Abstract
Multicellular organisms evolved sophisticated defense systems to confer protection against pathogens. An important characteristic of these immune systems is their ability to act both locally at the site of infection and at distal uninfected locations1-4. In insects, such as Drosophila melanogaster, RNA interference (RNAi) mediates antiviral immunity5-7. However, the antiviral RNAi defense in flies is thought to be a local, cell-autonomous process, since flies are considered unable to generate a systemic RNAi response8. Here we show that a recently defined double-stranded RNA (dsRNA) uptake pathway9 is essential for effective antiviral RNAi immunity in adult flies. Mutant flies defective in this dsRNA uptake pathway were hypersensitive to infection with Drosophila C virus (DCV) and Sindbis virus. Mortality in dsRNA-uptake defective flies was accompanied by 100-to 105-fold increases in viral titers and higher levels of viral RNA. Furthermore, inoculating naked dsRNA into flies elicited a sequence specific antiviral immune response that required an intact dsRNA uptake pathway. These findings suggest that spread of dsRNA to uninfected sites is essential for effective antiviral immunity. Strikingly, infection with Sindbis-GFP suppressed expression of host-encoded GFP at a distal site. Thus, similar to protein-based immunity in vertebrates, the antiviral RNAi-response in flies also relies on the systemic spread of a virus-specific immunity signal.
Based on the recent identification of a dsRNA uptake pathway in flies9, 10, we hypothesized that dsRNA produced and released from infected cells can be taken up locally, and perhaps at distal sites, to establish systemic pre-existing immunity in uninfected cells (Fig. 1). We thus examined whether naked dsRNA can mediate systemic RNAi spread by inoculating flies with dsRNA corresponding to two different regions of the Sindbis virus genome (dsSin1 and dsSin2, Supplementary Fig. S1a and Fig.2a). Two days after dsRNA inoculation, flies were infected with a recombinant Sindbis virus expressing GFP (Sindbis-GFP virus, Supplementary Fig. S1a). Strikingly, inoculation with dsSin1 and dsSin2 dramatically reduced accumulation of GFP as determined by fluorescence microscopy and immunoblotting (Fig 2b and 2c, lanes 7-11 and 18-22); control buffer had no effect on virus replication (Fig 2b and 2c, lanes 2-6 and Supplementary Fig S1b). This inhibitory response was sequence specific because flies inoculated with dsRNA corresponding to Drosophila C virus (DCV) genome showed no effect on Sindbis virus replication (Fig 2b and 2c, lanes 13-17). Further, inoculation of dsRNA corresponding to DCV (dsDCV) efficiently protected wild type flies against Drosophila C virus infection, but not against Sindbis (Supplementary Fig. S2a). The antiviral effect of exogenous dsRNA inoculation required a functional RNAi machinery as Dicer2 and Ago2 null mutant flies (dicer-2-/- and ago-2-/-) were unable to mount an effective antiviral response (Fig. 2d and Supplementary Fig. S2a). In addition, wild type flies accumulated siRNAs derived from injected dsRNA (Supplementary Fig. S2c). We conclude that inoculation of dsRNA initiates a bona-fide, specific RNAi response that protects flies against virus infection.
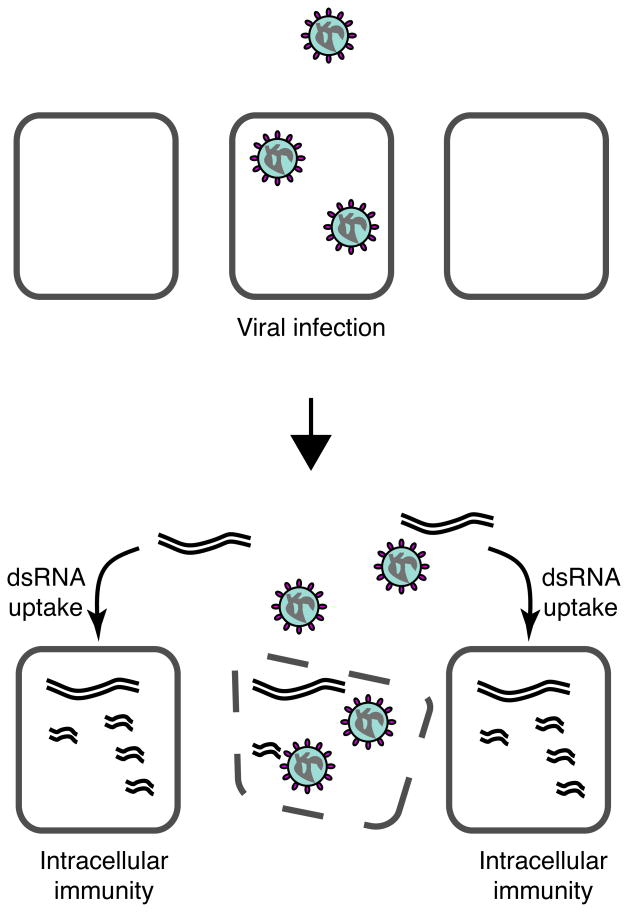
Figure 1.
