BRAF-V600E expression is identified in hematopoietic progenitor and precursor myeloid dendritic cells in patients with high-risk LCH, and enforced expression of BRAF-V600E in CD11c+ cells recapitulates a high-risk LCH-like phenotype in mice.
Abstract
Langerhans cell histiocytosis (LCH) is a clonal disorder with elusive etiology, characterized by the accumulation of CD207+ dendritic cells (DCs) in inflammatory lesions. Recurrent BRAF-V600E mutations have been reported in LCH. In this study, lesions from 100 patients were genotyped, and 64% carried the BRAF-V600E mutation within infiltrating CD207+ DCs. BRAF-V600E expression in tissue DCs did not define specific clinical risk groups but was associated with increased risk of recurrence. Strikingly, we found that patients with active, high-risk LCH also carried BRAF-V600E in circulating CD11c+ and CD14+ fractions and in bone marrow (BM) CD34+ hematopoietic cell progenitors, whereas the mutation was restricted to lesional CD207+ DC in low-risk LCH patients. Importantly, BRAF-V600E expression in DCs was sufficient to drive LCH-like disease in mice. Consistent with our findings in humans, expression of BRAF-V600E in BM DC progenitors recapitulated many features of the human high-risk LCH, whereas BRAF-V600E expression in differentiated DCs more closely resembled low-risk LCH. We therefore propose classification of LCH as a myeloid neoplasia and hypothesize that high-risk LCH arises from somatic mutation of a hematopoietic progenitor, whereas low-risk disease arises from somatic mutation of tissue-restricted precursor DCs.
Langerhans cell histiocytosis (LCH) is characterized by inflammatory lesions that include pathological langerin+ DCs. LCH has pleotropic clinical presentations ranging from single lesions cured by curettage to potentially fatal multi-system disease. The first descriptions of LCH, including Hand-Schüller-Christian disease and Letter-Siwe disease, were based on anatomical location and extent of the lesions (Arceci, 1999). The diagnosis of high-risk LCH, defined by involvement of “risk organs” which include BM, liver, and spleen, conferred mortality rates >20%, where patients with disease limited to non-risk organs (low-risk LCH) had nearly 100% survival, regardless of the extent of disease burden (Gadner et al., 2008). Despite clinical heterogeneity, LCH lesions are generally indistinguishable by histology, which led to the notion that the spectrum of clinical manifestations represents a single disorder, histiocytosis X (Lichtenstein, 1953). The designation “Langerhans cell histiocytosis” was subsequently proposed with discovery of cytoplasmic Birbeck granules in the pathological infiltrating DCs in histiocytosis X lesions, a feature shared by epidermal Langerhans cells (Nezelof et al., 1973). Birbeck granules are intracytoplasmic organelles whose role has remained poorly understood since their first identification in 1961 (Birbeck et al., 1961). Recent data revealed that the formation of the Birbeck granules is a consequence of the antigen capture function of a C–type II lectin receptor called langerin, recently named CD207 (Valladeau et al., 2000; Kissenpfennig et al., 2005; Verdijk et al., 2005). Langerin was initially described specifically on human and mouse epidermal Langerhans cells and subsequently found on histiocytosis X lesions, further supporting the epidermal Langerhans cell origin of the disease (Chikwava and Jaffe, 2004).
However, recent discoveries question the model of LCH arising from transformed or pathologically activated epidermal Langerhans cells. The cell-specific gene expression signature in langerin+ DCs within LCH lesions has been shown to be more consistent with immature myeloid DC precursors than epidermal Langerhans cells (Allen et al., 2010). Furthermore, mouse studies demonstrate that langerin is more promiscuous than previously appreciated (Ginhoux et al., 2007). In addition to epidermal Langerhans cells, langerin is also expressed on a subset of DC expressing the integrin CD103 in non-lymphoid tissues (Merad et al., 2008) and its expression is modulated by the tissue environment in which DCs reside (Chang et al., 2010).
The first recurrent somatic genetic mutation in LCH, BRAF-V600E, was recently reported in 57% of LCH lesions (Badalian-Very et al., 2010). Subsequently, recurrent BRAF-V600E mutations were reported in LCH as well as the related disorder Erdheim-Chester disease (ECD; Sahm et al., 2012; Satoh et al., 2012; Haroche et al., 2013). Case reports of two other LCH patients describe a potential activating BRAF-V600D somatic mutation and a novel germline BRAF mutation (Satoh et al., 2012; Kansal et al., 2013).
In this study, we investigate the clinical significance of the BRAF-V600E molecular signature and identify cells carrying the mutation to further define the cellular origins of LCH. We found that the presence of BRAF-V600E in pathological DCs within LCH lesions was associated with higher risk of refractory or recurrent disease. Importantly, we found that BRAF-V600E expression in circulating cells was also associated with disease severity in patients. Moreover, we demonstrate that BRAF-V600E expression in DC precursors is sufficient to induce an LCH-like phenotype in mice with risk organ involvement, whereas BRAF-V600E expression in differentiated DCs induces an attenuated phenotype. These results support a pivotal functional role of the mutation in LCH pathogenesis. We propose a model in which somatic mutation of BRAF-V600E in hematopoietic progenitors versus differentiated hematopoietic cells defines clinical risk in LCH.
RESULTS
BRAF genotype in LCH patients: frequency and clinical correlations
LCH lesions (n = 130) from 100 patients with LCH were analyzed for the presence of the BRAF-V600E mutation (Table S1). Patients were identified retrospectively by availability of tissue biopsies and informed consent, and the cohort largely represents patients seen by the Texas Children’s Histiocytosis Program or collaborators—including Cook Children’s Medical Center—over the past decade. Clinical characteristics of the patients represent a range of age, extent of disease, and clinical risk categories. Median follow-up for data from time of diagnosis was 2.3 yr (range, 0–9.3 yr).
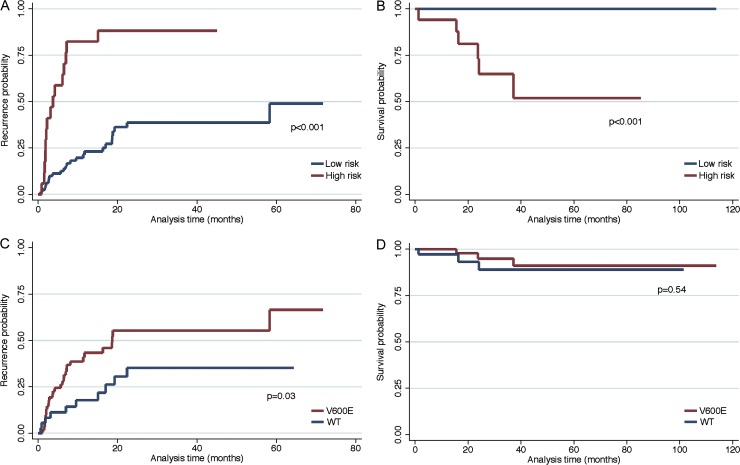
Genotyping was determined by high-sensitivity quantitative PCR (qPCR) of whole-lesion genomic DNA (gDNA) and/or cell-specific Sanger sequencing of cDNA from purified langerin+ cells. Overall, 64% (64/100) of the LCH patients had lesions expressing the BRAF-V600E mutation. The clinical significance of the BRAF mutation was analyzed by comparing BRAF genotype with categorical clinical variables including gender, age, single lesion versus multifocal lesions, high-risk versus low-risk clinical assessment, CNS risk location (skull base and orbit lesion are associated with increased risk of LCH-associated neurodegeneration; Grois et al., 1994), and diabetes insipidus versus no diabetes insipidus. Survival and recurrence/relapse categories were analyzed by Kaplan-Meier estimates. BRAF genotype did not correlate with gender, age, extent of lesions, clinical risk status, CNS risk, or development of diabetes insipidus (Table 1). High-risk clinical status, regardless of BRAF genotype, was associated with significantly higher frequency of relapse and lower survival (Fig. 1, A and B). The BRAF-V600E mutation was associated with approximately twofold increased risk of relapse/recurrence (hazard ratio 2.17, 95% CI: 1.06–4.46). Although both BRAF-V600E and high-risk status were associated with increased risk of recurrence, BRAF-V600E was not significantly associated with high-risk disease (Table 1). Despite the increased risk of recurrence from BRAF-V600E, the mutation was not associated with any difference in overall survival (Fig. 1, C and D).
Table 1.
Clinical and demographic characteristics by BRAF-V600E status
| Characteristic | V600E | WT | P-value |
| Gender | Number (%) | Number (%) | |
| Female | 23 (35.9) | 17 (47.2) | 0.27 |
| Male | 41 (64.1) | 19 (52.8) | |
| Age | |||
| <2 yr | 21 (32.8) | 10 (27.8) | 0.69 |
| 2–8 yr | 28 (43.8) | 19 (52.8) | |
| >8 yr | 15 (23.4) | 7 (19.4) | |
| Clinical risk category | |||
| Low | 52 (81.3) | 31 (86.1) | 0.53 |
| High | 12 (18.7) | 5 (13.9) | |
| Single/multifocal | |||
| Single | 27 (42.2) | 18 (50.0) | 0.45 |
| Multifocal | 37 (57.8) | 18 (50.0) | |
| CNS risk lesions | |||
| No | 36 (56.3) | 24 (66.7) | 0.31 |
| Yes | 28 (43.8) | 12 (33.3) | |
| Diabetes insipidus | |||
| No | 58 (90.6) | 34 (94.4) | 0.50 |
| Yes | 6 (9.4) | 2 (5.6) |
Figure 1.
Clinical status, BRAF genotype, and clinical outcomes. (A) Estimates for refractory or recurrent disease in the study population by disease risk (red: high-risk, blue: low-risk). (B) Estimates for survival based on disease risk (red: high-risk, blue: low-risk). (C) Estimates for refractory or recurrent disease by lesion BRAF genotype (red: wild-type, blue: BRAF-V600E). (D) Estimates for survival based on lesion BRAF genotype (blue: wild-type, red: BRAF-V600E).
