Abstract
A number of genome analyses and searches using programs that focus on the RNA-specific bulge-helix-bulge (BHB) motif have uncovered a wide variety of disrupted tRNA genes. The results of these analyses have shown that genetic information encoding functional RNAs is described in the genome cryptically and is retrieved using various strategies. One such strategy is represented by circularly permuted tRNA genes, in which the sequences encoding the 5′-half and 3′-half of the specific tRNA are separated and inverted on the genome. Biochemical analyses have defined a processing pathway in which the termini of tRNA precursors (pre-tRNAs) are ligated to form a characteristic circular RNA intermediate, which is then cleaved at the acceptor-stem to generate the typical cloverleaf structure with functional termini. The sequences adjacent to the processing site located between the 3′-half and the 5′-half of pre-tRNAs potentially form a BHB motif, which is the dominant recognition site for the tRNA-intron splicing endonuclease, suggesting that circularization of pre-tRNAs depends on the splicing machinery. Some permuted tRNAs contain a BHB-mediated intron in their 5′- or 3′-half, meaning that removal of an intron, as well as swapping of the 5′- and 3′-halves, are required during maturation of their pre-tRNAs. To date, 34 permuted tRNA genes have been identified from six species of unicellular algae and one archaeon. Although their physiological significance and mechanism of development remain unclear, the splicing system of BHB motifs seems to have played a key role in the formation of permuted tRNA genes. In this review, current knowledge of circularly permuted tRNA genes is presented and some unanswered questions regarding these species are discussed.
Keywords: tRNA gene, circular gene permutation, BHB motif, tRNA-splicing endonuclease, intron
Introduction
The cloverleaf structure of a single polynucleotide tRNA molecule is universally conserved among organisms. However, tRNA genes are often divided into parts on the chromosome; in bacteria, archaea, eukarya, and organelles, several tRNA genes are interrupted by various types of introns, which are removed by RNA splicing after transcription (Thompson and Daniels, 1988; Kjems et al., 1989; Westaway and Abelson, 1995; Belfort and Weiner, 1997; Marck and Grosjean, 2002, 2003; Jühling et al., 2009; Abe et al., 2011). Introns in nuclear and archaeal tRNAs are generally cleaved by tRNA-splicing endonuclease (Reyes and Abelson, 1988; Abelson et al., 1998; Calvin and Li, 2008), while those in eubacteria and organelle tRNAs are encoded as self-splicing group I or II introns (Kuhsel et al., 1990; Xu et al., 1990a; Reinhold-Hurek and Shub, 1992; Biniszkiewicz et al., 1994; Jacquier, 1996; Bonen and Vogel, 2001). In addition to these well-known cis-spliced tRNA genes (intron-containing tRNAs), recently developed software has enabled the identification of additional distinct types of disrupted tRNA genes. Use of the Split-tRNA-Search (Randau et al., 2005a), SPLITS and SPLITSX (Sugahara et al., 2006, 2007) packages, in combination with the widely used tRNAscan-SE program (Lowe and Eddy, 1997), has led to the discovery of a variety of disrupted tRNA genes from the archaeal lineage, such as trans-spliced tRNAs (split tRNAs) that are joined at several positions in the cloverleaf structure (Randau et al., 2005a,b; Fujishima et al., 2009; Chan et al., 2011) and cis-spliced tRNAs containing one or multiple introns at non-canonical positions (Sugahara et al., 2008, 2009; Chan and Lowe, 2009). These newly identified tRNAs commonly harbor a characteristic bulge-helix-bulge (BHB) motif, which comprises two 3-nucleotide bulges separated by a single 4-base pair stem and was originally identified around the intron-exon junction of eukaryal and archaeal tRNAs (Thompson and Daniels, 1988; Kjems et al., 1989; Belfort and Weiner, 1997; Fabbri et al., 1998; Marck and Grosjean, 2003).
Nuclear tRNA introns are generally short, comprise a relaxed form of the BHB motif denoted as a hBH or BHB-like (BHL) motif, and are located exclusively between positions 37 and 38 (37/38), which is 3′ adjacent to the anticodon (the canonical position) (Marck and Grosjean, 2002, 2003; Jühling et al., 2009). This limited location of the BHB motif in the cloverleaf structure is crucial for the precise recognition of precursor tRNAs (pre-tRNAs) by eukaryal tRNA-splicing endonucleases (Greer et al., 1987; Reyes and Abelson, 1988; Westaway and Abelson, 1995; Di Nicola Negri et al., 1997; Trotta et al., 1997, 2006; Xue et al., 2006; Calvin and Li, 2008). However, recent analyses of the nuclear and nucleomorph genomes of unicellular algae identified a number of non-canonical BHB-mediated disrupted tRNA genes, including circularly permuted and atypical intron-containing genes (Kawach et al., 2005; Soma et al., 2007, 2013; Landweber, 2007; Maruyama et al., 2010; Chan et al., 2011). Analysis of the processing intermediates of permuted tRNAs revealed a new strategy for post-transcriptional processing of genetic information by inversion of RNA fragments and relocation of the termini via circularization of pre-RNA molecules (Soma et al., 2007, 2013; Maruyama et al., 2010). A further analysis also identified permuted tRNA genes in an archaeal lineage (Chan et al., 2011), highlighting the considerable diversity and wide distribution of tRNA gene disruption among organisms. While BHB motifs and the tRNA-intron splicing system must have been a prerequisite for the development of permuted tRNA genes (Soma et al., 2007; Sugahara et al., 2009; Maruyama et al., 2010; Tocchini-Valentini and Tocchini-Valentini, 2012; Kanai, 2013), their detailed mechanisms and physiological relevance remain unclear.
Here, the structure, expression, and phylogeny of circularly permuted tRNA genes are summarized. Discussions of their possible physiological relevance and method of development in correlation with the tRNA expression system and other disrupted non-coding RNA genes are also provided.
Identification and distribution of circularly permuted tRNA genes
Circularly permuted tRNA genes were initially identified in the nuclear genome of Cyanidioschyzon merolae 10D (Soma et al., 2007), an ultra-small unicellular red alga that inhabits an extreme environment (pH 1–3, 40–50°C) and represents one of the most ancestral forms of eukaryote (Kuroiwa, 1998; Nozaki et al., 2003, 2007; Matsuzaki et al., 2004). A primary search of the complete 16.5 Mbp C. merolae nuclear genome sequence was performed using tRNAscan-SE, the most well-known and widely used software, which identifies tRNA genes without or with introns canonically located at position 37/38 in the anticodon-loop (Lowe and Eddy, 1997). This analysis identified a total of 30 predicted tRNA genes, which is insufficient to decode the 61 sense codons utilized in the nuclear genome of C. merolae (Matsuzaki et al., 2004). Therefore, to discover unidentified C. merolae nuclear tRNA genes, a genome-wide analysis was performed using the SPLITS and SPLITSX programs (Sugahara et al., 2006, 2007), which were developed to detect tRNA genes harboring BHB motifs, including cis-spliced tRNAs with introns inserted at various positions and split tRNAs that are joined at several positions in the cloverleaf structure. In addition, a BLAST search of conserved sequences in the TΨC-arm or anticodon-arm was also performed. This approach identified a total of 43 tRNA genes for 42 anticodons, which is sufficient to decode the 61 codons (Soma et al., 2007, 2013).
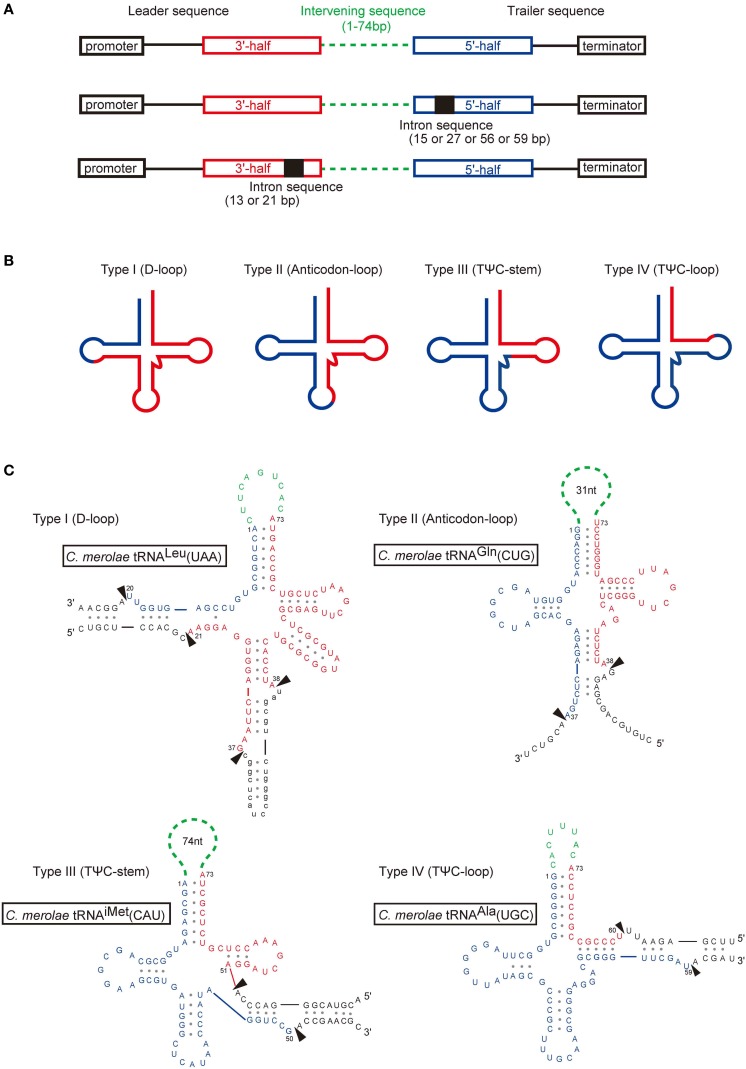
Notably, for 11 of the 43 tRNA genes identified in C. merolae, the sequence encoding the 3′-half of the tRNA is positioned upstream of the sequence encoding the 5′-half in the genome (Figure 1A), and the two halves are interrupted by an intervening sequence that corresponds to the boundary connecting the 5′- and 3′-ends of the acceptor-stem of a mature tRNA. This arrangement is termed the circular gene permutation model (Heinonen et al., 1987; Pan et al., 1991; Keiler et al., 2000); hence, these genes were named “circularly permuted tRNA genes” (Soma et al., 2007). The study by Soma et al. (2007) was the first report of the existence of permuted genes encoding tRNAs or eukaryal nuclear-encoded non-coding RNAs. A TATA-like sequence was identified within the region 50 bp upstream of the 3′-half of most of the permuted tRNA genes as well as the non-permuted tRNA genes in C. merolae (Matsuzaki et al., 2004; Soma et al., 2013), indicating its importance for the transcription of tRNA genes. A T-stretch corresponding to a termination signal for RNA polymerase III (RNAPIII) was identified downstream of the 5′-half of these genes (Sprague, 1995; Hamada et al., 2000; Nielsen et al., 2013), but no promoter or termination signals were identified in the intervening region between the 3′- and 5′-halves, which varies in length from 7 to 74 nucleotides (Table 1). These observations suggest that the 3′- and 5′-halves of the putative tRNA genes are transcribed as a linear RNA. The exon sequences of both the permuted and non-permuted C. merolae tRNA genes show ordinary characteristics of eukaryal tRNAs and contain consensus elements found in eukaryotic tRNAs (Marck and Grosjean, 2002; Jühling et al., 2009), including U8, the R15:Y48 tertiary base pairing, G18G19, and U33; in addition, the U54U55C56 for elongator tRNAs and the A54U55C56 for initiator tRNAMet are also conserved. The 3′-terminal CCA sequence, to which an amino acid is conjugated, is not encoded in the C. merolae genome, as found in other eukaryotes.
Figure 1.
Gene organization and structures of permuted tRNAs. (A) Schematic representations of the structures of permuted tRNA genes with or without an intron. The 5′-half (blue) and the 3′-half (red) of the mature tRNA, the intron sequence (black), and the intervening sequence (green) of the pre-tRNA are shown. (B) Most permuted tRNAs can be classified into four types based on the location of the junction between the 3′-end of the 5′-half (blue) and the 5′-end of the 3′-half (red) in the secondary structure. (C) Inferred secondary structures of pre-tRNAs representing the four types of permuted tRNA genes in C. merolae. The arrowheads indicate the positions to be processed. The intron sequence is shown in lower case. The tRNA positions are numbered according to Marck and Grosjean (2002). The figures are partially identical to the Figure 1 of Soma et al. (2007).
Table 1.