Model for systemic RNAi viral immunity in Drosophila melanogaster. Upon viral infection, virus-specific dsRNAs (eg., replication intermediates) are generated during the initial rounds of virus replication. Following cell death or lysis, dsRNAs are taken up and processed by uninfected cells to protect them from subsequent infection, thereby preventing virus spread.
Figure 2.
in vivo dsRNA immunization provides sequence-specific antiviral protection in D. melanogaster. a, Immunization protocol. b and c, Wild type flies infected with Sindbis-GFP virus two days after intrathoracical injection of dsRNA against Drosophila C virus (DCV, 442 base-pair in length, corresponding to the viral polymerase between nucleotides 5589 to 6030), dsRNA against Sindbis virus non-structural proteins 1 and 2 (dsSin1, 901 base-pairs in length, corresponding to nucleotides 1211 to 2112) or dsRNA Sindbis virus corresponding to the non-structural proteins 3 and 4 (dsSin2, 954 base-pairs long, length, corresponding to nucleotides 5485 to 6439). Buffer: control injection. d.p.i.: days post infection. Sindbis-GFP virus replication was monitored by GFP production. b, Fluorescence images. c, Western blot with an anti-GFP antibody. d, Sindbis-GFP virus challenge in wild type, homozygous Dcr2L811fsX (Dcr2-/-) and homozygous Ago414 (Ago2-/-) flies. e, dsRNA immunization protects in a dose-dependent manner. Flies were inoculated with dsRNA, dsSin2, directed against Sindbis-GFP (5ng, 0.5 ng, 50 pg, and 5 pg). Virus replication over time (d.p.i.: 2 to 5) was monitored by westernblotting using and an anti-GFP antibody.
Serial dilutions of dsSin2 indicated that very low concentrations of injected dsRNA sufficed to mount a very strong response (Fig. 2e). Accordingly, we observed a reduction on viral replication even after inoculation of 5 pg of dsRNA (equivalent to 1.5×105 molecules of dsSin2, Fig. 2e, lanes 17 to 20). Of note, while the maximal dose of dsSin2 (5 ng) elicited an inhibitory response that lasted 5 days (Fig. 2e, lanes 5 to 8), inoculation of a lower dose produced a shorter period of immunity (Fig. 2e, compare lanes 5 to 8 with 9 to 12, 13 to 16, and 17 to 20). This observation underscores the efficiency and persistence of the dsRNA mediated antiviral immunity in Drosophila, and supports the idea that exogenous dsRNA can initiate an RNA silencing response in flies, albeit rather without the RNAi amplification mechanism observed in plants and worms11, 12.
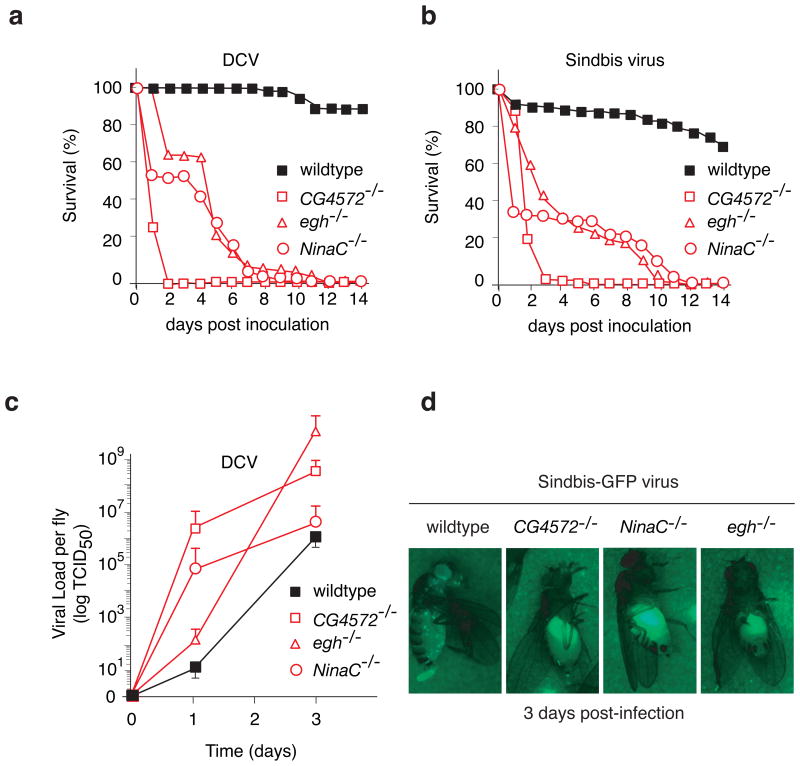
We previously described that dsRNA is taken up in Drosophila S2 cells by an active pathway, involving receptor-mediated endocytosis9. To examine whether this pathway is involved in the antiviral RNAi response mechanism we selected three genes implicated by the previous analysis in dsRNA uptake: egghead (egh) encoding a seven-transmembrane domains glycosyltransferase; NinaC, coding for a protein involved in vesicle transport; and a gene of unknown function CG4572 (Supplementary Fig. S2b). While viability and fertility of homozygous eghEP804, NinaC3 and CG4572c05963 mutant flies did not differ significantly from wild type, all three mutants were hypersensitive to DCV or Sindbis virus infection. In these dsRNA uptake-defective flies, we observed an earlier onset of disease (Fig. 3a and 3b). After infection, median survival of homozygous eghEP804, NinaC3 and CG4572c05963 flies was approximately 5 to 8 days, compared with more than 14 days in wild type flies, and the 50% lethal dose (LD50) in the CG4572c05963 was 9-fold lower than in wild type flies (not shown).
