Abstract
The hookworm Necator americanus is the predominant soil-transmitted human parasite. Adult worms feed on blood in the small intestine, causing iron deficiency anaemia, malnutrition, growth and development stunting in children, and severe morbidity and mortality during pregnancy in women. Characterization of the first hookworm genome sequence (244 Mb, 19,151 genes) identified genes orchestrating the hookworm's invasion of the human host, genes involved in blood feeding and development, and genes encoding proteins that represent new potential drug targets against hookworms. N. americanus has undergone a considerable and unique expansion of immunomodulator proteins, some of which we highlight as potential novel treatments against inflammatory diseases. We also utilize a protein microarray to demonstrate a post-genomic application of the hookworm genome sequence. This genome provides an invaluable resource to boost ongoing efforts towards fundamental and applied post-genomic research, including the development of new methods to control hookworm and human immunological diseases.
Keywords: nematodes, hookworm, necatoriasis, blood-feeding, SCP/TAPS protein, immunoregulation, anti-inflammation, genome, RNA-Seq, protein microarray
INTRODUCTION
Soil transmitted helminths (STHs), including Ascaris, Trichuris and hookworms, cause neglected tropical diseases (NTDs) affecting >1 billion people worldwide1,2. Hookworms alone infect approximately 700 million people (primarily in disadvantaged communities in tropical and subtropical regions), causing a disease burden of 1.5-22.1 million disability-adjusted life years (DALYs)3. Necator americanus represents ~85% of all hookworm infections4 and causes necatoriasis, characterized clinically by anaemia, malnutrition in pregnant women and an impairment of cognitive and/or physical development in children5.
The life cycle of N. americanus commences with eggs being shed in the faeces of infected people. Eggs embryonate in soil under favorable conditions, and then the first-stage larvae hatch, feed on environmental microbes and moult twice to reach the infective third-stage larvae (iL3). These larvae infect the human host by skin penetration, enter subcutaneous blood and lymph vessels and travel via the circulation to the lungs. The iL3 break into the alveoli and migrate via the trachea to the oropharynx, after which they are swallowed and travel to the small intestine, where they develop to become dioecious adults. The adult worms (~1 cm long) attach to the mucosa, where they feed on blood (up to 30 μl per day per worm), and can survive in the human host for up to a decade. The pre-patent period of N. americanus is 4-8 weeks and a female worm can produce up to 10,000 eggs per day.
New methods to control hookworm disease are urgently needed. Presently, the treatment of hookworm disease relies mainly on mass treatment with albendazole6, but its repeated and excessive use has the potential to lead to treatment failures7 and drug resistance8. Recent indications of reduced cure rates in infected humans9 imply an urgent need for new interventions strategies. Early attempts to utilize bioinformatic approaches for the discovery of immunogens were hampered by a lack of understanding of the molecular biology of N. americanus and other hookworms4, and the absence of genome and proteome sequences. A recent study10 demonstrated that comparative genomics facilitates the characterization and prioritization of anthelmintic targets which results in a higher hit rate compared with conventional approaches.
In addition to a need for anti-hookworm vaccines in countries with high rates of hookworm infections, hookworms and other helminths are being explored as treatments (probiotics) against immunological diseases in humans in many industrialized countries where hookworm infections are not endemic11. Recent studies12-14 indicate that hookworms suppress the production of pro-inflammatory molecules and promote anti-inflammatory and wound healing properties, suggesting a mechanism by which worms reside for long periods in humans and suppress autoimmune and allergic diseases. Indeed hookworm recombinant proteins have been tested in clinical trials for non-infectious diseases15.
Herein we characterized the N. americanus genome and compared it with those of other nematodes and the human host. Bioinformatic analyses of the protein-coding genes identified salient molecular groups, some of which may represent new intervention targets. The production and screening of a hookworm protein microarray reveals novel findings on the immune response to the parasite and demonstrates a post-genomic exploration of the genome sequence, including identification of molecules with low similarity to proteins in other species but recognized by all infected individuals, therefore demonstrating high diagnostic potential.
RESULTS
Genome features
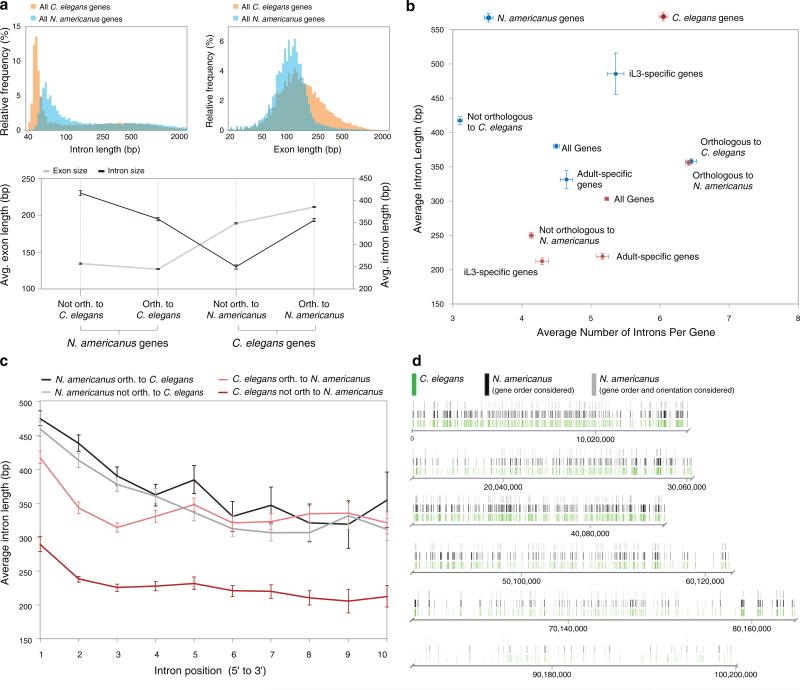
The nuclear genome of N. americanus (244 Mb) was assembled, with 11.4% (1,336) of the supercontigs (≥1 kb) comprising 90% of the genome. The 244 Mb sequence was estimated to represent 92% of the N. americanus genome (Table 1; Supplementary Fig. 1, 2 and 3; Supplementary Note). The GC content is 40.2%, the amino acid composition is comparable to other species (including 5 nematodes, the host and two outgroups; Supplementary Table 1) and the repeat content is 23.5%. In total, 669 repeat families were predicted and annotated (Supplementary Table 2, Supplementary Note). The protein-encoding genes predicted (n = 19,151) represent 33.7% of the genome at an average density of 78.5 genes per Mb and a GC content of 45.8%. Compared to C. elegans, N. americanus exons were shorter and the introns were longer (Fig. 1a), but the average intron length and count for genes orthologous between the two species was not significantly different (Fig. 1a and 1b; Supplementary Note). However, introns in C. elegans genes that are orthologous to N. americanus genes are significantly longer than introns in non-orthologous C. elegans genes (Fig. 1c), which may indicate a diversity of function for these genes, since longer introns are thought to contain functional elements that are present in addition to what might be regarded as ‘normal’ intron structure16. In addition, N. americanus iL3-overexpressed genes had longer introns than adult-overexpressed genes (Fig. 1b), which may indicate a greater diversity of regulation for these gene sets16. Positional bias was observed for intron length, which was comparable to C. elegans position-specific intron lengths for orthologous genes (Fig. 1c; Supplementary Note). Most genes (82.6%) were confirmed using RNA-Seq data from the iL3 and adult stages of N. americanus (two biological replicates per stage), and 6.5% and 3.7% were overexpressed in these stages, respectively (Supplementary Figs. 4 and 5, Supplementary Table 3). Alternative splicing was detected for 24.6% (4,712) of the genes, of which ~68.3% have orthologs in C. elegans. Among N. americanus genes with C. elegans orthologs, the alternatively spliced genes were more likely than other genes to belong to orthologous groups for which more than half of the CE genes were also alternatively spliced (p = 0.037, binomial distribution test). As expected, genes associated with alternative splicing had a higher number of exons than those without (p<10−15 and 2×10−7 for N. americanus and C. elegans, respectively). A total of 3,223 N. americanus genes were predicted to be trans-spliced, of which 818 had conserved gene order and orientation with 373 C. elegans operons (Fig. 1d; Supplementary Figs. 6 and 7; Supplementary Table 4; Supplementary Note). The genes within the operons had significantly more similar expression profiles to one another than to random subsets of non-operon genes (p<0.0001), supporting that they are co-transcribed under the similar regulatory control17.
