Abstract
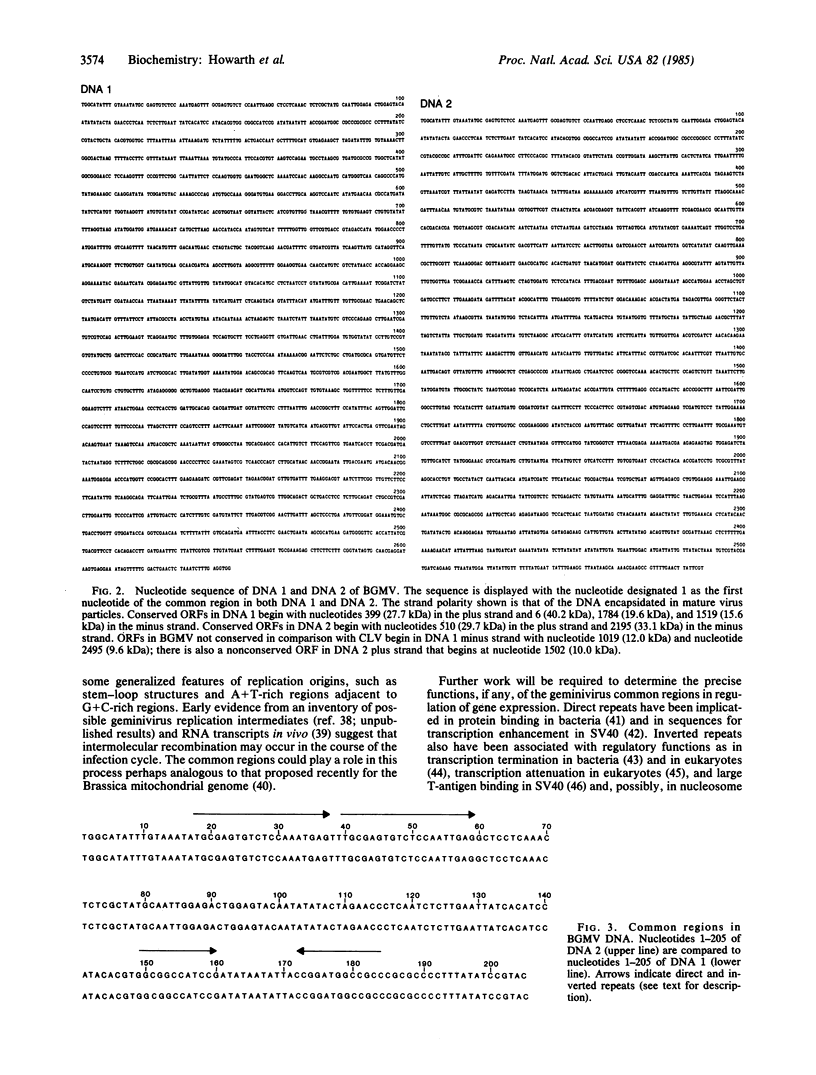
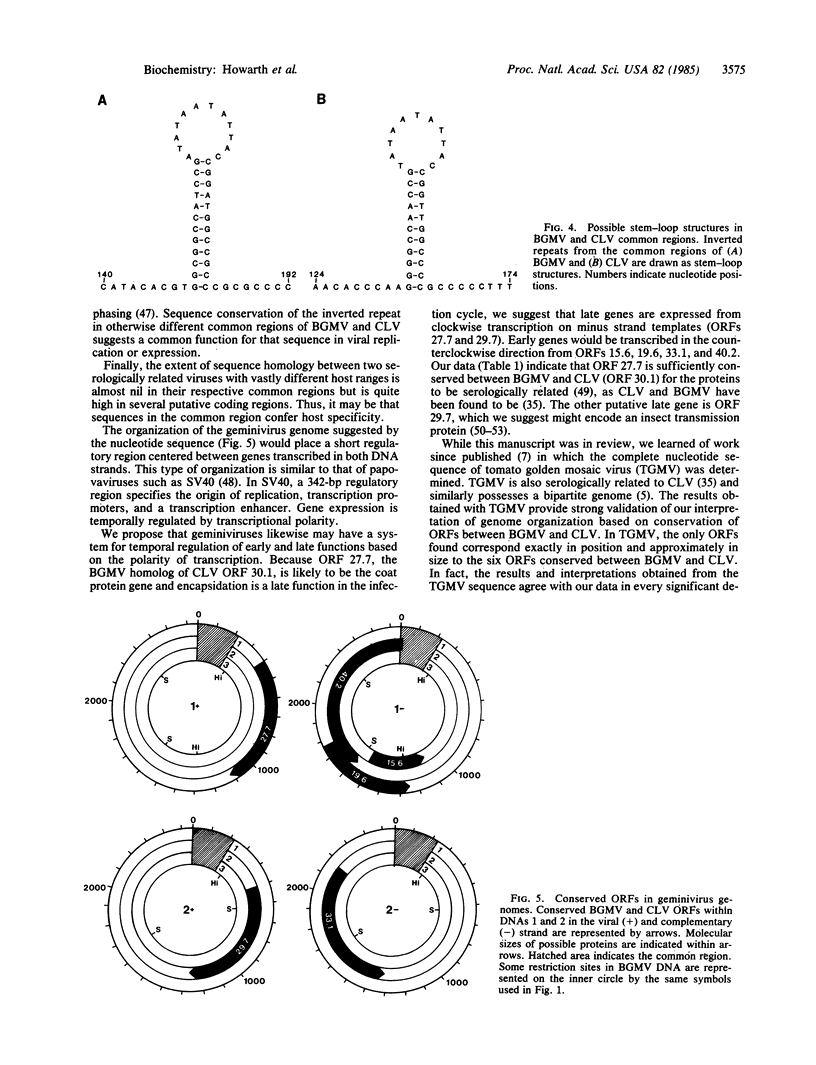
We have sequenced the genome of bean golden mosaic virus, which comprises two circular single-stranded DNA molecules (2646 and 2587 nucleotides long) of mostly unique sequence. Comparison of the sequences of bean golden mosaic virus and of cassava latent virus, which share serological relationship but are very different in host range and geographical origin, shows that each virus has a unique 200-nucleotide sequence (common region) on each 2.6-kilobase molecule of its genome. The common regions of the two viruses have no sequence homology except for a short inverted repeat near the 3′ end. Six open reading frames were identified that possess considerable sequence homology between the two viruses and, in bean golden mosaic virus, may encode proteins of 15.6, 19.6, 27.7, 29.7, 33.1, and 40.2 kDa. Conserved open reading frames are found in both the viral strand and the complementary strand, are approximately the same size, and are in the same orientation with respect to the common region in both viruses. We propose that temporal regulation in geminiviruses depends on the polarity of transcription and that the common region represents a replication origin and contains elements that serve to modulate gene expression.