BRAF genotype in LCH lesions
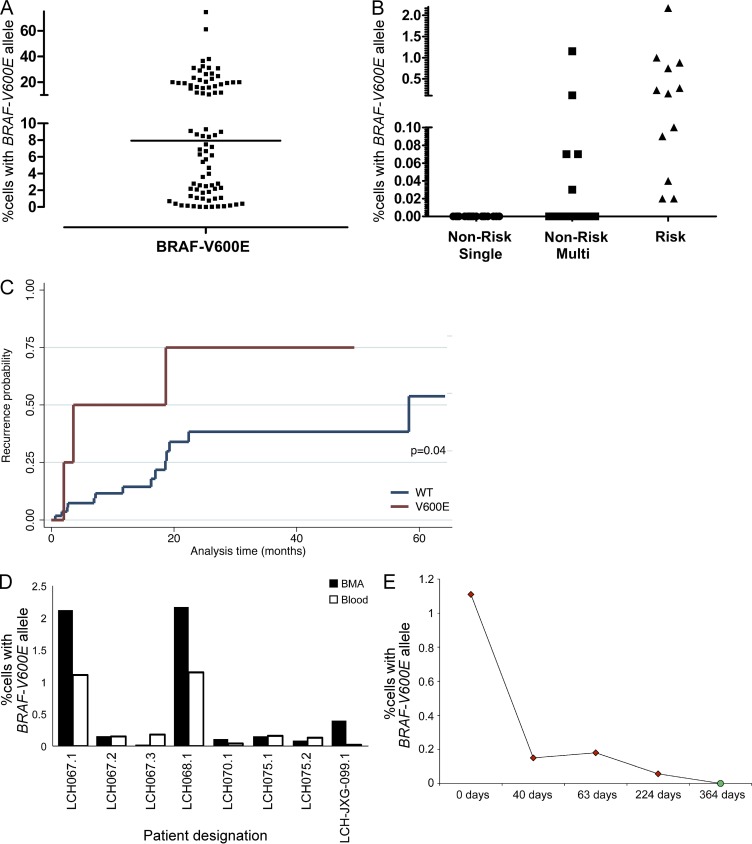
In every case where there were multiple lesions, either synchronous or recurrent, from the same patient (n = 16 patients), the BRAF genotype remained fixed, consistent with acquisition of BRAF-V600E as an early event in LCH pathogenesis (Table S2). Concordance between cell-specific Sanger sequencing of cDNA and qPCR of whole lesion gDNA was observed in 47 out of 48 cases. In the single discordant case (Sanger, wild-type; qPCR, V600E), it is possible that decreased allelic expression or incomplete transcriptome coverage of cDNA amplified from RNA may result in occasional false-negative tests with cDNA sequencing. The percentage of BRAF-V600E cells in LCH lesions varied widely from 0.01–74.5% of lesion cells, with a median of 8.0% (Table S2 and Fig. 2 A). The level of detection of commercially available deep sequencing–based diagnostic strategies requires ∼5% cells with the BRAF-V600E mutation (Verma et al., 2012). In this series, 42% (34/82) of the LCH lesion samples would fall below that threshold, suggesting that LCH will require more sensitive diagnostic approaches, as BRAF genotype becomes part of clinical risk stratification. In a subset of samples (n = 7), cDNA from CD3+ T cells purified from LCH lesions was analyzed along with purified langerin+ cells. Where 4/7 langerin+ cDNA samples had detectable BRAF-V600E, only wild-type BRAF was identified in all of the matched lesion CD3+ cDNA.
Figure 2.
Identification of cells with BRAF-V600E mutation in LCH lesions, circulating cells, and BM aspirates. (A) gDNA was isolated from biopsies of LCH lesions, and the percentage of cells with the BRAF-V600E allele was estimated by qPCR (line = median, 8.0%). (B) gDNA was isolated from PBMCs of patients with active LCH before initial therapy or at a time of recurrence before salvage chemotherapy. The percentage of circulating cells with the BRAF-V600E allele in patients from different clinical risk categories was estimated by qPCR. (C) Estimate of recurrence by circulating BRAF status among those who were clinically classified with low-risk LCH (blue = no circulating cells with BRAF-V600E detected, red = circulating cells with BRAF-V600E detected). (D) gDNA was isolated from gradient-separated leukocytes from synchronous peripheral blood and BM aspirate collected from patients with active LCH. The percentage of cells with the BRAF-V600E allele was determined by qPCR. (Every bar represents a single blood sample; technical duplicates were used in all experiments.) (E) The percentage of circulating cells with the BRAF-V600E allele was determined by qPCR of serial PBMC samples in one patient with disease refractory to multiple salvage therapies (red), and finally cured with clofarabine (green). (Every data point represents a single blood sample; technical duplicates were used in all experiments.)
This series included three patients with a biopsy reported with features with LCH, and also with a history of other histiocytic disorders. In two cases, LCH-JXG-098 and LCH-JXG-099, the patients had lesions with mixed populations of histiocytes described as LCH (CD207+, CD1a+, and S100A+) and juvenile xanthogranuloma (JXG; fascin+, Factor XIIIa+, CD207−, and CD1a−). BRAF-V600E was detectable in lesions from one of the two JXG/LCH hybrid patients (LCH-JXG099). Although serial and hybrid JXG and LCH have been reported previously (Stover et al., 2008), to our knowledge this is the first case of BRAF-V600E associated with a histiocytic lesion including features of JXG. In another case (LCH-ECD100), a patient with a history of ECD had a new bone lesion with histology consistent with LCH (CD207+, CD1a+, and S100A+) with the BRAF-V600E genotype. The presence of BRAF-V600E allele in ECD and the observation of serial ECD and LCH lesions have been reported previously (Pineles et al., 2011, 2012; Haroche et al., 2013). In this series, BRAF-V600E was not identified in lesions from additional patients with ECD (n = 2), JXG (n = 7), or Rosai-Dorfman disease (RDD; n = 5). Additionally, BRAF-V600E was not identified in several biopsies from patients evaluated for possible LCH, with subsequent histology consistent with diagnoses other than histiocytic disorders (n = 17; Table S3).
BRAF genotype in circulating blood and BM in LCH patients: frequency and clinical correlations
From the 100 patients with known BRAF genotype, peripheral blood samples were collected from 77 patients with clinically detectable active disease: 67 patients at the time of diagnosis before chemotherapy and 10 patients at time of relapse before salvage chemotherapy (Table S1). Circulating cells with BRAF-V600E were identified in 17 of the patients (0.02–2.2% of peripheral blood mononuclear cells), all of whom had BRAF-V600E identified in biopsy samples (Table S4 and Fig. 2 B). Clinical correlation with risk group shows that among patients with BRAF-V600E lesions, circulating BRAF-V600E was identified in 100% (12/12) of patients with high-risk disease (clinically documented liver, spleen, or BM involvement by LCH cells) and in 13% (5/39) of patients with low-risk disease. Circulating BRAF-V600E cells were not detected in peripheral blood gDNA from any of the patients with wild-type BRAF lesion (0/26). The presence of BRAF-V600E in circulating cells in patients with active LCH with BRAF-V600E lesions therefore correlates with 100% sensitivity and 87% specificity for clinically defined high-risk disease. All of the clinical low-risk patients with circulating BRAF-V600E cells (n = 5) had multifocal lesions in more than one organ system (including skin, bone, gastrointestinal mucosa, and pituitary), and this group had significantly higher frequency of recurrent/refractory disease than clinical low-risk patients without circulating BRAF-V600E cells detected (hazard ratio 3.31, 95% CI: 0.94–11.70; Fig. 2 C). Clinical risk categories were assigned according to the medical record. It is possible that these patients were incompletely evaluated or had subclinical liver/spleen/BM involvement. For example, patient LCH068 had biopsy-proven skin and bone lesions, but splenic lesions noted on abdominal ultrasound were not biopsied and the patient was regarded as multi-system low-risk by the treating team. Circulating BRAF-V600E cells were identified in this patient in this study. Because the initial therapy is the same for low-risk and high-risk LCH patients, the risks of proving liver or spleen involvement in cases where imaging is equivocal may not be warranted. Interestingly, the described splenic lesions resolved after successful therapy. This example highlights the limitations of conventional staging strategies.
Similarly, BRAF-V600E was detected in BM aspirate from 100% (7/7) of patients with BRAF-V600E lesions with active high-risk LCH (Table S5). qPCR estimated 0.2–2.1% of BM cells in this series with BRAF-V600E mutation. Interestingly, pathology reports describe abnormal findings in only 43% (3/7) of these patients, and one with mixed phenotype peripheral lesions was documented as JXG. In most cases, percentage of BRAF-V600E cells in BM and peripheral blood were similar (Fig. 2 D).
Serial blood samples were analyzed from patients with circulating cells with BRAF-V600E identified at diagnosis or at an early episode of relapse/refractory disease. In 100% (14/14) of episodes where there was no clinical, imaging, or laboratory evidence of active LCH documented in the medical record, the peripheral blood had no detectable cells with BRAF-V600E. In 97% (29/30) of episodes of patients with clinically defined active LCH, BRAF-V600E was detected in peripheral blood cells. In patients with circulating BRAF-V600E cells in peripheral blood at a time of active disease, analysis of circulating blood for BRAF-V600E defined active LCH with 97% sensitivity and 100% specificity (Table S4). In one case of a patient who developed symptoms of high-risk LCH at 6 mo of age (LCH075), BRAF-V600E cells were identified in multiple lesion samples and peripheral blood at the time of diagnosis; however, BRAF-V600E was not identified in a cord blood sample harvested at birth, suggesting that a precursor population was too rare to identify or that the somatic mutation was acquired at a later time (see Fig. 3 B and Fig. 4 B). These data demonstrate that the BRAF-V600E “bar code” may be used not only to diagnose high-risk LCH but also may potentially be tested in serial samples over time as a measure of residual disease (Fig. 2 E).
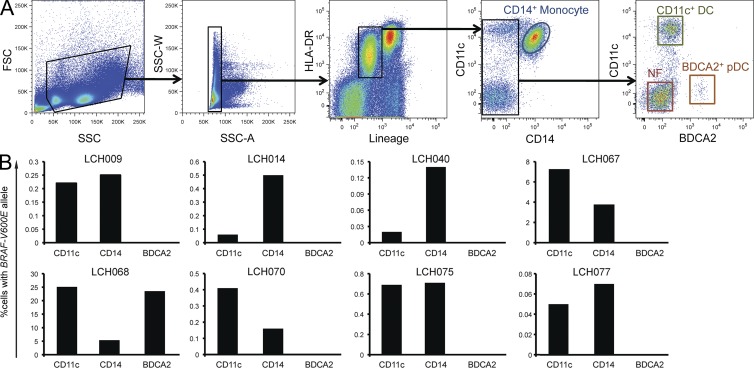
Figure 3.
