Summary
GATA transcription factors regulate transcription during development and differentiation by recognizing distinct GATA sites with a tandem of two conserved zinc fingers and by mediating long-range DNA looping. However, the molecular basis of these processes is not well understood yet. Here, we determined three crystal structures of the full DNA binding domain (DBD) of human GATA3 protein, which contains both zinc fingers, in complex with different DNA sites. In one structure, both zinc fingers wrap around a palindromic GATA site, cooperatively enhancing the binding affinity and kinetic stability. Strikingly, in the other two structures, the two fingers of GATA DBD bind GATA sites on different DNA molecules, thus bridging two separate DNA fragments, which is confirmed in solution by an in-gel FRET analysis. These findings not only provide new insights into the structure and function of GATA proteins, but also shed light on the molecular basis of long-range gene regulation.
Introduction
The GATA binding proteins are a group of structurally related transcription factors that bind to the DNA consensus sequence GATA. Members of the GATA protein family (GATA1-6) function as lineage-specific transcription factors for a number of cell types in the hematopoietic system (Molkentin, 2000; Patient and McGhee, 2002; Weiss and Orkin, 1995). For example, GATA1 is essential for erythroid and megakaryocytic development (Ferreira et al., 2005; Shivdasani et al., 1998); GATA2 plays an essential role in regulating transcription of genes involved in the development and proliferation of hematopoietic (Tsai and Orkin, 1997) and endocrine cell lineages (Dasen et al., 1999); GATA3 is an important regulator of T-cell development including Th2 (George et al., 1994; Ho et al., 1991; Zheng and Flavell, 1997) and regulatory T cells (Wang et al., 2011), and plays an important role in endothelial cell biology (Song et al., 2009); GATA4 regulates genes involved in embryogenesis and in myocardial differentiation and function (Watt et al., 2004).
The function of GATA proteins depends critically on two highly conserved zinc fingers and nearby basic regions (Shimizu et al., 2001). The C-terminal “C-finger” and its adjacent basic region are necessary and sufficient for GATA to bind its cognate sequence WGATAR (W=A/T, R=A/G) (Ko and Engel, 1993; Merika and Orkin, 1993; Omichinski et al., 1993; Visvader et al., 1995). The N-terminal “N-finger” can also bind DNA independently, but has a preference for GATC core motifs (Martin and Orkin, 1990; Newton et al., 2001; Pedone et al., 1997). Both fingers participate in binding the palindromic GATA motif ATCWGATA (W=A/T), resulting in markedly increased affinity (Trainor et al., 1996). Recent ChIP-seq experiments on GATA1, GATA2, and GATA3 have shown that GATA proteins mostly bind to a single GATA site ((A/T)GATAA) or a palindromic site (catctGATAAG) (Fujiwara et al., 2009; Horiuchi et al., 2011; Wei et al., 2011; Yu et al., 2009).
One mechanism by which GATA proteins contribute to transcriptional regulation is by facilitating chromosome looping, thereby mediating long-range control of gene expression in the nucleus. For example, GATA1 and its cofactor FOG-1 directly occupy looped enhancers as well as target gene promoters at the β–globin locus (Vakoc et al., 2005). Similarly, GATA3 has been shown to be important for the establishment and/or maintenance of long-range chromatin interactions at the Th2 cytokine locus (Spilianakis et al., 2005). Moreover, exchange of GATA1 and GATA2 has been shown to mediate transitions in looped chromatin organization of the KIT gene that is expressed during early erythropoiesis (Jing et al., 2008). However, the molecular mechanism of how GATA and FOG proteins mediate loop formation remains unclear.
Structural studies based on both NMR spectroscopy and X-ray crystallography have characterized the DNA binding mechanisms of the GATA C-finger (Bates et al., 2008; Omichinski et al., 1993; Starich et al., 1998). In addition, the structure of the GATA1 N-finger bound to the FOG-1 zinc finger 1 has been characterized using NMR (Liew et al., 2005). However, the DNA binding mechanisms of the GATA N-finger, the mechanisms by which the GATA double fingers bind to palindromic GATA sites, and the mechanisms by which GATA proteins loop DNA are not yet fully understood.
Here, we present the crystal structures of the full DNA-binding domain of GATA3 bound to different DNA sequences. These structures reveal that GATA zinc fingers can bind DNA using two different modes: the “wrapping ” mode wherein the two zinc fingers wrap around a single palindromic GATA site, and the “bridging” mode wherein the two zinc fingers bridge two distant sites on DNA, and in our case sites on different DNA molecules. Our results reveal the structural basis of how GATA fingers can bind distinct cis-regulatory elements in different conformations and how GATA proteins may participate in chromosome looping by bridging separated DNA molecules. These insights will help to understand gene regulation by GATA proteins and provide a basis for further studies of the complex functions of GATA proteins in vivo.
Results
Crystallographic study of GATA3 tandem zinc fingers bound to DNA
To better understand the mechanism by which GATA proteins recognize and bind DNA, we carried out crystallographic studies of the DNA binding domain (DBD) of the human GATA3 protein (amino acids 260–370) bound to DNA. GATA DBD contains two zinc fingers that are highly conserved within the GATA family (GATA1-6). Recent studies show that 40% of the GATA1 ChIP-seq peaks are palindromic GATA sites (Yu et al., 2009). Given the fact that GATA family members share a highly conserved DBD, it is reasonable to assume that all GATA proteins could bind palindromic GATA sites, although the binding preference and affinity may vary among different GATA members. To understand how GATA proteins bind these palindromic GATA sites, we tested different palindromic sequences with variable spacers between the two GATA sites in our crystallization attempts. Two DNA fragments crystallized successfully with the GATA3 DBD. In one DNA fragment, the two GATA sites are separated by one base pair. This fragment, which is derived from the mouse GATA1 promoter, contains the consensus palindromic motif defined by ChIP-seq, CATCTGATAAG. This complex (Complex 1) was solved at 2.8 Å resolution. In the other DNA fragment, the two GATA sites are separated by three base pairs. This DNA fragment crystallized with the GATA3 DBD in two different crystal shapes, one with a bar-shaped morphology and the other in plate shape. The bar-shaped crystal belongs to the P21 space group (Complex 2), while the plate-shaped crystal belongs to the C2 space group (Complex 3). The bar-shaped crystal structure was solved at 2.65 Å, while the plate-shaped crystal structure was solved at 1.6 Å. In total, we have solved three crystal structures of GATA3 DBD/DNA complexes. The structures of complexes 2 and 3 were solved by SAD phasing using zinc anomalous signal. We also found molecular replacement solution using previously published coordinates of the GATA3 C-finger (PDB 3DFX) as a partial search model. The structure of Complex 1 was solved by molecular replacement using the N-finger and C-finger determined in Complex 3 as partial search models. The X-ray crystallography statistics for all complexes are presented in Supplementary Table 1. Crystal packing and related analyses of each complex are described in Supplementary Figure 1.
