, Germany) and DNA samples were frozen at −80°C. Genotyping of CTLA-4 -1722T/C polymorphism was carried out using the polymerase chain reaction-ligase detection reactions (PCR-LDR) method [12]. The Shanghai Biowing Applied Biotechnology Company provides technical support for genotyping. One hundred and sixty samples were randomly selected and reciprocally tested with directly sequencing for quality control, and the reproducibility were 100%. The primers of directly sequencing used for CTLA-4 -1722T/C genotyping were as follows: F: 5' GCAATAACAACCTAATGGGCAC 3'; R: 5' ACTTCCACAGGCTGAACCACT 3' (Figure S1).
Statistical analysis
Chi-square test (χ 2) was conducted to measure the differences in the distributions of genotypes, demographic characteristics and selected variables between esophageal cancer cases and controls. Genotype frequencies of CTLA-4 -1722T/C polymorphism among the controls were tested for Hardy–Weinberg equilibrium (HWE) using an internet-based calculator (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl). The associations between CTLA-4 -1722T/C locus and the risk of ESCC were analyzed by unconditional logistic regression for crude ORs and adjusted ORs when it was appropriate. Statistical analyses were implemented in SAS 9.1.3 software (SAS Institute, Cary, NC). A P<0.05 (two-tailed) was defined as the criterion of statistical significance.
Meta analysis
The meta-analysis is reported on the basis of the Preferred Reporting Items for Meta-analyses (PRISMA) guideline (Checklist S1) [13].
Embase, PubMed, and CBM (Chinese BioMedical Disc), as well as CNKI (China National Knowledge Infrastructure) database were searched up to August 1st, 2013 for publications investigating the association of CTLA-4 -1722T/C polymorphism with cancer risk. The combination terms were ‘cancer’ or ‘tumor’ or ‘carcinoma’ or ‘neoplasm’ and ‘cytotoxic T-lymphocyte antigen 4′ or ‘CTLA-4′ or ‘CD152’, annexed with ‘mutation’ or ‘variant’ or ‘SNP’ or ‘polymorphism’. In addition, the publication language was restricted to English and Chinese, and all studies performed in human subjects were identified. The search results were supplemented by checking all references listed in these studies and published reviews. Included studies were qualified if they met the major included criteria: (1) designed as a retrospective or nested case-control study, (2) evaluated the CTLA-4 -1722T/C polymorphism and cancer risk, (3) provide genotype counts of CTLA-4 -1722T/C polymorphism between cancer cases and controls, and (4) control genotype distributions consistent with HWE. The major excluded criteria were: (1) not case-control studies, (2) review publications and (3) overlapping data. Information was carefully and independently extracted by three reviewers (W. Tang, H. Qiu, and H. Jiang). In case of conflicting evaluations, differences were resolved by further discussion among all authors. The following data was extracted: first author, year of publication, cancer type, country, ethnicity, number of cases and controls, genotype method, allele and genotype frequency, and HWE in controls.
In this meta-analysis, the crude odds ratio (OR) with the corresponding 95% confidence intervals (95% CI) was used to assess the strength of association between the CTLA-4 -1722T/C polymorphism and cancer risk. The Z-test and P-value (two-tailed) was used to measure the significance of the pooled OR, and statistical significance was defined as P<0.05 (two-tailed). Heterogeneity among studies was evaluated by a Chi-square-based I2 test, I2<25% indicated low heterogeneity, 25%≤I2≤50% indicated moderate heterogeneity, and I2>50% indicated large heterogeneity [14]. If I2>50% or P<0.10, the pooled ORs were calculated by the random-effects model (the DerSimonian–Laird method), otherwise the fixed-effects model was implemented (the Mantel–Haenszel method). Subgroup analyses were implemented to measure ethnicity-specific, cancer type-specific and system-specific effects according to ethnicity, cancer type (if any cancer type evaluated by less than three individual investigations, it was combined into "other cancers") and system. The funnel plot and Egger's test were carried out to measure publication bias, which was evaluated by visual inspection of an asymmetric plot. For heterogeneity, funnel plot and Egger's test, statistical significance was considered at P<0.1. In this meta-analysis, all statistical analyses were conducted by STATA software (version 12.0).
Results
Baseline characteristics
The demographics and risk factors of all subjects are presented in Table 1 . The results indicated that cases and controls were fully matched by age and gender. However, there was significant difference on drinking status and smoking between patients and controls (P<0.001). The primary information of CTLA-4 -1722T/C polymorphism was showed in Table 2 . For this SNP, the genotyping success rate was 96.43% in all samples. Minor allele frequency (MAF) of controls in our study, was similar to the database of Chinese for this SNP ( Table 2 ). The genotypic frequencies for CTLA-4 -1722T/C polymorphism among controls were used to evaluated deviation from the HWE, and the result was in HWE (P = 0.284) ( Table 2 ).