Table 1.
Summary of N. americanus genomic features
| Estimated genome size (Mega bases) | 244 |
|---|---|
| Assembly statistics | |
| Total number of supercontigs (>=1 kb) | 11,713 |
| Total number of base pairs (bp) in supercontigs | 244,009,025 |
| Number of N50 supercontigs | 283 |
| N50 supercontig length (bp) | 213,095 |
| Number N90 supercontigs | 1,336 |
| N90 supercontig length (bp) | 29,214 |
| GC content of whole genome (%) | 40.20% |
| Repetitive sequences (%) | 23.50% |
| Protein-coding loci | |
| Total number of protein coding genes | 19151 |
| Avg. gene loci footprint (bp) | 4289 |
| Avg. # exons per gene | 6.4 |
| Avg. exon size (bp) | 125 |
| Avg. intron size (bp) | 642 |
| Avg. intergenic space (bp) | 6631 |
N50: number-50% of all nucleotides in the assembly are in 283 supercontigs, length-50% of the genome is in supercontigs with a minimum length of 213kb; N90: number-90% of all nucleotides in the assembly are within 1,336 supercontigs, length-90% of the genome is in supercontigs with a minimum length of 29kb.
Figure 1. N. americanus gene feature organization compared to C. elegans.
a, The average exon of N. americanus genes is significantly shorter and the average intron length is significantly larger than for C. elegans genes. b, Orthologous genes have significantly more introns than non-orthologous genes in both species. c, Introns are longer for orthologous genes in C. elegans at every intron position (compared to non-orthologous genes). In a-c, Error bars indicate standard error values. d, N. americanus genes in operons and conserved with C. elegans shown on the C. elegans chromosomes.
The N. americanus predicted secretome (classical 1,590 and non-classical secretion 4,785 proteins) represents 33% of the deduced proteome. Functional annotation of predicted proteins based on sequence comparisons identified 4,961 unique domains and 1,411 gene ontology (GO) terms for 57% and 44% of the N. americanus genes, respectively, and annotations are provided for 68% of the predicted N. americanus proteins (Supplementary Table 5).
Transcriptional differences between infective and parasitic stages
Hookworms spend a considerable amount of time as free-living larvae in the external environment before transitioning to parasitism. Gene expression differences between these stages reflect this developmental progression (Supplementary Table 3; Supplementary Fig. 5). Of the 1,948 differentially expressed genes, 36% were significantly overexpressed in iL3-, and 64% in adult-. Compared to iL3-overexpressed genes, nearly twice as many of the adult-overexpressed genes were N. americanus-specific (58% compared to 32%, p<10−15), suggesting that species-specific genes are more likely to be related to parasitism rather than to non-parasitic iL3 stage18.
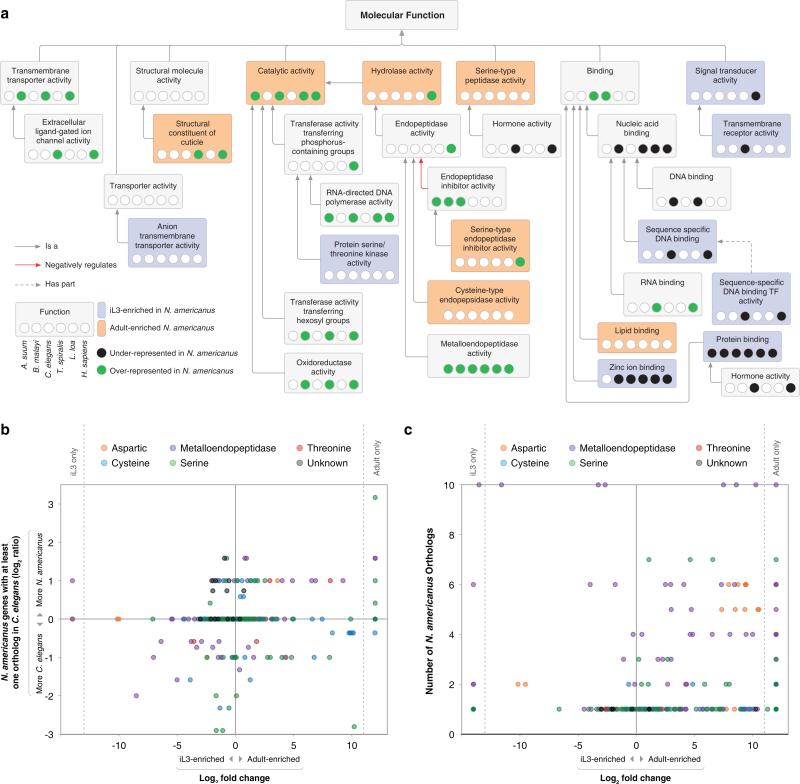
The iL3-overexpressed genes are over-represented (p<0.01) for eight molecular functions, including signal transduction, transmembrane receptor activity, and anion transporter activity, reflecting the ability of iL3 to adapt to a complex environment and infect a suitable host (Fig. 2a, Supplementary Table 6; Supplementary Note). This finding is supported by the enrichment of genes encoding G-protein coupled receptor proteins among iL3-overexpressed genes, (p=5.1×10−8), and not among adult-overexpressed genes (p=4.1×10−7) (Supplementary Fig. 8). Consistent with other parasitic nematodes19, serine/threonine protein kinase activity is also enriched among iL3-overexpressed genes (p=0.008). The complexity of transcription regulatory activities is likely to be high in iL3, as evidenced by the enrichment of sequence-specific DNA binding transcription factor activity genes (p=1.7×10−14) and genes with alternative splicing (p<2×10−13), and the fact that most (92.5%) of the differentially expressed transcription factors are iL3-overexpressed (Supplementary Note). This iL3-stage enrichment of transcription factor-related activity might indicate that transcription factors (TFs) are poised for rapid gene expression after host invasion (i.e.gene expression is not active but is likely to be primed, as observed in arrested stages of C. elegans20).
Figure 2. Molecular functions enriched among N. americanus genes, stage-enriched genes and the N. americanus degradome.
a, “Molecular Function” Gene Ontology (GO) terms enriched in life-cycle stages and in N. americanus compared to other species. Included are (i) categories enriched in the iL3 or adult life cycle stages in N. americanus (ii) categories significantly over-represented or depleted in N. americanus compared to at least two of the comparison species, and (iii) second-order root nodes. b, Expression profiling of N. americanus proteases with C.-elegans orthologs. c, Expression profiling of N. americanus proteases with no C. elegans orthologs.
In contrast, in the adult stage, a broad spectrum of enzymes, such as proteases, hydrolases and catalases (Supplementary Table 6) are detected, emphasizing nutritional adaptation of adult worms that demands of a high-protein diet (i.e. blood21) (Fig. 2, Supplementary Fig. 9, Supplementary Note). Proteins with a signal-peptide (SP) for secretion had transcripts which were enriched among adult-overexpressed genes (p<10−15), whereas transmembrane domain-containing proteins (p=1.2×10−8) had transcripts which were enriched among iL3-overexpressed genes. SP-containing genes are enriched for proteases and protease inhibitors, the former contributing substantially to the predicted secretome (Supplementary Table 6, Supplementary Note), with 55% of all proteases (325/592) predicted as secreted. Proteases (particularly N. americanus-specific proteases with no orthologs in C. elegans) are overexpressed more often in adult compared to iL3 (p<10−15 for all both comparisons; Fig. 2b,c; Supplementary Note, Supplementary Table 7). Serine-type endopeptidase inhibitor activity, required to protect the adult stage from the digestive and immunologically hostile environment in the host22, was adult-enriched (p=1.6×10−4). The adult enrichment of transcription pertaining to structural constituents of the cuticle (p=1.7×10−5) also relates to the importance of protection of the parasite from the host23.