Characteristics of permuted tRNA genes in unicellular algae and archaea.
| Organism | Permuted tRNA (anticodon) | Type (junction) | Intervening seq. (bp) | Intron (BHB, position, bp) |
|---|---|---|---|---|
| RED ALGA | ||||
| C. merolae | Leu(TAA) | I (20/21) | 10 | hBHBh', 37/38, 21 |
| Gln(CTG) | II (37/38) | 31 | No | |
| Leu(CAG) | II (37/38) | 9 | No | |
| Thr(CGT) | II (37/38) | 31 | No | |
| Thr(AGT) | II (37/38) | 24 | No | |
| Gly(CCC) | II (37/38) | 7 | no h, 55/56, 13 | |
| Gly(CCC) | II (37/38) | 7 | no h, 55/56, 13 | |
| iMet(CAT) | III (50/51) | 74 | No | |
| Ala(TGC) | IV (59/60) | 8 | No | |
| Arg(CCT) | IV (59/60) | 23 | hBHBh', 14/15, 27 | |
| Lys(TTT) | IV (59/60) | 38 | No | |
| GREEN ALGA | ||||
| O. lucimarinus | Leu (TAA) | II (37/38) | 25 | No |
| Ser(CGA) | II (37/38) | 12 | No | |
| Ser(TGA) | II (37/38) | 13 | no H, 27/28, 56 | |
| Cys(GCA) | II (37/38) | 26 | No | |
| O. tauri | Ser (CGA)* | V (15/16) | 17 | No |
| Ser(CGA) | II (37/38) | 28 | No | |
| Ser(TGA) | II (37/38) | 33 | No | |
| Arg(TCT) | II (37/38) | 11 | No | |
| Asn(GTT) | II (37/38) | 22 | No | |
| Cys(GCA) | II (37/38) | 38 | No | |
| M. pusilla | Ser (GCT) | II (37/38) | 15 | No |
| Ser(CGA) | II (37/38) | 19 | No | |
| Ser(AGA) | II (37/38) | 64 | No | |
| iMet(CAT) | II (37/38) | 47 | No | |
| Leu(TAA) | II (37/38) | 20 | No | |
| Cys(GCA) | II (37/38) | 12 | No | |
| M.sp RCC299 | Ser (CGA) | II (37/38) | 16 | No |
| Ser(TGA) | II (37/38) | 12 | No | |
| iMet(CAT) | II (37/38) | 12 | No | |
| CHLORARACHNIOPHYTE ALGA | ||||
| B. natans | Ser (AGA) | II (37/38) | 5 | No |
| (nucleomorph) | Ser(CGA) | II (37/38) | 5 | No |
| CRENARCHAEA | ||||
| T. pendens | iMet (CAT) | IV (59/60) | 7 | no H, 37/38, 15 |
| Tyr(GTA) | IV (59/60) | 1 | no H, 37/38, 59 | |
Each gene is identified by the amino acid with which the tRNA is charged. Classification of permuted tRNAs (types I–V) is based on the location of the junction between the 5 ′-half and the 3 ′-half in the inferred secondary structure for pre-tRNAs. The length (bp, base pair) of sequence encoding for the intervening sequence and the presence of an intron are indicated. In C. merolae, two distinct genes that produce identical mature tRNA sequences encode tRNAGly(CCC). iMet and eMet mean initiator and elongator tRNAMet(CAT), respectively. The asterisk indicates that the sequence of this tRNA is a typical, and experimental analysis would be preferable.
As shown in Figure 1B, C. merolae permuted tRNA genes can be classified into four types (I–IV) based on the location of the junction between the 3′-end of the 5′-half and the 5′-end of the 3′-half in the inferred secondary structures of the pre-tRNAs. The junctions are located at position 20/21 in the D-loop (type I), position 37/38 in the anticodon-loop (type II), position 50/51 in the TΨ C-stem (type III), or position 59/60 in the TΨ C-loop (type IV). In C. merolae, one type I, six type II, one type III, and three type IV candidate tRNAs have been identified (Figure 1C, Table 1). The sequences adjacent to the junctions in the pre-tRNAs are predicted to form a BHB motif that is generally found around the intron-exon junctions of nuclear and archaeal pre-tRNAs (Figure 1C).
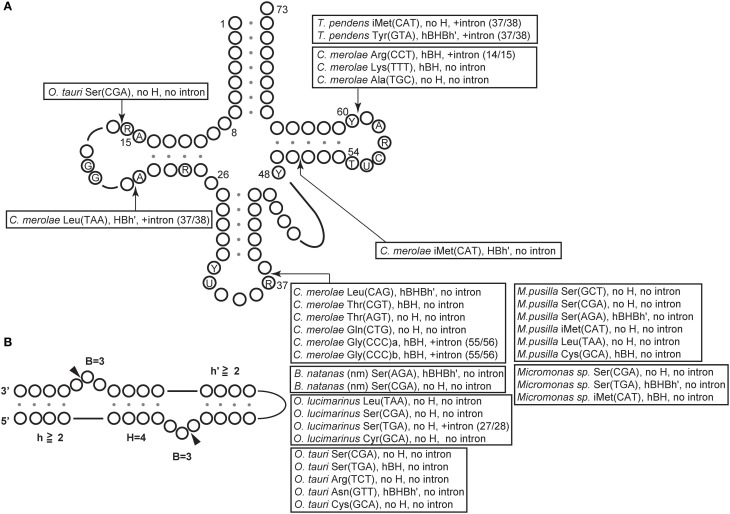
To date, 34 permuted tRNA genes have been identified in unicellular algae and archaea (Table 1), including 11 genes from the nuclear genome of the red alga C. merolae (Soma et al., 2007); 19 genes from the nuclear genome of four green algae (Ostreococcus lucimarimus, Ostreococcus tauri, Micromonas Pusilla, and Micromonas sp. RCC299) (Maruyama et al., 2010); two genes from the nucleomorph genome of the chlorarachiniophyte alga Bigelowiella natans (Maruyama et al., 2010), which is a remnant of a green algal nuclear DNA that developed as a secondary endosymbiont (Douglas et al., 2001; Archibald, 2007; Archibald and Lane, 2009); and two genes from the genome of the crenarchaeon Thermofilum pendens (Chan et al., 2011). In the nucleomorph and the nucleus of green algae, the junctions of the permuted pre-tRNAs are located most commonly at position 37/38 in the anticodon-loop (type II), while they are located at position 59/60 in the TΨ C-loop (type IV) in archaea (Figure 2A, Table 1). This tendency contrasts with that in the red alga C. merolae, in which the junctions are found at various positions in the cloverleaf structure. The intervening sequence varies from 1 to 74 bp among organisms (Table 1); tRNATyr(GTA) from T. pendens contains the shortest intervening sequence, while tRNAiMet(CAT) from C. merolae contains the longest intervening sequence. The species of amino acid or anticodon in the tRNAs encoded by permuted genes are not conserved among organisms. Interestingly, permuted tRNAiMet(CAT) exists in each lineage of red algae, green algae, and crenarchaea. In addition, tRNASer, tRNALeu, and tRNATyr, which are classified as class II tRNAs and have long variable-arms (Rich and Rajbhandary, 1976; Dirheimer et al., 1995), tend to be encoded as permuted genes. This observation may imply that the evolution of the long variable-arm, which is the dominant element required for recognition by corresponding aminoacyl-tRNA synthetases (Asahara et al., 1993; Himeno et al., 1997a; Soma et al., 1999), is correlated with that of the tRNA gene structure. Indeed, the long variable-arm is suggested to have arisen from an intron (Kjems et al., 1989). Further analyses of the sequences of disrupted tRNAs will aid identification of the types of tRNA genes that tend to be permuted.
Figure 2.
Distribution of BHB motifs at the junctions of permuted tRNAs in algae and archaea. (A) The tRNA nucleotides are numbered according to Marck and Grosjean (2002) and the arrows indicate the positions of the BHB motifs. The type of BHB motif and the presence or absence of an intron is indicated for each tRNA. For example, “C. merolae Leu(TAA), HBh', +intron, (37/38)” at the position between nucleotides 20 and 21 means that the HBh' is from the junction of the C. merolae permuted tRNALeu(UAA), which contains an intron at 37/38. (B) The BHB motif is classified by one or two 3-nucleotide (nt) bulges (denoted as “B = 3”) separated by a central 4-base pair (bp) helix (denoted as “H = 4”) and flanked by two helices (denoted as h or h′), each with more than two base pairings. “hBHBh′” is the strict form of the BHB motif, which contains two bulges, a central helix and two flanking helices. The relaxed form of the BHB motif (BHL) lacking some of these bulges or helices is denoted as “hBH” or “HBh'.” The term “no H” represents motifs that do not contain a central 4-bp helix.
Transcription of permuted tRNA genes
Northern blotting and aminoacylation analyses of C. merolae total RNA verified that tRNA molecules expressed from permuted genes are aminoacylated and are thus likely to participate in protein synthesis (Soma et al., 2007). Expression of some permuted tRNA genes from the nucleomorph and the nucleus of green algae has also been confirmed by northern blotting or reverse transcription polymerase chain reaction analyses (Maruyama et al., 2010). However, the function of mature tRNAs in the nucleomorph is unclear because protein synthesis in these structures has not yet been observed experimentally (Archibald and Lane, 2009; Curtis et al., 2012). The two permuted tRNAs in the archaea T. pendens are both encoded by single-copy genes for a unique anticodon that cannot be supplemented by other isoacceptors (Chan et al., 2011); therefore, they must be expressed and produce functional tRNA molecules.
The identification of unusual (permuted and atypical intron-containing) tRNA genes in eukaryotes raised an intriguing question about the mechanism of transcription. In eukaryotes, transcription of tRNA genes is generally performed by RNAPIII and it relies on an intragenic bipartite promoter consisting of an A box and a B box, which correspond to the highly conserved sequences in the D-arm (positions 8–19) and TΨ C-arm (positions 52–62), respectively, (Galli et al., 1981; Ciliberto et al., 1983; Sprague, 1995; Guffanti et al., 2006; Marck et al., 2006). The protein factors that bind to these motifs have been well-characterized in yeast (Willis, 1993; Paule and White, 2000; Geiduschek and Kassavetis, 2001; Huang and Maraia, 2001; Schramm and Hernandez, 2002). Polymerase III C (TFIIIC), a multi-subunit complex of transcription factors that is essential for transcription by RNAPIII, binds to the A and B boxes simultaneously and promotes binding of the TFIIIB complex, which includes the TATA-box binding protein, to the region upstream of the tRNA sequence, followed by recruitment of RNAPIII. The dependency of transcription of tRNA genes on the A and B boxes is predominantly conserved; however, the additional requirements for transcription are diverse among organisms and the upstream region sometimes contributes to the efficiency of the initiation step (Choisne et al., 1998; Yukawa et al., 2000; Hamada et al., 2001; Giuliodori et al., 2003; Dieci et al., 2006).
In permuted tRNA genes, the A box and B box are located inversely and are interrupted by an intervening sequence of variable length. This positional relationship is unsuitable for TFIIIC binding; therefore, the A and B boxes may not be uniformly bound by TFIIIC and the intragenic promoter may be dispensable for transcription of these genes. Instead, an upstream TATA-like sequence and a downstream T-stretch, which are probably the promoter and termination signal, respectively, (Sprague, 1995; Hamada et al., 2000, 2001; Nielsen et al., 2013), are located close to most permuted tRNA genes in C. merolae (Soma et al., 2007). This genomic arrangement also occurs for non-permuted tRNA genes in C. merolae, and the A and B boxes in the promoters of these genes may not be recognized by TFIIIC because they are often interrupted by a single or multiple (up to three) introns of various lengths (11–69 bp) (Matsuzaki et al., 2004; Soma et al., 2013). Homologs of TFC1 and TFC3, the TFIIIC components that are responsible for binding to the A and B boxes, have not been identified in C. merolae (Matsuzaki et al., 2004; Nozaki et al., 2007). Taken together, these findings suggest that C. merolae employs a non-canonical transcription system that is independent of TFIIIC and directs recruitment of TFIIIB to the upstream TATA-box, thereby enabling the transcription of various types of tRNA genes. An ambiguous AT-rich region is also located upstream of some permuted tRNA-encoding sequences in the B. natans nucleomorph and the nucleus of green algae (Maruyama et al., 2010). Therefore, TATA-like sequence-dependent transcription of tRNA genes may predominate in algae. This possibility is supported by the fact that an upstream TATA box is well conserved and functionally important for transcription of tRNA genes in some plants and fungi (Choisne et al., 1997, 1998; Yukawa et al., 2000; Hamada et al., 2001; Dieci et al., 2006). In addition, transcription of Saccharomyces cerevisiae tRNA genes harboring an upstream TATA box proceeds without TFIIIC in vitro (Dieci et al., 2000).
In archaea, transcription of a stable RNA depends on the upstream promoter including BRE (TFB response element) and TATA box (Wich et al., 1986; Thomm and Wich, 1988; Palmer and Daniels, 1995; Reeve, 2003), and on a downstream poly T sequence, which contributes to transcription termination (Santangelo et al., 2009). In T. pendens, which harbors two permuted tRNA genes, a predicted AT-rich promoter is located upstream of most of its tRNA genes (Chan et al., 2011), suggesting that various types of tRNA genes are potentially expressed. Consistent with this notion, T. pendens contains a large number of tRNA genes that are disrupted by various introns (Sugahara et al., 2009; Chan et al., 2011; Fujishima et al., 2011).