BRAF-V600E localizes to circulating myeloid DCs and monocytes in peripheral blood. (A) Sorting strategy for peripheral blood mononuclear cells into monocyte (CD14+), myeloid DC (lineage− HLADR+ CD11c+ BDCA2+), and plasmacytoid DC (lineage− HLADR+ BDCA2+) fractions. (B) Percentage of BRAF-V600E cells in circulating monocyte and DC fractions from the indicated LCH patients. (Every bar represents a single sorted blood sample; technical duplicates were used in all experiments.)
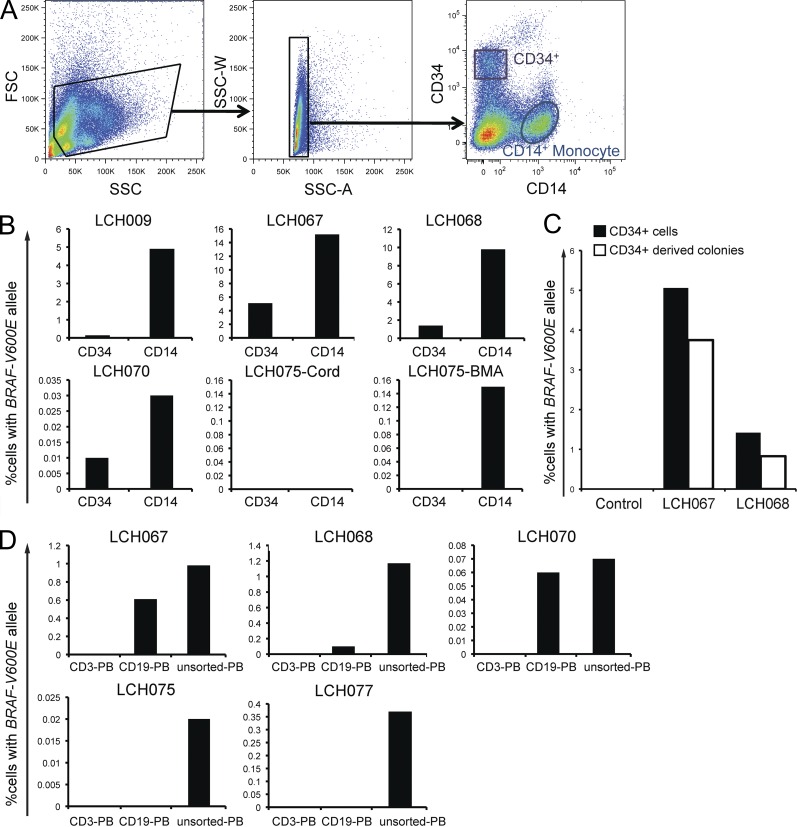
Figure 4.
BRAF-V600E localizes to CD34+ hematopoietic progenitor cells in BM. (A) Sorting strategy for BM aspirate cells into HSPCs (CD34+) and monocytes (CD14+). (B) CD34+ and CD14+ cells were sorted from BM aspirate fractions, gDNA was purified and amplified, and then the BRAF-V600E allele was detected by qPCR. (C) Percentage of CD34+ cells with BRAF-V600E in BM aspirate and the percentage of BRAF-V600E cells obtained from CD34+ HSPC-derived colonies generated in CFU Assay in two LCH patients. (D) PBMCs were sorted by FACS into CD3+ and CD19+ fractions. Percentage of peripheral B and T cells with BRAF-V600E in different LCH patients. (B–D: every bar represents a single sorted blood sample; technical duplicates were used in all experiments.)
BRAF-V600E may be expressed in CD34+ hematopoietic stem and progenitor cells (HSPCs) in LCH patients
To identify the circulating cells harboring the BRAF-V600E mutation, peripheral blood and BM aspirate fractions were analyzed. Samples were selected from available aliquots of viably stored peripheral blood mononuclear cells and BM aspirates from samples with BRAF-V600E detected in unsorted blood and BM aspirate (Table 2). Peripheral blood was first sorted into CD14+ monocytes, HLA-DR+CD11c+ classical DCs, BDCA2+ plasmacytoid DCs, and CD3−CD19−CD56−CD14−CD11c− negative fraction (Fig. 3 A). gDNA was then amplified and tested for the presence of BRAF-V600E. In all cases where BRAF-V600E was identified in the whole peripheral blood sample, it was also detected in CD11c+ DCs (0.2–25.2%) and CD14+ monocytes (0.07–5.4%). One case, LCH068, was initially categorized as low-risk multisystem by the treating team but with splenic lesions of uncertain etiology as described above. This patient was found to have 1.2% circulating BRAF-V600E cells, developed aggressive disease, and ultimately required multiple rounds of salvage therapy. In this patient, BRAF-V600E was detected in BDCA2+ fraction (23.5%), in addition to CD11c+ (25.2%) and CD14+ (5.4%) fractions (Fig. 3 B). BRAF-V600E was undetectable in the negative fraction in all cases (unpublished data).
Table 2.
Analysis of BRAF-V600E in peripheral blood and BM aspirate populations
| Sample | BRAF lesion | Circulating BRAF | CD11c+ PB | CD14+ PB | BDCA2+ PB | CD3+ PB | CD19+ PB | CD34+ BM | CD14+ BM | V600E in CFU assays | Cord blood |
| LCH009 | V600E | Yes | + | + | 0 | + | + | ||||
| LCH014 | V600E | Yes | + | + | 0 | ||||||
| LCH040 | V600E | Yes | + | + | 0 | 0 | 0 | ||||
| LCH067 | V600E | Yes | + | + | 0 | 0 | + | + | + | + | |
| LCH068 | V600E | Yes | + | + | + | 0 | + | + | + | + | |
| LCH070 | V600E | Yes | + | + | 0 | 0 | + | + | + | ||
| LCH075 | V600E | Yes | + | + | 0 | 0 | 0 | 0 | + | 0 | |
| LCH077 | V600E | Yes | + | + | 0 | 0 | 0 |
PB, peripheral blood; BM, BM aspirate.
It remains possible that the circulating cells identified in this series could represent “leaking” lesion or tumor cells rather than LCH precursors. However, the statistically significant frequency in patients with high-risk LCH and failure to identify any circulating cells in patients with large single-lesion tumor burden favor a precursor versus a lesional origin of these cells. To further explore the possibility that circulating cells could represent leaking tumor cells, we performed FACS analysis on a series of blood samples with detectable circulating BRAF-V600E and consistently failed to identify circulating CD207+ cells above background (<0.01%; not depicted).
BM aspirate samples were also analyzed to determine the cellular origins of the BRAF-V600E mutation (Fig. 4 A). They were sorted into CD34+ HSPCs, differentiated CD14+ monocytes, and a negative fraction, which included myeloid and lymphoid precursors as well as CD14-negative leukocytes (Fig. 4 B). In 4/5 BM aspirate samples from patients with BRAF-V600E cells in lesions and high-risk organ involvement (including patient LCH068 who was clinically classified as low-risk but with undefined splenic lesions that resolved with therapy), the BRAF-V600E mutation was identified in CD34+ cells, consistent with somatic mutation of the hematopoietic stem progenitor cell. Lineage− CD34+ HSPCs isolated from BM aspirate from two patients with high-risk LCH were used in CFU assays. The percentage of colonies with BRAF-V600E was similar to the percentage of CD34+ cells from BM aspirate carrying the mutation in both cases (Fig. 4 C). Due to the small number of cells, colonies were pooled to perform BRAF genotyping. BRAF-V600E colonies were identified in the macrophage/monocyte colonies from the LCH068 sample. The colony sizes were smaller with insufficient DNA to perform BRAF genotyping on phenotype-specific colonies for LCH067 (unpublished data). Together, these data suggest that BRAF-V600E mutations may arise in hematopoietic stem cells and promote stem cell differentiation into myelomonocytic precursor cells in patients with high-risk LCH.
Purified circulating CD3+ T lymphocytes and CD19+ B lymphocytes were also analyzed for the expression of BRAF-V600E in parallel aliquots from samples where BRAF-V600E was identified in peripheral monocyte/DC populations. BRAF was wild-type in CD3+ T cells from all samples. In the CD19+ fractions, BRAF-V600E was identified in some cases (LCH067, LCH068, and LCH070) but was absent from others (LCH040, LCH075, and LCH077). In all of the samples where BRAF-V600E was identified in peripheral CD19+ B lymphocytes, the mutation was also identified in CD34+ HSPCs from BM aspirate. In LCH075, BRAF-V600E was identified in neither CD34+ HSPC from BM aspirates nor circulating CD19+ B lymphocytes (Table 2 and Fig. 4 D). These observations are consistent with the potential of hematopoietic progenitors with BRAF hyper-activation to develop into B cells as well as monocytes and myeloid DCs. The absence of BRAF-V600E from T cells may be due to preferential commitment of the BRAF-V600E–expressing precursor to the myeloid and B cell lineages or to T cell–specific toxicity.
BRAF-V600E expression in the DC lineage drives formation of LCH-like lesions in mice
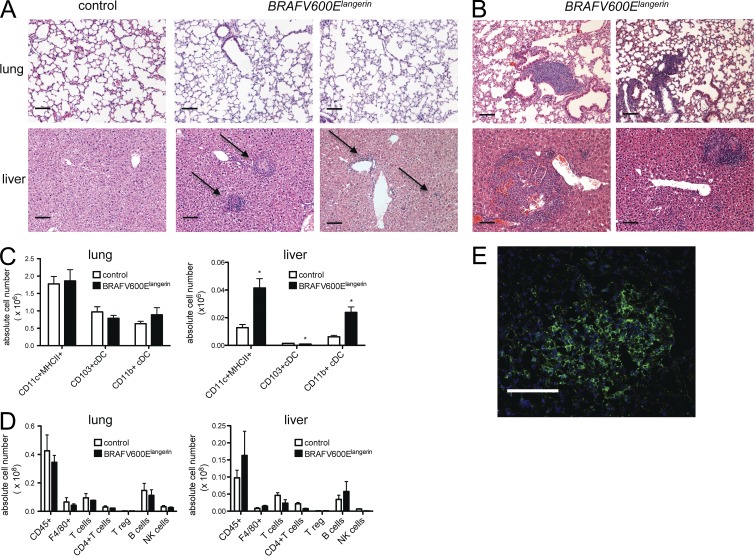
To determine if the BRAF-V600E expression identified in LCH lesions is not only a marker but also an essential driver of LCH pathogenesis, we generated a new mouse model in which we enforced the expression of conditional BRAF-V600E allele under the langerin promoter (BRAFV600Elangerin). In mice, langerin is absent from DC progenitors and is expressed only in differentiated tissue-resident DCs including epidermal Langerhans cells and a subset of tissue DCs that also express the integrin CD103 (Merad et al., 2008). Expression of BRAF-V600E in this mouse model remains under the control of the constitutive BRAF promoter similar to what occurs in patients with a somatic BRAF mutation in exon 15 (Badalian-Very et al., 2010). BRAFV600Elangerin mice were viable and developed normally without gross phenotypical alterations compared with control littermates. However, by 12 wk of age, BRAFV600Elangerin mice developed small inflammatory infiltrates restricted mainly to the liver (Fig. 5 A) that progressively increased in size with age (Fig. 5 B, bottom), and by 20 wk of age, all mice analyzed also displayed small lesions in the lung (Fig. 5 B, top). Flow cytometry analysis revealed an increased number of mainly MHC II+CD11c+ DCs (Fig. 5 C) in the liver of BRAF600Elangerin mice, whereas the number of DCs in the lung and total numbers of macrophages, NK cells, B cells, and T cells in the tissues were unaffected (Fig. 5, C and D). Immunofluorescence (IF) staining further confirmed that a significant proportion of liver tissue–infiltrating cells also expressed langerin (Fig. 5 E). Importantly, we failed to detect anemia or alteration of circulating blood DC numbers in 12- or 20-wk-old BRAFV600Elangerin mice (see Fig. 7, B and C).