Table 2. Primary information for CTLA4 -1722T/C (rs733618) polymorphism.
| Genotyped SNPs | CTLA4 -1722T/C (rs733618) |
| Chromosome | 2 |
| Function | nearGene-5 |
| Chr Pos (Genome Build 36.3) | 204439189 |
| Regulome DB Scorea | No Data |
| TFBSb | Y |
| Splicing (ESE or ESS) | — |
| miRNA (miRanda) | — |
| nsSNP | — |
| MAFc for Chinese in database | 0.390 |
| MAF in our controls (n = 686) | 0.414 |
| P value for HWEd test in our controls | 0.701 |
| Genotyping methode | LDR |
| % Genotyping value | 96.43% |
http://www.regulomedb.org/;
TFBS: Transcription Factor Binding Site (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm);
MAF: minor allele frequency;
HWE: Hardy–Weinberg equilibrium;
LDR: Ligation Detection Reaction.
Single-locus analysis
In the single locus analyses, the genotype frequencies of CTLA-4 -1722T/C were 16.53% (CC), 49.10% (TC) and 34.37% (TT) in the patients, and 17.50% (CC), 47.79% (TC) and 34.70% (TT) in the controls, and the difference was no statistically significant (P = 0.862) ( Table 3 ). In this case-control study, logistic regression analyses showed that the CTLA-4 -1722T/C SNP was not associated with the risk of ESCC. Tobacco use and alcohol consumption are two strong environmental factors, we examined the association in a stratified analysis by these two factors and the results were null association ( Table 4 ).
Table 3. Logistic regression analyses of associations between CTLA4 -1722T/C (rs733618) polymorphisms and risk of ESCC.
| Genotype | Cases (n = 629) | Controls (n = 686) | Crude OR (95%CI) | P | Adjusted OR a (95%CI) | P | ||
| n | % | n | % | |||||
| CTLA4 rs733618T/C | ||||||||
| TT | 210 | 34.37 | 228 | 34.70 | 1.00 | 1.00 | ||
| TC | 300 | 49.10 | 314 | 47.79 | 1.04 (0.81–1.33) | 0.770 | 1.06 (0.83–1.37) | 0.625 |
| CC | 101 | 16.53 | 115 | 17.50 | 0.95 (0.69–1.32) | 0.776 | 0.97 (0.69–1.35) | 0.846 |
| CC vs. TC vs. TT | 0.862 | |||||||
| TC+CC | 401 | 65.63 | 429 | 65.30 | 1.02 (0.81–1.28) | 0.901 | 1.04 (0.82–1.32) | 0.755 |
| TT+TC | 510 | 83.47 | 542 | 82.50 | 1.00 | 1.00 | ||
| CC | 101 | 16.53 | 115 | 17.50 | 0.93 (0.70–1.25) | 0.645 | 0.93 (0.69–1.26) | 0.649 |
| T allele | 720 | 58.92 | 770 | 58.60 | 0.99 (0.84–1.16) | 0.870 | ||
| C allele | 502 | 41.08 | 544 | 41.40 | ||||
Adjusted for age, sex, smoking and drinking status; Bold values are statistically significant (P<0.05).
Table 4. Stratified analyses between CTLA4 -1722T/C (rs733618) polymorphism and ESCC risk by sex, age, smoking status and alcohol consumption.
| Variable | CTLA4 rs733618 T/C (case/control)a | Adjusted ORb (95% CI); P | |||||||
| TT | TC | CC | TC+CC | TT | TC | CC | TC+CC | CC vs. (TC+TT) | |
| Sex | |||||||||
| Male | 150/154 | 209/214 | 70/76 | 279/290 | 1.00 | 1.04 (0.77–1.40); P: 0.815 | 0.96 (0.64–1.43); P: 0.828 | 1.02 (0.76–1.35); P: 0.916 | 0.94 (0.65–1.35); P: 0.723 |
| Female | 60/74 | 91/100 | 31/39 | 122/139 | 1.00 | 1.10 (0.70–1.72); P: 0.676 | 1.02 (0.57–1.83); P: 0.955 | 1.08 (0.71–1.64); P: 0.731 | 0.96 (0.57–1.63); P: 0.888 |
| Age | |||||||||
| <63 | 102/125 | 139/162 | 60/60 | 199/222 | 1.00 | 1.05 (0.74–1.51); P: 0.773 | 1.24 (0.79–1.96); P: 0.353 | 1.11 (0.79–1.55); P: 0.559 | 1.21 (0.80–1.82); P: 0.371 |
| ≥63 | 108/103 | 161/152 | 41/55 | 202/207 | 1.00 | 1.05 (0.73–1.49); P: 0.807 | 0.73 (0.45–1.20); P: 0.214 | 0.96 (0.69–1.35); P: 0.820 | 0.71 (0.46–1.11); P: 0.136 |
| Smoking status | |||||||||
| Never | 108/171 | 185/218 | 54/85 | 239/303 | 1.00 | 1.31 (0.96–1.80); P: 0.092 | 0.99 (0.65–1.52); P: 0.963 | 1.22 (0.91–1.65); P: 0.190 | 0.84 (0.58–1.24); P: 0.380 |
| Ever | 102/57 | 115/96 | 47/30 | 162/126 | 1.00 | 0.71 (0.46–1.10); P: 0.123 | 0.91 (0.51–1.62); P: 0.749 | 0.76 (0.50–1.14); P: 0.187 | 1.11 (0.66–1.86); P: 0.693 |
| Alcohol consumption | |||||||||
| Never | 145/178 | 208/231 | 63/91 | 271/322 | 1.00 | 1.17 (0.87–1.58); P: 0.300 | 0.89 (0.59–1.33); P: 0.563 | 1.09 (0.82–1.45); P: 0.548 | 0.81 (0.56–1.17); P: 0.257 |
| Ever | 65/50 | 92/83 | 38/24 | 130/107 | 1.00 | 0.81 (0.50–1.32); P: 0.399 | 1.20 (0.63–2.29); P: 0.577 | 0.90 (0.57–1.42); P: 0.648 | 1.36 (0.76–2.43); P: 0.296 |
The genotyping was successful in 611 (97.1%) ESCC cases, and 657 (95.8%) controls for CTLA4 -1722T/C (rs733618);
Adjusted for age, sex, smoking status and alcohol consumption (besides stratified factors accordingly) in a logistic regression model.