Maturation of permuted pre-tRNAs via a circular RNA intermediate
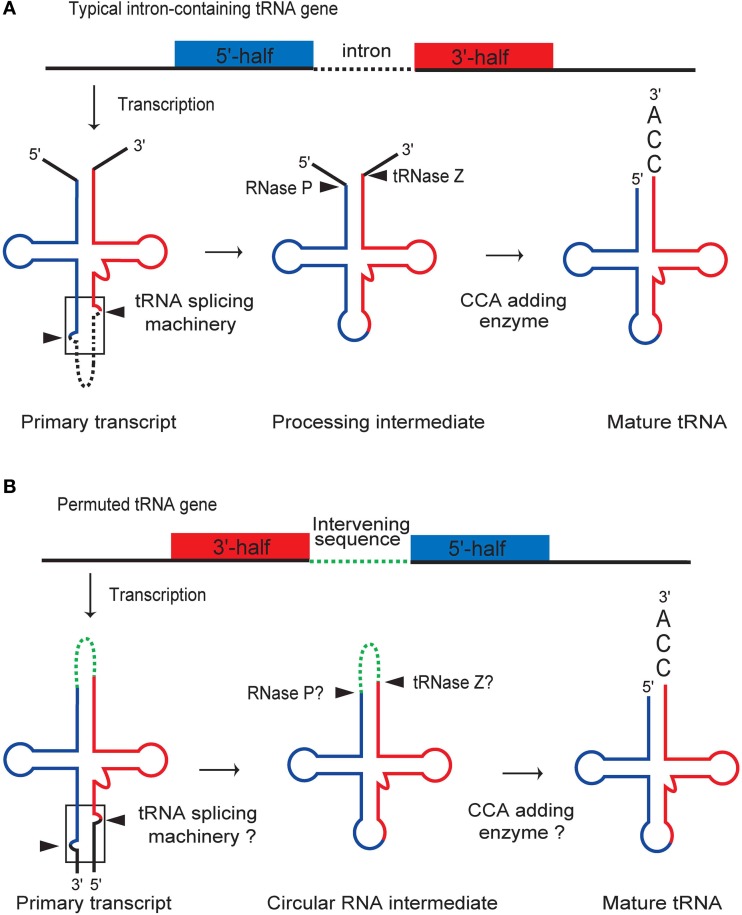
Processing of a pre-tRNA typically involves intronic splicing, maturation of the 5′- and 3′-ends at the acceptor stem, and nucleotide modification (Figure 3A) (Deutscher, 1995; Hopper and Phizicky, 2003). Biochemical analyses have shown that permuted pre-tRNAs in unicellular algae are maturated by a processing pathway that utilizes a circular RNA intermediate to exchange the location of the 5′- and 3′-halves of the tRNA (Soma et al., 2007, 2013; Maruyama et al., 2010). Reverse transcription polymerase chain reaction and sequencing analyses identified the following processing intermediates derived from algal permuted tRNAs: a circularly permuted pre-tRNA, the sequence of which aligns in the order of the leader sequence, the 3′-half of tRNA, the intervening sequence, the 5′-half of tRNA, and then the trailer sequence; and a circular RNA intermediate, in which the leader and trailer sequences are removed and the resulting ends are ligated, while the intervening sequence is retained. Furthermore, a consistent PCR product was also observed in these analyses, suggesting that two rounds of reverse transcription occur around a circular intermediate, thereby confirming the presence of the circular RNA molecule. Terminal sequences were also verified for a mature tRNA, in which the extra sequences are removed and the CCA sequence is added post-transcriptionally to the 3′-terminus of the acceptor-stem, as occurs in other eukaryotes.
Figure 3.
Comparison of the processing pathways for typical intronic and permuted pre-tRNAs. (A) Maturation of a typical intronic pre-tRNA involves intronic splicing, processing of the 5′- and 3′-ends by RNase P and tRNAse Z, and addition of the 3′-terminal CCA sequence. (B) Maturation of a permuted pre-tRNA starts with processing of the BHB motif (boxed) by the tRNA-splicing machinery, resulting in the formation of a circular RNA intermediate. The intervening sequence is then removed by RNase P and tRNase Z, followed by CCA addition. The sequential processing of permuted pre-tRNAs in the proposed pathway may be accomplished using processing machineries that are commonly used for typical pre-tRNAs, because the recognition elements for each processing enzyme are conserved in the permuted pre-tRNA and circular intermediate. The figure is partially identical to the Figure 3C of Soma et al. (2007).
As summarized in the model presented in Figure 3B, maturation of permuted pre-tRNAs in algal cells probably starts with processing of the junction of the termini to form a circular RNA intermediate in which the termini are joined by the intervening sequence. The intervening sequence in the acceptor-loop of the circular RNA intermediate is then removed, possibly by RNase P (Altman et al., 1995; Jarrous and Gopalan, 2010; Altman, 2011) and tRNase Z (Deutscher, 1995; Schürer et al., 2001; Schiffer et al., 2002; Späth et al., 2007), which are universal endoribonucleases. Finally, the 3′-terminal CCA sequence is added (Weiner, 2004) to generate the functional acceptor-stem of the tRNA. Because the circular RNA intermediate has been detected in red and green algae, this model is likely to be common to permuted tRNAs of both types of algae.
Cleavage of the leader and trailer sequences at the junction of permuted pre-tRNAs is most likely performed by the tRNA-intron splicing machinery, because the sequences adjoining the processing sites potentially form a BHB motif, which is the dominant recognition element for nuclear and archaeal tRNA-splicing endonucleases (Figures 1C, 2B). After excision of the BHB motifs at the junction, subsequent ligation of the 5′- and 3′-termini of the exons is required and is probably carried out by tRNA-splicing ligase (Xu et al., 1990b; Westaway and Abelson, 1995; Englert et al., 2011, 2012; Popow et al., 2011). It is intriguing that various positions in the cloverleaf structure of tRNAs, even the core region of the L-shaped tertiary structure, can serve as termini for permuted pre-RNA molecules that are recognized by the splicing machinery.
RNase P (McClain et al., 1987; Christian et al., 2002; Zahler et al., 2003; Kirsebom, 2007; Reiter et al., 2010; Altman, 2011) and tRNase Z (Nashimoto et al., 1999; Li de la Sierra-Gallay et al., 2006; Späth et al., 2007; Minagawa et al., 2008) generally recognize the top half of the L-shaped tertiary structure of a tRNA corresponding to the acceptor-stem and the TΨ C-arm, and do not require the mature body of the tRNA. Therefore, these enzymes may also perform endonucleolytic cleavage of the acceptor-loop of a circular RNA intermediate. Although some endoribonucleases require the linear ends of substrates to function (Mackie, 1998; Suzuki et al., 2006), it is not known whether this condition holds for RNase P and tRNase Z. The intron in the D- and TΨ C-arm, which inhibits folding of the tertiary structure of a tRNA, should be removed before processing of the acceptor-loop by RNase P and tRNase Z. Consistent with this requirement, the intron in the TΨ C-loop of a circular intermediate of C. merolae tRNAGly, which harbors both intronic and permuted structures, is removed before the intervening sequence at the acceptor-loop is processed (Soma et al., 2013). This finding can be explained by the fact that the top half of substrates for C. merolae tRNase Z must form a canonical tertiary structure, and circular pre-tRNAs without an intron would be able to fold into the canonical tertiary structure, which agrees with the previous discovery that artificially permuted tRNA molecules can fold into correct tertiary structures (Pan et al., 1991).
In eukaryotes, each tRNA processing step occurs at a different location in the cell, and the cellular distribution of processing enzymes is not conserved among organisms. In animal cells, the tRNA-splicing endonuclease and ligase are localized to the nucleus (Westaway and Abelson, 1995; Paushkin et al., 2004). By contrast, in budding yeast, the endonuclease is present on the surface of mitochondria (Huh et al., 2003; Yoshihisa et al., 2003) and the ligase is present in the cytosol (Huh et al., 2003). RNase P and tRNase Z are found in the nucleus and/or cytoplasm in eukaryotic cells (Späth et al., 2007; Canino et al., 2009; Gobert et al., 2010; Pinker et al., 2013). Accordingly, the order of the processing steps of a permuted pre-tRNA in algal cells will likely be governed by the location of the enzymes required.
It is unclear whether maturation of archaeal permuted pre-tRNAs involves the formation of a circular RNA intermediate. In a recent study, an in vitro transcript simulating a permuted pre-tRNA, which was composed of a tandem repeat of intron-containing tRNA, was cleaved at the BHB motif by a recombinant splicing endonuclease from the euryarchaeon Methanococcus jannaschii (Tocchini-Valentini and Tocchini-Valentini, 2012), suggesting that archaeal permuted pre-tRNAs can be processed in a similar pathway to that found in algae. Analysis of permuted pre-tRNA processing in T. pendens may also help to clarify whether the physiological role of permuted tRNA genes is ascribed to the formation of the circular RNA intermediate. With the exception of Nanoarchaeum equitans (Randau et al., 2008; Heinemann et al., 2010), RNase P and tRNase Z generally contribute to the end maturation of tRNAs in archaea (Späth et al., 2007; Jarrous and Gopalan, 2010). The 3′-terminal CCA sequence of two permuted tRNAs is encoded in the genome sequence of T. pendens (Chan et al., 2011) and one of these genes contains a short intervening sequence of only one nucleotide. It will be intriguing to clarify how such a short intervening sequence in the acceptor-loop is removed.
Processing of an intron in permuted pre-tRNAs
Four tRNA genes from the red alga C. merolae (Soma et al., 2007), one tRNA gene from the green alga O. lucimarinus (Maruyama et al., 2010), and two tRNAs genes from the crenarchaeon T. pendens (Chan et al., 2011) contain an intron in the 5′- or 3′-half of the gene (Figure 1A, Table 1), meaning that their pre-tRNAs require splicing of an intron in addition to swapping of the 5′- and 3′-halves. The position of the intron is not conserved among these organisms; in the four C. merolae tRNA genes, the introns are inserted at various positions (the D-loop, the anticodon-loop, and the TΨ C-loop), while those in the O. lucimarinus and T. pendens tRNA genes are inserted at specific positions: 27/28 in the anticodon-stem and 37/38 in the anticodon-loop, respectively. In C. merolae and T. pendens, the intron-exon junction and the termini of permuted pre-tRNAs harboring an intron can each form an independent BHB motif. The two BHB motifs are not nested; therefore, processing of one BHB motif can be preceded by processing of the other. Using C. merolae tRNAGly(CCC), which possesses both permuted (with the junction at position 37/38 in the anticodon-loop) and intronic (inserted at position 55/56 in the TΨ C-loop) structures, it was determined that the BHB motif in the intron is processed before the BHB motif in the termini of permuted pre-tRNAGly(CCC) (Soma et al., 2013). The theoretical ΔG of the BHB motif in the intron was calculated to be slightly lower than that of the BHB motif in the termini. The same phenomenon was also observed for precursors transcribed from multiple intron-containing (but not permuted) tRNA genes in C. merolae, in which the BHB motifs in the pre-tRNAs were removed in the order dictated by the theoretical free energy of each motif (Soma et al., 2013). These findings indicate that multiple BHB motifs in permuted and/or intronic pre-tRNAs in C. merolae are processed sequentially, even when each BHB motif can fold independently. This feature may be attributable to the stability of each BHB motif and their accessibility to the splicing endonuclease. Alternatively, it may depend on the position of the BHB motifs, because the BHB motif at the canonical position 37/38 is always the final substrate and has a relatively high ΔG. BHB motifs at 37/38, even those that form the junction of the permuted pre-tRNA or the intron, may be recognized by C. merolae endonuclease only after BHB motifs at the other positions have been processed. This procedure contrasts with the processing of multimeric introns in some archaeal pre-tRNAs, in which the introns are nested and the last intron can form a BHB motif only after the other introns are processed (Sugahara et al., 2007; Tocchini-Valentini et al., 2009).
Correlation between the BHB motif at the junction of permuted pre-tRNAs and the substrate specificity of splicing endonucleases
The BHB motif is the dominant recognition element for all known nuclear and archaeal tRNA-splicing endonucleases (Fruscoloni et al., 2001; Marck and Grosjean, 2003; Tocchini-Valentini et al., 2005a; Xue et al., 2006; Calvin and Li, 2008) and processing by these enzymes should have been pivotal for the development and maintenance of permuted tRNA genes in the genome. Archaeal endonucleases exhibit symmetrical architectures, and recognition of the splice sites of intronic pre-tRNAs by these enzymes is largely dependent on the BHB motif (Figure 4A) (Thompson and Daniels, 1988; Diener and Moore, 1998; Tocchini-Valentini et al., 2005a,b; Calvin and Li, 2008). In most archaeal tRNAs, the BHB motifs develop a relaxed form (hBH, as shown in Figure 2B) and are located at position 37/38 in the anticodon-loop, while several species from Crenarchaeota contain introns at non-canonical positions, such as the anticodon-arm, D-arm, TΨ C-arm, variable-arm, or acceptor-stem, with strict (hBHBh') or relaxed (hBH, BHh', or no H) forms of the BHB motif (Marck and Grosjean, 2003; Tocchini-Valentini et al., 2005a; Sugahara et al., 2007, 2008, 2009). In addition to tRNAs, BHB-mediated introns are also found in rRNAs and mRNAs in some archaea (Kjems and Garrett, 1988; Tang et al., 2002; Watanabe et al., 2002; Yoshinari et al., 2006). Furthermore, the combination of RNA fragments during maturation of split tRNAs depends on the processing of the BHB motifs by the tRNA-splicing machinery in N. equitans (Randau et al., 2005c), indicating that BHB-mediated disruption of genetic information and its processing by splicing endonucleases is widespread in archaea. Four different types of endonuclease have been identified in archaea (Tocchini-Valentini et al., 2005a; Calvin and Li, 2008; Fujishima et al., 2011; Hirata et al., 2011); the subunit architecture of these endonucleases seems to have co-evolved, by “subfunctionalization,” with their substrate specificity (Tocchini-Valentini et al., 2005b, 2007). T. pendens contains a heterotetrameric endonuclease (α2β 2) that can recognize both strict (hBHBh') and relaxed (BHL) motifs, and the junction of its two permuted tRNAs comprises no H or hBHBh' motif, and is located at position 59/60 in the TΨ C-loop (Figure 2, Table 1). The broad substrate specificity of the T. pendens endonuclease would have allowed the development and maintenance of permuted tRNAs during evolution.