Overall structure of GATA bound to a palindromic DNA site
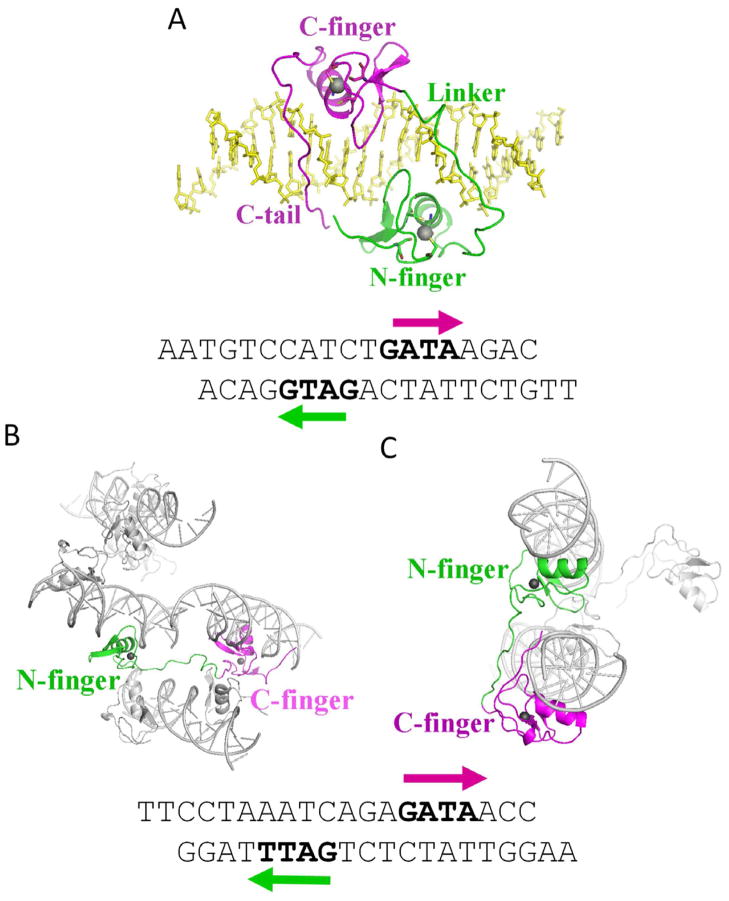
In Complex 1, GATA3 DBD binds to a 20-mer palindromic GATA site, AATGTCCATCTGATAAGACG (binding sites are underlined; Figure 1A lower panel). The crystal belongs to space group P21, wherein each asymmetric unit contains one GATA3 molecule bound to a double-stranded DNA (dsDNA) molecule (Figures 1A). The N-finger (amino acids 264–288, green) binds to the GATG site (CATC in complementary strand). The peptide region immediately following the N-finger (amino acids 289–317, green), which also serves as the linker to the C- finger (amino acids 318–342, magenta), tracks along one strand of the phosphodiester backbone and connects to the C-finger bound to the GATA site. The C-terminal basic region (amino acids 343–368, magenta) of the C-finger inserts deeply into the minor groove of the GATA site. Interestingly, the N- and C-terminus of the GATA3 DBD come close in space in this palindrome-bound form, rendering the protein in a shape of a parallelogram (Figure 1A). The DNA is in essentially straight B-form with a slightly widened major groove at the center of the palindromic site and an accompanying compression of the minor groove at the GATA site, presumably due to the insertion of the basic region of the C-terminal tail of the C-finger into the minor groove (Figure 1A). Our analysis demonstrates that the narrow minor groove correlates with enhanced negative electrostatic potential (Supplementary Figure 2). This effect of DNA shape-dependent electrostatic potential stabilizes the interaction with C-terminal arginines (Rohs et al., Nature 2009). Together, the two fingers and their respective C-terminal extensions form a C-shaped clamp that wraps around the DNA along the major groove of the palindromic site, which leads to an extensive shape and electrostatic potential complementarity between the GATA DBD and its target site (Harris et al., 2012).
Figure 1.
Overall structures of GATA3/DNA complexes. (A) The structure of Complex 1. The N-finger and the linker are colored in green, the C-finger and C-tail are colored in magenta, and the DNA is colored in yellow. (B) The structure of Complex 2. Complexes of GATA3 N-/C-fingers bound to DNA from neighbouring asymmetric units are shown to illustrate one GATA3 protein bridging two pieces of DNA. One N-finger is colored in green, the C-finger of the same GATA protein is colored in magenta, while all the other molecules are colored in gray. (C) on the structure of Complex 3. Again, complexes of GATA3 N-/C-fingers bound to DNA from neighbouring asymmetric units are shown to illustrate DNA bridging by GATA3 protein, and the same color scheme is used as described in (B). The sequences of the DNA are listed below. (See also Supplementary Figure S1 and Supplementary Table S1).
Structures of GATA bridging two separate DNA molecules
In an effort to crystallize GATA3 bound to palindromic GATA sites separated by different spacers, we unexpectedly solved two crystal structures of the GATA3 DBD bridging two double-stranded (ds) DNA molecules. In both complexes, the GATA3 DBD was co-crystallized with a palindromic site with two GATA binding sites separated by three base pairs, TTCCTAAATCAGAGATAACC (binding sites underlined; Figures 1B and 1C). One complex belongs to the P21 space group, where the asymmetric unit contains two GATA3 DBDs and two dsDNA molecules (Complex 2). In contrast to the one-spacer palindromic site, here the N-finger (green) and C-finger (magenta) of one GATA molecule bind to their cognate sites on two separate dsDNA molecules, which are parallel to each other in the crystal lattice (Figure 1B). Specifically, the C-finger binds to the GATA site of one dsDNA molecule, while the N-finger binds to the GATT site of another dsDNA molecule. The two DNA molecules are separated by 35 Å (axis to axis). In another complex, the protein and DNA were co-crystallized in the C2 space group, with the asymmetric unit containing one copy of GATA3 DBD and one copy of dsDNA molecule (Complex 3). Here again the C-finger of the GATA3 DBD binds to the GATA site of one dsDNA molecule, while its N-finger binds to the GATT site on a separated, symmetry-related dsDNA molecule (Figure 1C). The two dsDNA molecules are arranged in a relative angle of approximately 30° in the crystal lattice. The minimal axis-to-axis distance between the two DNA molecules is 29 Å. In Complexes 2 and 3, the relative orientations of the N-finger and C-finger are very different, reflecting the large conformational flexibility of the linker between the N-finger and C-finger and the different arrangement of the DNA molecules that can be accommodated by GATA3-mediated bridging.