Figure 3.
Increased viral susceptibility of dsRNA uptake deficient mutants. a, b, Survival of dsRNA uptake mutant flies after virus infection. Homozygous eghEP804 (egh-/-), NinaC3 (NinaC-/-), CG4572c05963 (CG4572-/-), and wild type flies were injected with 500 TCID50 DCV (a) or 500 PFU Sindbis-GFP virus (b) and monitored daily for survival. c, DCV replicates at higher levels in dsRNA uptake mutant flies. Flies were injected with 500 TCID50 DCV, and virus production was monitored over time. At each time point, three pools of five flies were homogenized, and the viral titer in the homogenate was determined by end-point dilution. The error bars report the average +/- s.d. for at least 3 independent experiments.d, Sindbis-GFP virus replicates at higher levels in dsRNA uptake mutant flies as shown by increased GFP expression in the fat body after 3 days post-infection when compared to wild type flies.
An important consideration when studying viral sensitivity in animals defective for components of a major cellular pathway, such as endocytosis or intracellular transport, is that enhanced death following viral infection may be caused by a decrease in fitness or general health of the mutant animal, and not by a direct antiviral activity of the deleted component. To establish whether the increased mortality of egh, NinaC and CG4572 mutants stems from their inability to control virus replication, we determined viral loads (Fig. 3c). Even at early time points after infection, before the onset of disease, DCV titers were 100- to 105-fold higher in homozygous eghEP804, NinaC3 and CG4572c05963 flies compared to wild type controls (Fig. 3c). The increase in viral titers in mutant flies was mirrored by a dramatic increase in viral RNA levels. While viral RNA was barely detected in wild type flies before day 5, it was clearly observed at 24 h after infection in homozygous NinaC3 and CG4572c05963 mutant flies, and by 48 h it accumulated at much higher levels in these mutants than in wild type flies (Supplementary Fig. S3a). We further examine the role of the dsRNA uptake pathway on virus replication by monitoring Sindbis-GFP virus tissue tropism. In wild type flies, GFP fluorescence was barely detected 3 days after infection and accumulated in discrete puncta throughout the fly. In contrast, in homozygous eghEP804, NinaC3 and CG4572c05963 mutants, GFP accumulated within a large structure in the abdomen of the animal and at much higher levels than in wild type flies (Fig. 3d and Supplementary Fig. S3b). These results indicate that the enhanced viral susceptibility of egh, NinaC and CG4572 mutant flies is due to their inability to control virus replication.