Eligible articles for meta-analysis
The initial search yielded a total of 345 potentially relevant publications. After applying additional filters, 12 case-control studies in 11 publications and our study were eligible for inclusion. The detailed process of selecting and excluding articles is presented in Figure 1 .
Figure 1. Flow diagram of articles selection process for CTLA-4 -1722T/C (rs733618) polymorphism and cancer risk meta-analysis.
Study characteristics
There were two groups in an article conducted by Hadinia et al. [15], we treated them separately. In total 12 separate studies plus our case-control study involving a total of 3420 cancer cases and 3675 controls were included in this meta-analysis. Among the 13 case-control studies, three investigated breast cancer [16]–[18], three investigated gastric cancer [15], [19], [20], and the other studies investigated cervical cancer, lung cancer, esophageal cancer, colorectal cancer, and oral cancer [6], [15], [21]–[24]. As for subjects in these studies, 8 were Asians [6], [17]–[21], [24] and 5 were Caucasians[15], [16] [22], [23]. Characteristics of each included study are presented in Table 5 . The detailed distribution of the CTLA-4 -1722T/C polymorphism and allele among cases and controls is presented in Table 6 .
Table 5. Characteristics of populations and cancer types of the individual studies included in the meta-analysis.
| study | year | country | ethnicity | cancer type | No. of cases/controls | Genotype Method |
| Bharti et al. | 2013 | India | Asians | oral cancer | 130/180 | PCR-RFLP |
| Li et al. | 2012 | China | Asians | breast cancer | 581/566 | PCR-RFLP |
| Qi et al. | 2012 | China | Asians | gastric cancer | 118/96 | PCR-RFLP |
| Jiang et al. | 2011 | China | Asians | cervical cancer | 100/100 | MALDI-TOF-MS |
| Khaghanzadeh et al. | 2010 | Iran | Caucasians | lung cancer | 127/124 | PCR-RFLP, PCR-ARMS |
| Rahimifar et al. | 2010 | Iran | Caucasians | cervical cancer | 55/110 | PCR-RFLP, PCR-ARMS |
| Li et al. | 2008 | China | Asians | breast cancer | 328/327 | PCR-RFLP |
| Sun et al. | 2008 | China | Asians | lung cancer | 765/800 | PCR-RFLP, MALDI-TOF MS |
| Hadinia et al. | 2007 | Iran | Caucasians | gastric cancer | 46/190 | RFLP, PCR-ARMS |
| Hadinia et al. | 2007 | Iran | Caucasians | colorectal cancer | 109/190 | RFLP, PCR-ARMS |
| Song et al. | 2006 | China | Asians | gastric cancer | 183/116 | PCR-RFLP |
| Erfani et al. | 2006 | Iran | Caucasians | breast cancer | 283/245 | PCR-CTPP |
| Our study | 2013 | China | Asians | esophageal cancer | 629/686 | PCR-LDR |
MALDI–TOF–MS: Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry.
PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism.
PCR-LDR: polymerase chain reaction-ligase detection reaction.
PCR-ARMS: AmplificationRefractory Mutation System-Polymerase Chain Reaction.
Table 6. Distribution of CTLA-4 -1722T/C (rs733618 T/C) polymorphisms genotype and allele among multiple cancer patients and controls.