Figure 5.
BRAFV600Elangerin mice spontaneously developed progressive accumulation of inflammatory infiltrates in peripheral tissue. (A and B) BRAFV600Elangerin mice were generated as described in Materials and methods and sacrificed at 12 wk (A) or 20 wk (B). Representative lung and liver sections were stained by H&E (in total at least 4–5 mice per group were analyzed; arrows indicate inflammatory infiltrates; bars, 100 µm). (C and D) Lung and liver tissue of BRAFV600Elangerin mice and control littermates were harvested at 12 wk of age and single cells suspensions were analyzed by multicolor flow cytometry analysis after digestion with collagenase. Absolute numbers of total MHCII+ CD11c+ DCs, MHCII+ CD11c+ CD103+ DCs, and MHCII+ CD11c+ CD11b+ DCs are presented (C, all cells are pregated on singlets, viable CD45+ cells, n = 3 per group, representative of two independent experiments). Absolute number of tissue infiltrating total hematopoietic CD45+ cells, F4/80+ macrophages, total CD3+ NK1.1− T cells, total CD3+ NK1.1− CD4+ CD8− T cells, total CD3+ NK1.1− CD4+ Foxp3+ T reg cells, total CD19+ B220+ B cells, and total CD3− NK1.1+ NK cells are depicted in D (all cells are pregated on singlets, viable CD45+ cells, n = 3 per group, representative of two independent experiments). (E) Representative picture of IF staining for langerin in liver lesion of 12-wk-old BRAFV600Elangerin mouse. Bars, 100 µm. Data are shown as mean ± SEM. *, P < 0.05.
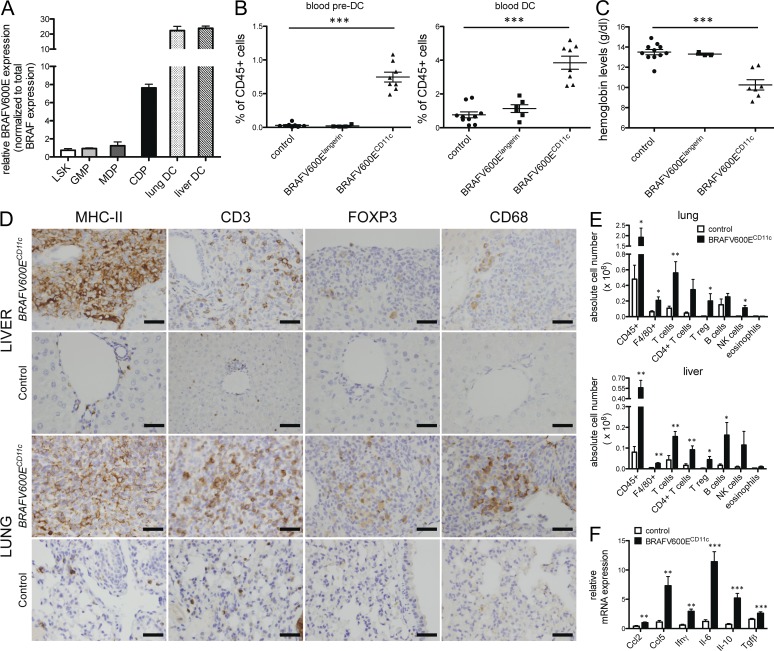
Figure 7.
Expression of BRAF-V600E in early BM-resident DC progenitors in BRAFV600ECD11c mice results in multi-systemic high-risk disease and is associated with high local cytokine expression and the recruitment of additional inflammatory cells. (A) Relative mRNA expression of BRAF-V600E in sorted BM lineage negative sca1+ c-kit+ (LSK), lin− sca1− c-kit+ CD34+ CD16/32+ granulocyte myeloid progenitors (GMPs), lin− sca1− CD135+ c-kithigh CD115+ monocyte and DC progenitors (MDPs), and lin− sca1− CD135+ c-kitlow CD115+ common DC progenitors (CDPs) and lung/liver CD45+ MHCII+ CD11c+ DCs in BRAFV600ECD11c mice as assessed by mutation-specific qPCR (data normalized to total BRAF expression, no BRAF-V600E expression detected in corresponding cells of control mice, n = 3, pooled data of three independent isolations). (B and C) Relative numbers of viable singlet CD11b+ CD115+ Flt3+ pre-DCs and CD11c+ MHCII+ DCs among circulating viable, singlet CD45+ cells (B) and hemoglobin levels (C) in BRAFV600ECD11c mice, BRAFV600Elangerin mice, and control mice (n = 4–10 per group, pooled data of four independent experiments, each data point represents one mouse). (D) Immunohistochemical analysis of liver and lung tissue sections stained with indicated mAb in BRAFV600ECD11c mice (top) and control littermates (bottom; bars, 50 µm). (E) Absolute numbers of tissue-infiltrating total hematopoietic CD45+ cells, F4/80+ macrophages, total CD3+ NK1.1− T cells, total CD3+ NK1.1− CD4+ CD8− T cells, total CD3+ NK1.1− CD4+Foxp3+ T reg cells, total CD19+ B220+ B cells, total CD3− NK1.1+ NK cells, and total SiglecF+ CD11c− eosinophils in lung and liver of BRAFV600ECD11c mice and control littermates as assessed by multicolor flow cytometry (n = 4–5 per group, representative of two independent experiments). (F) Local expression of indicated chemokines and cytokines in peripheral liver tissue was assessed by qPCR (data normalized to Gapdh expression, n = 4–5 per group). All data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
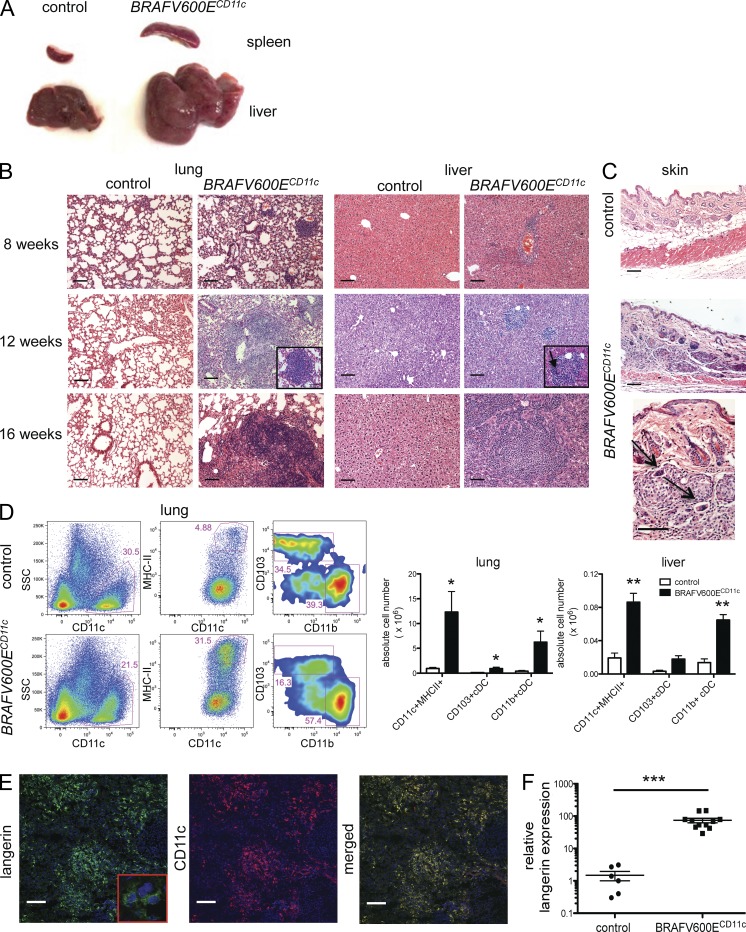
Our results in patients showing that BRAF-V600E expression in early hematopoietic precursors in human patients correlate with high-risk LCH and early involvement of risk organs prompted us to assess whether BRAF-V600E expression at earlier stages of DC development can affect LCH phenotype. Thus, we generated another mouse model in which the BRAF-V600E conditional allele was expressed under the CD11c promoter. In mice, CD11c is expressed on committed DC progenitors and remains expressed throughout DC differentiation (Merad et al., 2013). In striking contrast to BRAFV600Elangerin mice, BRAFV600ECD11c mice rapidly developed an aggressive LCH-like disease phenotype with a penetrance of 100% associated with a decreased lifespan. Along with severe hepatosplenomegaly and lymphadenopathy, organized histiocytic infiltrates were identified in the skin, liver, spleen, and lungs of all mice by 8 wk of age, resulting in a broad destruction of tissue architecture by 16 wk of age (Fig. 6, A–C). Histologically, the histiocytic lesions observed in BRAFV600ECD11c mice exhibited classical granulomatous organization, including multinucleated giant cell formation which is frequently observed in human LCH (da Costa et al., 2005; Fig. 6 B). These lesions were associated with a massive increase of MHC II+CD11c+ DCs (Fig. 6 D) with a classical DC shape (Fig. 6 E, higher magnification). The vast majority of the CD11c+ cells accumulating in the tissues also expressed high levels of langerin protein (Fig. 6 E) associated with a dramatic increase of langerin mRNA transcripts in tissues compared with control littermates (Fig. 6 F). As in human LCH lesions, mitoses were not observed in the granulomatous lesions observed in BRAFV600ECD11c mice (Senechal et al., 2007; Fig. 6 B).