Protein-DNA interactions
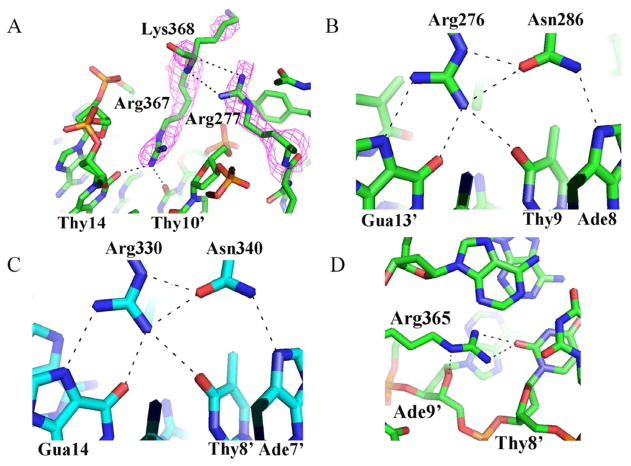
The GATA DBD contains two zinc fingers that can each bind its respective target site in a modular fashion. In Complex 1, the two fingers interact to bind adjacent sites in the palindromic motif cooperatively. In Complexes 2 and 3, the two fingers bind their target sites on separated dsDNA molecules inependent. Although the relative spatial arrangements of N-finger and C-finger vary substantially in different DNA binding mode, the detailed interactions between each zinc finger and its cognate DNA sites are largely conserved in diferent complexes. In all three complexes, the C-finger binds the consensus GATA site in a highly conserved mode that is also seen in the isolated C-finger/DNA complex (Bates et al., 2008). The conservation of the DNA binding mechanisms by the C-finger not only includes the detailed interactions of the core zinc module (amino acids 316–349) with the major groove but also the binding of the C-terminal basic tail (amino acids 350–366) to the minor groove. The detailed interactions in the minor groove, however, vary slightly between different complexes. For instance, in Complexes 2 and 3, Arg367 exhibits very weak density and is therefore not included in the final model; however, in Complex 1, Arg367 shows clear density and forms a hydrogen bond with both Thy14 and Thy10′ (Figure 2A). This difference could be due to the interactions with the N-finger that stabilize the conformation of the basic tail of the C-finger and facilitate its binding to DNA in the minor groove. As shown in Figure 2A, Arg277 of the N-finger interacts with the main chain of Lys368, while Met260 forms a van der Waals contact with Thr363 (not shown). In contrast, in Complexes 2 and 3, where the N-finger is located on a separate DNA molecule, the basic tail of the C-finger is less well ordered in the tip region including residue Arg367.
Figure 2.
DNA recognition by the GATA3 DBD. (A) Arg367 binds DNA only in the wrapping mode (Complex 1). These interactions are stabilized through the interaction between the Arg277 guanidinium group and the Lys368 backbone, reducing the conformational flexibility of the C-terminal tip. The electron density is calculated from a composite omit map. (B) Hydrogen bonding interactions among Arg276, Asn286 (N-finger) and three base pairs (GAT) of the binding site at their major groove core. These hydrogen bonds, in particular the bidentate hydrogen bond of the Arg276 guanidinium group with guanine, lead to the specific recognition of the base pair identity. (C) A network of hydrogen bonding interactions, similar to the one shown in (B), including a bidentate hydrogen bond, is formed between Arg330, Asn340 (C-finger), and three base pairs (GAT) of the binding site at their major groove core. (D) Arg365 forms hydrogen bonds with the minor groove edge of a base. Since these base readout interactions in the minor groove are not as specific as in the major groove, the negative electrostatic potential enhanced through the narrow minor groove stabilizes interactions with argine residues (shape readout; see also Supplementary Figure S2).
Although the N-finger structure has been solved in complex with a zinc finger of the FOG1 protein by NMR spectroscopy (Liew et al., 2005), the mechanism by which the N-finger recognizes DNA has not been elucidated structurally yet. In all three structures, the zinc core module of the N-finger binds to the first three nucleotides (GAT) in a similar manner as observed for the C-finger core. For example, Arg276 and Asn286 interact with Gua13′, Thy9, and Ade8 (Figure 2B), in an almost identical manner to Arg330 and Asn340 in the C-finger (Figure 2C).
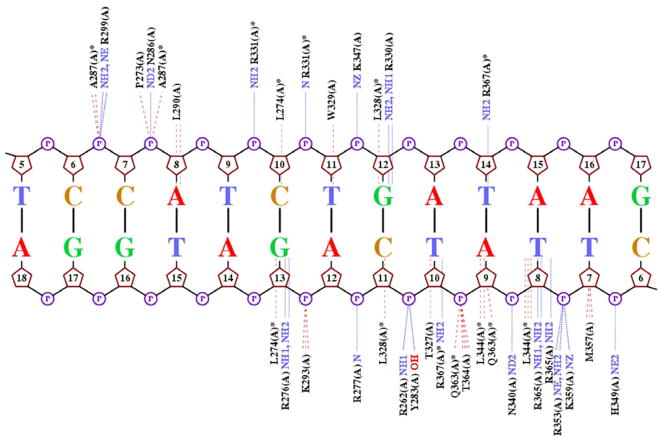
The major difference of DNA binding between the N-finger and the C-finger lies in the recognition of the fourth and variable position in their binding sites GATN (N refers to any nucleotide). The C-terminal basic tail of the C-finger inserts into the minor groove and, most importantly, Arg365 forms a hydrogen bond with the carbonyl of Thy8′, and also engages in extensive van der Waals contacts with neighboring bases and sugar moieties (Figure 2D). In addition, the enhanced negative electrostatic potential in this region of the minor groove attracts the positive charge located in the arginine’s guanidinium group (Supplementary Figure 2). The electrostatic potential at this position is about −8.5 kT/e due to the electrostatic focusing effect in narrow minor groove regions (Rohs et al., Nature 2009). Similar interactions are observed for the Arg367 of the C-finger in Complex 1, including hydrogen bonds with Thy14 and Thy10′ and an electrostatic potential of about −9.1 kT/e (Supplementary Figure 2A). Thus both Arg365 and Arg367 play a key role in DNA binding both through base-specific hydrogen bonding (base readout) and shape-dependent electrostatic interactions (shape readout) (Rohs et al., 2010; Rohs et al., 2009). Most of the minor groove interactions by the C-finger described above are missing in the N-finger. The detailed DNA-binding interactions by the N-finger and C-finger in Complex 1 are showon in Figure 3, which also represent most of the DNA-binding interactions by individual zinc fingers in Complexes 2 and 3 (not shown). Overall, compared to the C-finger, the N-finger makes fewer contacts to DNA both with the bases and phosphodiester backbone (Figure 3), and these differences may explain their difference in DNA binding affinity and specificity (Bates et al., 2008).
Figure 3.
Schematic representation of DNA contacts with GATA3 amino acids in Complex 1 on the palindromic site. The hydrogen bonds are indicated by blue dotted lines, while hydrophobic contacts are indicated by red dash lines. The protein/DNA contact diagram is generated by NUCPLOT (Luscombe et al., 1997). The contact map demonstrates that GATA3 contacts the DNA in both the major and minor groove and employs base and shape readout mechanisms.