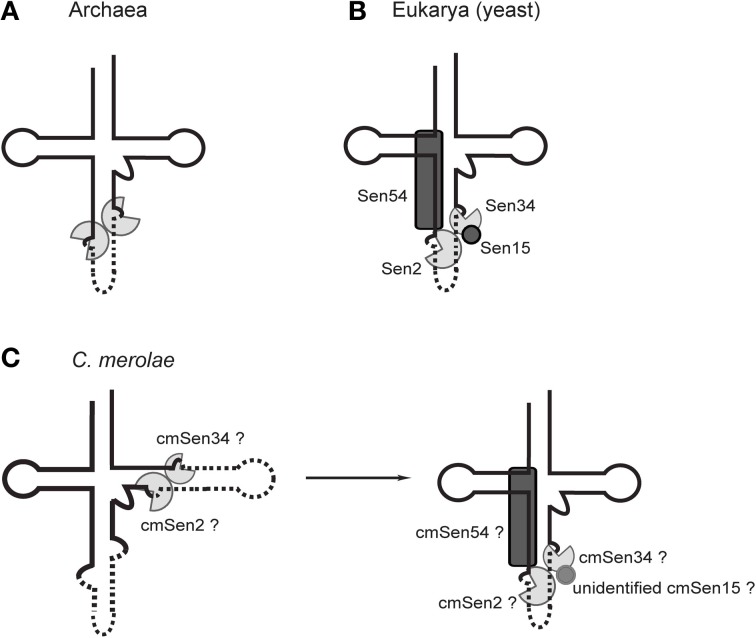
Figure 4.
Schematic representations of the tRNA-splicing endonuclease in archaea and eukaryotes. (A) An archaeal dimeric endonuclease comprising two catalytic subunits. (B) The yeast heterotetrameric endonuclease comprising two catalytic subunits (Sen2 and Sen34) and two accessory subunits (Sen15 and Sen54). (C) The possible heterodimeric (cmSen2 and cmSen34), heterotrimeric (cmSen2, cmSen34, and cmSen54), and heterotetrameric (cmSen2, cmSen34, cmSen54, and unidentified cmSen15) forms of the C. merolae endonuclease.
The S. cerevisiae splicing endonuclease forms a heterotetrameric structure (αβδε) comprised of two catalytic subunits (Sen2 and Sen34) and two accessory subunits (Sen15 and Sen54) (Rauhut et al., 1990; Westaway and Abelson, 1995; Trotta et al., 1997; Calvin and Li, 2008). Interactions between Sen2 and Sen54, and between Sen15 and Sen34, were identified by a yeast two-hybrid experiment (Trotta et al., 1997). These four subunits function cooperatively to recognize cleavage sites via “a ruler mechanism,” in which the endonuclease measures a specified distance to the site at which the cuts should be made in a pre-tRNA (Figure 4B) (Greer et al., 1987; Reyes and Abelson, 1988; Westaway and Abelson, 1995; Fabbri et al., 1998; Calvin and Li, 2008). In addition to the typical hBH motif at the canonical 37/38 position, yeast endonuclease recognizes the mature domain of pre-tRNA and the base pairs between the anticodon and the intron (A·I base pairs) (Mattoccia et al., 1988; Baldi et al., 1983, 1992; Di Nicola Negri et al., 1997; Trotta et al., 2006; Xue et al., 2006). Similarly, wheat germ endonuclease recognizes some specific nucleotides in the D-stem, and the mature tRNA domain is required for adequate binding to the endonuclease (Stange et al., 1992). Coordination between all four subunits of eukaryal endonucleases would stabilize the enzyme to place its active site at a specific position in the cloverleaf structure of pre-tRNA, namely position 37/38. Thus, it is likely that the recognition system and asymmetric subunit architecture of eukaryal endonucleases have co-evolved strictly with the BHB motifs at position 37/38.
The junctions of permuted pre-tRNAs in the B. natans nucleomorph and the nucleus of green algae comprise a hBH motif and are located at position 37/38 in the anticodon-loop (type II), which are the characteristics for recognition by the eukaryal splicing endonuclease (Table 1, Figure 2). In addition, B. natans and green algae contain almost no tRNA genes harboring atypical introns (Palenik et al., 2007; Maruyama et al., 2010). On the contrary, in the red alga C. merolae, the junctions of permuted pre-tRNAs and introns comprise various types of BHB motifs and are scattered along the cloverleaf structure (Figure 2A). This arrangement suggests that the C. merolae splicing endonuclease recognizes a wide variety of BHB motifs and employs a recognition strategy that is different from that of the known eukaryotic endonucleases.
A search of the C. merolae genome identified homologs of three of the yeast endonuclease subunits (cmSen2, cmSen34, and cmSen54) (Soma et al., 2013); however, no apparent homolog of the Sen15 accessory subunit was identified by homology searching or yeast two-hybrid analyses, which conflicts with the notion that all four subunits are essential for functional multimerization of the endonuclease. In yeast, Sen15 interacts with Sen34 to aid the proper positioning of the 3′-splice site (Westaway and Abelson, 1995; Di Nicola Negri et al., 1997; Trotta et al., 1997; Fabbri et al., 1998; Xue et al., 2006). The C. merolae endonuclease may contain an unidentified subunit or may comprise a novel heterotrimeric complex (Figure 4C). However, the C. merolae endonuclease containing accessory subunits is not likely to interact with pre-tRNAs that are disrupted at positions other than 37/38, because yeast Sen54 probably interacts with the D-arm and the acceptor-stem that are located in the core region of the L-shaped tertiary structure of a pre-tRNA (Di Nicola Negri et al., 1997; Xue et al., 2006). Thus, the C. merolae endonuclease may act on these pre-tRNAs as a dimer composed of catalytic subunits only (cmSen2 and cmSen34), via a tRNA mature domain-independent recognition mechanism. It is also tempting to speculate that the subunit composition of the C. merolae endonuclease depends on the positions or types of BHB motifs in the substrates. A feasible model may be that BHB motifs at positions other than 37/38 are removed by cmSen2-cmSen34, making the BHB motif at position 37/38 accessible to cmSen2-cmSen54-cmSen34 or cmSen2-cmSen54-cmSen34-cmSen15(unidentified), which interacts with the mature domain of the pre-tRNA, as occurs in yeast (Figure 4C). A previous study showing that the BHB motif at the canonical 37/38 position is always the final substrate during tRNA processing in C. merolae cells (Soma et al., 2013) may support this hypothesis. These observations imply that processing of the BHB motif in eukaryal tRNAs is more divergent among species than previously thought. Various types of BHB-mediated disrupted tRNA genes and splicing endonucleases may be present in other eukaryotes. In fact, ectopic intron-containing tRNA genes have been discovered in the nucleomorph of the cryptomonad Guillardia theta (Kawach et al., 2005), although many of these introns do not form a defined BHB motif. Furthermore, the absence of an accessory subunit (Sen15) homolog in Arabidopsis thaliana (Akama et al., 2000) implies that plant endonucleases have evolved various patterns of subunit architectures. On the other hand, A. thaliana contains only a few species of canonical intron-containing tRNA genes and does not contain any other disrupted tRNA genes; therefore, its endonuclease has not been adapted to process non-canonically disrupted pre-tRNAs.
Implications for the physiological relevance of permuted tRNA genes
To date, circular gene permutation of non-coding RNAs other than tRNA has been reported for the LSU rRNA from Tetrahymena mitochondria (Heinonen et al., 1987) as well as bacterial and organellar tmRNAs (Keiler et al., 2000; Mao et al., 2009), the latter of which are involved in the trans-translation system that rescues stalled ribosomes and maintains quality control of proteins in the cell (Keiler et al., 1996; Himeno et al., 1997b; Muto et al., 1998). However, permuted tRNAs show some substantial differences to permuted rRNAs and tmRNAs. A pre-tRNA of a permuted tRNA gene is processed and re-ligated at the junction of the 5′- and 3′-halves. The resultant tRNA molecule is composed of a continuous single-stranded RNA that can form a canonical cloverleaf structure, which is equipped with a functional acceptor-stem and an anticodon in the proper position. By contrast, the corresponding breaks between the 5′- and 3′-halves of rRNAs and tmRNAs encoded by permuted genes are not ligated and they function in a two-piece form. In the case of tmRNA, this form has been suggested to have a beneficial function, perhaps by solving topological problems on the ribosome (Williams, 2002; Sharkady and Williams, 2004). This idea is supported by the independent evolution of a similar two-piece form of tmRNA, encoded as a permuted gene in different lineages of bacteria (Sharkady and Williams, 2004; Williams, 2004). Additionally, the location of the junction of the 5′- and 3′-halves differs between permuted tmRNAs and permuted tRNAs. The two-piece form of tmRNA is adapted to its functional advantage, and the corresponding breakage between the 5′-half and the 3′-half is located at a unique position downstream of the tag peptide coding region. By contrast, the junctions of permuted tRNAs are located at various positions in the cloverleaf structure because breakage at any position is ultimately ligated to produce a typical tRNA molecule. Consequently, permutation of genes encoding tRNAs does not seem to affect the authentic function of the mature tRNA or confer any physiological benefit or restriction.
In C. merolae, disrupted tRNA genes that exhibit permuted (7/43), intron-containing (23/43), or both types of structures (4/43) account for 79.1% (34/43) of all nuclear tRNA genes (Soma et al., 2013), whereas only a few protein-encoding genes have spliceosomal introns (Matsuzaki et al., 2004). The conservation of a large number of permuted tRNAs, in addition to intronic tRNAs, which require more extensive processing, in the streamlined genome of C. merolae, implies that BHB-mediated disruption of tRNA genes has some physiological meaning. It is well known that while some tRNA introns are dispensable (Mori et al., 2011) others are involved in post-transcriptional modification (Johnson and Abelson, 1983; Szweykowska-Kulinska and Beier, 1992; Björk, 1995), quality control to ensure the supply of precisely processed tRNA molecules to the cytosol (Arts et al., 1998; Lund and Dahlberg, 1998; Takano et al., 2005; Hopper, 2013), and regulation of the cell cycle in response to DNA damage (Ghavidel et al., 2007; Weinert and Hopper, 2007). Therefore, permuted tRNA genes may contribute to essential cell functions. Alternatively, the circular RNA intermediate may be preferable because of its resistance to degradation in the cell.
From a physiological point of view, a possible explanation for the maintenance of disrupted tRNA genes is protection against mobile elements (Randau and Söll, 2008). Fragmentation of tRNA genes is thought to prevent the integration of mobile elements because tRNA gene sequences are sometimes used as conventional target sites in the genome (Devine and Boeke, 1996; Hani and Feldmann, 1998; Mou et al., 2006). This direct and valuable strategy would have functioned as a selective pressure at some point during evolution to increase the number of permuted tRNA genes. This possibility may be supported by the fact that almost no recognizable transposons or viruses are found in the contemporary genomes of C. merolae and M. pusilla, which harbor permuted tRNA genes (Matsuzaki et al., 2004; Worden et al., 2009). By contrast, Ostreococcus species, which contain some permuted tRNA genes and cis-spliced tRNA genes, have many transposons (Worden et al., 2009; Maruyama et al., 2010). Genome-wide analyses and studies focusing on the relationship between mobile elements and disrupted tRNA genes should further our understanding of this concept.
The eukaryal tRNA processing system has proofreading functions to ensure that only mature tRNAs are supplied for translation, and yeast cells possess multiple pathways to degrade inappropriately processed and folded tRNAs (Arts et al., 1998; Lund and Dahlberg, 1998; Kadaba et al., 2004; Takano et al., 2005; Whipple et al., 2011; Hopper, 2013; Kramer and Hopper, 2013). In Xenopus laevis oocytes, intron-containing pre-tRNAs are exported from the nucleus less efficiently than intron-spliced tRNAs, and nucleotide modifications and removal of the 5′- and 3′-flanking sequences at the acceptor-stem are monitored before transport of tRNAs into the cytosol (Arts et al., 1998). Therefore, the BHB motifs at various positions of permuted pre-tRNAs and the acceptor-loop of the circular RNA intermediate inhibit their exportin-dependent transport from the nucleus, and the sequential processing of permuted pre-tRNAs would contribute to the discrimination of immature tRNAs, providing a selective pressure to retain them in the genome. C. merolae cells use a small repertoire of tRNAs; hence, the quality of tRNA molecules must be checked to guarantee translational fidelity. Furthermore, elimination of incorrectly processed tRNA molecules might be more important for organisms that harbor a splicing endonuclease with relaxed substrate specificity.