| case | control | case | control | HWE,P value | ||||||||
| study | year | TT | TC | CC | TT | TC | CC | c | T | C | T | |
| Qi et al. | 2012 | 40 | 69 | 9 | 37 | 45 | 14 | 87 | 149 | 73 | 119 | 0.957723 |
| Li et al. | 2012 | 184 | 276 | 114 | 207 | 256 | 88 | 504 | 644 | 432 | 670 | 0.552314 |
| Jiang et al. | 2011 | 37 | 49 | 14 | 43 | 39 | 18 | 77 | 123 | 75 | 125 | 0.092957 |
| Rahimifar et al. | 2010 | 46 | 8 | 1 | 90 | 20 | 0 | 10 | 100 | 20 | 200 | 0.294266 |
| Khaghanzadeh et al. | 2010 | 106 | 19 | 1 | 98 | 16 | 1 | 21 | 231 | 18 | 212 | 0.702320 |
| Sun et al. | 2008 | 719 | 43 | 3 | 762 | 37 | 1 | 49 | 1481 | 39 | 1561 | 0.435355 |
| Li et al. | 2008 | 125 | 163 | 40 | 111 | 168 | 48 | 243 | 413 | 264 | 390 | 0.224758 |
| Hadinia et al.(colorectal) | 2007 | 97 | 12 | 0 | 165 | 24 | 0 | 12 | 206 | 24 | 354 | 0.351131 |
| Hadinia et al.(gastric) | 2007 | 42 | 4 | 0 | 165 | 24 | 0 | 4 | 88 | 24 | 354 | 0.351131 |
| Erfani et al. | 2006 | 225 | 54 | 3 | 204 | 41 | 0 | 60 | 504 | 41 | 449 | 0.152921 |
| Bharti et al. | 2013 | 92 | 25 | 6 | 131 | 46 | 3 | 37 | 209 | 52 | 308 | 0.648604 |
| Song et al. | 2006 | 62 | 113 | 8 | 45 | 54 | 17 | 129 | 237 | 88 | 144 | 0.902590 |
| Our study | 2013 | 210 | 300 | 101 | 228 | 314 | 115 | 502 | 720 | 544 | 770 | 0.700586 |
HWE: Hardy–Weinberg equilibrium.
Meta-analysis results
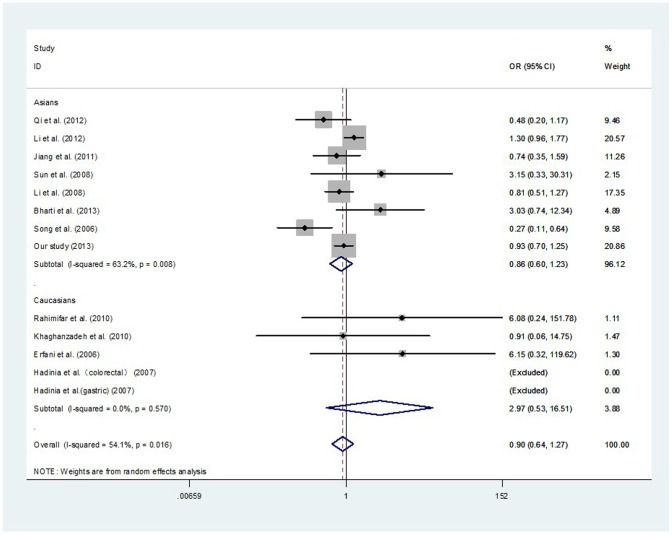
After combining all qualified studies, a total of 3420 cancer cases and 3675 controls from 13 eligible case–control studies were included for meta-analysis of the association between the CTLA-4 -1722T/C polymorphism and cancer risk. There was null association of CTLA-4 -1722T/C polymorphism with overall cancer risk in all genetic models ( Table 7 , Table 8 , Table 9 , Figure 2 , and Figure 3 ). In a stratified analysis by ethnicity, the similar results were observed in both Asians and Caucasians ( Table 7 ). In a stratified analysis by cancer type, there was a decreased risk of gastric cancer in two genetic models: CC vs. TC+TT (OR, 0.36; 95% CI, 0.19–0.66; P = 0.001) and CC vs. TT (OR, 0.45; 95% CI, 0.23–0.86; P = 0.016) ( Table 8 ). In a stratified analysis by system, null association was also observed ( Table 9 ).
Table 7. Summary of results of the meta-analysis from different comparative genetic models in the subgroup analysis by ethnicity.
| Polymorphism | Genetic comparison | Population | OR(95%CI); P | Test of heterogeneity | |
| (p -Value, I2) | Model | ||||
| CC+TC vs. TT | All | 1.09(0.97–1.22);0.159 | 0.762,0.0% | F | |
| Asians | 1.09(0.97–1.24);0.160 | 0.494,0.0% | F | ||
| Caucasians | 1.04(0.78–1.41);0.773 | 0.767,0.0% | F | ||
| CC vs. TC+TT | All | 0.90(0.64–1.27);0.553 | 0.016,54.1% | R | |
| Asians | 0.86(0.60–1.23);0.400 | 0.008,63.2% | R | ||
| Caucasians | 3.27(0.65–16.32);0.149 | 0.570,0.0% | F | ||
| CTLA-4 -1722T/C | CC vs. TT | All | 0.98(0.70–1.37);0.906 | 0.050,45.3% | R |
| Asians | 0.94(0.66–1.33);0.719 | 0.028,55.4% | R | ||
| Caucasians | 3.29(0.66–16.46);0.146 | 0.575,0.0% | F | ||
| TC vs. TT | All | 1.09(0.97–1.23);0.154 | 0.641,0.0% | F | |
| Asians | 1.11(0.97–1.26);0.124 | 0.358,9.3% | F | ||
| Caucasians | 1.01(0.74–1.36);0.970 | 0.792,0.0% | F | ||
| C vs. T | All | 1.04(0.95–1.13);0.383 | 0.577,0.0% | F | |
| Asians | 1.03(0.95–1.13);0.460 | 0.301,16.4% | F | ||
| Caucasians | 1.08(0.82–1.43);0.575 | 0.744,0.0% | F | ||
F indicates fixed model; R indicates random model.