The linker region
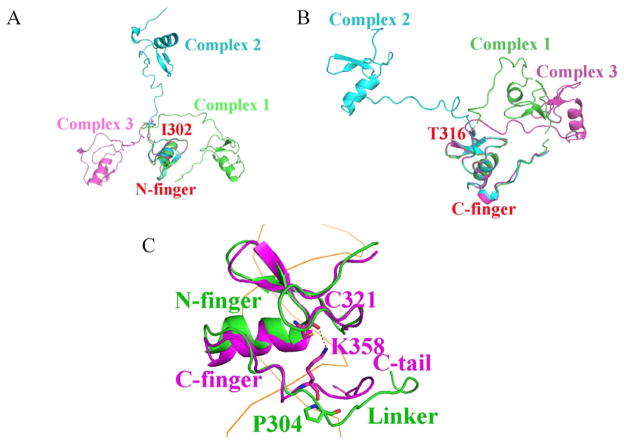
The linker region in all three crystal structures shows a large flexibility as evident by high B-factors and a weak electron density. Although the electron density for the main chain of the linker region is visible in Complex 1, the side chains are not defined (Supplementary Figure S3A). This observation suggests that the linker region makes little contribution to DNA binding in the “wrapping” model. In Complexes 2 and 3, there are three unique copies of the linker region in total, all of which show densities for the backbone but not the side chains (Supplementary Figures S3B and S3C). A superposition of the N-finger of all three molecules shows the linker region of these structures diverges at Ile302 and extends in different directions (Figure 4A). Similarly, superposition of the C-finger shows different paths of the linker region converging at Thr316, immediately in front of the C-finger (Figure 4B). These structural analyses suggest that the linker region between the N-finger and the C-finger in GATA3 does not appear to have any conformational limitations. One of its functional roles appears to be providing a constraint on the spacing of the palindromic sites and the geometric distance between the DNA molecules bridged by GATA, while the flexibility of the linker region allows a wide variety of orientations of the bridged DNA molecules.
Figure 4.
Superposition of zinc finger motifs displaying the varying linker conformations. In one representation (A), the N-fingers from the three complexes are superimposed, while in a different representaion (B), the C-fingers from the three complexes are superimposed. (C) Superposition of the N-finger and C-finger in Complex 1. P304 push the linker region away from the minor groove, while K358 pull the C-tail close to the minor groove throuth its interaction with C321. The DNA is visualized as orange ribbon. (see also Supplementary Figures S3 and S4).
It is somewhat surprising that the linker region does not bind to DNA in the minor groove in Complex 1 as the C-terminal tail of the C-finger (C-tail) does. The linker region is similar to the C-tail at sequence level as it has multiple basic residues (Supplementary Figure S4). The major difference lies between P304 and K358. In the C-tail, the side chain of K358 interacts with the main chain of C321, and this interaction pulls the tail close to the minor groove; while in the linker region, P304 cannot interact with C267, and pushes the linker away from the minor groove (Figure 4C).
DNA binding analyses
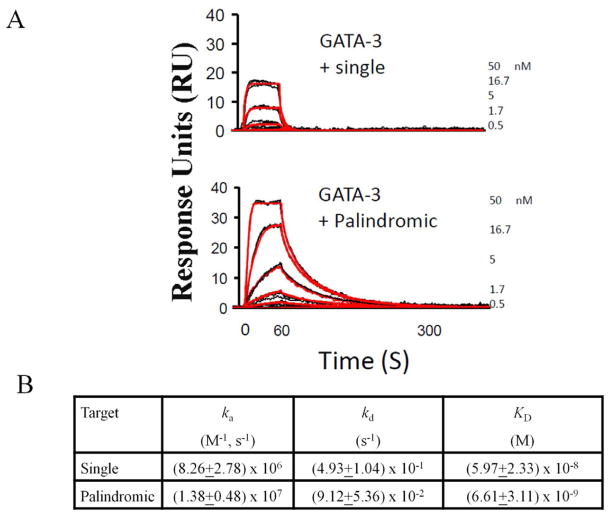
Although the GATA N-finger and C-finger alone can bind DNA, a previous study showed that both fingers of GATA1 participate in binding palindromic GATA sites, resulting in markedly increased affinity (Trainor et al., 1996). Consistent with this biochemical finding, a recent ChIP-seq study shows that the palindromic GATA site accounts for 40% of GATA1 enrichment peaks and that the palindromic GATA site has an increased peak height compared to the peak average (Yu et al., 2009). Our structure (Complex 1) shows that the two fingers of GATA3 bind opposite faces of the DNA and that the N-finger interacts with the C-terminal basic tail of the C-finger, thereby enhancing GATA/DNA interactions. These structural features suggest that the higher affinity of GATA DBD for the palindromic site compared to the single site is likely a result of more extensive protein/DNA contacts and the clamping mode of DNA binding by the two fingers of GATA, which could lead to enhanced kinetic stability of the protein-DNA complex (Stroud et al., 2002). To see if GATA3 also binds palindromic sites with higher affinity than single sites, we further analyzed the binding of GATA3 to DNA. We performed binding studies using surface plasmon resonance (SPR) on a Biacore 2000 instrument. DNA probes containing either a single or a palindromic GATA site were immobilized to the same low density on the surface of a biosensor chip. GATA3 protein was injected over the sensor chip surfaces to examine its interaction with the different DNA sites. A 1:1 interaction model with mass transport correction was applied to obtain the association (ka) and dissociation (kd) rates. The affinity was calculated from these values (KD = kd/ka). Biacore analyses revealed that GATA3 binds to DNA containing a single GATA site at relatively low affinity with a fast off rate (Figure 5A, top panel). In contrast, the binding of GATA3 to DNA containing the palindromic GATA site shows a markedly increased affinity, largely due to enhanced kinetic stability as evident by the slower off rate (p< 0.05) (Figure 5A, bottom panel). In addition, the response units at equilibrium binding of GATA3 to a palindromic GATA site was two-fold higher than the response when binding to a single GATA site, further confirming that GATA3 has a stronger affinity for the palindromic GATA site over the single site (Figure 5A). Fitting of the data indicates that the GATA3 DBD binds the palindromic site with a KD of about 6 nM, while it binds the single site with a KD of about 60 nM (Figure 5B). Therefore, the binding affinity of GATA3 DBD for the palindromic site is about 10-fold higher than that of the single site.
Figure 5.
Biacore analyses. (A) The sensorgrams showing kinetic analysis of GATA3 DBD with either single GATA site (top) or a palindromic site (bottom). Black lines represent triplicate injections performed in random order over the indicated DNA surface. A 1-min association was followed by a 5-min dissociation phase. Red lines represent the global fit of data sets using CLAMP. (B) Association rate constants ka, dissociation rate constants kd, and equilibrium constants KD=ka/kd demonstrate the kinetics of GATA3 DBD binding to either a single or palindromic GATA site. Standard error of the mean is indicated. The dissociation rates of single and palindromic sites are significantly different (p<0.05).