A different perspective is that permuted tRNA genes might have been formed as a remnant of genome dynamics under relatively neutral selective pressure. Even if such tRNA genes were acquired, most of them could not be retained because of the failure of transcription or subsequent RNA processing. In some organisms, including early-rooted algae and archaea, permuted tRNA genes could have persisted in the genome because of the upstream promoter-dependent transcription system and the capacity of the splicing machinery to process disrupted pre-tRNAs into the canonical cloverleaf structure. An expression system adapted to the wide variety of tRNA genes might have been preferable for organisms attempting to reduce redundantly duplicated tRNA genes, thereby enabling disruption of tRNA genes in various ways while maintaining the repertoire of those essential for protein synthesis. It has been suggested that permuted tRNA genes might have contributed to the maintenance of genome integrity during the reduction of the B. natans nucleomorph genome, which is the smallest eukaryotic genome (Gilson et al., 2006; Maruyama et al., 2010). Thus, plasticity of tRNA gene structure and expression systems may be more important than permuted tRNA genes.
Scenarios for the development of permuted tRNA genes
There are two hypotheses for the development of permuted tRNA genes: the “ancient origin” hypothesis, which is related to the origin of the cloverleaf structure of tRNA (Di Giulio, 2008; Fujishima et al., 2008); and the “recent origin” hypothesis, which assumes that permuted tRNA genes arose from existing tRNAs in a relatively late stage of evolution (Randau and Söll, 2008; Sugahara et al., 2008; Maruyama et al., 2010; Chan et al., 2011).
The cloverleaf structure of tRNA is thought to have originated from mini-hairpins (Weiner and Maizels, 1987; Di Giulio, 1992, 2006; Schimmel and Ribas De Pouplana, 1995; Widmann et al., 1995), and tRNA sequences sometimes form a double hairpin structure flanked by the anticodon sequence (Tanaka and Kikuchi, 2001). The dominant localization of introns at position 37/38, which divide tRNAs into two hairpins, may be a remnant of the boundary connecting the hairpins, and disrupted tRNAs may represent plesiomorphic forms produced during the development of the modern cloverleaf structure (Di Giulio, 2008; Fujishima et al., 2008). The results of archaeal genome analyses have consistently suggested that modern tRNAs evolved through the combination of 5′-half and 3′-half fragments (Fujishima et al., 2008). Based on this concept, it was proposed that permuted tRNA genes arose from an event in which the two hairpin-like structures encoding the 5′- and 3′-halves of a tRNA were brought together in an inverted configuration on the genome (Di Giulio, 2008). However, there is some debate surrounding this idea. First, some permuted tRNAs are intervened at positions other than 37/38, which conflicts with the assumptions of the hairpin model (Di Giulio, 2008). Second, it is questionable whether the ancient forms of tRNA genes are preserved in the modern genome (Randau and Söll, 2008).
Based on comparative genome analyses, another hypothesis suggests that BHB-mediated disrupted tRNA genes were gained by gene transfer as apomorphies or were developed from extant tRNA genes (Di Giulio, 2008; Randau and Söll, 2008; Sugahara et al., 2009, 2012; Fujishima et al., 2010; Maruyama et al., 2010; Chan et al., 2011). Given that permuted tRNAs are present in early-rooted algae (Nozaki et al., 2003, 2007; Matsuzaki et al., 2004) and deep-branching Crenarchaeota from which eukarya might have derived (Lake et al., 1984; Cox et al., 2008), the algal genome may retain permuted tRNAs as a vestigial trait inherited from archaea. In fact, C. merolae tRNAs exhibit some characteristics that are found in archaea but not eukaryotes. For example, a number of C. merolae tRNAs contain ectopic and multiple introns, and C. merolae tRNAIle has the anticodon GAU (Matsuzaki et al., 2004), which has been identified in prokaryotes but not eukaryotes. However, sequence and structural similarities of the disrupted tRNAs in C. merolae and archaea have not been identified. Moreover, archaeal permuted tRNA genes encode the terminal CCA sequence, which is not encoded in the eukaryal genome, indicating that they have not simply been exchanged between archaea and algae (Chan et al., 2011). Therefore, permuted tRNA genes might have arisen independently in each lineage. This possibility is supported by the fact that BHB-mediated disrupted tRNA genes exhibit a discontinuous and patchy distribution in eukaryotes and archaea (Maruyama et al., 2010; Chan et al., 2011; Soma et al., 2013). An evolutionary relationship between cis-spliced tRNAs and split tRNAs has been suggested, because the leader sequences of some split tRNAs show a high degree of homology to the intronic sequence of tRNAs in correlated archaea (Fujishima et al., 2010). In addition, continuous transcripts corresponding to read-through of adjacently encoded 5′- and 3′-halves of split tRNAs are produced, albeit at very low levels, suggesting that they represent a transition state between a split tRNA and a cis-spliced tRNA in the genome (Chan et al., 2011). Thus, it is possible that permuted tRNAs emerged from extant tRNA genes.
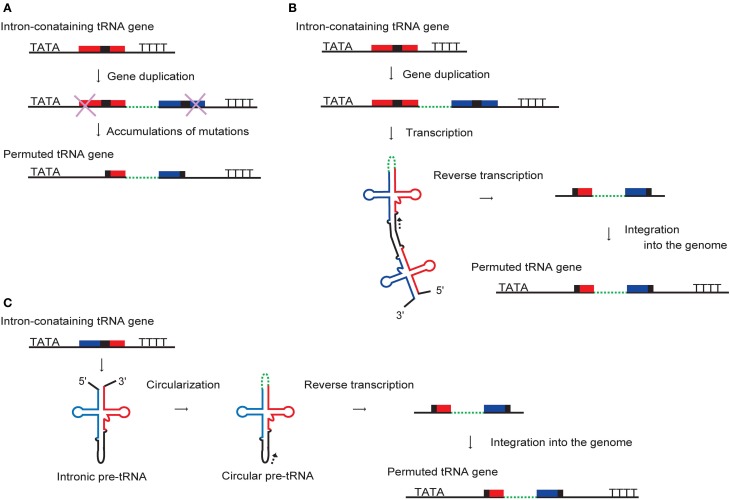
A plausible description of the emergence of permuted tRNA genes via convergent evolution can be inferred from the model proposed for permuted rRNAs and tmRNAs, which function as a two-piece form as described earlier. These species are hypothesized to have been established by a gene duplication event that formed a tandem repeat of the RNA genes, followed by the loss of the outer segment of each copy (Heinonen et al., 1987; Williams, 2002). Similarly, permuted tRNA genes might have originated from duplication of an intronic tRNA gene, followed by the loss of the outer exons to leave the 3′-half of the upstream tRNA gene and the 5′-half of the downstream tRNA gene (Figure 5A) (Soma et al., 2007; Di Giulio, 2008; Maruyama et al., 2010). In algae and archaea, these rearranged tRNA genes could have persisted in the genome because of the use of the upstream promoter-dependent transcription system and the tRNA maturation system that allows processing of permuted pre-tRNAs. In this context, the high frequency of permutation with the junction at position 37/38 (type II) can be ascribed to the overall dominance of introns located at the corresponding position in both eukaryotes and archaea. It is noteworthy that some tandem repeats of tRNA genes composed of single tRNA species containing an intron have been found in the nuclear genomes of green algae, namely the prasinophyte O. lucimarinus and the chlorophycea Chlamydomonas reinhardtii, which contain some and no permuted tRNAs, respectively, (Table 1) (Maruyama et al., 2010). Furthermore, an additional 5′-half is located downstream of the 5′-half of the permuted tRNACys(GCA) gene in the nuclear genome of O. lucimarinus. These duplicated tRNA genes may be structurally identical to the plausible intermediate stage of permuted tRNA evolution shown in the proposed model.
Figure 5.
Proposed models for the development of permuted tRNA genes. (A) Permuted tRNA genes might have formed by gene duplication and loss of the outer segments. A tandem repeat of an intron-containing tRNA gene could be arranged into a permuted tRNA gene by the accumulation of mutations at both ends. Alternative models are based on reverse transcription of permuted or circularized pre-tRNA molecules. (B) A permuted pre-tRNA formed by an interaction between the 3′-half of the tRNA encoded by the initial gene and the 5′-half of the duplicated tRNA derived from the tandemly-repeated gene may have been reverse transcribed from the point indicated by an arrowhead and integrated back into the genome. (C) A circular intron-containing pre-tRNA produced by ligation of an intron-containing pre-tRNA at the acceptor-stem may also have been reverse transcribed and integrated into the genome.
An alternative scenario is that the formation of permuted or circularized tRNA molecules preceded that of the corresponding permuted genes. Canonical pre-tRNAs co-transcribed from two tandemly-repeated intronic tRNA genes might be able to form a permuted pre-tRNA via the combination of the 3′-half of the initial tRNA and the 5′-half of the duplicated tRNA (Figure 5B). In support of this concept, a recent study showed that an artificial transcript simulating a tandemly-repeated intron-containing pre-tRNA could form a permuted tRNA structure in vitro (Tocchini-Valentini and Tocchini-Valentini, 2012). Furthermore, a circular pre-tRNA may be produced by ligation of an intron-containing (non-permuted) pre-tRNA at the acceptor-stem (Figure 5C). Indeed, many kinds of circularized non-coding RNAs have been identified in archaeal cells, indicating that the circularization of RNA is fairly prevalent, although the significance of this feature is still unknown (Danan et al., 2012). The resulting permuted or circular pre-tRNA molecules might have been reverse transcribed and integrated back into the genome to generate permuted tRNA genes (Figures 5B,C). Therefore, it is plausible that circular permutation has contributed to the evolution of the tRNA-like structures that are prevalent in nature (Pan et al., 1991; Pan and Uhlenbeck, 1993; Florentz and Giegé, 1995), and the cloverleaf structure of tRNA might have developed as a circularly permuted RNA isomer.
Regardless of the mechanism(s) by which permuted tRNA genes originated, the BHB motifs must have played a pivotal role during their development. The existence of a number of BHB-mediated cis-spliced tRNAs in algae and archaea may reflect a background that has accelerated the production of permuted tRNA genes. If so, permuted tRNA genes could have occurred frequently in archaea, especially in Crenarchaeota, whose splicing machinery can process various types of BHB motifs. However, only two permuted tRNA genes have been identified from one crenarchaeon (T. pendens) (Table 1), which harbors plenty of intron-containing tRNA genes (Fujishima et al., 2011). In eukaryotes, tRNA genes contain an intron at the canonical 37/38 position and tRNA genes of plants and yeast can be transcribed depending on the upstream promoter; therefore, it is plausible that eukaryotes could possess permuted tRNA genes with the junction at the canonical 37/38 position. However, most eukaryotes do not contain permuted tRNAs. These observations may indicate that the background for the development of permuted tRNA genes is intrinsically different among organisms. Moreover, even if permuted tRNA genes did once emerge in archaeal and eukaryotic species, they may not have been maintained in the genome due to their instability or harmful influence. For example, an inverted tRNA gene structure might have been lost easily, or the BHB motifs may have been associated with a specific adverse effect on the genome or organism.
The phylogenic distributions of BHB-mediated disruptions of tRNA genes are biased and an organism harboring all three types of disrupted tRNAs has not yet been identified. Some archaea, including N. equitans and Caldiviga maquilingensis, harbor split tRNAs but only a few cis-spliced tRNAs and no permuted tRNAs (Chan et al., 2011; Fujishima et al., 2011). Other archaea, including the Pyrobaculum and Thermofilum genera, harbor a number of intronic tRNAs that are disrupted at various positions, although Pyrobaculum have no split or permuted tRNAs (Fujishima et al., 2011). Similarly, green algae contain some permuted tRNAs but almost no ectopic intron-containing tRNAs (Kawach et al., 2005; Maruyama et al., 2010). Hence, C. merolae is unique because it possesses a number of permuted tRNAs and various intron-containing tRNAs. C. merolae might be permissive for the absorption and retention of various tRNA genes, or some characteristics of C. merolae may have accelerated the development and preservation of permuted tRNA genes during evolution. Considering that C. merolae has a compact genome, it is possible that the successive genome size reduction put pressure on redundantly duplicated tRNA genes to be arranged into a single permuted tRNA gene. Split tRNAs have not been identified in C. merolae, despite its potential ability to express them. Formation of a split tRNA may be a less efficient strategy to reduce the genome size because it requires two sets of promoter and terminator sequences to produce one species of tRNA. To date, there has been no report of an organism in which split tRNAs coexist with permuted tRNAs; therefore, the individual mechanisms and requisite elements required for the acquisition or maintenance of each disrupted tRNA gene could be substantially different, as suggested previously (Chan et al., 2011). This hypothesis supports a non-monophyletic origin of BHB-mediated disrupted tRNA genes, which may have arisen and disappeared multiple times independently in various organisms. The next challenge will be to identify the specific characteristics and fundamental background that led to the BHB-mediated disruption of tRNA genes, and to clarify the method of formation of each type of tRNA gene.