Blood feeding in adult hookworms is facilitated by an anticoagulation process and degradation of blood proteins by proteases. Known hookworm anticoagulants24 are dominated by single-domain serine protease inhibitors (SPIs). We annotated 87 serine protease inhibitors (SPIs) in N. americanus, accounting for 8 of 17 protease inhibitor clans. Given that serine proteases in humans are involved in diverse physiological functions (including blood coagulation and immunomodulation) the diversity of SPIs in N. americanus are likely critical not only for anticoagulation during blood-feeding, but also for long-term survival in the host. Specifically, SPIs are also likely to protect adult worms from enzymes in the small intestine where serine proteases, including trypsin, chymotrypsin, and elastase are prominent 25, therefore mediating hookworm-associated growth delay22. SPIs are enriched among the adult-overexpressed genes (p=3.9×10−8), but not among the iL3-overexpressed genes (p=0.35). Most of the SPIs characterized in hookworms are Kunitz-type molecules (Supplementary Note), but our findings suggest that multiple types of SPIs are produced by adult N. americanus in the human host. A mass spectrometry-based proteomics analysis was also performed using whole adult N. americanus worms (Full Methods Online), and the proteins detected (Supplementary Table 7, Supplementary Fig. 10) were also enriched for proteases (p=4.9×10−7), SPIs (p=1.8×10−4), as well as proteins with signal peptides (p=4.7×10−11) and a wide range of GO terms, many of which were related to proteolysis (Supplementary Table 6; Supplementary Note).
Pathogenesis and immunobiology of hookworm disease
N. americanus causes chronic disease and does not usually induce sterile immunity in the host. Adult hookworms live in the host for several years due to their ability to modulate and evade host immune defenses13 with their E/S products that sustain development and create a site of immune privilege26. Comparing the N. americanus genome with genomes from other nematodes, its host, and distant species, resulted in identification of molecules that facilitate parasitism. Sixty percent of N. americanus genes share an ortholog with other species (Supplementary Table 8; Supplementary Fig. 11, Supplementary Note). Comparative analysis identified metalloendopeptidases as the most prominent N. americanus protease (Fig. 2a), which is likely associated with the cleavage of eotaxin and inhibition of eosinophil recruitment27, in addition to tissue penetration28 and haemoglobinolysis29. N. americanus is the only blood-feeding nematode included in the comparison, and the hierarchical structure for enriched molecular functions (Fig. 2a) reveals shared and unique patterns and subsequent functional relationships.
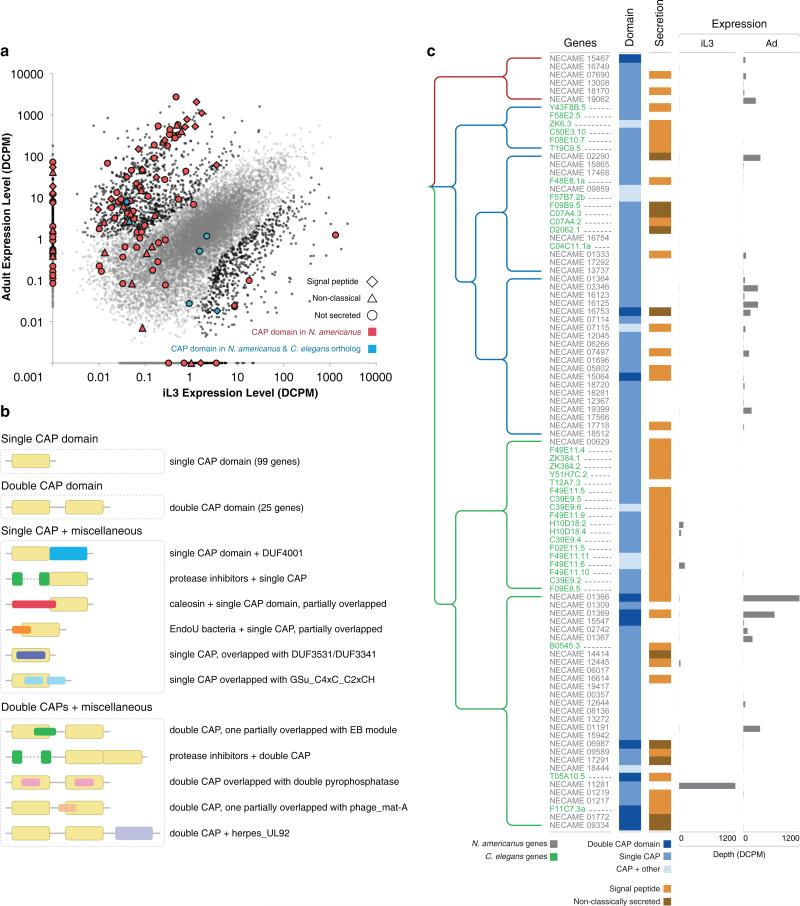
SCP/Tpx-1/Ag5/PR-1/Sc7 (SCP/TAPS; IPR014044; Supplementary Table 5) is a protein family inferred to be involved in host-parasite interactions (Supplementary Note). There are 137 SCP/TAPS proteins in N. americanus, a 4-fold expansion of this protein family compared to other nematodes. More than half (69/137) of the N. americanus SCP/TAPS proteins are adult-overexpressed (p<10−15; Fig. 3a), and only 6 of the 137 N. americanus SCP/TAPS proteins have orthologs in C. elegans (according to the MCL clustering; see Methods). The presence of a limited repertoire of orthologs in C. elegans suggests that nematode SCP/TAPS proteins may have originated prior to parasitism. Primary sequence similarity classified SCP/TAPS proteins into multiple groups (Fig. 3b, c; Supplementary Fig. 12), which do or do not contain C. elegans members, suggesting independent expansion of SCP/TAPS proteins after parasite speciation. The large expansion of SCP/TAPS proteins in N. americanus suggests multiple, possibly distinct roles in host-parasite interactions. SCP/TAPS proteins have been studied extensively as potential hookworm drug/vaccine candidates30 or as therapeutics for human inflammatory diseases15 or stroke31 (Supplementary Note). The 96 N. americanus-specific SCP/TAPS identified might serve as candidates for selective drug or vaccine targets32 (Supplementary Table 5).
Figure 3. SCP/TAPS (SCP/Tpx-1/Ag5/PR-1/Sc7) gene family expansion in N. americanus.
a, SCPs/TAPS are enriched in the Adult stage of N. americanus. b, A schematic representation of gene structure from SCP/TAPS family members. All SCP/TAPS proteins are grouped according to the number of CAP domains and regions outside the CAP domains: single CAP domain, double CAP domain, single CAP+miscellaneous and double CAP+miscellaneous. c, Neighbor joining clustering of the all C. elegans and ungapped N. americanus SCP/TAPS genes based on their full-length primary sequence similarity of the CAP domain. Data on domain representation, secretion type and stage of expression is included.
A total of 336 N. americanus genes that are orthologous to previously-predicted immunogenic/immunomodulatory proteins in A. suum24 were identified, along with three homologs to of transforming growth factor beta (TGF-β), an important protein in modulation of inflammation and the evolution of nematode parasitism33 (Supplementary Table 5). Additional protein-coding genes in N. americanus inferred to be involved in host-parasite immunomodulatory interactions include macrophage migration inhibitory factors (MIF), neutrophil inhibitor factor (NIF), hookworm platelet inhibitor (HPI), galectins, C-type lectins (C-TL), peroxiredoxins (PRX), glutathione S-transferases (GST), etc (Supplementary Note).