DNA looping by GATA in solution
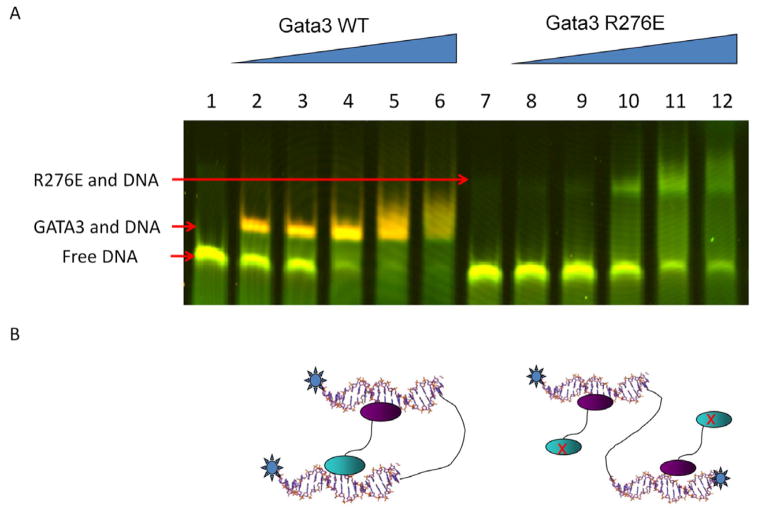
Our structures of Complexes 2 and 3 reveal that GATA3 DBD can bridge two DNA molecules. This mode of DNA binding may have important implications for long-range gene regulation by GATA proteins (Jing et al., 2008; Spilianakis et al., 2005; Vakoc et al., 2005). However, in order to exclude potential crystal packing effects, it is important to test if GATA can bind two segments of DNA in solution. For this purpose, we used FRET (Fluorescence or Förster Resonance Energy Transfer) to monitor the proximity of DNA ends in solution. We synthesized a 40-mer DNA that contains the binding site for the C-finger (GATA) at one end and the N-finger (GATC) at the other end. The two ends of the DNA were labeled by fluorophore Cy3 (donor) and Cy5 (acceptor), respectively. The central region of the DNA is a stretch of single-stranded DNA made of poly(dT). This design was chosen to reduce non-specific binding and increase DNA flexibility. Based on our crystal structures (Figures 1B and 1C), simultaneous binding of GATA3 to both sites on the DNA would bring the two DNA ends together, thus allowing FRET to occur when the donor is excited. However, detection of FRET signal in solution does not necessarily indicate DNA looping because two different DNA molecules may be brought together by non-specific interactions. To address this issue, we developed an in-gel FRET assay so that we can measure the FRET signal for a given complex with a well-defined mobility shift. As shown in Figure 6A (lanes 1–6), the FRET signal as indicated by the color change in the GATA3/DNA complex is reproducibly and significantly stronger than that in free DNA, suggesting that the DNA ends are indeed closer in the GATA3-bound complex. The tight band and fast mobility suggest that the FRET signal most likely arises from GATA3-mediated looping rather than binding of multiple DNA molecules. To further test this, we mutated the N-finger (R276E) to disrupt its DNA-binding activity. Compared with the wild-type protein, the mutant showed lower DNA binding affinity, presumably due to the loss of avidity; and more importantly, the mutant complex had similar FRET signal as free DNA, suggesting an open DNA structure (Figure 6A, lanes 7–12). These results are consistent with a model wherein the mutant GATA3 binds only one site with the C-finger in an open conformation (Figure 6B). Interestingly, the mutant-bound complex also showed slower mobility, consistent with a less compact structure and/or binding of two molecules of GATA3 as depicted in Figure 6B. The above studies strongly suggest that GATA3 can indeed loop DNA in solution.
Figure 6.
In-gel FRET analysis of DNA looping by GATA3 in solution. (A) GATA3 N-/C-fingers can bringe two binding sites into close proximity despite their separation in sequence. Pseudo-colored image showing a superposition of the fluorescence of the Cy3-donor (green) and the fluorescence of the Cy5-acceptor (red) fluorophores. Labels to the left indicate the respective protein/DNA species. Lanes 1–6 show the binding of wild-type GATA3 to DNA, while lanes 7–12 show the binding of a GATA3 mutant R276E, which abolishes N-finger DNA binding. (B) Cartoons represent models of the distinct overall architecture corresponding to the two protein shifts. The N-finger is displayed as a cyan oval and the C-finger as a purple oval.
Discussion
Here we present a comprehensive analysis of the DNA recognition mechanisms by the GATA protein in the context of its full DNA-binding domain, i.e. with both zinc fingers. Compared with previous studies with isolated GATA zinc fingers (Bates et al., 2008; Liew et al., 2005; Omichinski et al., 1993; Starich et al., 1998), the present study reveals novel aspects of DNA binding by GATA that have important implications for the function of GATA in transcriptional regulation.
It is known that GATA can bind palindromic sites with high affinity and that such binding may be functionally important (Trainor et al., 1996; Yu et al., 2002). However, it was not known until recently how prevalent this DNA binding mode of GATA is in vivo. Genome-wide analysis of GATA1 binding sites using ChIP-seq revealed that 40% of GATA1 binding sites follow the palindromic consensus motif. Moreover, ChIP-seq peaks from palindromic sites are significantly higher than those from isolated GATA1 sites (Yu et al., 2009), consistent with the higher affinity binding of GATA1 to palindromic sites (Trainor et al., 1996). These observations suggest that binding of GATA1 to palindromic sites plays a significant role in vivo (Yu et al., 2009). Although similar analysis on GATA3 has not yet been reported, it is possible that GATA3 also binds palindromic GATA sites. Our structure of the GATA3 DBD bound to the palindromic site presents the first example of such an important DNA binding mode by GATA proteins, which is, more generally, also a novel mode of DNA binding by a zinc finger protein. Unlike classical Cys2His2 zinc finger proteins, such as Zif268 and TFIIIA, the GATA proteins have a longer linker region allowing more complex and versatile DNA binding patterns (Stroud and Chen, 2003). Zif268 and TFIIIA have shorter linkers and use multiple zinc finger domains to wrap around DNA and track along the major groove (Elrod-Erickson et al., 1998; Wuttke et al., 1997). Our structure is also different from most other Cys4 zinc finger proteins, such as nuclear receptor proteins. Although most nuclear receptor proteins also have two Cys4 zinc finger motifs, they use one for DNA binding and the other for dimerization. In addition, nuclear receptors form dimers to bind the same side of the DNA (Bain et al., 2007). In our structure, which exhibits the “wrapping” mode (Complex 1), the N-finger binds to one side of DNA in the major groove, then tracks along the minor groove without DNA binding; the C-finger binds to the major groove on the other side of the double helix, tracks along the minor groove with extensive DNA binding; and the end of the C-tail reaches the N-finger and completes the “wrapping” architecture. Several structural features may account for the enhanced binding affinity of GATA to the palindromic sites. First, the N-finger and C-finger bind to adjacent sites on the DNA, making extensive protein-DNA contacts. Second, the binding of a zinc finger induces conformational changes of the DNA, and such changes may facilitate the binding of the second zinc finger to the adjacent site. Third, the N-finger and C-finger interact directly on palindromic sites, and these protein-protein interactions may lead to cooperative binding of the two fingers to DNA, similar to the cooperative binding of GATA C-fingers to adjacent sites (Bates et al., 2008). Finally, the N-finger and C-fingers, in conjunction with their C-terminal extensions, wrap around DNA in the palindromic complex, and such encirclement of DNA by the protein usually leads to enhanced kinetic stability of the protein/DNA complex (Stroud et al., 2002). Our Biacore analysis suggests that GATA3 DBD indeed binds the palindromic site with a significantly slower off rate than the single site.