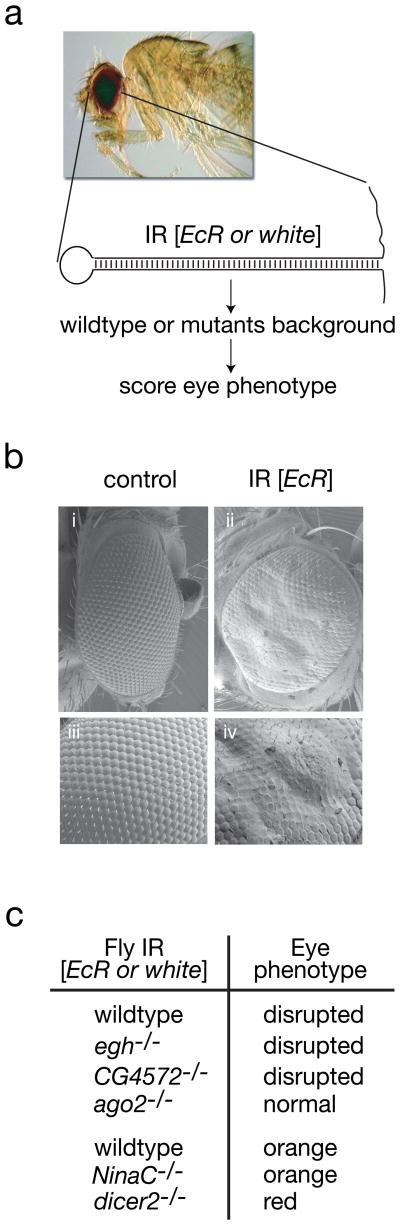
The hyper-susceptibility to virus infection of flies defective in dsRNA uptake is strikingly similar to that previously seen in ago-2 defective flies6. We thus examined whether the RNAi core function is impaired in egh, NinaC and CG4572 mutant flies. Eye-specific silencing of the Ecdysone Receptor gene (EcR) by an endogenously expressed EcR hairpin dsRNA13 leads to abnormal eye structure resulting from impaired corneal lens formation (Fig. 4a and b). Under these conditions, disruption of the core RNAi machinery in homozygous ago414 mutant suppressed EcR RNAi and restored normal eye structure. In contrast, efficient EcR RNAi was observed in homozygous eghEP804 and CG4572c05963 flies. Similar experiments monitored RNAi in homozygous NinaC3 flies using the expression of a hairpin dsRNA targeting the white gene, that causes a decrease of eye pigmentation and orange eye colour in control flies (Supplementary Fig. S4a and b)14. Silencing of white was suppressed in homozygous Dcr2L811fsX mutant flies while it was fully maintained in homozygous NinaC3 flies (Fig. 4c and Supplementary Fig. S4a and b). We further confirmed this conclusion by injecting dsRNA against the fushi tarazu gene (ftz)15 into syncitial embryos before cellularization. Injection of ftz dsRNA in wild-type embryos resulted in the expected segmentation defects, namely loss of denticle belts in the cuticule of pre-hatching larvae (ftz phenotype) (Supplementary Fig. S4c). Injection of ftz dsRNA in homozygous eghEP804, NinaC3 and CG4572c05963 embryos induced the same defects, indicating that RNA silencing proceeded normally in these mutants. In contrast, homozygous ago414 control embryos were unable to silence ftz expression and thus hatched with a wild type cuticule (Supplementary Fig. S4c). These results indicate that mutant flies support efficient RNAi silencing if dsRNA uptake is bypassed through expression of dsRNA hairpins intracellularly or by injecting dsRNA into syncytial embryos.
Figure 4.
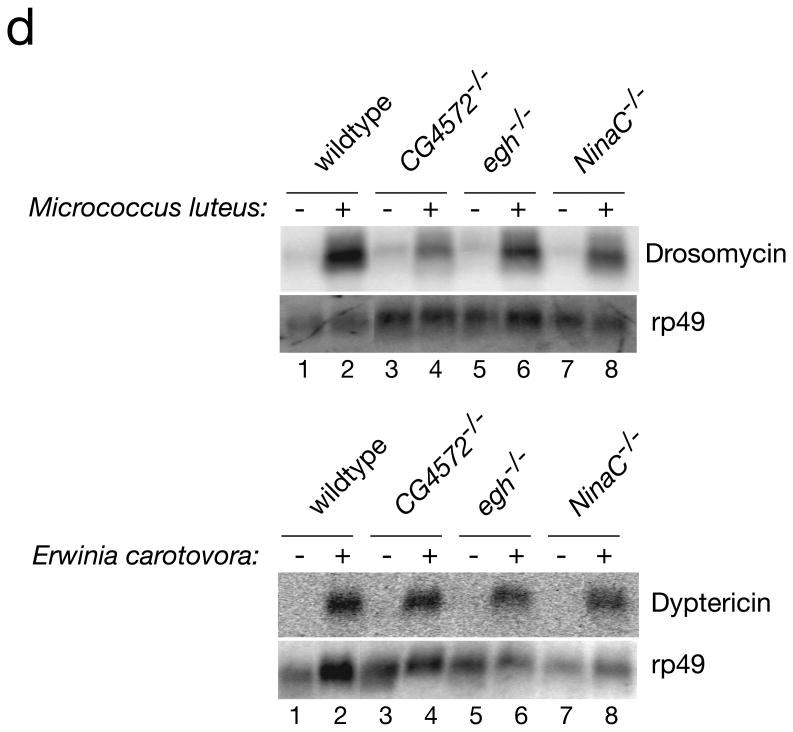
Core RNAi machinery and antibacterial immunity are intact in dsRNA uptake mutants. a, Schematic to test the core RNAi machinery integrality. b, RNAi processing of an inverted repeat IR [Ecr] induced by the GMR-GAL4 driver prevents the formation of the corneal lens (EM picture). c, Monitoring corneal lens formation and eye color in transgenic flies deficient in the dsRNA uptake pathway. d, Susceptibility of dsRNA uptake mutant flies to infection is specific to the viruses, as the dsRNA uptake mutant flies are able to produce antimicrobial peptides in response to an infection by pathogenic gram + and gram − bacteria.