Prospects for new interventions
Historically, anthelmintic drugs have been discovered using in vivo and in vitro compound screens34. Recent comparative ‘omics’ studies (accompanied by experimental screening) in multiple nematode species10 demonstrate that genomic and transcriptomic data can be used to prioritize targets, with a higher hit rate compared with conventional approaches. Hence, the availability of the N. americanus genome is expected to enable comparative genomic and chemogenomic studies for the prediction and prioritization of therapeutic targets. Since more than half (53%) of all current drug targets35 consist of rhodopsin-like G-protein-coupled receptors (GPCRs), nuclear receptors (NRs), ligand-gated ion channels (LGICs), kinases and voltage-gated ion channels (VGICs, these protein groups were investigated in the N. americanus genome to identify potential therapeutic targets (Supplementary Table 9, Supplementary Note). GPCRs are attractive drug targets due to their importance in signal transduction35. We identified 272 GPCR genes, whereas there are nearly 1,700 GPCR genes in C. elegans. Although GPCRs are challenging to characterize at the primary sequence level (and the N. americanus genome is in a draft state), there may be a biological explanation for this difference in the number of GPCRs identified, including frequent amplifications of several subfamilies of GPCRs in C. elegans relative to the closely-related C. briggsae36. Three of the 5 GRAFS families (glutamate, rhodopsin, and frizzled, but not adhesion or secretin) are found in N. americanus. The putative GPCRs are enriched for iL3-overexpression (30 genes, p=5.1×10−8), with only one gene being adult-overexpressed (p=4.1×10−7 for under-representation). N. americanus encodes members of both major ion channel categories (LGICs and VGICs); 224 LGICs belonging to two of the three subfamilies of LGIC (Cys-loop family and glutamate-activated cation channels) were identified, compared with 159 LGIC-encoding genes in C. elegans37. Genes encoding nicotinic acetylcholine receptor subunits (nAChR) of cys-loop family members are also found. Nematodes have a much larger number of nAChR alpha subunits than examined vertebrates (17 nAChR-encoding genes in mammals and birds vs. 29 nAChR subunits in C. elegans38), and several anthelmintics such as levamisole39 and monepantel40 have been developed to exploit these differences. Ivermectin41 targets a subunit of glutamate-gated chloride channels that are present in N. americanus (eight genes; IPR015680); three of these genes clustered with six C. elegans glutamate-gated chloride channel genes (avr 14/15 and glc 1-442). The lack of a clear ortholog of the ivermectin-sensitive genes within the N. americanus genome and the underlying sequence diversity at a position correlated with direct activation by ivermectin may explain the relative ivermectin insensitivity of N. americanus43 (Supplementary Note; Supplementary Fig. 13) compared to other nematodes44.
VGICs include sodium, potassium and calcium channels, and are anthelmintic targets (e.g., emodepside inhibiting SLO-1 in C. elegans45 and parasitic nematodes such as A. suum46). N. americanus encodes 48 VGICs (less than C. elegans), including members from the major families such as 6-transmembrane (6TM) potassium channels, voltage-gated calcium channels, and voltage-gated chloride channels (Supplementary Note). Consistent with other nematodes47, voltage-gated sodium channels are not present in N. americanus.
Protein kinases are involved in numerous signal transduction pathways that regulate biological processes, and have been exploited major focus for drug discovery48. Of the 274 N. americanus genes encoding kinases, 15 and 12 are overexpressed in iL3 and adults, respectively. Gene expression, tissue expression, conservation among nematodes and dissimilarity to human ortholog was used for prioritization10 of candidate targets (Supplementary Table 10). To evaluate current drugs and inhibitors that target homologous kinases, compounds from a publicly available database were also prioritized (Full Methods Online). The highest scoring compound is an approved tyrosine kinase inhibitor for treating chronic myelogenous leukemia (CML)49. A total of 233 other compounds had the second-highest score of 5 (Supplementary Table 11), indicating that these existing drugs might be repurposed for treating neglected tropical diseases, thus minimizing development time and cost50.
Chokepoints in metabolic pathways51 were analyzed and prioritized to identify further drug targets. N. americanus encodes at least 3,976 protein-coding genes associated with 3,265 KEGG orthology (KO) terms (Supplementary Table 7), 938 (24%) of which are involved in metabolic pathways (Supplementary Fig. 14), representing 32 potentially complete modules. A total of 34% of the metabolic pathway genes are classified as a chokepoint (Supplementary Table 12), of which 120 are conserved among nematodes and non-nematode species used in the comparative analysis. Chokepoint prioritization, along with a requirement for a chokepoint to be an expression bottleneck in N. americanus and to display a lethal RNAi phenotype of the C. elegans orthologous gene prioritized 8 enzymes encoded by 10 distinct genes (Supplementary Table 12-14, Supplementary Note). Among the prioritized chokepoints is adenylosuccinate lyase (ASL) (EC 4.3.2.2) (Supplementary Figs. 15-17), an enzyme involved in the purine metabolism pathway (ko00230) and a chokepoint in the adenine ribonucleotide biosynthesis module (M00049). To identify chokepoint inhibitors for repurposing, compounds from publicly available databases (449 target-compound pairs) were assessed using the same method as for kinase inhibitors. The highest ranked candidates include compounds such as azathioprine (DB00993), a pro-drug that is converted into mercaptopurine (DB01033) to inhibit purine metabolism and DNA synthesis (Supplementary Fig. 18, Supplementary Table 14, Supplementary Note).
Post-genomic exploration using the N. americanus immunome
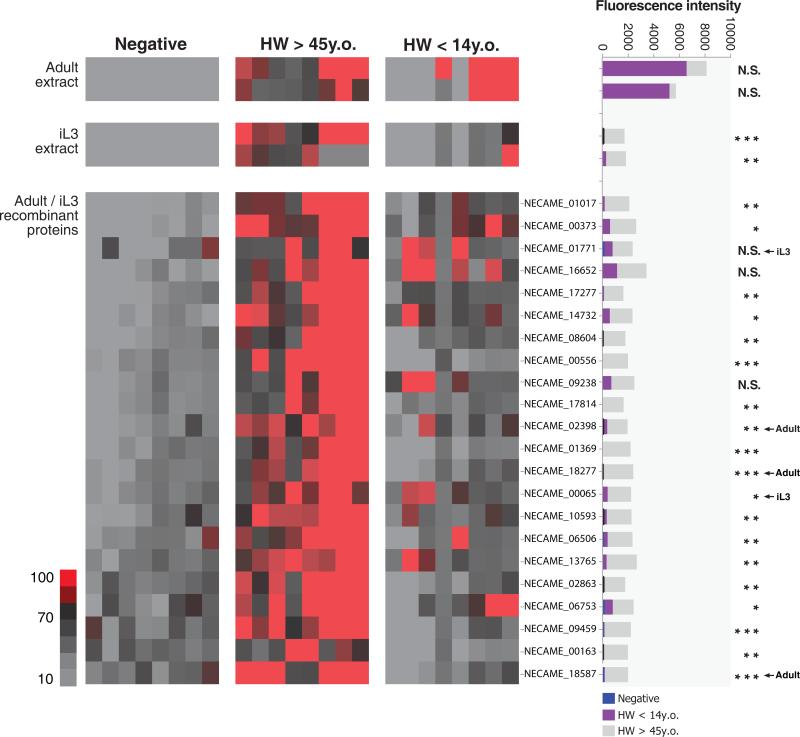
The N. americanus genome enables development of post-genomic tools to address the immuno-biology of human hookworm disease and accelerate antigen discovery for the development of vaccines and diagnostics. We developed a protein microarray containing 564 N. americanus recombinant proteins inferred from the genome (Supplementary Table 15, Supplementary Note). The microarray was probed with sera from individuals aged 4 to 66 years residents in an N. americanus-endemic area of northeastern Minas Gerais state in Brazil. This pilot study based on 200 individuals from the youngest (<14 years of age) and the oldest age strata (>45 years of age), resulted in identification of 22 antigens that were significant targets of anti-hookworm immune responses (Fig. 4). Older individuals showed stronger IgG responses to a larger number of secreted antigens, but these antibodies appear to play no role in killing the parasite or protecting against heavy infection. Hence, unlike other STHs of humans, protective immunity to N. americanus does not seem to develop in most individuals during adolescence. This is consistent with knowledge that, in Necator endemic areas, older human individuals often harbour the heaviest-intensity infections1,52,53. Younger individuals showed IgG responses against fewer antigens, usually with lower intensity. Thus, while antibodies are a key feature of the immune response to N. americanus and increase with host age, they fail to protect individuals from infection over time. The absence of overall protective immunity to hookworm infection as opposed to age-acquired protective immunity observed with other STH infections is likely multifactorial. Detailed kinetic studies of the IgG subclasses and IgE responses to hookworm antigens represented on our protein microarray will be required to better understand the roles of these antibodies in the acquisition of immunity against hookworm13. The protein microarray can be probed with sera from individuals with different genetic backgrounds and different histories of exposure to hookworm54, as well as animals rendered immunologically resistant to hookworm infection by vaccination with irradiated iL355, thereby facilitating efforts to develop an efficacious vaccine against hookworm disease. Furthermore, secreted proteins recognized by most or all the infected individuals and with weak or no homologies to other nematode species, indicate identification of antigens that might form the basis of sensitive and specific serodiagnostic tests (Supplementary Note; e.g. Supplementary Fig. 19).