The GATA family of proteins contains two zinc fingers that can each bind to a separate target site. Although Complex 1 reveals that the two fingers can function jointly to recognize the palindromic sites, the majority (60%) of GATA1 response elements is comprised of single GATA sites (Yu et al., 2009). Binding of these isolated GATA sites by the N-finger and C-finger of GATA proteins could lead to chromosome looping or inter-chromosome interactions. This is a very attractive model of transcriptional regulation by GATA proteins given the increasing evidence implicating a role of GATA proteins in long-range gene regulation (Jing et al., 2008; Spilianakis et al., 2005; Vakoc et al., 2005). Complexes 2 and 3 reveal that GATA3 DBD can indeed bridge two dsDNA molecules, thereby providing a direct mechnism of chromosome looping. These structures, to our knowledge, are the first crystal structures of a single protein bridging two separate DNA fragments. Several lines of evidence support that the DNA bridging by GATA protein is not an artifact. First, we observed the DNA bridging in two crystal structures with different overall architecture; second, in-gel FRET experiments provided further evidence that the GATA DBD could bridge DNA in solution. We would like to provide functional relevance of this DNA bridging in cells, however, there is no method, to our knowledge, that can provide direct evidence that a GATA protein can bridge two separate DNA molecules in cells. Given the fact that it is well known that GATA proteins mediate long-range gene regulation, we believe that our model, represents at leaast one of the mechanisms for GATA proteins to loop DNA in cells. In addition to the direct DNA bridging, GATA proteins may loop DNA through indirect mechanisms, for instance via their co-factors, such as FOG proteins, which have also been shown to play important roles in mediating DNA loop formation (Vakoc et al., 2005). We have recently shown that FOXP3, a transcription factor critical to the function of regulatory T cells, also bridges DNA to mediate DNA looping (Bandukwala et al., 2011). However, the mechanisms of DNA bridging by FOXP3 and GATA3 differ in two aspects. First, DNA bridging by FOXP3 occurs through a domain-swapped dimer whereas GATA3 bridges DNA through two covalently linked DNA-binding motifs of the same protein. Second, the domain-swapped dimer of FOXP3 has a rigid structure which bridges two DNA molecules in a fixed orientation, whereas the flexible linker between the N- and C-finger in GATA allows different orientations of bridged DNA molecules, as seen in Complexes 2 and 3. These structural differences may reflect differences in long-range gene regulation by FOXP3 and GATA3. In spite of these differences, DNA bridging appears to be a common property of many transcription factors, including FOXP2 (Stroud et al., 2006), MEF2 (Guo et al., 2007), FOXP3 (Bandukwala et al., 2011), and GATA3 discussed here. Notably these transcription factors are all implicated in lineage control during cellular differentiation. We thus propose that one mechanism to control epigenetic expression patterns of a given cell is through specific folding of the 3D structure of the genome by transcription factors that control a given lineage (Kalhor et al., 2012).
We have characterized two distinct DNA binding modes of GATA transcription factors: the “wrapping” mode, in which both zinc fingers synergistically enhance the binding affinity and kinetic stability; the “bridging” mode, in which a single GATA DBD bridges two pieces of DNA. Since the DBDs of GATA proteins are highly conserved, the structural features described here likely hold true for all six GATA family members. The fact that GATA can bind DNA in two distinct modes with different binding affinity/kinetic stability and conformations has important implications for transcriptional regulation by this family of proteins. For example, varying protein concentrations of GATA proteins during development may not just affect their occupancy of DNA, but also could switch their DNA binding mode and affected transcriptional networks (Georgescu et al., 2008). At low expression levels, the GATA proteins may preferentially bind to the palindromic sites; at high expression levels, the GATA proteins might be able to bind both palindromic sites and single sites, leading to more intra-chromosomal and inter-chromosomal interaction, either by themselves or through their interaction with cofactors, such as the FOG protein. The different DNA-bound conformations of GATA proteins may recruit different cofactors or change local chromosomal conformation, leading to different biological functions.
Materials and methods
Sample Preparation and Crystallization
The sequence containing both the N- and C-terminal zinc finger of human GATA3 (amino acids 260–370) was cloned into the pET-28a vector as a 6× His-tagged fusion protein and was expressed in Rosetta (DE3) pLysS cells. The protein was first purified by Ni-NTA beads and then digested by thrombin protease to remove the His-tag. The protein was further purified by Mono S cation exchange and Superdex 75 size exclusion column (Amersham Biosciences, Piscataway, NJ). The protein was then concentrated to approximately 40 mg/ml in 10 mM HEPES (pH 7.63), 5 mM β-mercaptoethanol, 0.5 μ ZnAcetate, 100 mM NaCl, 200 mM NH4Acetate, and 20% glycerol and stored at −80°C. DNA was synthesized by Integrated DNA Technologies (Coralville, IA). The DNA sequences are listed in Figure 1.
The protein/DNA complex was prepared by mixing protein and DNA at 1:1 molar ratio. Crystals were grown by the hanging drop method at 18°C using a reservoir buffer of either 400 mM NH4(OAc), 50 mM Acetate (pH 4.7), and 18% PEG 4K (for Complex 1), or 400 mM NH4(OAc),10 mM Mg(OAc)2, 50mM Cacodylic acid (pH 6.33), and 15% PEG 4K (for Complexes 2 and 3).
Data Collection and Structure Determination
Crystals were stabilized in the crystallization buffer with 25% (w/v) glycerol and flash frozen with liquid nitrogen for cryocrystallography. Data were collected at the ALS BL8.2.1, BL8.2.2 beamline at the Lawrence Berkeley National Laboratory. Data were reduced using HKL2000 (Otwinowski and Minor, 1997). Molecular replacement solutions for Complexes 2 and 3 were found using the coordinates of the C-finger (PDB 3DFX) (Bates et al., 2008) as a partial search model. Independent phases for Complexes 2 and 3 were also obtained using SAD phasing by Zinc anomalous signal, which cross-validated the molecular replacement solutions. Complex 1 was determined solely by molecular replacement, using the N-finger and C-finger obtained from Complex 3 as partial search models. Refinement were done using CNS refine (Brunger et al., 1998), CCP4 refmac5 (CCP4, 1994), Phenix.refine (Adams et al., 2002). Model building and analysis were carried out using Phenix.autobuild (Adams et al., 2002) and O (Jones et al., 1991). The statistics of the crystallographic analysis are presented in Supplementary Table 1. Graphical representations of structure were prepared using PyMol (DeLano Scientific, San Francisco, CA).