We next examined whether other arms of the immune system were affected in egh, NinaC and CG4572 mutants. Insects produce a number of antimicrobial peptides, which are secreted into the hemolymph, in response to immune challenge. These peptides are effective against Gram-negative and Gram-positive bacteria as well as fungi16. We determined whether egh, NinaC and CG4572 mutant flies can support production of the antimicrobial peptides drosomycin and dyptericin in response to septic injury with Gram-positive and Gram-negative bacteria. Drosomycin production was measured after septic injury with Micrococcus luteus, a Gram-positive bacterium that signals through the Toll pathway. Production of dyptericin was measured after septic injury with Erwinia carotovora, a Gram-negative bacterium that induces the Imd pathway. Homozygous eghEP804 , NinaC3 and CG4572c05963 flies were able to efficiently respond to bacterial infection, (Fig 4d). Similarly, the JAK/STAT signaling pathway17 appears unimpaired in eghEP804and CG4572c05963 flies as DCV infection induced normal vir-1 expression in these mutants (Supplementary Fig. S5). Thus, defects in cellular components that abrogate dsRNA uptake and its ensuing antiviral immunity do not generally impair other arms of the fly innate immune system.
We hypothesize that the dsRNA uptake pathway underlies systemic antiviral immunity, which is required to control virus replication. We thus examined whether dsRNA inoculation in egh, NinaC and CG4572 mutant flies was able to elicit the protective immunity observed in wild type flies (Fig. 2). Indeed, while inoculation of DCV dsRNA dramatically reduced DCV replication in wild type flies (Fig 5a, lanes 1-9), homozygous eghEP804, NinaC3 and CG4572c05963 mutant flies were unable to mount an antiviral response upon DCV dsRNA inoculation (Fig 5a, lanes 10-30). Similarly, the dsRNA uptake pathway was required for protection against Sindbis virus infection by naked dsSin1 inoculation (Supplementary Fig. S6). Furthermore, while wild type flies efficiently processed inoculated dsRNA into siRNAs, eghEP804, NinaC3 and CG4572c05963 mutant flies accumulated siRNAs at much lower levels (Supplementary Fig. S2b).
Figure 5.
Systemic spread of dsRNA follows virus infection and it is essential for effective antiviral immunity. a, Drosophila C virus infection in wild type, and homozygous eghEP804 (egh-/-), NinaC3 (NinaC-/-) and CG4572c05963 (CG4572-/-) mutant flies treated with the inoculated with dsRNA. DCV replication was monitored by westernblotting using an antibody directed against DCV capsid protein VP1. (b-d) dsRNA produced during virus replication can spread and silence endogenous GFP expressed at a distal site of infection. Flies expressing eGFP (Tub-eGFP) inoculated with Sindbis-GFP (b, c) or Sindbis:Luciferase virus (d) by intrathoracical inoculation. b, Viral replication monitored by RT-PCR using primers that amplify NSP1-2 virus genes. c, Expression of endogenous GFP monitored by Western blot with an anti-GFP antibody. d, same as (c) except that flies were infected with Sindbis-Luciferase virus.
Our model is that infected cells released viral dsRNA that is subsequently taken up by uninfected cells through the dsRNA uptake pathway thereby eliciting an antiviral RNAi response. A direct prediction of this model is that during infection, viral-derived dsRNA spread to induce systemic silencing. To test this prediction we examined whether infection with a Sindbis virus carrying the GFP gene could silence an ubiquitously expressed endogenous GFP at a distal site. Following intrathoraxic inoculation, Sindbis-GFP virus RNA was readily detected in the thorax and in the abdomen of the fly starting at one day post infection (Fig. 5b, lanes 5 to 8). In contrast, the viral RNA was not detectable in the head until day 5 post infection (Fig. 5b, lanes 1 to 4). Strikingly, endogenous GFP expression in the head was significantly reduced already at day 2, despite the absence of any detectable viral replication in this organ (Fig.5c, lane 3). In contrast, infection with control Sindbis virus carrying firefly luciferase gene did not silence GFP expression (Fig.5d). These results indicate that a virus specific derived RNAi signal spread from the thorax to the head early after infection.