Keywords: genome organization, stem-loop, comparative virology, viral replication
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birchmeier C., Grosschedl R., Birnstiel M. L. Generation of authentic 3' termini of an H2A mRNA in vivo is dependent on a short inverted DNA repeat and on spacer sequences. Cell. 1982 Apr;28(4):739–745. doi: 10.1016/0092-8674(82)90053-8. [DOI] [PubMed] [Google Scholar]
- Bisaro D. M., Hamilton W. D., Coutts R. H., Buck K. W. Molecular cloning and characterisation of the two DNA components of tomato golden mosaic virus. Nucleic Acids Res. 1982 Aug 25;10(16):4913–4922. doi: 10.1093/nar/10.16.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. C., Howarth A. J., Messing J., Shepherd R. J. Cloning and sequencing of restriction fragments generated by Eco RI*. DNA. 1982;1(2):109–115. doi: 10.1089/dna.1.1982.1.109. [DOI] [PubMed] [Google Scholar]
- Gay N. J., Walker J. E. Homology between human bladder carcinoma oncogene product and mitochondrial ATP-synthase. Nature. 1983 Jan 20;301(5897):262–264. doi: 10.1038/301262a0. [DOI] [PubMed] [Google Scholar]
- Hamilton W. D., Bisaro D. M., Buck K. W. Identification of novel DNA forms in tomato golden mosaic virus infected tissue. Evidence for a two component viral genome. Nucleic Acids Res. 1982 Aug 25;10(16):4901–4912. doi: 10.1093/nar/10.16.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton W. D., Stein V. E., Coutts R. H., Buck K. W. Complete nucleotide sequence of the infectious cloned DNA components of tomato golden mosaic virus: potential coding regions and regulatory sequences. EMBO J. 1984 Sep;3(9):2197–2205. doi: 10.1002/j.1460-2075.1984.tb02114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell S. W., Byrne B. J., Subramanian K. N. Mapping of the late promoter of simian virus 40. Proc Natl Acad Sci U S A. 1984 Jan;81(1):23–27. doi: 10.1073/pnas.81.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidecker G., Messing J., Gronenborn B. A versatile primer for DNA sequencing in the M13mp2 cloning system. Gene. 1980 Jun;10(1):69–73. doi: 10.1016/0378-1119(80)90145-6. [DOI] [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Ikegami M., Haber S., Goodman R. M. Isolation and characterization of virus-specific double-stranded DNA from tissues infected by bean golden mosaic virus. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4102–4106. doi: 10.1073/pnas.78.7.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson R., Messing J. Apple II computer software for DNA and protein sequence data. DNA. 1983;2(1):31–35. doi: 10.1089/dna.1.1983.2.31. [DOI] [PubMed] [Google Scholar]
- Larson R., Messing J. Apple II software for M13 shotgun DNA sequencing. Nucleic Acids Res. 1982 Jan 11;10(1):39–49. doi: 10.1093/nar/10.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Murotsu T., Matsubara K., Sugisaki H., Takanami M. Nine unique repeating sequences in a region essential for replication and incompatibility of the mini-F plasmid. Gene. 1981 Nov;15(2-3):257–271. doi: 10.1016/0378-1119(81)90135-9. [DOI] [PubMed] [Google Scholar]
- Nickol J., Martin R. G. DNA stem-loop structures bind poorly to histone octamer cores. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4669–4673. doi: 10.1073/pnas.80.15.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Queen C., Korn L. J. A comprehensive sequence analysis program for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):581–599. doi: 10.1093/nar/12.1part2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M., Illmensee R., Summers J. Efficeint transcription of RNA into DNA by avian sarcoma virus polymerase. Biochim Biophys Acta. 1976 Sep 6;442(3):324–330. doi: 10.1016/0005-2787(76)90307-5. [DOI] [PubMed] [Google Scholar]
- Tenen D. G., Livingston D. M., Wang S. S., Martin R. G. Effect of a stem-loop structure within the SV40 replication origin upon SV40 T antigen binding to origin region sequences. Cell. 1983 Sep;34(2):629–639. doi: 10.1016/0092-8674(83)90395-1. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tischer I., Gelderblom H., Vettermann W., Koch M. A. A very small porcine virus with circular single-stranded DNA. Nature. 1982 Jan 7;295(5844):64–66. doi: 10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- Toh H., Hayashida H., Miyata T. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. 1983 Oct 27-Nov 2Nature. 305(5937):827–829. doi: 10.1038/305827a0. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Augereau P., Chambon P. The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell. 1983 Feb;32(2):503–514. doi: 10.1016/0092-8674(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Wilkie N. M., Clements J. B., Boll W., Mantei N., Lonsdale D., Weissmann C. Hybrid plasmids containing an active thymidine kinase gene of Herpes simplex virus 1. Nucleic Acids Res. 1979 Oct 25;7(4):859–877. doi: 10.1093/nar/7.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolston C. J., Covey S. N., Penswick J. R., Davies J. W. Aphid transmission and a polypeptide are specified by a defined region of the cauliflower mosaic virus genome. Gene. 1983 Jul;23(1):15–23. doi: 10.1016/0378-1119(83)90212-3. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]