Figure 4. The serum responses to Necator americanus antigens vary with age and infection intensity.
The heat map shows the immunoreactivity of 22 antigens to the IgG antibodies from groups of uninfected individuals, infected children (HW <14 y.o.) and infected adults (HW >45 y.o.)[n=8 in each group]. Duplicate crude somatic extracts from iL3 and adult stage were included as control naive antigens; Every other row represents an individual recombinant in vitro translation product. The stacked bar plot represents the mean immunoreactivity for each antigen for the three groups based on mean fluorescence intensity. Highlighted “iL3” or “Adult” indicates corresponding stage-specific expression based on RNA-seq data. Significant differences in antibody responses between adult and children group were detected using student t test [P<0.05 (*); P<0.01 (**); P<0.001 (***); N.S., no significant difference].
DISCUSSION
N. americanus is responsible for causing more disease worldwide than any other STH. The characterization of the first genome of a human hookworm is expected to significantly facilitate future fundamental explorations of the epidemiology and evolutionary biology of hookworms as well as efforts toward the development of therapeutics to combat hookworm disease. Since N. americanus is the first hookworm whose genome has been sequenced, the data presented provide a first insight into blood-feeding nematodes of major human and animal health importance. Our post-genomic exploration of inferred proteomic information highlights the utility of the draft genome sequence for understanding the immuno-biology of human hookworm disease and accelerating the development of vaccines and diagnostics. It is also pertinent to note that hookworms are garnering interest for their therapeutic properties against a range of non-infectious inflammatory diseases of humans. The genome sequence, therefore, represents a veritable pharmacopoeia – indeed, recombinant hookworm molecules have already undergone clinical trials for stroke and deep vein thrombosis15. Clearly the N. americanus genome sequence will have broad implications and provides many exciting opportunities to establish post-genomic methods in the quest to develop improved interventions against this ancient and neglected parasite, as well as inflammatory diseases that are reaching epidemic proportions in industrialized societies.
Online Methods
Parasite material
The Anhui strain of N. americanus was maintained56 in Golden Syrian Hamster (3-4 weeks, male) from Harlan under the George Washington University IACUC approved protocol 053-12,2, and in accordance with all Animal Welfare guidance. Adult worms were collected from intestines of hamsters infected subcutaneously with N. americanus iL3 for 8 weeks57. DNA was extracted with the QIAamp DNA Mini Kit according to manufacturer's instruction (Qiagen). For transcriptome sequencing, two key developmental stages from a host-parasite interaction perspective, the infective L3 (iL3; environmental) and adult (parasitic) worm stages, were collected.
Sequencing, assembly and annotation
Fragment, paired-end whole-genome shotgun libraries (3kb and 8 kb insert sizes) were sequenced using Roche/454 platform and assembled with Newbler58. A repeat library was generated (Repeatmodeler) and repeats characterized (CENSOR59 v. 4.2.27 against RepBase release 17.0360). Ribosomal RNA genes (RNAmmer61) and transfer RNAs (tRNAscan-SE62) were identified. Other non-coding RNAs were identified by a sequence homology search against the Rfam database63. Repeats and predicted RNAs were then masked using RepeatMasker. Protein-coding genes were predicted using a combination of ab initio programs64,65 and the annotation pipeline tool MAKER66. A consensus high confidence gene set from the above prediction algorithms was generated (Supplementary Note). The size and number of exons and introns in N. americanus were determined by parsing exon sizes from gff-format annotations (considering only exon features tagged as “coding_exon”) and calculating intron sizes and compared to the C. elegans genes (WS230). Significant differences in exon and intron lengths and numbers were tested between species and orthologous and non-orthologous gene groups using two-tailed T-tests with unequal variance (Supplementary Note). Two separate approaches were used to identify putative operons in N. americanus (Supplementary Note). Gene product naming was determined by BER (JCVI) and functional categories of deduced proteins were assigned67-69. Orthologous groups were built from 13 species using OrthoMCL70 and genes not orthologous to the other 12 species were classified as N. americanus-specific.
RNA-seq
RNA was extracted18, DNAse treated and used to generate both Roche/454 and Illumina cDNA libraries (Supplementary Note) that were sequenced using a Genome Sequencer Titanium FLX (Roche Diagnostics) and Illumina (Illumina Inc, San Diego, CA), with slight modification (Supplementary Note). The 454 cDNA reads were analyzed as previously described18. The Illumina RNA-seq data were processed71 and low-compositional complexity bases were masked72. RNA-Seq reads were aligned73 to the predicted gene set and genes with a breadth of coverage ≥50% across the gene sequence (i.e., “expressed”) were used for further downstream analysis. Expression was quantified using expression values normalized to the depth of coverage per 100 million mapped bases (DCPM). Expressed genes were subject to further differential expression analysis using EdgeR74 (false discovery rate <0.05), in order to identify stage-overexpressed genes (Supplementary Note).
Deduced Proteome Functional Annotation and Enrichment
Proteins were searched against KEGG75 using KAAS68 (cut-off 35 bits) and InterProScan69 was used to get InterPro76 domain matches and Gene Ontology67 (GO) annotations. Proteins with signal peptides77, non-classical secretion78 and transmembrane topology77 were identified. The degradome was identified by comparison to the MEROPS79 protease unit database using WU-BLAST (identifying the best hit with E≤e-10). Enrichment of different protease groups among different gene sets (based on similarity to C. elegans) was detected based on False Discovery Rate (FDR)-corrected binomial distribution probability tests80. GO enrichment significance comparing the iL3 and adult-overexpressed gene sets was calculated using FUNC81 at a 0.01 significance threshold after Family-Wise Error Rate (FWER) population correction81. QuickGO82 was used to analyze the hierarchical structure of significant GO categories.
Proteomic analysis of somatic worm extract
Whole worms were ground under liquid nitrogen before solubilisation in lysis buffer, total protein was precipitated, and established methods83 were used to reduce, alkyate and tryptic-digest two 1.5 mg samples of total somatic protein. Peptide fractions were prepared before LC and mass spectral analysis (Supplementary Note). Only proteins confirmed with at least two peptides and a confidence of p≤0.05 were considered identified. GO functional enrichment among the genes supported by proteomics was calculated81, using all of the genes without proteomics support as a background for comparison.