It was previously thought that Drosophila is unable to systemically spread an RNAi response, based on observations that endogenously expressed RNA hairpins did not spread from cell to cell8. However, we demonstrate that, upon virus infection infected cells spread systemically a silencing signal that elicits protective RNAi-dependent immunity through out the organism. While uninfected Drosophila cells appear to lack a constitutive mechanism for RNAi systemic spread, unlike plant and worm cells, they do have an active and highly efficient mechanism for dsRNA uptake, which we here show is essential for antiviral immunity.
Accordingly, dsRNA is normally not released from uninfected cells, but virus infection may induce dsRNA release either through lysis of infected cells or through a virally induced shedding mechanism. We propose that these virally derived dsRNAs are taken up into uninfected cells to generate virus-specific intracellular immunity that prevents virus spread (Fig. 1). In support of this idea, this specific antiviral response in flies requires both the RNAi core machinery and the recently described dsRNA uptake pathway. Furthermore, simple inoculation of even very low amounts of dsRNA, in the absence of virus infection, can by itself promote a potent antiviral immunity, which is similarly dependent on the RNAi core machinery and the dsRNA uptake pathway. Our previous results indicated that while dsRNA is readily uptake by S2 Drosophila cells, siRNAs are not efficiently taken up9. We thus conclude that systemic spread of a specific antiviral RNAi activity, most likely mediated by large viral dsRNAs or intramolecular base-pairing structures released from infected cells, is an essential component of the immune response elicited by virus infection in flies. The precise nature of the RNAi spread intermediate reminds to be further defined.
It is remarkable that blocking the spread of the RNAi signal has such a profound effect on antiviral immunity. This suggests that the cell autonomous RNAi response is insufficient to control a viral infection. In striking parallel to vertebrates, flies also rely on systemic immunity, albeit in this case the virus-specific signal is dsRNA-based. These observations speak to the evolutionarily conserved principles of immunity in multicellular organisms, requiring both cell-autonomous responses as well as systemic mechanisms to create pre-existing immunity to protect uninfected cells.
Methods Online
Cells, plasmids and viruses
Drosophila S2 cells (Invitrogen) were cultured at 25°C in Schneider's Drosophila medium (GIBCO), supplemented with 10% heat inactivated foetal calf serum, 2mM L-glutamine, 100 U/ml penicillin, and 100 mg/ml streptomycin. Firefly (Photinus pyralis) and Renilla reniformis luciferase sequences from the plasmids pGL3 and pRL-CMV (Promega) were cloned into pMT/V5-HisB (Invitrogen), generating pMT-Luc and pMT-Ren allowing Copper-inducible expression from a metallothein promoter.
Transfections were performed using Effectene transfection reagent (Qiagen) according to the manufacturer's recommendations. Luciferase expression was assayed using the Dual-Luciferase Reporter Assay System (Promega) and analyzed on a Tecan Ultra-evolution platereader. Double stranded RNA was generated by in vitro transcription from T7 promoter flanked PCR products. Viral stocks were prepared on low passage S2 cells and titered by end point dilution. Briefly, 25.000 S2 cells per well in a 96 well plate were inoculated with 10-fold dilutions of viral stocks. Cells were transferred to fresh medium at day 7 and CPE was monitored visually over 14 days. Viral titers were calculated according to the method of Reed and Muench.
Recombinant Sindbis virus expressing GFP during viral replication was generated by cloning enhanced GFP into the XbaI site of the double subgenomic Sindbis vector pTE3′2J (kindly provided by C. Rice)1. The resulting plasmid was linearized and in vitro transcribed using the mMessage machine kit (Ambion). RNA was purified and electroporated into BHK-21 cells and supernatant was harvested and virus title determined by plaque assay on BHK cells.
Fly stocks
dsRNA uptake mutant stocks were obtained from the Bloomington Drosophila Stock Center. The genomic structure of these mutant alleles stocks was confirmed by inverse PCR and sequencing. The eghEP804 allele is a P-element insertion in the coding sequence of the egh first exon. The NinaC3 allele is a replacement of the K1078 codon by a stop codon2. The CG4572c05963 allele is a PiggyBac insertion in the open reading frame of CG4572c05963. The UAS>IR[EcR] transgene producing EcR dsRNA3 and the P{GAL4-ninaE.GMR}12 GAL4 driver were recombined on chromosome 2 before genetic crosses with eghEP804 and CG4572c05963 mutant stocks. The GMR>IR[white] inverted repeat transgene has been previously described4. The Tub-eGFP transgenic line was obtained from Steve Cohen5.