Table 8. Summary of results of the meta-analysis from different comparative genetic models in the subgroup analysis by cancer type.
| Polymorphism | Genetic comparison | Cancer type | OR(95%CI); P | Test of heterogeneity | |
| (p -Value, I2) | Model | ||||
| CC+TC vs. TT | All | 1.09(0.97–1.22);0.159 | 0.762,0.0% | F | |
| Gastric cancer | 1.15(0.81–1.62);0.430 | 0.571,0.0% | F | ||
| Breast cancer | 1.10(0.83–1.47);0.514 | 0.100,56.5% | R | ||
| Other cancers | 1.05(0.89–1.24);0.589 | 0.903,0.0% | F | ||
| CC vs. TC+TT | All | 0.90(0.64–1.27);0.553 | 0.016,54.1% | R | |
| Gastric cancer | 0.36(0.19–0.66);0.001 | 0.347,0.0% | F | ||
| Breast cancer | 1.10(0.68–1.77);0.689 | 0.121,52.7% | R | ||
| Other cancers | 0.98(0.76–1.28);0.903 | 0.374,6.6% | F | ||
| CTLA-4-1722T/C | CC vs. TT | All | 0.98(0.70–1.37);0.906 | 0.050,45.3% | R |
| Gastric cancer | 0.45(0.23–0.86);0.016 | 0.412,0.0% | F | ||
| Breast cancer | 1.15(0.60–2.22);0.672 | 0.046,67.6% | R | ||
| Other cancers | 1.04(0.78–1.39);0.798 | 0.496,0.0% | F | ||
| TC vs. TT | All | 1.09(0.97–1.23);0.154 | 0.641,0.0% | F | |
| Gastric cancer | 1.34(0.94–1.91);0.107 | 0.392,0.0% | F | ||
| Breast cancer | 1.09(0.90–1.31);0.383 | 0.259,25.9% | F | ||
| Other cancers | 1.04(0.88–1.24);0.637 | 0.741,0.0% | F | ||
| C vs. T | All | 1.04(0.95–1.13);0.383 | 0.577,0.0% | F | |
| Gastric cancer | 0.90(0.70–1.15);0.406 | 0.833,0.0% | F | ||
| Breast cancer | 1.09(0.85–1.41);0.504 | 0.044,68.0% | R | ||
| Other cancers | 1.02(0.90–1.16);0.733 | 0.931,0.0% | F | ||
F indicates fixed model; R indicates random model.
Table 9. Summary of results of the meta-analysis from different comparative genetic models in the subgroup analysis by system.
| Polymorphism | Genetic comparison | Cancer type | OR(95%CI); P | Test of heterogeneity | |
| (p -Value, I2) | Model | ||||
| CC+TC vs. TT | All | 1.09(0.97–1.22);0.159 | 0.762,0.0% | F | |
| Digestive system cancer | 1.02(0.86–1.22);0.797 | 0.839,0.0% | F | ||
| Reproductive and breast cancer | 1.12(0.95–1.32);0.186 | 0.275,22.0% | F | ||
| Respiratory system cancer | 1.22(0.84–1.78);0.288 | 0.697,0.0% | F | ||
| CC vs. TC+TT | All | 0.90(0.64–1.27);0.553 | 0.016,54.1% | R | |
| Digestive system cancer | 0.71(0.33–1.53);0.381 | 0.008,74.5% | R | ||
| Reproductive and breast cancer | 1.11(0.88–1.40);0.395 | 0.171,37.5% | F | ||
| Respiratory system cancer | 1.99(0.37–10.85);0.425 | 0.498,0.0% | F | ||
| CTLA-4-1722T/C | CC vs. TT | All | 0.98(0.70–1.37);0.906 | 0.050,45.3% | R |
| Digestive system cancer | 0.79(0.41–1.52);0.476 | 0.056,60.3% | R | ||
| Reproductive and breast cancer | 1.18(0.91–1.53);0.217 | 0.111,46.7% | F | ||
| Respiratory system cancer | 2.02(0.37–10.99);0.417 | 0.499,0.0% | F | ||
| TC vs. TT | All | 1.09(0.97–1.23);0.154 | 0.641,0.0% | F | |
| Digestive system cancer | 1.06(0.88–1.27);0.529 | 0.386,4.8% | F | ||
| Reproductive and breast cancer | 1.10(0.92–1.31);0.289 | 0.392,2.6% | F | ||
| Respiratory system cancer | 1.19(0.81–1.75);0.367 | 0.791,0.0% | F | ||
| C vs. T | All | 1.04(0.95–1.13);0.383 | 0.577,0.0% | F | |
| Digestive system cancer | 0.96(0.85–1.09);0.569 | 0.966,0.0% | F | ||
| Reproductive and breast cancer | 1.09(0.96–1.23);0.168 | 0.175,37.0% | F | ||
| Respiratory system cancer | 1.24(0.87–1.78);0.232 | 0.595,0.0% | F | ||
F indicates fixed model; R indicates random model.
