Abstract
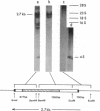
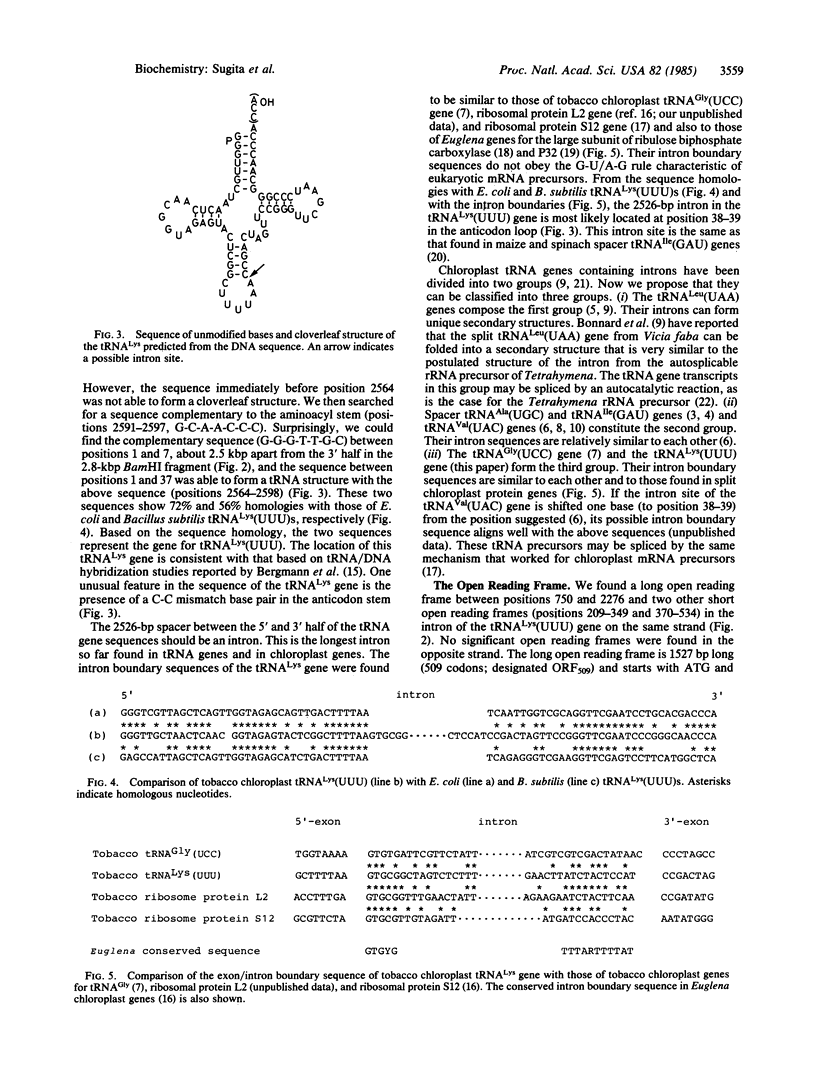
The nucleotide sequence of a tRNALys(UUU) gene on tobacco (Nicotiana tabacum) chloroplast DNA has been determined. This gene is located 215 base pairs upstream from the gene for the 32,000-dalton thylakoid membrane protein on the same DNA strand and has a 2526-base-pair intron in the anticodon loop. The intron boundary sequence does not follow the G-U/A-G rule but is similar to those of tobacco chloroplast split genes for tRNAGly(UCC) and ribosomal proteins L2 and S12. The intron contains one major open reading frame of 509 codons. The codon usage in the open reading frame resembles those observed in the genes for tobacco chloroplast proteins so far analyzed. The primary transcript of this tRNA gene is 2.7 kilobases long.