RNAi in S2 cells
The effect of down regulating NinaC, CG4572, and Egghead on dsRNA uptake was analyzed in a silencing of luciferase expression assay. S2 cells were transfected with corresponding dsRNA (NinaC, Egghead and CG4572). Cells were pre-treated with approximately 500 nt long dsRNA targeting either egh (nucleotides 488 to 1103 – 616 bp product), NinaC (nucleotides 161 to 761 – 601 bp product), cg4572 (nucleotides 61 to 731 – 671 bp product), or ago2 (nucleotides 214 to 865 – 652 bp product), or with dsRNA targeting GFP as a negative control. Three days after knock down of these gene products, the cells were co-transfected with an RNAi dual reporter system, consisting of firefly luciferase and Renilla luciferase expression plasmids. Then, dsRNA directed against firefly luciferase (nucleotides 66 to 658 – 592 bp product) was either added to the culture supernatant ‘soaking’) or directly introduced into cells by co-transfection with the dual reporter plasmids (‘transfection’). Twenty four hours after dsRNA luciferase treatment, expression of luciferase was induced by adding CuSO4 to the culture supernatant, and cell lysates were generated after an additional 18 hour incubation.
Microbial infection
The bacteria Erwinia carotorova and Micrococcus luteus were precultured in LB medium. Pellets taken when the cultures were in the log phase of growth were resuspended in a small amount of culture medium, and sharpened needles dipped into these suspensions. Flies were harvested at 6 and 36 h after septic injury. Total RNA extraction and Northern blots were performed following standard procedures.
Fly infections
Flies were reared on standard medium at 25°C. Ago-2414 and Dcr-2L811fsX flies have been described previously 4[Okamura, 2004 #164], w1118 flies were used as a wildtype controls. UAS-B2 flies will be described in more detail elsewhere. Two to three day old female flies were injected with 50 nl of the appropriate virus dilution in 10 mM Tris.Cl (pH 7.5) as described previously [Cherry, 2004 #170], using a Drummond nanoject injector. Fly mortality at day one was attributed to damage invoked by the injection procedure, and these flies were excluded from further analyses. Mortality was monitored daily for 14 days, and every three to four days the flies were transferred to fresh food. In all experiments 40-60 flies per genotypic group were injected. Unless noted otherwise, female flies were used. No significant difference in survival was observed between flies after injection of buffer (data not shown). For northern blots, RNA was isolated from 25 flies using Trizol reagent. Viral titers in the flies were determined by end-point dilution of fly homogenate of three pools of five flies. At the indicated time points, flies were harvested and stored at -70°C until further processing. We confirmed the absence of endogenous virus in fly stocks by titration of uninfected fly homogenate on S2 cells (data not shown).
dsRNA preparation and injection into adult flies
dsRNA was generated by in vitro transcription using T7 RNA-polymerase using as template PCR products corresponding to nucleotides 1211 to 2112 (NSP1/2), nucleotides 5485 to 6439 (NSP3/4) of Sindbis virus genome, or nucleotides 5589 to 6030 of the DCV genome. Five-day-old female flies were CO2-anesthetized and injected in the thorax with 50 nl of the appropriate dsRNA (5mg/ml) using a nanoinjector (Nanoject II, Drummond Scientific). Two days later flies were CO2-anesthetized and injected in the opposite side of the thorax with the appropriate virus dilution in 10mMTris-Cl (pH 7.5). Injection of the same volume of 10 mM Tris-HCl, pH 7.5 was used as a control. Age of the flies and amount of dsRNA injected was determined according to Goto et al.6. Virus infection was previously described7.
Western-Blot analysis
For protein analysis, equal amounts of proteins from total fly extracts were boiled in Laemmli buffer and loaded on 10% SDS-PAGE. After transfer nitrocellulose membranes were blocked in 5% milk, 1X PBS, 0.1% Tween, and incubated overnight with rabbit polyclonal anti-GFP (Santa Cruz Biotechnology) or rabbit polyclonal anti VP1 (custom made). For normalization a monoclonal antibody anti-alpha tubulin (Sigma Aldrich) was used. Detection was performed using Supersignal West Pico Chemiluminescent Substrate (Pierce).