Figure 2. Meta-analysis with a fixed-effects model for the association between the risk of cancer and the CTLA-4 -1722T/C polymorphism (C vs. T).
Figure 3. Meta-analysis with a random-effects model for the association between the risk of cancer and the CTLA-4 -1722T/C polymorphism (CC vs. TC+TT).
Tests for publication bias, sensitivity analyses, and heterogeneity
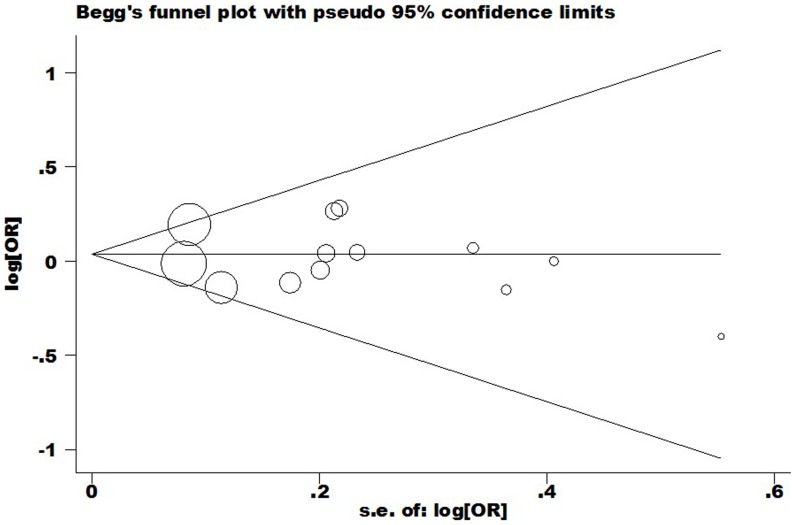
In this meta-analysis, potential publication bias was detected by Begg's Funnel plot and Egger's test ( Figure 4 ), and the shape of funnel was symmetry in all genetic model. It suggested that there were no publication bias for overall cancer in this meta-analysis (C vs. T: Begg's test P = 0.855, Egger's test P = 0.675; CC vs. TT: Begg's test P = 0.350, Egger's test P = 0.709; TC vs. TT: Begg's test P = 0.583, Egger's test P = 0.702; CC+TC vs. TT: Begg's test P = 0.161, Egger's test P = 0.576; CC vs. TT+TC: Begg's test P = 0.533, Egger's test P = 0.845).
Figure 4. Begg's funnel plot of meta-analysis of between the CTLA-4 -1722T/C polymorphism and the risk of cancer (fixed–effects estimates) (C vs. T compare genetic model).
Sensitivity analyses were carried out to detect the influence of each individual dataset on the pooled OR, with each study dataset set dropped at a time. The outcomes did not change when any individual study was omitted, suggesting the stability of our results ( Figure 5 ) (data not shown).
Figure 5. Sensitivity analysis of the influence of C vs. T in overall cancer meta–analysis (fixed–effects estimates).
Large heterogeneities among the studies were indentified in the recessive model and homozygous model. Since tumor origin, ethnicity and system can influence the results from meta–analyses, we carried out subgroup analyses and the results were presented in Table 7 , Table 8 and Table 9 . The results indicated that breast cancer, digestive system cancer and Asian population subgroup may contribute to the major heterogeneity. As shown in Table 7 , heterogeneity was significant in the recessive model. Further analysis was conducted by Galbraith radial plot in the recessive model ( Figure 6 ), and the result showed one outlier might contribute to the major sources of heterogeneity. From the forest plot in the recessive model ( Figure 2 ), one can identify that a case-control study conducted by Erfani et al.[16] contributes the main heterogeneity.
Figure 6. Galbraith radial plot of meta–analysis (CC vs. TC+TT compare genetic model).
Discussion
Of late, several studies have investigated the association between CTLA-4 -1722T/C polymorphism and multiple cancers, a decisive answer is lacking. In this study, a case-control study in Han Chinese population, along with a meta-analysis on overall cancer, attempted to derive a comprehensive evaluation and the results were non-significance. To the best of our knowledge, this is the first case-control study investigating the association between CTLA-4 -1722T/C polymorphism and esophageal cancer risk.
Cancer and autoimmune disease are both multifactorial disorders that results from complex interactions between genetic backgrounds and environmental factors. The CTLA-4 -1722T/C polymorphism (T→C) would reduce a transcription factor binding site for nuclear factor 1 and weaken the expression of cell surface CTLA-4 [11], [25], which might play an important role in cancer and autoimmune disease susceptibility. Several meta-analyses showed that CTLA-4 -1722T/C polymorphism might be a risk factor for systemic lupus erythematosus susceptibility [26]–[29]. However, the association between this locus and cancer risk was inconclusive. With a growing interest in the associations of genetic polymorphisms and cancer, several studies have examined the hypothesis that CTLA-4 -1722T/C polymorphism is relevant to the risk of a number of cancers; however, the results remain elusive. Considering the fact that most common SNPs usually make low penetrance cancer susceptibility, this study includes 13 case-control studies with relatively large sample sizes to obtain a precise evaluation between CTLA-4 -1722T/C genetic variation and cancer risk. One individual study has reported positive signal of CTLA-4 -1722T/C polymorphism with cancer [18]; the other individual study has reported negative signal [20]; however, as demonstrated in our overall genetic model results among 7098 subjects, there were non-significance, even in different population subgroups and different system. In a stratified analysis by cancer type, the protective effect conferred by the recessive model and homozygous model was appreciably obvious in gastric cancer subgroup. Considering only three case-control studies were conducted in gastric cancer subgroup and these studies were small sample sizes, which might restrict power to confirm a real influence or generate a fluctuated assessment. All results should be interpreted with very caution. It is also possible that the potential function of this polymorphism is diluted or covered by other genetic background or environment factors, and these important factors should not be ignored. Considering only 13 case-control studies were recruited in this meta-analysis and most of these studies were small sample sizes, in the future, further investigations with large sample sizes should be carried out to confirm or refute these results.