Figure 6.
BRAFV600ECD11c mice develop an aggravated phenotype with massive infiltration of MHCII+ CD11c+ langerin-expressing cells in peripheral tissues. (A) Representative pictures of the spleen and liver of BRAFV600ECD11c mice (right) and control littermates (left) at 12 wk of age (n = 16 per group). (B) Representative H&E staining of liver and lung tissue sections of BRAFV600ECD11c mice and corresponding controls at the indicated ages (arrow marks multinucleated giant cells; bars, 100 µm; n = 5–11 animals per group). (C) H&E staining of back skin tissue sections of 12-wk-old BRAFV600ECD11c mice and controls (arrows indicate multinucleated giant cells; bars, 100 µm; n = 4 per group). (D) Absolute numbers and percentages of MHC II+ CD11c+ DC among total CD45+ lung and liver cells in 12-wk-old BRAFV600ECD11c mice and control littermates were determined by multicolor flow cytometry (all cells pregated on singlets, viable cells, n = 3–5 per group, representative of three independent experiments). (E) Representative image of frozen liver tissue section isolated from the liver of 16-wk-old BRAFV600ECD11c mice stained with anti-langerin mAb (left) and anti-CD11c mAb (right) or with both langerin and CD11c mAb (merged) and analyzed by confocal microscopy (bars, 100 µm). (F) Langerin mRNA expression in whole liver tissue of BRAFV600ECD11c mice and corresponding controls was analyzed by qPCR (normalized to Gapdh mRNA expression, n = 6–11 per group). All data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We confirmed that lesional MHC II+CD11c+ DCs isolated from the liver of BRAFV600ECD11c mice specifically expressed the BRAF-V600E mutation as assessed by mutation-specific qPCR (Fig. 7 A). Importantly, and consistent with the data in patients with high-risk multi-systemic LCH, BRAF-V600E was also present in BM-resident common DC progenitors (CDPs) in BRAFV600ECD11c mice (Fig. 7 A), whereas it was absent from BM progenitors in BRAFV600Elangerin mice (not depicted). The BRAF-V600E mutation was restricted to the DC lineage and was absent from early myeloid precursors or multipotent progenitors in BRAFV600ECD11c mice. Expression of the mutation in DC-restricted precursors was associated with severe anemia, confirming BM involvement in these mice (Fig. 7 C), together with a dramatic expansion of blood circulating DC precursors (pre-DCs) and blood circulating DCs (Fig. 7 B), whereas blood pre-DCs and DC numbers, as well as hemoglobin levels, remained unaffected in BRAFV600Elangerin mice.
The granuloma lesions observed in BRAFV600ECD11c mice contained, in addition to massive CD11c+ langerin+ infiltrates, a large number of macrophages, NK cells, B cells, and T cells, with a specific accumulation of regulatory T cells (Fig. 7, D and E) closely resembling LCH lesions in humans (Hicks and Flaitz, 2005; Senechal et al., 2007). Importantly, these granuloma-like lesions were also associated with a significant increase in several chemokines and cytokines (Ccl2, Ccl5, Il-6, Il-10, IFN-γ, and TGF-β; Fig. 7 F), consistent with the “local cytokine storm” that characterizes human LCH (Allen et al., 2010). Additionally, a local fibrotic response occasionally observed in human LCH granuloma and likely due to high cytokines expression—for example, TGF-β and IFN-γ (de Graaf et al., 1996)—was detected within the granulomas and the surrounding stroma (not depicted).
DISCUSSION
The etiology of LCH has intrigued and eluded the medical community for the past century as a paradox of a collection of highly variable clinical manifestations connected by common histopathology. The resulting debate on LCH pathogenesis has recently focused on aberrant activation versus malignant transformation of the epidermal Langerhans cell. Previous studies demonstrated that pathological DCs in LCH are clonal (Willman et al., 1994; Yu et al., 1994) and that the BRAF-V600E point mutation occurs with significant frequency in LCH lesions (Badalian-Very et al., 2010; Haroche et al., 2012; Sahm et al., 2012; Satoh et al., 2012). In this study, we demonstrate that BRAF status of pathological DCs in LCH lesions is fixed in synchronous lesions as well as serial relapse samples, consistent with the BRAF-V600E mutation as an early or initiating event in LCH pathogenesis, tipping the scales to classify LCH as a neoplastic disorder. We also demonstrate that BRAF-V600E mutation in patients with high-risk LCH is a somatic event that seems to arise in very early myelomonocytic precursors or in stem cells that may be subsequently driven to myelomonocytic differentiation, whereas BRAF-V600E mutation in patients with low-risk LCH was not routinely identified in circulating cells, suggesting it may be restricted to differentiated tissue-resident DCs in these patients. We cannot entirely exclude that the BRAF-V600E–expressing cells in the blood and BM of high-risk patients might represent circulating lesional cells; however, the statistically significant presence of circulating cells in high-risk patients and infrequent detection in low-risk patients, lack of langerin+ cells in the blood of patients with circulating BRAF-V600E–expressing cells, and the identification of the mutation in the CD34+ fraction in the BM render this hypothesis unlikely. CD34 is a major positive marker of human HSPCs and is not expressed by mature blood cells or tissue-restricted myeloid cells (Kondo et al., 2003). Furthermore, the progenitor nature of BRAF-V600E–expressing CD34+ cells is greatly supported by the CFU assay performed with sorted CD34+ BM cells of two high-risk patients, which yield a percentage of colonies with BRAF-V600E similar to the percentage of CD34+ cells from BM aspirates. This shows the high proliferative potential of mutation-bearing CD34+ BM cells, defining them as colony-forming cells/progenitor cells. As lesional cells display only low proliferation rate (Senechal et al., 2007), contamination of the assay with circulating LCH cells seems implausible.
Using two novel mouse models, we established that the BRAF-V600E mutation in myeloid cells results in the formation of LCH-like lesions in mice. Induction of physiological expression levels of BRAF-V600E in BM DC progenitors induced a severe LCH-like phenotype with multi-organ involvement and reduced lifespan, whereas expression of BRAF-V600E in differentiated DCs resulted in an attenuated phenotype. Consistent with identification of BRAF-V600E mutation in patients with high-risk LCH, BRAF-V600E expression in hematopoietic progenitors in mice resulted in BM involvement, indicated by severe anemia and expansion of circulating DC precursors and circulating DC, whereas anemia or alteration in circulating DCs was not detectable when the mutation was restricted to the more differentiated DCs. These findings implicate the BRAF-V600E mutation as a driver of LCH-like disease in mice. Although these mouse models recapitulate several features of human LCH, the extent to which these results can be translated to human LCH pathology remains a matter for investigation.
Based on our results, we propose a new hypothetical model of LCH in which the initiating cell defines the extent of the disease. The mixed inflammatory lesion that defines LCH clinically is indistinguishable between different LCH clinical risk groups. However, LCH lesions may represent only a superficial downstream manifestation of pathogenesis. If BRAF-V600E is the activating event, somatic mutation in a hematopoietic stem cell or myeloid DC restricted precursors may determine progression to high-risk disease, potentially with lesions in BM, liver, spleen, or virtually any organ system. Neoplastic transformation of an early multipotent myeloid precursor could explain the occurrence of coexistent histiocytic disorders (JXG/LCH and ECD/LCH) in the same patients in this study, as well as in others, and the common BRAF-V600E mutation in both LCH and ECD (Stover et al., 2008; Pineles et al., 2011; Haroche et al., 2012). Moreover, immature myeloid origins of high-risk LCH are consistent with effectiveness of acute myelogenous leukemia–based salvage chemotherapy with cytarabine/cladribine and clofarabine for patients with recurrent/refractory high-risk LCH (Bernard et al., 2005; Rodriguez-Galindo et al., 2008).
We further hypothesize that if the initiating mutation occurs in tissue-restricted DC progenitors, multifocal low-risk disease could result. Finally, if an initiating mutation arises in a differentiated DC with restricted tissue tropism, low-risk LCH with a single lesion could result. In this model, as pathological DCs from circulating or tissue-restricted precursors migrate to sites of lesion formation, they may acquire CD1a and langerin expression and recruit and activate “innocent bystander” inflammatory cells, including macrophages and lymphocytes, resulting in the formation of characteristic granulomatous LCH lesions mediated, at least in part, by an altered expression of chemotactic cytokines. Based on the misguided differentiation of the pathological DC and their ability to recruit and activate additional inflammatory cells, we propose that the answer to the neoplasia versus inflammation debate is that both processes may be essential to LCH pathogenesis.
The results from this study also revealed some innovative diagnostic tools that will need to be validated in future prospective clinical studies. The qPCR assay was highly sensitive and specific for diagnosis of LCH. In the absence of fresh tissue on which to perform cell sorting, qPCR on whole tissue samples appears to be a robust strategy compared with sequencing strategies for unmanipulated biopsy samples, many of which have pathological DC content <5%. The BRAF-V600E mutation is clinically relevant, as LCH patients with BRAF-V600E expression within the lesions were approximately twice as likely to have recurrent or refractory disease as patients with wild-type lesions, and detection of BRAF-V600E in peripheral blood was 100% sensitive for clinical high-risk disease associated with significant mortality. Patients with clinical low-risk multisystem LCH but evidence of circulating BRAF-V600E–expressing cells also had a significantly higher chance of recurrent/refractory disease compared with low-risk multisystem LCH patients without detectable circulating BRAF-V600E cells. It is possible that our findings differ with previous reports of absent circulating BRAF-V600E cells due to the relative insensitivity of deep sequencing and/or lack of inclusion of patients with BRAF-V600E and active high-risk disease in the study cohorts (Sahm et al., 2012; Satoh et al., 2012). Future multi-center prospective studies will determine if multifocal clinical low-risk disease with circulating BRAF-V600E cells represents an independent clinical risk category or simply reflects the limitation of existing clinical scoring strategies to accurately categorize low-risk versus high-risk disease.
In addition to potential clinical utility in risk stratification, BRAF mutation status might also be useful for disease surveillance. In patients with circulating BRAF-V600E cells, detection of BRAF-V600E in peripheral blood was 97% sensitive to active disease. The potential value of surveillance strategies in LCH is indicated by the published very high rates of treatment failure or disease reactivation, which may represent persistence of LCH precursor cells. In the Histiocyte Society trial LCH-I, 57–65% of high-risk patients were either nonresponders or had progressive disease at the sixth week of treatment with vinblastine or etoposide, and reactivations were observed in 55–61% (Gadner et al., 2001). The LCH-II study reported similar results with 51–64% nonresponse or progressive disease in high-risk patients treated with prednisone and vinblastine ± etoposide, and reactivations occurred in 46% of patients (Gadner et al., 2008). In this study, we observed refractory or recurrent LCH in nearly 90% of our high-risk patients, with twofold increased risk across all risk groups in patients with the BRAF-V600E mutation. We question the wisdom of generally treating clinical high-risk, or even clinical low-risk patients, with BRAF-V600E lesions initially with vinblastine, prednisone, or etoposide with such high rates of failure. As we are now able to identify BRAF-V600E minimal residual disease, future clinical trials will test sustained responses to experimental therapies more efficiently.