Northern blots
Total RNAs were extracted from whole flies were extracted using Trizol (Invitrogen). 15 ug of total RNA was size fractionated on 1% (w/v) agarose gels containing 1.1 mM formaldehyde, After electrophoresis, the RNA was transferred overnight by capillarity to a nylon membrane (Nytran Supercharge; Schleicher and Schuell) and covalently bound to the membrane using a Stratalinker UV crosslinker . Northern blots were hybridized with DNA probes generated by a random- primed labelling reaction and [alpha-32P]dCTP. Membranes were exposed overnight to a PhosphorImager screen at room temperature. Viral RNA was detected by northern blot using standard procedures with a random primed DNA probe corresponding to nt 1947 to 2528 of DCV.
Oligonucleotide primers
All the primers used to produce dsRNA had a T7 promoter sequence (taatacgactcactatagggaga) at the 5′.
DCVpol Fwd: 5′ CAACGAATATGTCGCCTTGA 3′
DCVpol Rev: 5′ TTGGTTGTACGTCAAAATCTGAG 3′
SINnsp1 Fwd : 5′ TCTGCCGATCATAGCACAAG 3′
SINnsp2 Rev : 5′ CCTTCTTAACGCAACGCTTC 3′
SINnsp3 Fwd : 5′ GAGGATCAATTTTCGACGGAGA 3′
SINnsp4 Rev : 5′ GATTGAATGTCGCTGAGTCCAG 3′
Vir1-Fwd: 5′ TTCGATTCCTCAGACGATGA 3′
Vir1-Rev: 5′ GGTCAATGGGCACAAAGTTC 3′
Rp49-Fwd: 5′ AAGGGTATCGACAACAGAGTGC 3′
Rp49-Rev: 5′ ACAAATGTGTATTCCGACCACG 3′
Supplementary Material
Acknowledgments
We are grateful to members of the Andino and the O'Farrell lab for discussion, technical support and advice on fly work. We thank Drs. Judith Frydman, Pat O'Farrell and Marco Vignuzzi for useful discussions and comments on the manuscript. We also thank Tiffany Cook for advice on the IR Ecr eye phenotype, Richard Carthew for providing the GMR-wIR and Dcr2 flies stock and Mikiko Siomi for the Ago2 fly stock. BG is a Manlio Cantarini fellow. BB is a Lebanese CNRSL Fellow. CJ is a University of Paris VI and Ministère de la Recherche fellow. This work was financially supported by NIH grants AI40085 and AI064738 to R.A, .the Institut Pasteur to M-C.S and C.A., the CNRS, ANR and ARC grants to C.A.
Footnotes
Competing interests statement: The authors declare that they have no competing financial interests.
References
- 1.Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–63. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 2.Dorner T, Radbruch A. Antibodies and B cell memory in viral immunity. Immunity. 2007;27:384–92. doi: 10.1016/j.immuni.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Roitt I, Brostoff J, Male D. Immunology. Mosby; 2001. [Google Scholar]
- 4.Voinnet O. Non-cell autonomous RNA silencing. FEBS Lett. 2005;579:5858–71. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 5.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7:590–7. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 6.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20:2985–95. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang XH, et al. RNA interference directs innate immunity against viruses in adult. Drosophila Science. 2006;312:452–4. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roignant JY, et al. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA. 2003;9:299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saleh MC, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulvila J, et al. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–5. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- 11.Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–67. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sijen T, et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–76. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 13.Schubiger M, Carre C, Antoniewski C, Truman JW. Ligand-dependent de-repression via EcR/USP acts as a gate to coordinate the differentiation of sensory neurons in the Drosophila wing. Development. 2005;132:5239–48. doi: 10.1242/dev.02093. [DOI] [PubMed] [Google Scholar]
- 14.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 15.Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–26. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426:33–8. doi: 10.1038/nature02021. [DOI] [PubMed] [Google Scholar]
- 17.Dostert C, et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat Immunol. 2005;6:946–53. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 18.Porter JA, Minke B, Montell C. Calmodulin binding to Drosophila NinaC required for termination of phototransduction. Embo J. 1995;14:4450–9. doi: 10.1002/j.1460-2075.1995.tb00124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen S. M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 20.Goto A, Blandin S, Royet J, Reichhart JM, Levashina EA. Silencing of Toll pathway components by direct injection of double-stranded RNA into Drosophila adult flies. Nucleic Acids Res. 2003;31:6619–23. doi: 10.1093/nar/gkg852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn CS, Hahn YS, Braciale TJ, Rice CM. Infectious Sindbis virus transient expression vectors for studying antigen processing and presentation. Proc Natl Acad Sci U S A. 1992;89:2679–83. doi: 10.1073/pnas.89.7.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.