Some merit of current study should be adequate consideration. First, this is to date the first case-control study detecting the association of CTLA-4 gene -1722T/C polymorphism with esophageal cancer. Second, the findings of our case-control study conform to that of the subsequent meta-analysis. Third, in our case-control study, control genotype distributions were consistent with HWE showed our results were less prone to selection bias, the shape of funnel plot indicated that there were no publication bias in current meta-analysis. Fourth, relatively low heterogeneity was observed between publications for CTLA-4 -1722T/C polymorphism.
In addition, some limitations in current study should be acknowledged when interpreting our results. First, in this case-control study, all cases and controls were recruited from two hospitals and might not fully represent the general Chinese populations. Second, all included case–control studies for meta-analysis were from Asians and Caucasians; thus, our findings might only be suitable for these two populations. Third, only published studies were recruited in this meta-analysis, publication bias might have inevitably occurred. Fourth, due to the lack of uniform background data for recruited studies, data were not further stratified by other factors (such as, age, gender, smoking, alcohol consumption, and other lifestyle factors). Fifth, in this study, we focused on only -1722T/C polymorphism in CTLA-4, and did not consider other susceptibility genes or polymorphisms. For the low penetrance cancer susceptibility gene effects from SNP, these important genetic and environmental factors should be adequately considered.
In summary, this case-control study along with a meta-analysis, failed to confirm the association between CTLA-4 -1722T/C polymorphism and cancer risk, even across different ethnic subgroups and different systems. In the future, further investigations with large sample sizes and detailed gene–environment data, should be carried out to confirm or refute these results.
Supporting Information
Direct sequencing analyses for genotypes of CTLA-4 -1722T/C SNP (The three charts represent three genotypes).
(TIF)
PRISMA checklist, Checklist of items to include when reporting a systematic review or meta-analysis (diagnostic review consisting of cohort studies).
(DOCX)
Funding Statement
This study was supported by Jiangsu University Clinical medicine science and technology development fund (JLY20120004), National Natural Science Foundation of China (81370001, 81101889, 81000028), Jiangsu Province Natural Science Foundation (BK2010333, BK2011481), Social Development Foundation of Zhenjiang (SH2010017), Changzhou Young Talents and Science-Technology Foundation of Health Bureau (QN201102) and Affiliated People's Hospital of Jiangsu University fund (Y200913). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Liu L, Zhong R, Wei S, Yin JY, Xiang H, et al. (2011) Interactions between genetic variants in the adiponectin, adiponectin receptor 1 and environmental factors on the risk of colorectal cancer. PLoS One 6: e27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reeves GK, Pirie K, Green J, Bull D, Beral V, et al. (2012) Comparison of the effects of genetic and environmental risk factors on in situ and invasive ductal breast cancer. Int J Cancer 131: 930–937. [DOI] [PubMed] [Google Scholar]
- 4. Zhang Y, Zhang J, Deng Y, Tian C, Li X, et al. (2011) Polymorphisms in the cytotoxic T-lymphocyte antigen 4 gene and cancer risk: a meta-analysis. Cancer 117: 4312–4324. [DOI] [PubMed] [Google Scholar]
- 5. Welsh MM, Applebaum KM, Spencer SK, Perry AE, Karagas MR, et al. (2009) CTLA4 variants, UV-induced tolerance, and risk of non-melanoma skin cancer. Cancer Res 69: 6158–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sun T, Zhou Y, Yang M, Hu Z, Tan W, et al. (2008) Functional genetic variations in cytotoxic T-lymphocyte antigen 4 and susceptibility to multiple types of cancer. Cancer Res 68: 7025–7034. [DOI] [PubMed] [Google Scholar]
- 7. Hu L, Liu J, Chen X, Zhang Y, Liu L, et al. (2010) CTLA-4 gene polymorphism +49 A/G contributes to genetic susceptibility to two infection-related cancers-hepatocellular carcinoma and cervical cancer. Hum Immunol 71: 888–891. [DOI] [PubMed] [Google Scholar]