In light of the findings of this study, we hypothesize that optimal therapy for LCH would specifically impact immature myeloid DC precursors. Targeted inhibition of BRAF-V600E is an obvious strategy with reported short-term responses in a pilot study of adults with features of LCH along with ECD (Haroche et al., 2013). Although potentially promising, the long-term efficacy of targeted therapy against BRAF in LCH remains to be proven. Even if effective, the relative mortality in most patients with LCH and the known toxicities of BRAF inhibition make optimal use of these agents an uncertain equation, especially in children where the effects of BRAF inhibition on development are unknown (Chapman et al., 2011). In addition to chemotherapy or BRAF inhibition, novel strategies could include inhibition of myeloid growth factors, inhibition of maturation or migration of pathological DCs, and disruption of cell–cell interactions that recruit and activate bystander inflammatory cells.
Until now, development and evaluation of novel therapies for patients have been limited by a lack of animal models of LCH. Although enforcement of BRAF-V600E expression across a DC lineage in mice is not completely parallel to development of somatic mutation in a single progenitor cell in humans, the BRAFV600ECD11c mouse is a promising model with significant similarity to high-risk human LCH. This model not only supports a model of pathogenesis in which LCH may arise through expression of BRAF-V600E in precursor cells and DCs but may also represent a valuable tool with which to develop and evaluate future therapeutic strategies. In summary, the observation of BRAF-V600E in BM progenitor cells and immature circulating cells in patients with high-risk LCH, coupled with the development of LCH-like lesions in mice expressing the BRAF mutation within the myeloid lineage, supports a causative role of BRAF-V600E in LCH pathogenesis and progression, consistent with classification of LCH as a bone fide myeloid neoplasia.
MATERIALS AND METHODS
Human studies
Subjects.
LCH diagnosis was established by the presence of CD1a+ or CD207+ histiocytes in clinical biopsy specimens along with characteristic histopathology. Clinical data were collected from approved chart reviews. Studies with patient tissue and clinical data were performed according to protocols approved by the Institutional Review Board of Baylor College of Medicine.
Analysis of clinical variables.
Frequency distributions of demographic and clinical characteristics were tabulated for those with and without the BRAF-V600E mutation. The crude odds ratio (OR) and 95% confidence interval (CI) were estimated using unconditional logistic regression for the association between each characteristic and mutation status. Kaplan-Meier survival estimates were calculated for recurrence/relapse and overall mortality. The sensitivity and specificity of circulating cells with BRAF-V600E were compared with the clinical assessment of disease risk (i.e., gold standard). All statistical analyses were performed using Intercooled Stata (version 12.1; StatCorp LP).
BRAF-V600E assays: LCH lesions.
gDNA was isolated from frozen whole lesions, or from frozen sections cut from biopsies embedded in OCT blocks using the QIAamp DNA mini and QIAamp DNA micro protocols (QIAGEN), including treatment with RNase A (QIAGEN). When gDNA isolated from scrolls of formalin fixed and paraffin embedded (FFPE) archived biopsy samples were used, the samples were processed with QIAamp DNA FFPE Tissue kit per manufacturer’s protocol (QIAGEN), including treatment with RNase A. Purified gDNA (30 ng/sample) was used in the BRAF-V600E qPCR mutation assay (Somatic Mutation Assay for BRAF_476; QIAGEN). BRAF mutation and reference primers were included in each reaction. Duplicate reactions were performed for each sample. All experiments were performed on an iQ5 iCycler (Bio-Rad Laboratories). The means of Ctmut and Ctref were calculated to determine ΔCt = Ctmut(ave) − Ctref(ave). The ΔCT was compared with a standard curve to estimate the percentage of cells with BRAF-V600E alleles. The standard curve was created by making 13 dilutions of gDNA from A375 cell line (American Type Culture Collection), which carries one allele of BRAF-V600E and one allele with wild-type BRAF per cell, with gDNA from HEK293 cell line (American Type Culture Collection), which carries two wild-type BRAF alleles per cell. The ΔCT was plotted against the percentage of cells from A375/HEK293 cell pools (0.1–100% BRAF-V600E) from which the gDNA was isolated. For each LCH gDNA sample, the percentage of cells with the BRAF-V600E mutation was then calculated based on the standard curve. Independent controls (wild type and 100% BRAF-V600E cell lines) were routinely tested along with standard curve and experimental samples.
Cell-specific mutation analysis was performed in langerin+ and CD3+ cells purified from viable LCH lesions. Cell sorting, RNA purification, and cDNA amplification was performed as described previously (Allen et al., 2010). BRAF sequence was analyzed in cDNA from purified cells by PCR amplification, followed by Sanger sequencing. BRAF-V600E was identified in cases where GAG peak was visible, along with wild-type GTG at codon 600. Primers: forward, 5′-AGCTCAATAGAGGCGAGAAT-3′; reverse, 5′-AGCTCAATAGAGGCGAGAAT-3′.
BRAF-V600E assays: peripheral blood and BM aspirate.
Buffy coat from fresh peripheral blood, BM aspirate, and cord blood samples was purified by spinning an equal volume over Histopaque-1077 (Sigma-Aldrich) at 400 g for 30 min. Samples were washed with PBS, and then viably stored long-term in Recovery Cell Culture Freezing Media (Gibco) until they were thawed for gDNA isolation, per protocol described above, or used for sorting experiments. gDNA from sorted cells was purified with the QIAamp DNA Micro kit (QIAGEN), and then amplified with the REPLI-g Midi kit (QIAGEN) per manufacturer’s instructions. qPCR was performed as described above on the amplified gDNA. BRAF-V600E:wild-type standard curves were created for amplified gDNA from 13 dilutions of amplified gDNA from A375 cells with amplified gDNA from HEK293 cells.
BRAF-V600E assays: sorting peripheral blood.
Purified leukocytes from peripheral whole blood were subjected to staining using anti–CD11c-PE, anti–HLADR-APC-Cy7, lineage antibodies (anti-CD3–Pacific blue, anti-CD19–Pacific blue, and anti-CD56–Pacific blue), anti–CD14-APC (BD), and anti–BDCA2-FITC (Miltenyi Biotec). Thawed samples were blocked for 30 min in FcR-block (Miltenyi Biotec) and stained for 30 min in ice-cold PBS supplemented with 2 mM EDTA and 5% BSA. For purified peripheral blood populations, isolation of lin−HLADR+CD14−CD11c+, lin−HLADR+CD14+, lin−HLADR+BDCA2+, and lin−HLADR+ precursor cells were achieved by six-color sorting on an Aria BSL2 (BD). B and T cell populations were isolated from select samples using anti–CD3-FITC and anti–CD19-PE (BD) on a MoFlo sorter (Beckman Coulter). Purified populations were sorted directly into ice-cold fetal bovine serum (Invitrogen), and gDNA was isolated using the QIAamp DNA Micro kit with RNase A treatment and amplified with the REPLI-g Midi kit.
BRAF-V600E assays: sorting BM and cord blood.
Purified cord blood and BM leukocyte populations were isolated by staining with anti–CD34-PE and anti–CD14-APC antibodies (BD) and subsequent three-color sorting for the CD34+CD14, CD34−CD14+, and double-negative populations. Populations were sorted into ice-cold fetal bovine serum (Invitrogen) and gDNA isolated using the QIAamp DNA Micro kit with RNase A treatment to remove endogenous RNA. All flow cytometry analysis was performed using FlowJo software (Tree Star).
BM CFU assays.
Frozen healthy or patient BM samples were stained with anti-CD123–Brilliant violet 421, anti–CD38-FITC, anti–CD90-PE, anti-lineage markers (CD3, CD4, CD8, CD11b, CD14, and GPA)–Tricolor, anti–CD34-PE-Cy7, anti–CD127-biotin, and anti–CD45RA-APC-Cy7 antibodies (BioLegend, BD, or Invitrogen). Subsequently, biotin was detected by streptavidin-APC (eBioscience). CD34+lin− cells were sorted into methylcellulose (H4230; STEMCELL Technologies) containing 20 ng/ml IL-3, 10 ng/ml IL-6, 10 ng/ml IL-11, 10 ng/ml SCF, 50 ng/ml TPO, 4 U/ml EPO, 50 ng/ml GM-CSF, and 10 ng/ml Flt3L (PeproTech), and cultured for 12–15 d, followed by colony classification and isolation of gDNA from single colonies with the QIAamp DNA Micro kit.
Mouse studies
Mouse models.
All animal experiments performed in this study were approved by the Institutional Animal Care and Use Committee (IACUC) of Mount Sinai School of Medicine. BRAFV600Elangerin mice were generated by crossing mice expressing cre recombinase under the control of the murine langerin promoter (C57BL/6 background; provided by B. Claussen, Erasmus University Medical Center, Rotterdam, Amsterdam; Zahner et al., 2011) with heterozygous mice carrying a cre-activated allele of BRAF-V600E (BRAFV600Eca/wt, C57BL/6 background) in which wild-type BRAF is expressed in both alleles in the absence of cre-mediated recombination (provided by M.W. Bosenberg, Yale University, New Haven, CT; Dankort et al., 2007). BRAFV600ECD11c mice were created by crossing BRAFV600Eca/wt mice with mice expressing cre recombinase under the control of the CD11c promoter (C57BL/6 background; The Jackson Laboratory). All animals were housed under specific pathogen-free (SPF) conditions and sacrificed at the indicated time points. All experiments were controlled using littermates negative for the cre recombinase transgene construct.
Histology, immunohistochemistry (IHC), and IF analysis.