Conclusions
The identification of circularly permuted tRNA genes has revealed a unique style of gene structure and RNA processing. Comparative genome analyses should be performed to identify more examples of permuted genes and to investigate the origin of the permuted tRNAs in correlation with other BHB-mediated disrupted tRNAs and mobile elements that target tRNA genes. Studies of the transcription and maturation systems for tRNAs that must have co-evolved with disrupted tRNA genes would help to clarify the physiological meaning and the mechanisms that govern the development and maintenance of permuted tRNA genes.
Author contributions
Akiko Soma wrote the manuscript, and prepared Figures 1–5 and Table 1.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author thanks A. Muto, S. Maruyama, S. Goto, Y. Sekine, A. Onodera, J. Sugahara, N. Yachie, and A. Kanai for valuable discussions; and T. Kuroiwa, K. Tanaka, O. Misumi, and M. Matsuzaki for technical advice; and M. Ohara and K. Nakayama for helping preparation of the figures. This work was supported by a grant from a Grant-in-Aid for Scientific Research C (25440003 to Akiko Soma), Career-Support Program for Woman Scientists at Chiba University (to Akiko Soma), and the Nakajima Foundation (to Akiko Soma).
References
- Abe T., Ikemura T., Sugahara J., Kanai A., Ohara Y., Uehara H., et al. (2011). tRNADB-CE 2011: tRNA gene database curated manually by experts. Nucleic Acids Res. 39, D210–D213 10.1093/nar/gkq1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abelson J., Trotta C. R., Li H. (1998). tRNA splicing. J. Biol. Chem. 273, 12685–12688 10.1074/jbc.273.21.12685 [DOI] [PubMed] [Google Scholar]
- Akama K., Junker V., Beier H. (2000). Identification of two catalytic subunits of tRNA splicing endonuclease from Arabidopsis thaliana. Gene 257, 177–185 10.1016/S0378-1119(00)00408-X [DOI] [PubMed] [Google Scholar]
- Altman S. (2011). Ribonuclease P. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366, 2936–2941 10.1098/rstb.2011.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman S., Kirsebom L., Talbot S. (1995). Recent studies of RNase P, in tRNA, eds Söll D., RajBhandary U. L. (Washington, DC: ASM Press; ), 67–78 [Google Scholar]
- Archibald J. (2007). Nucleomorph genomes: structure, function, origin and evolution. Bioessays. 29, 392–402 10.1002/bies.20551 [DOI] [PubMed] [Google Scholar]
- Archibald J., Lane C. E. (2009). Going, going, not quite gone: nucleomorphs as a case study in nuclear genome reduction. J. Hered. 100, 582–590 10.1093/jhered/esp055 [DOI] [PubMed] [Google Scholar]
- Arts G., Kuersten S., Romby P., Ehresmann B., Mattaj I. W. (1998). The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 17, 7430–7441 10.1093/emboj/17.24.7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara H., Himeno H., Tamura K., Hasegawa T., Watanabe K., Shimizu M. (1993). Recognition nucleotides of Escherichia coli tRNA(Leu) and its elements facilitating discrimination from tRNASer and tRNA(Tyr). J. Mol. Biol. 231, 219–229 10.1006/jmbi.1993.1277 [DOI] [PubMed] [Google Scholar]
- Baldi M., Mattoccia E., Bufardeci E., Fabbri S., Tocchini-Valentini G. P. (1992). Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science 255, 1404–1408 10.1126/science.1542788 [DOI] [PubMed] [Google Scholar]
- Baldi M., Mattoccia E., Tocchini-Valentini G. P. (1983). Role of RNA structure in splicing: excision of the intervening sequence in yeast tRNA3leu is dependent on the formation of a D stem. Cell 35, 109–115 10.1016/0092-8674(83)90213-1 [DOI] [PubMed] [Google Scholar]
- Belfort M., Weiner A. (1997). Another bridge between kingdoms: tRNA splicing in archaea and eukaryotes. Cell 89, 1003–1006 10.1016/S0092-8674(00)80287-1 [DOI] [PubMed] [Google Scholar]
- Biniszkiewicz D., Cesnaviciene E., Shub D. A. (1994). Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 13, 4629–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk G. R. (1995). Biosynthesis and function of modified nucleosides, in tRNA, eds Söll D., RajBhandary U. L. (Washington, DC: ASM Press; ), 165–206 [Google Scholar]
- Bonen L., Vogel J. (2001). The ins and outs of group II introns. Trends Genet. 17, 322–331 10.1016/S0168-9525(01)02324-1 [DOI] [PubMed] [Google Scholar]
- Calvin K., Li H. (2008). RNA-splicing endonuclease structure and function. Cell. Mol. Life Sci. 65, 1176–1185 10.1007/s00018-008-7393-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canino G., Bocian E., Barbezier N., Echeverría M., Forner J., Binder S., et al. (2009). Arabidopsis encodes four tRNase Z enzymes. Plant Physiol. 150, 1494–1502 10.1104/pp.109.137950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P., Cozen A., Lowe T. (2011). Discovery of permuted and recently split transfer RNAs in Archaea. Genome Biol. 12: R38 10.1186/gb-2011-12-4-r38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P., Lowe T. (2009). GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37, D93–D97 10.1093/nar/gkn787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choisne N., Carneiro V. T., Pelletier G., Small I. (1998). Implication of 5′-flanking sequence elements in expression of a plant tRNA(Leu) gene. Plant Mol. Biol. 36, 113–123 10.1023/A:1005988004924 [DOI] [PubMed] [Google Scholar]
- Choisne N., Martin-Canadell A., Small I. (1997). Transactivation of a target gene using a suppressor tRNA in transgenic tobacco plants. Plant J. 11, 597–604 10.1046/j.1365-313X.1997.11030597.x [DOI] [PubMed] [Google Scholar]
- Christian E., Zahler N. H., Kaye N. M., Harris M. E. (2002). Analysis of substrate recognition by the ribonucleoprotein endonuclease RNase P. Methods 28, 307–322 10.1016/S1046-2023(02)00238-4 [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Raugei G., Costanzo F., Dente L., Cortese R. (1983). Common and interchangeable elements in the promoters of genes transcribed by RNA polymerase III. Cell 32, 725–733 10.1016/0092-8674(83)90058-2 [DOI] [PubMed] [Google Scholar]
- Cox C., Foster P. G., Hirt R. P., Harris S. R., Embley T. M. (2008). The archaebacterial origin of eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 105, 20356–20361 10.1073/pnas.0810647105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis B., Tanifuji G., Burki F., Gruber A., Irimia M., Maruyama S., et al. (2012). Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492, 59–65 10.1038/nature11681 [DOI] [PubMed] [Google Scholar]
- Danan M., Schwartz S., Edelheit S., Sorek R. (2012). Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 40, 3131–3142 10.1093/nar/gkr1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M. P. (1995). Processing nucleases, in tRNA, eds Söll D., RajBhandary U. L. (Washington, DC: ASM Press; ), 51–66 [Google Scholar]
- Devine S., Boeke J. D. (1996). Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10, 620–633 10.1101/gad.10.5.620 [DOI] [PubMed] [Google Scholar]
- Dieci G., Percudani R., Giuliodori S., Bottarelli L., Ottonello S. (2000). TFIIIC-independent in vitro transcription of yeast tRNA genes. J. Mol. Biol. 299, 601–613 10.1006/jmbi.2000.3783 [DOI] [PubMed] [Google Scholar]
- Dieci G., Yukawa Y., Alzapiedi M., Guffanti E., Ferrari R., Sugiura M., et al. (2006). Distinct modes of TATA box utilization by the RNA polymerase III transcription machineries from budding yeast and higher plants. Gene 379, 12–25 10.1016/j.gene.2006.03.013 [DOI] [PubMed] [Google Scholar]
- Diener J., Moore P. B. (1998). Solution structure of a substrate for the archaeal pre-tRNA splicing endonucleases: the bulge-helix-bulge motif. Mol. Cell 1, 883–894 10.1016/S1097-2765(00)80087-8 [DOI] [PubMed] [Google Scholar]
- Di Giulio M. (1992). On the origin of the transfer RNA molecule. J. Theor. Biol. 159, 199–214 10.1016/S0022-5193(05)80702-7 [DOI] [PubMed] [Google Scholar]
- Di Giulio M. (2006). Nanoarchaeum equitans is a living fossil. J. Theor. Biol. 242, 257–260 10.1016/j.jtbi.2006.01.034 [DOI] [PubMed] [Google Scholar]
- Di Giulio M. (2008). Permuted tRNA genes of Cyanidioschyzon merolae, the origin of the tRNA molecule and the root of the Eukarya domain. J. Theor. Biol. 253, 587–592 10.1016/j.jtbi.2008.04.022 [DOI] [PubMed] [Google Scholar]
- Di Nicola Negri E., Fabbri S., Bufardeci E., Baldi M. I., Gandini Attardi D., Mattoccia E., et al. (1997). The eucaryal tRNA splicing endonuclease recognizes a tripartite set of RNA elements. Cell 89, 859–866 10.1016/S0092-8674(00)80271-8 [DOI] [PubMed] [Google Scholar]
- Dirheimer G., Keith G., Dumas P., Westhof E. (1995). Primary, secondary, and tertiary structure of tRNAs, in tRNA, eds Söll D., RajBhandary U. L. (Washington, DC: ASM Press; ), 93–126 [Google Scholar]
- Douglas S., Zauner S., Fraunholz M., Beaton M., Penny S., Deng L. T., et al. (2001). The highly reduced genome of an enslaved algal nucleus. Nature 410, 1091–1096 10.1038/35074092 [DOI] [PubMed] [Google Scholar]
- Englert M., Sheppard K., Aslanian A., Yates J. R., 3rd., Söll D. (2011). Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 108, 1290–1295 10.1073/pnas.1018307108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M., Xia S., Okada C., Nakamura A., Tanavde V., Yao M., et al. (2012). Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc. Natl. Acad. Sci. U.S.A. 109, 15235–15240 10.1073/pnas.1213795109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri S., Fruscoloni P., Bufardeci E., Di Nicola Negri E., Baldi M. I., Attardi D. G., et al. (1998). Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science 280, 284–286 10.1126/science.280.5361.284 [DOI] [PubMed] [Google Scholar]
- Florentz C., Giegé R. (1995). tRNA-like structures in plant viral RNAs, in tRNA, eds Söll D., RajBhandary U. L. (Washington, DC: ASM Press; ), 141–164 [Google Scholar]
- Fruscoloni P., Baldi M. I., Tocchini-Valentini G. P. (2001). Cleavage of non-tRNA substrates by eukaryal tRNA splicing endonucleases. EMBO Rep. 2, 217–221 10.1093/embo-reports/kve040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishima K., Sugahara J., Kikuta K., Hirano R., Sato A., Tomita M., et al. (2009). Tri-split tRNA is a transfer RNA made from 3 transcripts that provides insight into the evolution of fragmented tRNAs in archaea. Proc. Natl. Acad. Sci. U.S.A. 106, 2683–2687 10.1073/pnas.0808246106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishima K., Sugahara J., Miller C. S., Baker B. J., Di Giulio M., Takesue K., et al. (2011). A novel three-unit tRNA splicing endonuclease found in ultrasmall Archaea possesses broad substrate specificity. Nucleic Acids Res. 39, 9695–9704 10.1093/nar/gkr692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishima K., Sugahara J., Tomita M., Kanai A. (2008). Sequence evidence in the archaeal genomes that tRNAs emerged through the combination of ancestral genes as 5′ and 3′ tRNA halves. PLoS ONE 3: e1622 10.1371/journal.pone.0001622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujishima K., Sugahara J., Tomita M., Kanai A. (2010). Large-scale tRNA intron transposition in the archaeal order Thermoproteales represents a novel mechanism of intron gain. Mol. Biol. Evol. 27, 2233–2243 10.1093/molbev/msq111 [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. (1981). Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature 294, 626–631 10.1038/294626a0 [DOI] [PubMed] [Google Scholar]
- Geiduschek E., Kassavetis G. A. (2001). The RNA polymerase III transcription apparatus. J. Mol. Biol. 310, 1–26 10.1006/jmbi.2001.4732 [DOI] [PubMed] [Google Scholar]
- Ghavidel A., Kislinger T., Pogoutse O., Sopko R., Jurisica I., Emili A. (2007). Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell 131, 915–926 10.1016/j.cell.2007.09.042 [DOI] [PubMed] [Google Scholar]
- Gilson P. R., Su V., Slamovits C. H., Reith M. E., Keeling P. J., McFadden G. I. (2006). Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature's smallest nucleus. Proc. Natl. Acad. Sci. U.S.A. 103, 9566–9571 10.1073/pnas.0600707103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliodori S., Percudani R., Braglia P., Ferrari R., Guffanti E., Ottonello S., et al. (2003). A composite upstream sequence motif potentiates tRNA gene transcription in yeast. J. Mol. Biol. 333, 1–20 10.1016/j.jmb.2003.08.016 [DOI] [PubMed] [Google Scholar]
- Gobert A., Gutmann B., Taschner A., Gössringer M., Holzmann J., Hartmann R. K., et al. (2010). A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 17, 740–744 10.1038/nsmb.1812 [DOI] [PubMed] [Google Scholar]