- 8. Scheipers P, Reiser H (1998) Fas-independent death of activated CD4(+) T lymphocytes induced by CTLA-4 crosslinking. Proc Natl Acad Sci U S A 95: 10083–10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vandenborre K, Van Gool SW, Kasran A, Ceuppens JL, Boogaerts MA, et al. (1999) Interaction of CTLA-4 (CD152) with CD80 or CD86 inhibits human T-cell activation. Immunology 98: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ueda H, Howson JM, Esposito L, Heward J, Snook H, et al. (2003) Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 423: 506–511. [DOI] [PubMed] [Google Scholar]
- 11.Geng R, Song F, Yang X, Sun P, Hu J, et al.. (2013) Association between cytotoxic T lymphocyte antigen-4 +49A/G, -1722T/C, and -1661A/G polymorphisms and cancer risk: a meta-analysis. Tumour Biol. [DOI] [PubMed]
- 12. Chen ZJ, Zhao H, He L, Shi Y, Qin Y, et al. (2011) Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet 43: 55–59. [DOI] [PubMed] [Google Scholar]
- 13.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 151: : 264–269, W264. [PMC free article] [PubMed] [Google Scholar]
- 14. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hadinia A, Hossieni SV, Erfani N, Saberi-Firozi M, Fattahi MJ, et al. (2007) CTLA-4 gene promoter and exon 1 polymorphisms in Iranian patients with gastric and colorectal cancers. J Gastroenterol Hepatol 22: 2283–2287. [DOI] [PubMed] [Google Scholar]
- 16. Erfani N, Razmkhah M, Talei AR, Pezeshki AM, Doroudchi M, et al. (2006) Cytotoxic T lymphocyte antigen-4 promoter variants in breast cancer. Cancer Genet Cytogenet 165: 114–120. [DOI] [PubMed] [Google Scholar]
- 17. Li H, Fu ZK, Wang LH, Li DL, Wu N, et al. (2008) [Association of cytotoxic T lymphocyte antigen-4 gene polymorphisms with susceptibility to breast cancer]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 24: 282–284. [PubMed] [Google Scholar]
- 18. Li D, Zhang Q, Xu F, Fu Z, Yuan W, et al. (2012) Association of CTLA-4 gene polymorphisms with sporadic breast cancer risk and clinical features in Han women of northeast China. Mol Cell Biochem 364: 283–290. [DOI] [PubMed] [Google Scholar]
- 19. Qi YQ SL, Cui JY, Ji LS (2012) Polym orphism of CTLA-4 gene prom oter region and extron region in gastric cancer. Chinese Clinical Oncolog 17: 828–831. [Google Scholar]
- 20. Song JQ XB, Li R, Li C, Wu YH (2006) Association of the CTLA4 promoter region polymorphism with gastric cancer. Chinese Journal of Experimental Surgery 23: 1373–1374. [Google Scholar]
- 21. Jiang L, Luo RY, Zhang W, Wang LR, Wang F, et al. (2011) [Single nucleotide polymorphisms of CTLA4 gene and their association with human cervical cancer]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 28: 313–317. [DOI] [PubMed] [Google Scholar]
- 22. Khaghanzadeh N, Erfani N, Ghayumi MA, Ghaderi A (2010) CTLA4 gene variations and haplotypes in patients with lung cancer. Cancer Genet Cytogenet 196: 171–174. [DOI] [PubMed] [Google Scholar]
- 23. Rahimifar S, Erfani N, Sarraf Z, Ghaderi A (2010) ctla-4 gene variations may influence cervical cancer susceptibility. Gynecol Oncol 119: 136–139. [DOI] [PubMed] [Google Scholar]
- 24. Bharti V, Mohanti BK, Das SN (2013) Functional genetic variants of CTLA-4 and risk of tobacco-related oral carcinoma in high-risk North Indian population. Hum Immunol 74: 348–352. [DOI] [PubMed] [Google Scholar]
- 25. Jones KA, Kadonaga JT, Rosenfeld PJ, Kelly TJ, Tjian R (1987) A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell 48: 79–89. [DOI] [PubMed] [Google Scholar]
- 26. Zhu JM, Li BK, Chen GM, Feng CC, Cen H, et al. (2013) CTLA-4 -1722T/C polymorphism and systemic lupus erythematosus susceptibility: a meta-analysis involving ten separate studies. Immunol Invest 42: 91–105. [DOI] [PubMed] [Google Scholar]
- 27. Chen SL, Deng F, Jiang F, Liu JJ, Meng W (2012) [Correlation between CTLA-4 gene polymorphism and systemic lupus erythematosus: a meta-analysis]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 28: 1320–1323. [PubMed] [Google Scholar]
- 28. Liu J, Zhang H (2013) -1722T/C polymorphism (rs733618) of CTLA-4 significantly associated with systemic lupus erythematosus (SLE): a comprehensive meta-analysis. Hum Immunol 74: 341–347. [DOI] [PubMed] [Google Scholar]
- 29. Zhai JX, Zou LW, Zhang ZX, Fan WJ, Wang HY, et al. (2013) CTLA-4 polymorphisms and systemic lupus erythematosus (SLE): a meta-analysis. Mol Biol Rep 40: 5213–5223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Direct sequencing analyses for genotypes of CTLA-4 -1722T/C SNP (The three charts represent three genotypes).
(TIF)
PRISMA checklist, Checklist of items to include when reporting a systematic review or meta-analysis (diagnostic review consisting of cohort studies).
(DOCX)