Transcription Factors and the binding sites
Transcription factors in N. americanus were identified by extracting KEGG Orthology (KO) numbers from the KEGG transcription factor database (derived from TRANSFAC 7.084) and comparing to N. americanus KOs. Documented matrices of transcription factor binding sites were downloaded from the JASPAR database85. The corresponding protein accession numbers were extracted and converted to KOs, and were compared to N. americanus transcription factor KOs to define a subset of N. americanus transcription factors with available binding site information. The binding site matrices of this subset of N. americanus transcription factors were used to scan the sequences of up to 500 bp downstream and upstream of differentially expressed genes using Patser.
SCP/TAPS
Each protein was searched for the SCP/TAPS-representative protein domains86 IPR014044 (“CAP domain”) and PF00188 (“CAP”)86 using Interproscan69 and hmmpfam87. Phylogenetic relationship trees using full length primary sequences derived from ungapped genes were built using Bayesian inference88 and Neighbor Joining89 as previously described for other helminths32,86,90. Leaves of the tree were annotated with domain information, secretion mode and expression data, and then visualized using iTOL91.
Potential Drug Targets
GPCRs, LGICs and VGICs were identified with InterProScan69. Ion channels were identified using WU-BLASTP (E≤e-10) against the C. elegans proteome (WS230). Ivermectin Target Characterization: sequence alignments were obtained by MUSCLE92 for the C. elegans and N. americanus orthologs within two orthologous groups (NAIF1.5_00184 and NAIF1.5_06724). Homology models for the two N. americanus orthologs (NECAME_16744 and NECAME_16780) were built by MODELLER93 using the C. elegans crystal structure as template94. For each ortholog, five models were built and the one with the lowest total function score (energy) was chosen as the model shown. Sequence alignments are colored by Clustalx scheme in JalView95; protein structure models are rendered in PyMol (Schrodinger, LLC, The PyMOL Molecular Graphics System, Version 1.3r1. 2010).
Kinome and Chokepoints
N. americanus genes were screened against the collection of kinase domain models in the Kinomer96 and custom score thresholds applied for each kinase group and then adjusted until an hmmpfam search87 came as close as possible to identifying known C. elegans kinases. Those same cutoffs were then applied to the N. americanus gene set to identify putative kinases as previously described97. Kinase prioritization was done adapting the protocol as previously described10 (Supplementary Note).
Chokepoints of KEGG metabolic pathways were defined as a reaction that either consumes a unique substrate or produces a unique product. The reaction database from KEGG v5898 was used and the chokepoint were identified and prioritized as previously described99 (Supplementary Note). Metabolic module abundances were calculated (and normalized in DCPM) based on KAAS annotations68, and module bottlenecks were defined as reaction steps in the cascade that are both essential for the module completion and have that have low enzyme abundance that primarily constrains the overall module abundance. Homology models were aligned with their reference sequence using T-COFFEE100, constructed using MODELLER101 with default parameters using PDB structures with the highest sequence similarity, and docking was performed using AutoDock4.2102 using default parameters. Chemogenomic screening for compound prioritization was performed as previously described99 (Supplementary Note).
Protein microarray
In 2005, 1494 individuals between the ages 4 and 66 years (inclusive) were enrolled (with informed consent) into a cross-sectional study in an N. americanus-endemic area of Northeastern Minas Gerais state in Brazil, using protocols approved by the George Washington University IRB (117040 and 060605), the Ethics Committee of Instituto René Rachou, and the National Ethics Committee of Brazil (CONEP) (Protocol numbers 04/2008 and 12/2006). Venous blood (15 mL) was collected from individuals determined to be positive for N. americanus (Supplementary Note).
A total of 1,275 N. americanus open reading frames (ORFs) contained a classical signal peptide for secretion and had RNA-seq evidence for transcription in iL3 and/or adult worms. Of those, 623 corresponding cDNAs were successfully amplified, cloned, expressed and the protein extracts were contact-printed without purification onto nitrocellulose glass FAST® slides (Supplementary Note). The printed in vitro-expressed proteins were quality-checked using antibodies against incorporated N-terminal poly-histidine (His) and C-terminal hemaglutinin (HA) tags.
Protein arrays were blocked in blocking solution (Whatman) and probed with human sera overnight. Arrays were washed and isotype and subclass-specific responses were detected using biotinylated mouse monoclonal antibodies against human IgG1, IgG3, IgG4 (Sigma) and biotin-conjugated mouse monoclonal anti-human IgE Fc (Human Reagent Laboratory, Baltimore, MD). Microarrays were scanned using a GenePix microarray scanner (Molecular Devices). The data was analyzed using the “group average” method103, whereby the mean fluorescence was considered for analysis (Supplementary Note).
Supplementary Material
ACKNOWLEDGEMENTS
The genome sequencing and annotation work was funded by NIH-NHGRI grant U54HG003079 to R.K.W. The comparative genome analysis was funded by AI081803 and GM097435 to M.M. Funds from the Australian Research Council (ARC) and the National Health and Medical Research Council (NHMRC) to R.B.G. are gratefully acknowledged. P.W.S. is an investigator with the Howard Hughes Medical Institute. We thank the faculty and staff of The Genome Institute, who contributed to this study.
Footnotes
AUTHOR CONTRIBUTIONS: These authors contributed equally to this work: Y.T.T., X.G., B.A.R; Conceived and planned the project: M.M., R.B.G., P.W.S., R.K.W., S.R.; Led the project, analysis and manuscript preparation: M.M.; Provided material: B.Z., P.J.H., J.H., P.L.F., J.B., E.M.R.; Sequence data production, assembly construction, annotation, and submission: K.H.P., X.Z., V.B.P, P.M., W.W., J.M., S.A.; Genome-based comparative studies, differential transcription, host-parasite interaction analysis, proteomics, protein-array analysis: M.M., Y.T.T., X.G., B.A.R., R.T., Q.W., S.A., J.M., A.L., S.G., P.L.F., JM, JS, AD; Drafted, edited and wrote the manuscript: M.M., R.B.G, A.L., J.M.H.
URLS. NCBI SRA, http://www.ncbi.nlm.nih.gov/sra; RepeatModeler, http://www.repeatmasker.org/RepeatModeler.html; RNAmmer, http://www.cbs.dtu.dk/services/RNAmmer/; Rfam database, http://selab.janelia.org/software.html; RepeatMasker, http://repeatmasker.org; Fgenesh, www.softberry.com; BER, http://ber.sourceforge.net; Seqclean, http://compbio.dfci.harvard.edu/tgi/software/; Refcov, http://gmt.genome.wustl.edu/gmt-refcov; PyMol, www.pymol.org; KEGG transcription factor database, http://www.genome.jp/keggbin/get_htext?ko03000.keg; Jaspar database, jasper.genereg.net; Patser, stormo.wustl.edu/resourse.html; Kinomer, http://www.compbio.dundee.ac.uk/kinomer; SignalP, www.cbs.dtu.dk/services/SignalP/.
Accession codes. The whole-genome sequence of N. americanus has been deposited in DDBJ/EMBL/GenBank under the project accession ANCG00000000. The version described in this paper is the first version ANCG01000000. All short read data have been deposited in the Short Read Archive under the following accessions: SRR036799 - SRR036800 SRR036802 SRR036804 - SRR036811 SRR341459 - SRR341460 SRR609850 - SRR609895 SRR609951 SRR610281 - SRR610282 SRR611341 - SRR611350. RNA-Seq profiles have been deposited in Nematode.net and a browse-able genome is also available at Nematode.net and WormBase. The authors declare no competing financial interest.
COMPETING INTERESTS
The authors declare no competing financial interests.
Note: Supplementary information is available on the Nature Genetics website.