Biosensor Analysis
Binding experiments were performed on a BIACORE 2000 instrument (Biacore Inc., Piscataway, NJ, USA). Double-stranded DNA oligos containing either a palindromic GATA site (5′-Bi-CTCCCGCTCGCTATCAGATAAGGCCTTAT-3′ and 5′-ATAAGGCCTTATCTGATAGCGAGCGGGAG-3′) or a single GATA site (5′-Bi-CTCCCGCTCGCTCAGAGATAAGGCCTTAT-3′ and 5′-ATAAGGCTTTATCTCTGAGCGAGCGGGAG-3′) were synthesized by Integrated DNA Technologies (Coralville, IA). One strand of each pair carried a 5′-biotin tag (Bi) to allow coating on streptavidin-coated sensorchips (SA chip, GE Healthcare). Two comparable density surfaces were generated using DNA containing the palindromic GATA site and the single GATA site on two distinct flow cells of a single sensorchip. GATA3 was serially diluted in four three-fold steps from 50 nM to 0.2 nM. The five concentrations of protein samples were injected at 20°C over the chip surface, using 1-min injections followed by a 5-min dissociation. Samples with different concentrations of protein were injected in random order and every injection was performed in triplicate within each experiment. All experiments were done at least three times. Data were processed using Scrubber and analyzed using CLAMP XP (Myszka and Morton, 1998). The data was fit globally using a simple 1:1 Langmuir interaction model with a correction for mass transport (Myszka et al., 1998). The results for differential protein/DNA binding strengths were compared using the Student’s t-test. Equal or unequal variance of the samples was determined using the F-test. Mean association and dissociation rates were used to calculate the equilibrium binding constants, and the standard error of the mean values in the ka and kd were used to compute the error in the mean KD values reported in Figure 5B.
In-gel FRET Analysis
The ssDNAs labeled with either Cy3 (containing a GATA site) or Cy5 (containing a GATC site) were synthesized by IDT and purified by HPLC. A 40 base pair oligonucleotide, which anneals to both Cy3 and Cy5 oligos, was also synthesized by IDT. The DNA was annealed at a 1:1:1 ratio of DNAs. The resulting probe, labeled at the 5′ and 3′ ends with the FRET-pair Cy3 and Cy5, respectively, consisted of two double-stranded DNA regions at the ends and a 20 nucleotide poly(dT) region at the center. The DNA was incubated with GATA3 or R276E mutant protein for 25 minutes. A native 6% (w/v) polyacrylamide gel in 0.5×TBE buffer was used to resolve the free DNA from the protein/DNA complex. The gel was scanned with a Typhoon 8610 variable mode imager (Amersham Biosciences) at excitation wavelength of 532nm, which excited Cy3, and the fluorescence images were detected at emission wavelength of 580nm for Cy3 and 670nm for Cy5, respectively. The Cy3 image was assigned in green and Cy5 image in red. The two images were superpositioned resulting in the final image.
Computational analysis
Electrostatic potential was calculated with DelPhi (Honig and Nicholls, Science 1995) based on the non-linear Poisson-Boltzmann equation, which was solved in five focusing steps at physiologic ionic strength I=0.145 (Rohs et al., Nature 2009). The electrostatic potential of the protein was mapped on the molecular surface. Although the protein side chains could not be resolved in Complex 1 due to flexibility, these residues were modeled for the electrostatic potential calculation in order to mimic the correct charges. The potential of the DNA was shown as an isopotential surface and plotted as a function of sequence in reference points at the center of the minor groove (Rohs et al., Nature 2009). DNA minor groove geometry was analyzed with the CURVES algorithm (Lavery and Sklenar, J. Biomol. Struct. Dyn. 1989).
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Liang Guo, Dr. Aidong Han, Grace Kim for expeirmental assistance and discussion. The authors would also like to thank ALS BCSB staff member Corie Ralston, Peter Zwart, and Kevin Royal for help with data collection. L.C. is supported by NIH grants GM064642 and GM077320. R.R. is supported in part by grant #IRG-58-007-51 from the American Cancer Society. Y.C. is partly supported by an NIH postdoctoral fellowship.
Footnotes
Accession numbers
The Protein Data Bank accession numbers for the atomic coordinates and structural factors are 4HCA, 4HC7 and 4HC9.
Author Contribution
Y.C, D.L.B. and L.C. designed research; Y.C. and D.L.B. purified protein and DNA; Y.C. and D.L.B. grew the crystals and collected diffraction data; Y.C., D.L.B and R.D. solved and refined the structures; P.C. and I.A.L. conducted the Biacore experiments; D.L.B performed In-gel FRET; A.C.D.M and R.R. performed the computational work; Y.C., D.L.B. and L.C. wrote the manuscript. All authors commented on and revised the manuscript.
Conflict of Interest
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Bain DL, Heneghan AF, Connaghan-Jones KD, Miura MT. Nuclear Receptor Structure: Implications for Function. Annual Review of Physiology. 2007;69:201–220. doi: 10.1146/annurev.physiol.69.031905.160308. [DOI] [PubMed] [Google Scholar]
- Bandukwala Hozefa S, Wu Y, Feuerer M, Chen Y, Barboza B, Ghosh S, Stroud James C, Benoist C, Mathis D, Rao A, et al. Structure of a Domain-Swapped FOXP3 Dimer on DNA and Its Function in Regulatory T Cells. Immunity. 2011;34:479–491. doi: 10.1016/j.immuni.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates DL, Chen Y, Kim G, Guo L, Chen L. Crystal Structures of Multiple GATA Zinc Fingers Bound to DNA Reveal New Insights into DNA Recognition and Self-Association by GATA. Journal of molecular biology. 2008;381:1292–1306. doi: 10.1016/j.jmb.2008.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dasen JS, O’Connell SM, Flynn SE, Treier M, Gleiberman AS, Szeto DP, Hooshmand F, Aggarwal AK, Rosenfeld MG. Reciprocal Interactions of Pit1 and GATA2 Mediate Signaling Gradient-Induced Determination of Pituitary Cell Types. Cell. 1999;97:587–598. doi: 10.1016/s0092-8674(00)80770-9. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M, Benson TE, Pabo CO. High-resolution structures of variant Zif268 DNA complexes: implications for understanding zinc finger DNA recognition. 1998;6:451–464. doi: 10.1016/s0969-2126(98)00047-1. [DOI] [PubMed] [Google Scholar]
- Ferreira R, Ohneda K, Yamamoto M, Philipsen S. GATA1 Function, a Paradigm for Transcription Factors in Hematopoiesis. Mol Cell Biol. 2005;25:1215–1227. doi: 10.1128/MCB.25.4.1215-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering Hematopoietic Mechanisms through Genome-wide Analysis of GATA Factor Chromatin Occupancy. Molecular cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George KM, Leonard MW, Roth ME, Lieuw KH, Kioussis D, Grosveld F, Engel JD. Embryonic expression and cloning of the murine GATA-3 gene. Development (Cambridge, England) 1994;120:2673–2686. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- Georgescu C, Longabaugh WJR, Scripture-Adams DD, David-Fung ES, Yui MA, Zarnegar MA, Bolouri H, Rothenberg EV. A gene regulatory network armature for T lymphocyte specification. Proceedings of the National Academy of Sciences. 2008;105:20100–20105. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Han A, Bates DL, Cao J, Chen L. Crystal structure of a conserved N-terminal domain of histone deacetylase 4 reveals functional insights into glutamine-rich domains. Proceedings of the National Academy of Sciences. 2007;104:4297–4302. doi: 10.1073/pnas.0608041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RC, Mackoy T, Dantas Machado AC, Xu D, Rohs R, Fenley MO. Innovations in Biomolecular Modeling and Simulations. The Royal Society of Chemistry; 2012. Chapter 3 Opposites Attract: Shape and Electrostatic Complementarity in Protein-DNA Complexes; pp. 53–80. [Google Scholar]