Keywords: molecular cloning, DNA sequence, blot hybridization, precursor RNA, codon usage
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burke J. M., RajBhandary U. L. Intron within the large rRNA gene of N. crassa mitochondria: a long open reading frame and a consensus sequence possibly important in splicing. Cell. 1982 Dec;31(3 Pt 2):509–520. doi: 10.1016/0092-8674(82)90307-5. [DOI] [PubMed] [Google Scholar]
- Deno H., Kato A., Shinozaki K., Sugiura M. Nucleotide sequences of tobacco chloroplast genes for elongator tRNAMet and tRNAVal (UAC): the tRNAVal (UAC) gene contains a long intron. Nucleic Acids Res. 1982 Dec 11;10(23):7511–7520. doi: 10.1093/nar/10.23.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deno H., Sugiura M. Chloroplast tRNA gene contains a long intron in the D stem: Nucleotide sequences of tobacco chloroplast genes for tRNA (UCC) and tRNA (UCU). Proc Natl Acad Sci U S A. 1984 Jan;81(2):405–408. doi: 10.1073/pnas.81.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemaut P., Weil J. H. The nucleotide sequence of the maize and spinach chloroplast isoleucine transfer RNA encoded in the 16S to 23S rDNA spacer. Nucleic Acids Res. 1982 Mar 11;10(5):1653–1659. doi: 10.1093/nar/10.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabin G. D., Farley M., Hallick R. B. Chloroplast gene for Mr 32000 polypeptide of photosystem II in Euglena gracilis is interrupted by four introns with conserved boundary sequences. Nucleic Acids Res. 1984 Jul 25;12(14):5801–5812. doi: 10.1093/nar/12.14.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W., Edwards K., Kössel H. Sequencing of the 16S-23S spacer in a ribosomal RNA operon of Zea mays chloroplast DNA reveals two split tRNA genes. Cell. 1981 Jul;25(1):203–213. doi: 10.1016/0092-8674(81)90245-2. [DOI] [PubMed] [Google Scholar]
- Koller B., Gingrich J. C., Stiegler G. L., Farley M. A., Delius H., Hallick R. B. Nine introns with conserved boundary sequences in the Euglena gracilis chloroplast ribulose-1,5-bisphosphate carboxylase gene. Cell. 1984 Feb;36(2):545–553. doi: 10.1016/0092-8674(84)90247-2. [DOI] [PubMed] [Google Scholar]
- Kruger K., Grabowski P. J., Zaug A. J., Sands J., Gottschling D. E., Cech T. R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982 Nov;31(1):147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Lazowska J., Jacq C., Slonimski P. P. Sequence of introns and flanking exons in wild-type and box3 mutants of cytochrome b reveals an interlaced splicing protein coded by an intron. Cell. 1980 Nov;22(2 Pt 2):333–348. doi: 10.1016/0092-8674(80)90344-x. [DOI] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montandon P. E., Stutz E. Nucleotide sequence of a Euglena gracilis chloroplast genome region coding for the elongation factor Tu; evidence for a spliced mRNA. Nucleic Acids Res. 1983 Sep 10;11(17):5877–5892. doi: 10.1093/nar/11.17.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M., Gauss D. H. Compilation of tRNA sequences. Nucleic Acids Res. 1984;12 (Suppl):r1–57. [PMC free article] [PubMed] [Google Scholar]
- Steinmetz A., Gubbins E. J., Bogorad L. The anticodon of the maize chloroplast gene for tRNA Leu UAA is split by a large intron. Nucleic Acids Res. 1982 May 25;10(10):3027–3037. doi: 10.1093/nar/10.10.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaiwa F., Sugiura M. Nucleotide sequence of the 16S - 23S spacer region in an rRNA gene cluster from tobacco chloroplast DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2665–2676. doi: 10.1093/nar/10.8.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Bottomley W., Whitfeld P. R. Junctions of the large single copy region and the inverted repeats in Spinacia oleracea and Nicotiana debneyi chloroplast DNA: sequence of the genes for tRNAHis and the ribosomal proteins S19 and L2. Nucleic Acids Res. 1984 Aug 24;12(16):6547–6558. doi: 10.1093/nar/12.16.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Clegg M. T. The barley chloroplast DNA atpBE, trnM2, and trnV1 loci. Nucleic Acids Res. 1984 Mar 12;12(5):2549–2559. doi: 10.1093/nar/12.5.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]