- Greer C., Söll D., Willis I. (1987). Substrate recognition and identification of splice sites by the tRNA-splicing endonuclease and ligase from Saccharomyces cerevisiae. Mol. Cell. Biol. 7, 76–84 10.1128/MCB.7.1.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti E., Percudani R., Harismendy O., Soutourina J., Werner M., Iacovella M. G., et al. (2006). Nucleosome depletion activates poised RNA polymerase III at unconventional transcription sites in Saccharomyces cerevisiae. J. Biol. Chem. 281, 29155–29164 10.1074/jbc.M600387200 [DOI] [PubMed] [Google Scholar]
- Hamada M., Huang Y., Lowe T. M., Maraia R. J. (2001). Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast. Mol. Cell. Biol. 21, 6870–6881 10.1128/MCB.21.20.6870-6881.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M., Sakulich A. L., Koduru S. B., Maraia R. J. (2000). Transcription termination by RNA polymerase III in fission yeast. A genetic and biochemically tractable model system. J. Biol. Chem. 275, 29076–29081 10.1074/jbc.M003980200 [DOI] [PubMed] [Google Scholar]
- Hani J., Feldmann F. (1998). tRNA genes and retroelements in the yeast genome. Nucleic Acids Res. 26, 689–696 10.1093/nar/26.3.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann I., Söll D., Randau L. (2010). Transfer RNA processing in archaea: unusual pathways and enzymes. FEBS Lett. 584, 303–309 10.1016/j.febslet.2009.10.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen T., Schnare M. N., Young P. G., Gray M. W. (1987). Rearranged coding segments, separated by a transfer RNA gene, specify the two parts of a discontinuous large subunit ribosomal RNA in Tetrahymena pyriformis mitochondria. J. Biol Chem. 262, 2879–2887 [PubMed] [Google Scholar]
- Himeno H., Sato M., Tadaki T., Fukushima M., Ushida C., Muto A. (1997b). In vitro trans translation mediated by alanine-charged 10Sa RNA. J. Mol. Biol. 268, 803–808 10.1006/jmbi.1997.1011 [DOI] [PubMed] [Google Scholar]
- Himeno H., Yoshida S., Soma A., Nishikawa K. (1997a). Only one nucleotide insertion to the long variable arm confers an efficient serine acceptor activity upon Saccharomyces cerevisiae tRNA(Leu) in vitro. J. Mol. Biol. 268, 704–711 10.1006/jmbi.1997.0991 [DOI] [PubMed] [Google Scholar]
- Hirata A., Kitajima T., Hori H. (2011). Cleavage of intron from the standard or non-standard position of the precursor tRNA by the splicing endonuclease of Aeropyrum pernix, a hyper-thermophilic Crenarchaeon, involves a novel RNA recognition site in the Crenarchaea specific loop. Nucleic Acids Res. 39, 9376–9389 10.1093/nar/gkr615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. (2013). Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194, 43–67 10.1534/genetics.112.147470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Phizicky E. M. (2003). tRNA transfers to the limelight. Genes Dev. 17, 162–180 10.1101/gad.1049103 [DOI] [PubMed] [Google Scholar]
- Huang Y., Maraia R. J. (2001). Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 29, 2675–2690 10.1093/nar/29.13.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., et al. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Jacquier A. (1996). Group II introns: elaborate ribozymes. Biochimie 78, 474–487 10.1016/0300-9084(96)84754-7 [DOI] [PubMed] [Google Scholar]
- Jarrous N., Gopalan V. (2010). Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res. 38, 7885–7894 10.1093/nar/gkq701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P., Abelson J. (1983). The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature 302, 681–687 10.1038/302681a0 [DOI] [PubMed] [Google Scholar]
- Jühling F., Mörl M., Hartmann R. K., Sprinzl M., Stadler P. F., Pütz J. (2009). tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37, D159–D162 10.1093/nar/gkn772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S., Krueger A., Trice T., Krecic A. M., Hinnebusch A. G., Anderson J. (2004). Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18, 1227–1240 10.1101/gad.1183804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai A. (2013). Molecular evolution of disrupted transfer RNA genes and their introns in archaea, in Evolutionary Biology: Exobiology and Evolutionary Mechanisms, ed Pontarotti P. (Berlin: Springer-Verlag Press; ), 181–193 [Google Scholar]
- Kawach O., Voss C., Wolff J., Hadfi K., Maier U. G., Zauner S. (2005). Unique tRNA introns of an enslaved algal cell. Mol. Biol. Evol. 22, 1694–1701 10.1093/molbev/msi161 [DOI] [PubMed] [Google Scholar]
- Keiler K., Shapiro L., Williams K. P. (2000). tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc. Natl. Acad. Sci. U.S.A. 97, 7778–7783 10.1073/pnas.97.14.7778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler K., Waller P. R., Sauer R. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993 10.1126/science.271.5251.990 [DOI] [PubMed] [Google Scholar]
- Kirsebom L. (2007). RNase P RNA mediated cleavage: substrate recognition and catalysis. Biochimie 89, 1183–1194 10.1016/j.biochi.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Kjems J., Garrett R. A. (1988). Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell 54, 693–703 10.1016/S0092-8674(88)80014-X [DOI] [PubMed] [Google Scholar]
- Kjems J., Leffers H., Olesen T., Garrett R. A. (1989). A unique tRNA intron in the variable loop of the extreme thermophile Thermofilum pendens and its possible evolutionary implications. J. Biol. Chem. 264, 17834–17837 [PubMed] [Google Scholar]
- Kramer E., Hopper A. K. (2013). Retrograde transfer RNA nuclear import provides a new level of tRNA quality control in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 110, 21042–21047 10.1073/pnas.1316579110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhsel M. G., Strickland R., Palmer J. D. (1990). An ancient group I intron shared by eubacteria and chloroplasts. Science 250, 1570–1573 10.1126/science.2125748 [DOI] [PubMed] [Google Scholar]
- Kuroiwa T. (1998). The primitive red algae Cyanidium caldarium and Cyanidioschyzon merolae as model system for investigating the dividing apparatus of mitochondria and plastids. Bioessays 20, 344–354 [DOI] [Google Scholar]
- Lake J., Henderson E., Oakes M., Clark M. W. (1984). Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 81, 3786–3790 10.1073/pnas.81.12.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landweber L. F. (2007). Why genomes in pieces? Science 318, 405–407 10.1126/science.1150280 [DOI] [PubMed] [Google Scholar]
- Li de la Sierra-Gallay I., Mathy N., Pellegrini O., Condon C. (2006). Structure of the ubiquitous 3′ processing enzyme RNase Z bound to transfer RNA. Nat. Struct. Mol. Biol. 13, 376–377 10.1038/nsmb1066 [DOI] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Dahlberg J. E. (1998). Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282, 2082–2085 10.1126/science.282.5396.2082 [DOI] [PubMed] [Google Scholar]
- Mackie G. (1998). Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395, 720–723 10.1038/27246 [DOI] [PubMed] [Google Scholar]
- Mao C., Bhardwaj K., Sharkady S. M., Fish R. I., Driscoll T., Wower J., et al. (2009). Variations on the tmRNA gene. RNA Biol. 6, 355–361 10.4161/rna.6.4.9172 [DOI] [PubMed] [Google Scholar]
- Marck C., Grosjean H. (2002). tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8, 1189–1232 10.1017/S1355838202022021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C., Grosjean H. (2003). Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications. RNA 9, 1516–1531 10.1261/rna.5132503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C., Kachouri-Lafond R., Lafontaine I., Westhof E., Dujon B., Grosjean H. (2006). The RNA polymerase III-dependent family of genes in hemiascomycetes: comparative RNomics, decoding strategies, transcription and evolutionary implications. Nucleic Acids Res. 34, 1816–1835 10.1093/nar/gkl085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S., Sugahara J., Kanai A., Nozaki H. (2010). Permuted tRNA genes in the nuclear and nucleomorph genomes of photosynthetic eukaryotes. Mol. Biol. Evol. 27, 1070–1076 10.1093/molbev/msp313 [DOI] [PubMed] [Google Scholar]
- Matsuzaki M., Misumi O., Shin I. T., Maruyama S., Takahara M., Miyagishima S. Y., et al. (2004). Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657 10.1038/nature02398 [DOI] [PubMed] [Google Scholar]
- Mattoccia E., Baldi I. M., Gandini-Attardi D., Ciafrè S., Tocchini-Valentini G. P. (1988). Site selection by the tRNA splicing endonuclease of Xenopus laevis. Cell 55, 731–738 10.1016/0092-8674(88)90231-0 [DOI] [PubMed] [Google Scholar]
- McClain W., Guerrier-Takada C., Altman S. (1987). Model substrates for an RNA enzyme. Science 238, 527–530 10.1126/science.2443980 [DOI] [PubMed] [Google Scholar]
- Minagawa A., Ishii R., Takaku H., Yokoyama S., Nashimoto M. (2008). The flexible arm of tRNase Z is not essential for pre-tRNA binding but affects cleavage site selection. J. Mol. Biol. 381, 289–299 10.1016/j.jmb.2008.05.016 [DOI] [PubMed] [Google Scholar]
- Mori S., Kajita T., Endo T., Yoshihisa T. (2011). The intron of tRNA-TrpCCA is dispensable for growth and translation of Saccharomyces cerevisiae. RNA 17, 1760–1769 10.1261/rna.2851411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z., Kenny A. E., Curcio M. J. (2006). Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics 172, 2157–2167 10.1534/genetics.105.054072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Ushida C., Himeno H. (1998). A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem. Sci. 23, 25–29 10.1016/S0968-0004(97)01159-6 [DOI] [PubMed] [Google Scholar]
- Nashimoto M., Tamura M., Kaspar R. L. (1999). Minimum requirements for substrates of mammalian tRNA 3′ processing endoribonuclease. Biochemistry 38, 12089–12096 10.1021/bi9911942 [DOI] [PubMed] [Google Scholar]
- Nielsen S., Yuzenkova Y., Zenkin N. (2013). Mechanism of eukaryotic RNA polymerase III transcription termination. Science 340, 1577–1580 10.1126/science.1237934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki H., Matsuzaki M., Takahara M., Misumi O., Kuroiwa H., Hasegawa M., et al. (2003). The phylogenetic position of red algae revealed by multiple nuclear genes from mitochondria-containing eukaryotes and an alternative hypothesis on the origin of plastids. J. Mol. Evol. 56, 485–497 10.1007/s00239-002-2419-9 [DOI] [PubMed] [Google Scholar]
- Nozaki H., Takano H., Misumi O., Terasawa K., Matsuzaki M., Maruyama S., et al. (2007). A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga. Cyanidioschyzon merolae. BMC Biol. 5:28 10.1186/1741-7007-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B., Grimwood J., Aerts A., Rouzé P., Salamov A., Putnam N., et al. (2007). The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. U.S.A. 104, 7705–7710 10.1073/pnas.0611046104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. R., Daniels C. J. (1995). In vivo definition of an archaeal promoter. J. Bacteriol. 177, 1844–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T., Gutell R. R., Uhlenbeck O. C. (1991). Folding of circularly permuted transfer RNAs. Science 254, 1361–1364 10.1126/science.1720569 [DOI] [PubMed] [Google Scholar]
- Pan T., Uhlenbeck O. C. (1993). Circularly permuted DNA, RNA and proteins–a review. Gene 125, 111–114 10.1016/0378-1119(93)90317-V [DOI] [PubMed] [Google Scholar]
- Paule M., White R. J. (2000). Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 28, 1283–1298 10.1093/nar/28.6.1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S., Patel M., Furia B. S., Peltz S. W., Trotta C. R. (2004). Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 117, 311–321 10.1016/S0092-8674(04)00342-3 [DOI] [PubMed] [Google Scholar]
- Pinker F., Bonnard G., Gobert A., Gutmann B., Hammani K., Sauter C., et al. (2013). PPR proteins shed a new light on RNase P biology. RNA Biol. 10, 1457–1463 10.4161/rna.25273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., et al. (2011). HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 331, 760–764 10.1126/science.1197847 [DOI] [PubMed] [Google Scholar]
- Randau L., Calvin K., Hall M., Yuan J., Podar M., Li H., et al. (2005b). The heteromeric Nanoarchaeum equitans splicing endonuclease cleaves noncanonical bulge-helix-bulge motifs of joined tRNA halves. Proc. Natl. Acad. Sci. U.S.A. 102, 17934–17939 10.1073/pnas.0509197102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randau L., Münch R., Hohn M. J., Jahn D., Söll D. (2005a). Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature 433, 537–541 10.1038/nature03233 [DOI] [PubMed] [Google Scholar]