- Ho IC, Vorhees P, Marin N, Oakley BK, Tsai SF, Orkin SH, Leiden JM. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. The EMBO journal. 1991;10:1187–1192. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi S, Onodera A, Hosokawa H, Watanabe Y, Tanaka T, Sugano S, Suzuki Y, Nakayama T. Genome-Wide Analysis Reveals Unique Regulation of Transcription of Th2-Specific Genes by GATA3. The Journal of Immunology. 2011 doi: 10.4049/jimmunol.1100179. [DOI] [PubMed] [Google Scholar]
- Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Exchange of GATA Factors Mediates Transitions in Looped Chromatin Organization at a Developmentally Regulated Gene Locus. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotech. 2012;30:90–98. doi: 10.1038/nbt.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Molecular and cellular biology. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CK, Simpson RJY, Kwan AHY, Crofts LA, Loughlin FE, Matthews JM, Crossley M, Mackay JP. Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/Friend of GATA interaction. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:583–588. doi: 10.1073/pnas.0407511102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscombe NM, Laskowski RA, Thornton JM. NUCPLOT: A Program to Generate Schematic Diagrams of Protein-Nucleic Acid Interactions. Nucleic acids research. 1997;25:4940–4945. doi: 10.1093/nar/25.24.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DI, Orkin SH. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes & development. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Molecular and cellular biology. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- Myszka DG, Jonsen MD, Graves BJ. Equilibrium Analysis of High Affinity Interactions Using BIACORE. Analytical Biochemistry. 1998;265:326–330. doi: 10.1006/abio.1998.2937. [DOI] [PubMed] [Google Scholar]
- Myszka DG, Morton TA. CLAMP©: a biosensor kinetic data analysis program. Trends in Biochemical Sciences. 1998;23:149–150. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
- Newton A, Mackay J, Crossley M. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J Biol Chem. 2001;276:35794–35801. doi: 10.1074/jbc.M106256200. [DOI] [PubMed] [Google Scholar]
- Omichinski JG, Clore GM, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl SJ, Gronenborn AM. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science (New York, NY. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Current Opinion in Genetics & Development. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- Pedone PV, Omichinski JG, Nony P, Trainor C, Gronenborn AM, Clore GM, Felsenfeld G. The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. The EMBO journal. 1997;16:2874–2882. doi: 10.1093/emboj/16.10.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of Specificity in Protein-DNA Recognition. Annual Review of Biochemistry. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009;461:1248–1253. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu R, Takahashi S, Ohneda K, Engel JD, Yamamoto M. In vivo requirements for GATA-1 functional domains during primitive and definitive erythropoiesis. The EMBO journal. 2001;20:5250–5260. doi: 10.1093/emboj/20.18.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, McDevitt MA, Fujiwara Y, Orkin SH. Transcription factor GATA-1 in megakaryocyte development. STEM CELLS. 1998;16:79–83. doi: 10.1002/stem.5530160710. [DOI] [PubMed] [Google Scholar]
- Song H, Suehiro J-i, Kanki Y, Kawai Y, Inoue K, Daida H, Yano K, Ohhashi T, Oettgen P, Aird WC, et al. Critical Role for GATA3 in Mediating Tie2 Expression and Function in Large Vessel Endothelial Cells. Journal of Biological Chemistry. 2009;284:29109–29124. doi: 10.1074/jbc.M109.041145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Starich MR, Wikstrom M, Arst HN, Jr, Clore GM, Gronenborn AM. The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors. Journal of molecular biology. 1998;277:605–620. doi: 10.1006/jmbi.1998.1625. [DOI] [PubMed] [Google Scholar]
- Stroud JC, Chen L. Structure of NFAT Bound to DNA as a Monomer. Journal of Molecular Biology. 2003;334:1009–1022. doi: 10.1016/j.jmb.2003.09.065. [DOI] [PubMed] [Google Scholar]
- Stroud JC, Lopez-Rodriguez C, Rao A, Chen L. Structure of a TonEBP-DNA complex reveals DNA encircled by a transcription factor. Nat Struct Mol Biol. 2002;9:90–94. doi: 10.1038/nsb749. [DOI] [PubMed] [Google Scholar]
- Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006;14:159–166. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Trainor CD, Omichinski JG, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol Cell Biol. 1996;16:2238–2247. doi: 10.1128/mcb.16.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FY, Orkin SH. Transcription Factor GATA-2 Is Required for Proliferation/Survival of Early Hematopoietic Cells and Mast Cell Formation, But Not for Erythroid and Myeloid Terminal Differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among Distant Regulatory Elements at the [beta]-Globin Locus Requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Crossley M, Hill J, Orkin SH, Adams JM. The C-terminal zinc finger of GATA-1 or GATA-2 is sufficient to induce megakaryocytic differentiation of an early myeloid cell line. Molecular and cellular biology. 1995;15:634–641. doi: 10.1128/mcb.15.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Su Maureen A, Wan Yisong Y. An Essential Role of the Transcription Factor GATA-3 for the Function of Regulatory T Cells. Immunity. 2011;35:337–348. doi: 10.1016/j.immuni.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Abraham Brian J, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup Daniel L, Tang Q, Paul William E, et al. Genome-wide Analyses of Transcription Factor GATA3-Mediated Gene Regulation in Distinct T Cell Types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Experimental hematology. 1995;23:99–107. [PubMed] [Google Scholar]
- Wuttke DS, Foster MP, Case DA, Gottesfeld JM, Wright PE. Solution structure of the first three zinc fingers of TFIIIA bound to the cognate DNA sequence: determinants of affinity and sequence specificity. Journal of molecular biology. 1997;273:183–206. doi: 10.1006/jmbi.1997.1291. [DOI] [PubMed] [Google Scholar]
- Yu C, Niakan KK, Matsushita M, Stamatoyannopoulos G, Orkin SH, Raskind WH. X-linked thrombocytopenia with thalassemia from a mutation in the amino finger of GATA-1 affecting DNA binding rather than FOG-1 interaction. Blood. 2002;100:2040–2045. doi: 10.1182/blood-2002-02-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al. Insights into GATA-1-Mediated Gene Activation versus Repression via Genome-wide Chromatin Occupancy Analysis. Molecular cell. 2009;36:682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.