SUPPLEMENTARY INFORMATION
PDF files
The supplementary information PDF file (Supplementary Info.pdf; 2.7MB) contains Supplementary Figures 1 to 19, Supplementary Tables 1 and 2, and Supplementary Note.
Excel files
Supplementary tables 3 to 15 are provided as individual excel-format spreadsheets (total file size 9.6MB)
References
- 1.Bethony J, et al. Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet. 2006;367:1521–32. doi: 10.1016/S0140-6736(06)68653-4. [DOI] [PubMed] [Google Scholar]
- 2.Schneider B, et al. A history of hookworm vaccine development. Human vaccines. 2011;7:1234–44. doi: 10.4161/hv.7.11.18443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hotez PJ, Bethony JM, Diemert DJ, Pearson M, Loukas A. Developing vaccines to combat hookworm infection and intestinal schistosomiasis. Nature reviews. Microbiology. 2010;8:814–26. doi: 10.1038/nrmicro2438. [DOI] [PubMed] [Google Scholar]
- 4.Loukas A, et al. Vaccinomics for the major blood feeding helminths of humans. Omics : a journal of integrative biology. 2011;15:567–77. doi: 10.1089/omi.2010.0150. [DOI] [PubMed] [Google Scholar]
- 5.Diemert DJ, Bethony JM, Hotez PJ. Hookworm vaccines. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2008;46:282–8. doi: 10.1086/524070. [DOI] [PubMed] [Google Scholar]
- 6.Steinmann P, et al. Efficacy of single-dose and triple-dose albendazole and mebendazole against soil-transmitted helminths and Taenia spp.: a randomized controlled trial. PloS one. 2011;6:e25003. doi: 10.1371/journal.pone.0025003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keiser J, Utzinger J. Efficacy of current drugs against soil-transmitted helminth infections: systematic review and meta-analysis. JAMA. 2008;299:1937–48. doi: 10.1001/jama.299.16.1937. [DOI] [PubMed] [Google Scholar]
- 8.Jia TW, Melville S, Utzinger J, King CH, Zhou XN. Soil-transmitted helminth reinfection after drug treatment: a systematic review and meta-analysis. PLoS neglected tropical diseases. 2012;6:e1621. doi: 10.1371/journal.pntd.0001621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soukhathammavong PA, et al. Low efficacy of single-dose albendazole and mebendazole against hookworm and effect on concomitant helminth infection in Lao PDR. PLoS Negl Trop Dis. 2012;6:e1417. doi: 10.1371/journal.pntd.0001417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor CM, et al. Using Existing Drugs as Leads for Broad Spectrum Anthelmintics Targeting Protein Kinases. PLoS Pathog. 2013;9:e1003149. doi: 10.1371/journal.ppat.1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott DE, Weinstock JV. Helminth-host immunological interactions: prevention and control of immune-mediated diseases. Ann N Y Acad Sci. 2012;1247:83–96. doi: 10.1111/j.1749-6632.2011.06292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daveson AJ, et al. Effect of hookworm infection on wheat challenge in celiac disease--a randomised double-blinded placebo controlled trial. PloS one. 2011;6:e17366. doi: 10.1371/journal.pone.0017366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McSorley HJ, Loukas A. The immunology of human hookworm infections. Parasite immunology. 2010;32:549–59. doi: 10.1111/j.1365-3024.2010.01224.x. [DOI] [PubMed] [Google Scholar]
- 14.Ferreira I, et al. Hookworm excretory/secretory products induce interleukin-4 (IL-4)+ IL-10+ CD4+ T cell responses and suppress pathology in a mouse model of colitis. Infect Immun. 2013;81:2104–11. doi: 10.1128/IAI.00563-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navarro S, Ferreira I, Loukas A. The hookworm pharmacopoeia for inflammatory diseases. Int J Parasitol. 2013;43:225–31. doi: 10.1016/j.ijpara.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Bradnam KR, Korf I. Longer first introns are a general property of eukaryotic gene structure. PLoS One. 2008;3:e3093. doi: 10.1371/journal.pone.0003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lercher MJ, Blumenthal T, Hurst LD. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome research. 2003;13:238–43. doi: 10.1101/gr.553803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, et al. Characterizing Ancylostoma caninum transcriptome and exploring nematode parasitic adaptation. BMC Genomics. 2010;11:307. doi: 10.1186/1471-2164-11-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell BE, Hofmann A, McCluskey A, Gasser RB. Serine/threonine phosphatases in socioeconomically important parasitic nematodes--prospects as novel drug targets? Biotechnology advances. 2011;29:28–39. doi: 10.1016/j.biotechadv.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–4. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- 21.Williamson AL, Brindley PJ, Knox DP, Hotez PJ, Loukas A. Digestive proteases of blood-feeding nematodes. Trends in parasitology. 2003;19:417–23. doi: 10.1016/s1471-4922(03)00189-2. [DOI] [PubMed] [Google Scholar]
- 22.Chu D, et al. Molecular characterization of Ancylostoma ceylanicum Kunitz-type serine protease inhibitor: evidence for a role in hookworm-associated growth delay. Infection and immunity. 2004;72:2214–21. doi: 10.1128/IAI.72.4.2214-2221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page AP, Winter AD. Enzymes involved in the biogenesis of the nematode cuticle. Adv Parasitol. 2003;53:85–148. doi: 10.1016/s0065-308x(03)53003-2. [DOI] [PubMed] [Google Scholar]
- 24.Jex AR, et al. Ascaris suum draft genome. Nature. 2011;479:529–533. doi: 10.1038/nature10553. [DOI] [PubMed] [Google Scholar]
- 25.Whitcomb DC, Lowe ME. Human pancreatic digestive enzymes. Digestive diseases and sciences. 2007;52:1–17. doi: 10.1007/s10620-006-9589-z. [DOI] [PubMed] [Google Scholar]
- 26.Maizels RM, Yazdanbakhsh M. Immune regulation by helminth parasites: cellular and molecular mechanisms. Nature reviews. Immunology. 2003;3:733–44. doi: 10.1038/nri1183. [DOI] [PubMed] [Google Scholar]
- 27.Culley FJ, et al. Eotaxin is specifically cleaved by hookworm metalloproteases preventing its action in vitro and in vivo. J Immunol. 2000;165:6447–53. doi: 10.4049/jimmunol.165.11.6447. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Pritchard DI. Secretion of metalloproteases by living infective larvae of Necator americanus. The Journal of parasitology. 1992;78:917–9. [PubMed] [Google Scholar]
- 29.Ranjit N, et al. Proteolytic degradation of hemoglobin in the intestine of the human hookworm Necator americanus. The Journal of infectious diseases. 2009;199:904–12. doi: 10.1086/597048. [DOI] [PubMed] [Google Scholar]
- 30.Goud GN, et al. Expression of the Necator americanus hookworm larval antigen Na- ASP-2 in Pichia pastoris and purification of the recombinant protein for use in human clinical trials. Vaccine. 2005;23:4754–64. doi: 10.1016/j.vaccine.2005.04.040. [DOI] [PubMed] [Google Scholar]
- 31.Krams M, et al. Acute Stroke Therapy by Inhibition of Neutrophils (ASTIN): an adaptive dose-response study of UK-279,276 in acute ischemic stroke. Stroke. 2003;34:2543–8. doi: 10.1161/01.STR.0000092527.33910.89. [DOI] [PubMed] [Google Scholar]
- 32.Cantacessi C, Gasser RB. SCP/TAPS proteins in helminths--where to from now? Mol Cell Probes. 2012;26:54–9. doi: 10.1016/j.mcp.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Viney ME, Thompson FJ, Crook M. TGF-beta and the evolution of nematode parasitism. Int J Parasitol. 2005;35:1473–5. doi: 10.1016/j.ijpara.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Kotze AC. Target-based and whole-worm screening approaches to anthelmintic discovery. Veterinary parasitology. 2012;186:118–23. doi: 10.1016/j.vetpar.2011.11.052. [DOI] [PubMed] [Google Scholar]
- 35.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nature reviews. Drug discovery. 2006;5:993–6. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 36.Robertson HM, Thomas JH. The putative chemoreceptor families of C. elegans. WormBook : the online review of C. elegans biology. 2006:1–12. doi: 10.1895/wormbook.1.66.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Littleton JT, Ganetzky B. Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron. 2000;26:35–43. doi: 10.1016/s0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 38.Jones AK, Davis P, Hodgkin J, Sattelle DB. The nicotinic acetylcholine receptor gene family of the nematode Caenorhabditis elegans: an update on nomenclature. Invert Neurosci. 2007;7:129–31. doi: 10.1007/s10158-007-0049-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lionel ND, Mirando EH, Nanayakkara JC, Soysa PE. Levamisole in the treatment of ascariasis in children. British medical journal. 1969;4:340–1. doi: 10.1136/bmj.4.5679.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaminsky R, et al. Identification of the amino-acetonitrile derivative monepantel (AAD 1566) as a new anthelmintic drug development candidate. Parasitology research. 2008;103:931–9. doi: 10.1007/s00436-008-1080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell WC, Fisher MH, Stapley EO, Albers-Schonberg G, Jacob TA. Ivermectin: a potent new antiparasitic agent. Science. 1983;221:823–8. doi: 10.1126/science.6308762. [DOI] [PubMed] [Google Scholar]