Langerin and CD11c staining was performed on acetone-fixed frozen tissues, and all other staining was performed on FFPE tissue at the Histology Shared Resource Facility of Mount Sinai School of Medicine and at the Baylor College of Medicine Pathology Core according to standard operating procedures. For IF staining with anti-langerin and CD11c mAb, fresh tissue was cryopreserved in OCT (Sakura) and stored at 80°C. 8-µm-thick sections were air dried for 1 h and fixed in acetone for 20 min at −20°C. After incubation with 10% goat serum for 1 h, sections were stained overnight at 4°C with rat anti-langerin (clone eBioL31; eBioscience) or Armenian hamster anti-CD11c (clone N418; eBioscience) mAbs. After several washing steps, sections were incubated for 60 min with goat anti–rat antibody labeled with Alexa Fluor 488 (Invitrogen) and goat anti–Armenian hamster labeled with Alexa Fluor 594 (BioLegend) and embedded in ProLong Gold Antifade Reagent with DAPI (Life Technologies). Images were acquired with a confocal microscope (SP5 DM; Leica) and analyzed with ImageJ software (National Institutes of Health).
For the IHC analysis, 4-µm tissue sections were deparaffinized and rehydrated through xylene/absolute alcohol. Endogenous peroxidase activity was blocked by incubating the sections in methanol with 0.6% hydrogen peroxide for 10 min at room temperature. Heat-induced antigen retrieval was performed with citrate buffer, pH 6.0 (Diagnostic BioSystems), in a steamer for 15 min. To block nonspecific staining, Rodent Block M (Biocare Medical) was incubated with the section for 1 h at room temperature. Sections were stained with anti-CD3 (Abcam), anti–MHC II (BD), anti-FOXP3 (BioLegend), or anti-CD68 (Thermo Fisher Scientific) mAbs overnight at 4°C. Application of the primary antibodies was followed by a 30-min incubation with a Rabbit on Rodent/Mouse on mouse/rat on Rodent Polymer-HRP (BioCare Medical) and visualized with DAB (Diagnostics BioSystems) as a chromogen with CAT hematoxylin counterstaining (BioCare Medical).
Flow cytometry analysis.
Single cell suspension was obtained from indicated tissues after digestion with 0.4 mg/ml of type IV collagenase (Sigma-Aldrich) at 37°C for 45 min. For liver tissues, nonparenchymal cells were enriched by density gradient centrifugation with 40/70 Percoll (GE Healthcare) for 30 min at 1,100 rcf. BM and blood single cell suspensions were incubated with RBC lysis buffer (BioLegend) for 2 min at room temperature before staining. mAbs specific to mouse CD45 (clone 30F11; BioLegend), MHC II (I-A/I-E; clone M5/114.15.2; BioLegend), CD11c (clone N418; eBioscience), CD103 (clone 2E7; eBioscience), CD11b (clone M1/70; BioLegend), F4/80 (clone CI: A3-1; BioLegend), CD3 (clone 145-2C11; BioLegend), CD4 (clone L3T4; BioLegend), NK1.1 (clone PK136; eBioscience), FoxP3 (clone FJK-16s; eBioscience), Sca-1 (clone D7; eBioscience), c-kit (clone 2B8; eBioscience), CD16/32 (clone 2.4G2; BD), CD135 (clone A2F10; eBioscience), CD115 (clone AFS98; eBioscience), CD34 (clone RAM34; eBioscience), and Siglec F (clone E50-2440; BD) were purchased from the indicated vendors. For intracellular FoxP3 staining, cells were fixed overnight in Fixation/Permeabilization buffer (eBioscience). Before acquisition, cells were resuspended in PBS/BSA 0.5%/EDTA (2 mM) solution with 1 µg/ml of DAPI to exclude dead cells. For FoxP3 staining, cells were stained with live/dead fixable dead cell stain kit (Invitrogen) before fixation to assess viability. Multiparameter analysis was performed on the LSR II (BD) and analyzed with FlowJo software.
qPCR.
Total RNA was isolated from tissue using TRIzol reagent (Invitrogen) and transcribed with RNA to the cDNA EcoDry kit (Takara Bio Inc.). qPCR was performed and analyzed on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories) using SYBR Green PCR Master Mix (Invitrogen) and the following primers: IFN-γ forward, 5′-TCAAGTGGCATAGATGTGGAAGAA-3′, and reverse, 5′-TGGCTCTGCAGGATTTTCATG-3′; IL-10 forward, 5′-GGCTGAGGCGCTGTCATCG-3′, and reverse, 5′-TCATTCATGGCCTTGTAGACACC-3′; TGF-β forward, 5′-GCAAAGACCATCTGTCTCACA-3′, and reverse, 5′-GCAAAGACCATCTGTCTCACA-3′; IL-6 forward, 5′-CAGAGGATACCACTCCCAACA-3′, and reverse, 5′-TCCAGTTTGGTAGCATCCATC-3′; Ccl2 forward, 5′-ATTGGGATCATCTTGCTGGT-3′, and reverse, 5′-CCTGCTGTTCACAGTTGCC-3′; Ccl5 forward, 5′-TGCCCTCACCATCATCCTCACT-3′, and reverse, 5′-GGCGGTTCCTTCGAGTGACA-3′; and langerin forward, 5′-ACGCACCCCAAAGACCTGGTACAG-3′, and reverse, 5′-AGACACCCTGATATTGGCACAGTG-3′. All data were normalized to GAPDH expression using the following primers: forward, 5′-CAATGACCCCTTCATTGACC-3′; and reverse, 5′-GATCTCGCTCCTGGAAGATG-3′.
Assessment of mutation-specific BRAF-V600E mRNA expression in isolated cells.
To examine the presence of the mutation-specific BRAF-V600E mRNA in peripheral DC and distinct BM progenitors, single cell suspensions from BM and liver were stained for the appropriate surface markers, and individual cell subsets were purified using FACSAria (BD) directly into TRIzol reagent (Invitrogen). After RNA isolation and reverse transcription with RNA to cDNA EcoDry kit (Takara Bio Inc.), expression level of BRAF-V600E was assessed by probe-based mutation-specific qPCR using iTaq Universal Probes Supermix (Bio-Rad Laboratories) on a CFX384 Touch Real-Time PCR Detection system. In detail, reaction was performed with a final dilution of 500 nM of the reverse primer 5′-GTAGCTGGCCGGTCATCAGTTCG-3′, 500 nM of mutation-specific forward primer 5′-TAGGTGACTTTGGTCTAGCCACGGA-3′, which includes the specific nucleotide substitution of the T>A at the 3′ end and an additional mismatch nucleotide (A>G at the third position from the 3′ end), further enhancing the specific detection of mutated transcripts (strategy adapted from Schnittger et al., 2012), and 250 nM of the FAM-labeled hybridization probe 6-FAM-TCAGACGTGTATGCGTTTGGGATTG-MGBNFQ at an annealing temperature 68°C for 45 cycles. Data were normalized to total BRAF expression using the TaqMan Gene Expression Assay Mm01165837_m1 (Life Technologies) recognizing the common exon 13–14 of wild-type BRAF and BRAF-V600E.
Online supplemental material
Table S1 shows the BRAF-V600E status and clinical variables of LCH Patients. Table S2 shows the cell-specific BRAF-V600E status of LCH lesions. Table S3 shows the BRAF-V600E status of non-LCH lesions. Table S4 shows LCH peripheral blood analysis: detectable BRAF-V600E in circulating cells and disease activity. Table S5 shows LCH BM aspirate analysis: detectable BRAF-V600E and disease activity. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20130977/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank the flow cytometry and microscopy shared research facility at Mount Sinai School of Medicine, Munu Bilgi (Baylor College of Medicine, Texas Children’s Cancer Center) for data management support, and Billie Smith (Baylor College of Medicine, Pathology Core) for expert technical assistance. We also appreciate the support of shared resources by Dan L. Duncan Cancer Center support grant (P30CA125123). Finally, we would like to acknowledge this collaboration as a result of the Nikolas Symposia on the Histiocytoses, organized and supported by the Kontoyannis family.