- Randau L., Pearson M., Söll D. (2005c). The complete set of tRNA species in Nanoarchaeum equitans. FEBS Lett. 579, 2945–2947 10.1016/j.febslet.2005.04.051 [DOI] [PubMed] [Google Scholar]
- Randau L., Schröder I., Söll D. (2008). Life without RNase P. Nature 453, 120–123 10.1038/nature06833 [DOI] [PubMed] [Google Scholar]
- Randau L., Söll D. (2008). Transfer RNA genes in pieces. EMBO Rep. 9, 623–628 10.1038/embor.2008.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut R., Green P. R., Abelson J. (1990). Yeast tRNA-splicing endonuclease is a heterotrimeric enzyme. J. Biol. Chem. 265, 18180–18184 [PubMed] [Google Scholar]
- Reeve J. (2003). Archaeal chromatin and transcription. Mol. Microbiol. 48, 587–598 10.1046/j.1365-2958.2003.03439.x [DOI] [PubMed] [Google Scholar]
- Reinhold-Hurek B., Shub D. A. (1992). Self-splicing introns in tRNA genes of widely divergent bacteria. Nature 357, 173–176 10.1038/357173a0 [DOI] [PubMed] [Google Scholar]
- Reiter N., Osterman A., Torres-Larios A., Swinger K. K., Pan T., Mondragón A. (2010). Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468, 784–789 10.1038/nature09516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. (1988). Substrate recognition and splice site determination in yeast tRNA splicing. Cell 55, 719–730 10.1016/0092-8674(88)90230-9 [DOI] [PubMed] [Google Scholar]
- Rich A., Rajbhandary U. L. (1976). Transfer RNA: molecular structure, sequence, and properties. Annu. Rev. Biochem. 45, 805–860 10.1146/annurev.bi.45.070176.004105 [DOI] [PubMed] [Google Scholar]
- Santangelo T., Cubonová L., Skinner K. M., Reeve J. N. (2009). Archaeal intrinsic transcription termination in vivo. J. Bacteriol. 191, 7102–7108 10.1128/JB.00982-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer S., Rösch S., Marchfelder A. (2002). Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J. 21, 2769–2777 10.1093/emboj/21.11.2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P., Ribas De Pouplana L. (1995). Transfer RNA: from minihelix to genetic code. Cell 81, 983–986 10.1016/S0092-8674(05)80002-9 [DOI] [PubMed] [Google Scholar]
- Schramm L., Hernandez N. (2002). Recruitment of RNA polymerase III to its target promoters. Genes Dev. 16, 2593–2620 10.1101/gad.1018902 [DOI] [PubMed] [Google Scholar]
- Schürer H., Schiffer S., Marchfelder A., Mörl M. (2001). This is the end: processing, editing and repair at the tRNA 3′-terminus. Biol. Chem. 382, 1147–1156 10.1515/BC.2001.144 [DOI] [PubMed] [Google Scholar]
- Sharkady S., Williams K. P. (2004). A third lineage with two-piece tmRNA. Nucleic Acids Res. 32, 4531–4518 10.1093/nar/gkh795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma A., Onodera A., Sugahara J., Kanai A., Yachie N., Tomita M., et al. (2007). Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science 318, 450–453 10.1126/science.1145718 [DOI] [PubMed] [Google Scholar]
- Soma A., Sugahara J., Onodera A., Yachie N., Kanai A., Watanabe S., et al. (2013). Identification of highly-disrupted tRNA genes in nuclear genome of the red alga, Cyanidioschyzon merolae 10D. Sci. Rep. 3: 2321 10.1038/srep02321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma A., Uchiyama K., Sakamoto T., Maeda M., Himeno H. (1999). Unique recognition style of tRNA(Leu) by Haloferax volcanii leucyl-tRNA synthetase. J. Mol. Biol. 293, 1029–1038 10.1006/jmbi.1999.3219 [DOI] [PubMed] [Google Scholar]
- Späth B., Canino G., Marchfelder A. (2007). tRNase Z: the end is not in sight. Cell. Mol. Life Sci. 64, 2404–2412 10.1007/s00018-007-7160-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague K. (1995). Transcrption of eukaryotic tRNA genes, in tRNA, eds Söll D., RajBhandary U. L. (Washington, DC: ASM Press; ), 31–50 [Google Scholar]
- Stange N., Beier D., Beier H. (1992). Intron excision from tRNA precursors by plant splicing endonuclease requires unique features of the mature tRNA domain. Eur. J. Biochem. 210, 193–203 10.1111/j.1432-1033.1992.tb17408.x [DOI] [PubMed] [Google Scholar]
- Sugahara J., Fujishima K., Morita K., Tomita M., Kanai A. (2009). Disrupted tRNA gene diversity and possible evolutionary scenarios. J. Mol. Evol. 69, 497–504 10.1007/s00239-009-9294-6 [DOI] [PubMed] [Google Scholar]
- Sugahara J., Fujishima K., Nunoura T., Takai Y., Takami H., Takai K., et al. (2012). Genomic heterogeneity in a natural archaeal population suggests a model of tRNA gene disruption. PLoS ONE 7: e32504 10.1371/journal.pone.0032504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara J., Kikuta K., Fujishima K., Yachie N., Tomita M., Kanai A. (2008). Comprehensive analysis of archaeal tRNA genes reveals rapid increase of tRNA introns in the order thermoproteales. Mol. Biol. Evol. 25, 2709–2716 10.1093/molbev/msn216 [DOI] [PubMed] [Google Scholar]
- Sugahara J., Yachie N., Arakawa K., Tomita M. (2007). In silico screening of archaeal tRNA-encoding genes having multiple introns with bulge-helix-bulge splicing motifs. RNA 13, 671–681 10.1261/rna.309507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara J., Yachie N., Sekine Y., Soma A., Matsui M., Tomita M., et al. (2006). SPLITS: a new program for predicting split and intron-containing tRNA genes at the genome level. In Silico Biol. 6, 411–418 [PubMed] [Google Scholar]
- Suzuki H., Zuo Y., Wang J., Zhang M. Q., Malhotra A., Mayeda A. (2006). Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34, e63 10.1093/nar/gkl151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szweykowska-Kulinska Z., Beier H. (1992). Sequence and structure requirements for the biosynthesis of pseudouridine (psi 35) in plant pre-tRNA(Tyr). EMBO J. 11, 1907–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A., Endo T., Yoshihisa T. (2005). tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309, 140–142 10.1126/science.1113346 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Kikuchi Y. (2001). Origin of the cloverleaf shape of transfer RNA-the double-hairpin model: implication for the role of tRNA intron and the long extra loop. Viva Origino 29, 134–142 [Google Scholar]
- Tang T., Rozhdestvensky T. S., D'orval B. C., Bortolin M. L., Huber H., Charpentier B., et al. (2002). RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 30, 921–930 10.1093/nar/30.4.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomm M., Wich G. (1988). An archaebacterial promoter element for stable RNA genes with homology to the TATA box of higher eukaryotes. Nucleic Acids Res. 16, 151–163 10.1093/nar/16.1.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L., Daniels C. J. (1988). A tRNA(Trp) intron endonuclease from Halobacterium volcanii. Unique substrate recognition properties. J. Biol. Chem. 263, 17951–17959 [PubMed] [Google Scholar]
- Tocchini-Valentini G. D., Fruscoloni P., Tocchini-Valentini G. P. (2005a). Structure, function, and evolution of the tRNA endonucleases of Archaea: an example of subfunctionalization. Proc. Natl. Acad. Sci. U.S.A. 102, 8933–8938 10.1073/pnas.0502350102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini G. D., Fruscoloni P., Tocchini-Valentini G. P. (2005b). Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc. Natl. Acad. Sci. U.S.A. 102, 15418–15422 10.1073/pnas.0506750102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini G. D., Fruscoloni P., Tocchini-Valentini G. P. (2007). The dawn of dominance by the mature domain in tRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 104, 12300–12305 10.1073/pnas.0705537104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini G. D., Fruscoloni P., Tocchini-Valentini G. P. (2009). Processing of multiple-intron-containing pretRNA. Proc. Natl. Acad. Sci. U.S.A. 106, 20246–20251 10.1073/pnas.0911658106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchini-Valentini G. D., Tocchini-Valentini G. P. (2012). Avatar pre-tRNAs help elucidate the properties of tRNA-splicing endonucleases that produce tRNA from permuted genes. Proc. Natl. Acad. Sci. U.S.A. 109, 21325–21329 10.1073/pnas.1219336110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta C. R., Miao F., Arn E. A., Stevens S. W., Ho C. K., Rauhut R., et al. (1997). The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89, 849–858 10.1016/S0092-8674(00)80270-6 [DOI] [PubMed] [Google Scholar]
- Trotta C. R., Paushkin S. V., Patel M., Li H., Peltz S. W. (2006). Cleavage of pre-tRNAs by the splicing endonuclease requires a composite active site. Nature 441, 375–377 10.1038/nature04741 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Yokobori S., Inaba T., Yamagishi A., Oshima T., Kawarabayasi Y., et al. (2002). Introns in protein-coding genes in Archaea. FEBS Lett. 510, 27–30 10.1016/S0014-5793(01)03219-7 [DOI] [PubMed] [Google Scholar]
- Weiner A. (2004). tRNA maturation: RNA polymerization without a nucleic acid template. Curr. Biol. 14, R883–R885 10.1016/j.cub.2004.09.069 [DOI] [PubMed] [Google Scholar]
- Weiner A., Maizels N. (1987). tRNA-like structures tag the 3′ ends of genomic RNA molecules for replication: implications for the origin of protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 84, 7383–7387 10.1073/pnas.84.21.7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert T., Hopper A. K. (2007). tRNA traffic meets a cell-cycle checkpoint. Cell 131, 838–840 10.1016/j.cell.2007.11.014 [DOI] [PubMed] [Google Scholar]
- Westaway K., Abelson J. (1995). Splicing of tRNA precursor, in tRNA, eds Söll D., RajBhandary U. L. (Washington, DC: ASM Press; ), 79–92 [Google Scholar]
- Whipple J., Lane E. A., Chernyakov I., D'silva S., Phizicky E. M. (2011). The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 25, 1173–1184 10.1101/gad.2050711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wich G., Hummel H., Jarsch M., Bär U., Böck A. (1986). Transcription signals for stable RNA genes in Methanococcus. Nucleic Acids Res. 14, 2459–2479 10.1093/nar/14.6.2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann J., Di Giulio M., Yarus M., Knight R. (1995). tRNA creation by hairpin duplication. J. Mol. Evol. 61, 524–530 10.1007/s00239-004-0315-1 [DOI] [PubMed] [Google Scholar]
- Williams K. (2002). Descent of a split RNA. Nucleic Acids Res. 30, 2025–2030 10.1093/nar/30.9.2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. (2004). Evolutionary resealing of a split RNA: reversal of gene permutation. RNA 10, 555–557 10.1261/rna.5274104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I. M. (1993). RNA polymerase III. Genes, factors and transcriptional specificity. Eur. J. Biochem. 212, 1–11 10.1111/j.1432-1033.1993.tb17626.x [DOI] [PubMed] [Google Scholar]
- Worden A., Lee J. H., Mock T., Rouzé P., Simmons M. P., Aerts A. L., et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324, 268–272 10.1126/science.1167222 [DOI] [PubMed] [Google Scholar]
- Xu M. Q., Kathe S. D., Goodrich-Blair H., Nierzwicki-Bauer S. A., Shub D. A. (1990a). Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science 250, 1566–1570 10.1126/science.2125747 [DOI] [PubMed] [Google Scholar]
- Xu Q., Teplow D., Lee T. D., Abelson J. (1990b). Domain structure in yeast tRNA ligase. Biochemistry 29, 6132–6138 10.1021/bi00478a004 [DOI] [PubMed] [Google Scholar]
- Xue S., Calvin K., Li H. (2006). RNA recognition and cleavage by a splicing endonuclease. Science 312, 906–910 10.1126/science.1126629 [DOI] [PubMed] [Google Scholar]
- Yoshihisa T., Yunoki-Esaki K., Ohshima C., Tanaka N., Endo T. (2003). Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol. Biol. Cell. 14, 3266–3279 10.1091/mbc.E02-11-0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari S., Itoh T., Hallam S. J., Delong E. F., Yokobori S., Yamagishi A., et al. (2006). Archaeal pre-mRNA splicing: a connection to hetero-oligomeric splicing endonuclease. Biochem. Biophys. Res. Commun. 346, 1024–1032 10.1016/j.bbrc.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Yukawa Y., Sugita M., Choisne N., Small I., Sugiura M. (2000). The TATA motif, the CAA motif and the poly(T) transcription termination motif are all important for transcription re-initiation on plant tRNA genes. Plant J. 22, 439–447 10.1046/j.1365-313X.2000.00752.x [DOI] [PubMed] [Google Scholar]
- Zahler N., Christian E. L., Harris M. E. (2003). Recognition of the 5′ leader of pre-tRNA substrates by the active site of ribonuclease P. RNA 9, 734–745 10.1261/rna.5220703 [DOI] [PMC free article] [PubMed] [Google Scholar]