- 42.Hobert O. The neuronal genome of Caenorhabditis elegans. WormBook. 2013:1–106. doi: 10.1895/wormbook.1.161.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richards JC, Behnke JM, Duce IR. In vitro studies on the relative sensitivity to ivermectin of Necator americanus and Ancylostoma ceylanicum. Int J Parasitol. 1995;25:1185–91. doi: 10.1016/0020-7519(95)00036-2. [DOI] [PubMed] [Google Scholar]
- 44.Geary TG, et al. Haemonchus contortus: ivermectin-induced paralysis of the pharynx. Exp Parasitol. 1993;77:88–96. doi: 10.1006/expr.1993.1064. [DOI] [PubMed] [Google Scholar]
- 45.Bull K, et al. Effects of the novel anthelmintic emodepside on the locomotion, egg-laying behaviour and development of Caenorhabditis elegans. International journal for parasitology. 2007;37:627–36. doi: 10.1016/j.ijpara.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 46.Willson J, Amliwala K, Harder A, Holden-Dye L, Walker RJ. The effect of the anthelmintic emodepside at the neuromuscular junction of the parasitic nematode Ascaris suum. Parasitology. 2003;126:79–86. doi: 10.1017/s0031182002002639. [DOI] [PubMed] [Google Scholar]
- 47.Zakon HH. Adaptive evolution of voltage-gated sodium channels: the first 800 million years. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(Suppl 1):10619–25. doi: 10.1073/pnas.1201884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nature reviews. Drug discovery. 2002;1:309–15. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 49.Shah NP, et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 50.Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nature reviews. Drug discovery. 2004;3:673–83. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 51.Yeh I, Hanekamp T, Tsoka S, Karp PD, Altman RB. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome research. 2004;14:917–24. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Humphries DL, et al. The use of human faeces for fertilizer is associated with increased intensity of hookworm infection in Vietnamese women. Trans R Soc Trop Med Hyg. 1997;91:518–20. doi: 10.1016/s0035-9203(97)90007-9. [DOI] [PubMed] [Google Scholar]
- 53.Bethony J, et al. Emerging patterns of hookworm infection: influence of aging on the intensity of Necator infection in Hainan Province, People's Republic of China. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2002;35:1336–44. doi: 10.1086/344268. [DOI] [PubMed] [Google Scholar]
- 54.Quinnell RJ, et al. Genetic and household determinants of predisposition to human hookworm infection in a Brazilian community. J Infect Dis. 2010;202:954–61. doi: 10.1086/655813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller TA. Vaccination against the canine hookworm diseases. Adv Parasitol. 1971;9:153–83. doi: 10.1016/s0065-308x(08)60161-x. [DOI] [PubMed] [Google Scholar]
- 56.Jian X, et al. Necator americanus: maintenance through one hundred generations in golden hamsters (Mesocricetus auratus). I. Host sex-associated differences in hookworm burden and fecundity. Exp Parasitol. 2003;104:62–6. doi: 10.1016/s0014-4894(03)00094-8. [DOI] [PubMed] [Google Scholar]
- 57.Xiao S, et al. The evaluation of recombinant hookworm antigens as vaccines in hamsters (Mesocricetus auratus) challenged with human hookworm, Necator americanus. Experimental parasitology. 2008;118:32–40. doi: 10.1016/j.exppara.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 58.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenetic and genome research. 2005;110:462–7. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 61.Lagesen K, et al. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic acids research. 2007;35:3100–8. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic acids research. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic acids research. 2003;31:439–41. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korf I. Gene finding in novel genomes. BMC bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–44. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 66.Cantarel BL, et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome research. 2008;18:188–96. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic acids research. 2007;35:W182–5. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quevillon E, et al. InterProScan: protein domains identifier. Nucleic acids research. 2005;33:W116–20. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li L, Stoeckert CJ, Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome research. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hancock JM, Armstrong JS. SIMPLE34: an improved and enhanced implementation for VAX and Sun computers of the SIMPLE algorithm for analysis of clustered repetitive motifs in nucleotide sequences. Comput Appl Biosci. 1994;10:67–70. doi: 10.1093/bioinformatics/10.1.67. [DOI] [PubMed] [Google Scholar]
- 73.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic acids research. 2012;40:D109–14. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hunter S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic acids research. 2012;40:D306–12. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. Journal of molecular biology. 2004;338:1027–36. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 78.Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein engineering, design & selection : PEDS. 2004;17:349–56. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 79.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic acids research. 2010;38:D227–33. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 81.Prufer K, et al. FUNC: a package for detecting significant associations between gene sets and ontological annotations. BMC bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Binns D, et al. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045–6. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mulvenna J, et al. Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum. Molecular & cellular proteomics : MCP. 2009;8:109–21. doi: 10.1074/mcp.M800206-MCP200. [DOI] [PubMed] [Google Scholar]
- 84.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic acids research. 2006;34:D108–10. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bryne JC, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic acids research. 2008;36:15. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cantacessi C, et al. Insights into SCP/TAPS proteins of liver flukes based on large-scale bioinformatic analyses of sequence datasets. PLoS One. 2012;7:e31164. doi: 10.1371/journal.pone.0031164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eddy SR. Accelerated Profile HMM Searches. PLoS computational biology. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 89.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 90.Cantacessi C, et al. A portrait of the “SCP/TAPS” proteins of eukaryotes--developing a framework for fundamental research and biotechnological outcomes. Biotechnology advances. 2009;27:376–88. doi: 10.1016/j.biotechadv.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 91.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 92.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic acids research. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Eswar N, et al. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007;2 doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 94.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miranda-Saavedra D, Barton GJ. Classification and functional annotation of eukaryotic protein kinases. Proteins. 2007;68:893–914. doi: 10.1002/prot.21444. [DOI] [PubMed] [Google Scholar]
- 97.Mitreva M, et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 2011;43:228–35. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic acids research. 2010;38:30. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Taylor CM, et al. Discovery of anthelmintic drug targets and drugs using chokepoints in nematode metabolic pathways. PLoS Pathog. 2013;9:e1003505. doi: 10.1371/journal.ppat.1003505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. Journal of molecular biology. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 101.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. Journal of molecular biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 102.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of computational chemistry. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sundaresh S, et al. Identification of humoral immune responses in protein microarrays using DNA microarray data analysis techniques. Bioinformatics. 2006;22:1760–6. doi: 10.1093/bioinformatics/btl162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.