This work was performed with support from the HistioCure Foundation (Texas Children’s Cancer Center Histiocytosis Program). Grant support includes NIH R01 (CA154489; C.E. Allen and K. McClain), NIH SPORE in Lymphoma (P50CA126752; C.E. Allen), NIH R01 (CA154947A, AI10008, and AI089987; M. Merad), the German Research Association (Deutsche Forschungsgemeinschaft, BE 4818/1-1; M.-L. Berres), NIH K12 (CA090433; S.J. Simko), the Histiocytosis Association (C.E. Allen, K. McClain, T. Peters, and M. Merad), the Baylor College of Medicine Junior Faculty Seed Grant (C.E. Allen), the Texas Children’s Hospital Pediatric Pilot Award (C.E. Allen),the ASH Scholar Award (C.E. Allen), and the University of Zurich Clinical Research Program (M.G. Manz).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ECD
- Erdheim-Chester disease
- FFPE
- formalin fixed and paraffin embedded
- gDNA
- genomic DNA
- HSPC
- hematopoietic stem and progenitor cell
- IF
- immunofluorescence
- JXG
- juvenile xanthogranuloma
- LCH
- Langerhans cell histiocytosis
- qPCR
- quantitative PCR
- RDD
- Rosai-Dorfman disease
References
- Allen C.E., Li L., Peters T.L., Leung H.C., Yu A., Man T.K., Gurusiddappa S., Phillips M.T., Hicks M.J., Gaikwad A., et al. 2010. Cell-specific gene expression in Langerhans cell histiocytosis lesions reveals a distinct profile compared with epidermal Langerhans cells. J. Immunol. 184:4557–4567 10.4049/jimmunol.0902336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arceci R.J. 1999. The histiocytoses: the fall of the Tower of Babel. Eur. J. Cancer. 35:747–767 10.1016/S0959-8049(99)00039-8 [DOI] [PubMed] [Google Scholar]
- Badalian-Very G., Vergilio J.A., Degar B.A., MacConaill L.E., Brandner B., Calicchio M.L., Kuo F.C., Ligon A.H., Stevenson K.E., Kehoe S.M., et al. 2010. Recurrent BRAF mutations in Langerhans cell histiocytosis. Blood. 116:1919–1923 10.1182/blood-2010-04-279083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard F., Thomas C., Bertrand Y., Munzer M., Landman Parker J., Ouache M., Colin V.M., Perel Y., Chastagner P., Vermylen C., Donadieu J. 2005. Multi-centre pilot study of 2-chlorodeoxyadenosine and cytosine arabinoside combined chemotherapy in refractory Langerhans cell histiocytosis with haematological dysfunction. Eur. J. Cancer. 41:2682–2689 10.1016/j.ejca.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Birbeck M.S.C., Breathnach A.S., Everall J.D. 1961. An electron microscope study of basal melanocytes and high-level clear cells (Langerhans cell) in vitiligo. J. Invest. Dermatol. 37:51–64 10.1038/jid.1961.7 [DOI] [Google Scholar]
- Chang S.Y., Cha H.R., Chang J.H., Ko H.J., Yang H., Malissen B., Iwata M., Kweon M.N. 2010. Lack of retinoic acid leads to increased langerin-expressing dendritic cells in gut-associated lymphoid tissues. Gastroenterology. 138:1468–1478 10.1053/j.gastro.2009.11.006 [DOI] [PubMed] [Google Scholar]
- Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., et al. BRIM-3 Study Group. 2011. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364:2507–2516 10.1056/NEJMoa1103782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikwava K., Jaffe R. 2004. Langerin (CD207) staining in normal pediatric tissues, reactive lymph nodes, and childhood histiocytic disorders. Pediatr. Dev. Pathol. 7:607–614 10.1007/s10024-004-3027-z [DOI] [PubMed] [Google Scholar]
- da Costa C.E., Annels N.E., Faaij C.M., Forsyth R.G., Hogendoorn P.C., Egeler R.M. 2005. Presence of osteoclast-like multinucleated giant cells in the bone and nonostotic lesions of Langerhans cell histiocytosis. J. Exp. Med. 201:687–693 10.1084/jem.20041785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D., Filenova E., Collado M., Serrano M., Jones K., McMahon M. 2007. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 21:379–384 10.1101/gad.1516407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf J.H., Tamminga R.Y., Dam-Meiring A., Kamps W.A., Timens W. 1996. The presence of cytokines in Langerhans’ cell histiocytosis. J. Pathol. 180:400–406 [DOI] [PubMed] [Google Scholar]
- Gadner H., Grois N., Arico M., Broadbent V., Ceci A., Jakobson A., Komp D., Michaelis J., Nicholson S., Pötschger U., et al. ; Histiocyte Society. 2001. A randomized trial of treatment for multisystem Langerhans’ cell histiocytosis. J. Pediatr. 138:728–734 10.1067/mpd.2001.111331 [DOI] [PubMed] [Google Scholar]
- Gadner H., Grois N., Pötschger U., Minkov M., Aricò M., Braier J., Broadbent V., Donadieu J., Henter J.I., McCarter R., Ladisch S.; Histiocyte Society. 2008. Improved outcome in multisystem Langerhans cell histiocytosis is associated with therapy intensification. Blood. 111:2556–2562 10.1182/blood-2007-08-106211 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Collin M.P., Bogunovic M., Abel M., Leboeuf M., Helft J., Ochando J., Kissenpfennig A., Malissen B., Grisotto M., et al. 2007. Blood-derived dermal langerin+ dendritic cells survey the skin in the steady state. J. Exp. Med. 204:3133–3146 10.1084/jem.20071733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grois N., Tsunematsu Y., Barkovich A.J., Favara B.E. 1994. Central nervous system disease in Langerhans cell histiocytosis. Br. J. Cancer Suppl. 23:S24–S28. [PMC free article] [PubMed] [Google Scholar]
- Haroche J., Charlotte F., Arnaud L., von Deimling A., Hélias-Rodzewicz Z., Hervier B., Cohen-Aubart F., Launay D., Lesot A., Mokhtari K., et al. 2012. High prevalence of BRAF V600E mutations in Erdheim-Chester disease but not in other non-Langerhans cell histiocytoses. Blood. 120:2700–2703 10.1182/blood-2012-05-430140 [DOI] [PubMed] [Google Scholar]
- Haroche J., Cohen-Aubart F., Emile J.F., Arnaud L., Maksud P., Charlotte F., Cluzel P., Drier A., Hervier B., Benameur N., et al. 2013. Dramatic efficacy of vemurafenib in both multisystemic and refractory Erdheim-Chester disease and Langerhans cell histiocytosis harboring the BRAF V600E mutation. Blood. 121:1495–1500 10.1182/blood-2012-07-446286 [DOI] [PubMed] [Google Scholar]
- Hicks J., Flaitz C.M. 2005. Langerhans cell histiocytosis: current insights in a molecular age with emphasis on clinical oral and maxillofacial pathology practice. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 100:S42–S66 10.1016/j.tripleo.2005.06.016 [DOI] [PubMed] [Google Scholar]
- Kansal R., Quintanilla-Martinez L., Datta V., Lopategui J., Garshfield G., Nathwani B.N. 2013. Identification of the V600D mutation in Exon 15 of the BRAF oncogene in congenital, benign langerhans cell histiocytosis. Genes Chromosomes Cancer. 52:99–106 10.1002/gcc.22010 [DOI] [PubMed] [Google Scholar]
- Kissenpfennig A., Aït-Yahia S., Clair-Moninot V., Stössel H., Badell E., Bordat Y., Pooley J.L., Lang T., Prina E., Coste I., et al. 2005. Disruption of the langerin/CD207 gene abolishes Birbeck granules without a marked loss of Langerhans cell function. Mol. Cell. Biol. 25:88–99 10.1128/MCB.25.1.88-99.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Wagers A.J., Manz M.G., Prohaska S.S., Scherer D.C., Beilhack G.F., Shizuru J.A., Weissman I.L. 2003. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21:759–806 10.1146/annurev.immunol.21.120601.141007 [DOI] [PubMed] [Google Scholar]
- Lichtenstein L. 1953. Histiocytosis X; integration of eosinophilic granuloma of bone, Letterer-Siwe disease, and Schüller-Christian disease as related manifestations of a single nosologic entity. AMA Arch. Pathol. 56:84–102. [PubMed] [Google Scholar]
- Merad M., Ginhoux F., Collin M. 2008. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat. Rev. Immunol. 8:935–947 10.1038/nri2455 [DOI] [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., Mortha A. 2013. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31:563–604 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezelof C., Basset F., Rousseau M.F. 1973. Histiocytosis X histogenetic arguments for a Langerhans cell origin. Biomedicine. 18:365–371. [PubMed] [Google Scholar]
- Pineles S.L., Liu G.T., Acebes X., Arruga J., Nasta S., Glaser R., Pramick M., Fogt F., Roux P.L., Gausas R.E. 2011. Presence of Erdheim-Chester disease and Langerhans cell histiocytosis in the same patient: a report of 2 cases. J. Neuroophthalmol. 31:217–223 10.1097/WNO.0b013e31820a204e [DOI] [PubMed] [Google Scholar]
- Rodriguez-Galindo C., Jeng M., Khuu P., McCarville M.B., Jeha S. 2008. Clofarabine in refractory Langerhans cell histiocytosis. Pediatr. Blood Cancer. 51:703–706 10.1002/pbc.21668 [DOI] [PubMed] [Google Scholar]
- Sahm F., Capper D., Preusser M., Meyer J., Stenzinger A., Lasitschka F., Berghoff A.S., Habel A., Schneider M., Kulozik A., et al. 2012. BRAFV600E mutant protein is expressed in cells of variable maturation in Langerhans cell histiocytosis. Blood. 120:e28–e34 10.1182/blood-2012-06-429597 [DOI] [PubMed] [Google Scholar]
- Satoh T., Smith A., Sarde A., Lu H.C., Mian S., Trouillet C., Mufti G., Emile J.F., Fraternali F., Donadieu J., Geissmann F. 2012. B-RAF mutant alleles associated with Langerhans cell histiocytosis, a granulomatous pediatric disease. PLoS ONE. 7:e33891 10.1371/journal.pone.0033891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger S., Bacher U., Haferlach T., Wendland N., Ulke M., Dicker F., Grossmann V., Haferlach C., Kern W. 2012. Development and validation of a real-time quantification assay to detect and monitor BRAFV600E mutations in hairy cell leukemia. Blood. 119:3151–3154 10.1182/blood-2011-10-383323 [DOI] [PubMed] [Google Scholar]
- Senechal B., Elain G., Jeziorski E., Grondin V., Patey-Mariaud de Serre N., Jaubert F., Beldjord K., Lellouch A., Glorion C., Zerah M., et al. 2007. Expansion of regulatory T cells in patients with Langerhans cell histiocytosis. PLoS Med. 4:e253 10.1371/journal.pmed.0040253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover D.G., Alapati S., Regueira O., Turner C., Whitlock J.A. 2008. Treatment of juvenile xanthogranuloma. Pediatr. Blood Cancer. 51:130–133 10.1002/pbc.21523 [DOI] [PubMed] [Google Scholar]
- Valladeau J., Ravel O., Dezutter-Dambuyant C., Moore K., Kleijmeer M., Liu Y., Duvert-Frances V., Vincent C., Schmitt D., Davoust J., et al. 2000. Langerin, a novel C-type lectin specific to Langerhans cells, is an endocytic receptor that induces the formation of Birbeck granules. Immunity. 12:71–81 10.1016/S1074-7613(00)80160-0 [DOI] [PubMed] [Google Scholar]
- Verdijk P., Dijkman R., Plasmeijer E.I., Mulder A.A., Zoutman W.H., Mieke Mommaas A., Tensen C.P. 2005. A lack of Birbeck granules in Langerhans cells is associated with a naturally occurring point mutation in the human Langerin gene. J. Invest. Dermatol. 124:714–717 10.1111/j.0022-202X.2005.23645.x [DOI] [PubMed] [Google Scholar]
- Verma S., Greaves W.O., Ravandi F., Reddy N., Bueso-Ramos C.E., O’Brien S., Thomas D.A., Kantarjian H., Medeiros L.J., Luthra R., Patel K.P. 2012. Rapid detection and quantitation of BRAF mutations in hairy cell leukemia using a sensitive pyrosequencing assay. Am. J. Clin. Pathol. 138:153–156 10.1309/AJCPL0OPXI9LZITV [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willman C.L., Busque L., Griffith B.B., Favara B.E., McClain K.L., Duncan M.H., Gilliland D.G. 1994. Langerhans’-cell histiocytosis (histiocytosis X)—a clonal proliferative disease. N. Engl. J. Med. 331:154–160 10.1056/NEJM199407213310303 [DOI] [PubMed] [Google Scholar]
- Yu R.C., Chu C., Buluwela L., Chu A.C. 1994. Clonal proliferation of Langerhans cells in Langerhans cell histiocytosis. Lancet. 343:767–768 10.1016/S0140-6736(94)91842-2 [DOI] [PubMed] [Google Scholar]
- Zahner S.P., Kel J.M., Martina C.A., Brouwers-Haspels I., van Roon M.A., Clausen B.E. 2011. Conditional deletion of TGF-βR1 using Langerin-Cre mice results in Langerhans cell deficiency and reduced contact hypersensitivity. J. Immunol. 187:5069–5076 10.4049/jimmunol.1101880 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.