Two interleukin-7 receptor mutants identified in human early T cell precursor leukemia are sufficient to induce disease in mice when expressed in primitive, Arf-null thymocytes.
Abstract
Early T cell precursor acute lymphoblastic leukemia (ETP-ALL) exhibits lymphoid, myeloid, and stem cell features and is associated with a poor prognosis. Whole genome sequencing of human ETP-ALL cases has identified recurrent mutations in signaling, histone modification, and hematopoietic development genes but it remains to be determined which of these abnormalities are sufficient to initiate leukemia. We show that activating mutations in the interleukin-7 receptor identified in human pediatric ETP-ALL cases are sufficient to generate ETP-ALL in mice transplanted with primitive transduced thymocytes from p19Arf−/− mice. The cellular mechanism by which these mutant receptors induce ETP-ALL is the block of thymocyte differentiation at the double negative 2 stage at which myeloid lineage and T lymphocyte developmental potential coexist. Analyses of samples from pediatric ETP-ALL cases and our murine ETP-ALL model show uniformly high levels of LMO2 expression, very low to undetectable levels of BCL11B expression, and a relative lack of activating NOTCH1 mutations. We report that pharmacological blockade of Jak–Stat signaling with ruxolitinib has significant antileukemic activity in this ETP-ALL model. This new murine model recapitulates several important cellular and molecular features of ETP-ALL and should be useful to further define novel therapeutic approaches for this aggressive leukemia.
Early T cell precursor acute lymphoblastic leukemia (ETP-ALL) is a recently described subtype of acute lymphoblastic leukemia that occurs in both adults and children and has a relatively poor survival rate with current therapies (Coustan-Smith et al., 2009). The leukemic blasts in ETP-ALL have a unique phenotype characterized by cytoplasmic expression of CD3, a lack of expression of mature T cell markers such as CD4 and CD8, and aberrant expression of myeloid and stem cell markers. There has been no animal model for ETP-ALL, so the biology of the disease and identification of new therapeutics remains relatively unexplored.
The prevailing hypothesis is that ETP-ALL is caused by transformation of a primitive hematopoietic cell that retains the capacity to differentiate into both T cells and myeloid cells. The thymus is seeded by primitive thymic immigrants derived from the BM that then proceed through a series of maturational steps, ultimately generating CD4 and CD8 single-positive T cells (Rothenberg et al., 2010). The initial stages of thymocyte development are characterized by differentiation of cells that lack expression of CD4 or CD8. As these double negative (DN) cells differentiate, at least four distinct differentiation stages can be distinguished by differential expression of CD44 and CD25 (DN1, DN2, DN3, and DN4). The potential for myeloid, dendritic, and natural killer cell differentiation is retained at the DN1 stage and at the early DN2 stage (Bell and Bhandoola, 2008). The ability to adopt non–T cell fates is lost by the DN3 stage and most likely by the latter half of DN2 progression (Yui et al., 2010). Therefore, it seems plausible that the tumor-initiating cell in ETP-ALL could be derived from DN1 and/or DN2 thymocytes.
Whole-genome sequencing studies in ETP-ALL have discovered several recurrent mutations involving genes that participate in cytokine signaling, epigenetic control of gene expression, and hematopoietic transcriptional regulation. In particular, activating mutations in the IL7R pathway were seen in five of 54 cases of pediatric ETP-ALL (Zhang et al., 2012). Most of these IL7R mutants can confer cytokine independent proliferation in various cell lines (Shochat et al., 2011; Zenatti et al., 2011; Zhang et al., 2012); however, there is no proof that these signaling mutants are sufficient to initiate ETP leukemia in primary cells. Furthermore, it is unclear how these mutations can lead the specific phenotypic features of ETP-ALL and what other collaborative mutations may be required. Therefore, we tested Il7r mutant alleles homologous to those that have been identified in human ETP cases (Zhang et al., 2012) in a mouse thymocyte transplant assay to determine if they were sufficient to generate ETP-ALL.
Our experimental system is based on transducing CD4−CD8− thymocytes from Arf−/− mice with retroviral vectors expressing either of two constitutively active Il7r mutant receptors. These transduced thymocytes were then transplanted into sublethally irradiated recipients by tail vein injection. The use of Arf−/− thymocytes allows efficient thymic engraftment due to enhanced self-renewal associated with p19Arf loss (Treanor et al., 2011). Transplanted mice were followed over time for the development of leukemia and the resulting malignancies were characterized at a cellular and molecular level. Relevant molecular abnormalities seen in the murine model system were then evaluated in human ETP-ALL samples from pediatric cases identified at St. Jude Children’s Research Hospital to determine if they were present in both human and murine ETP-ALL.
RESULTS
Constitutively active, Il7r mutants block thymocyte differentiation at the DN2 stage and induce ETP-like ALL in transplanted mice
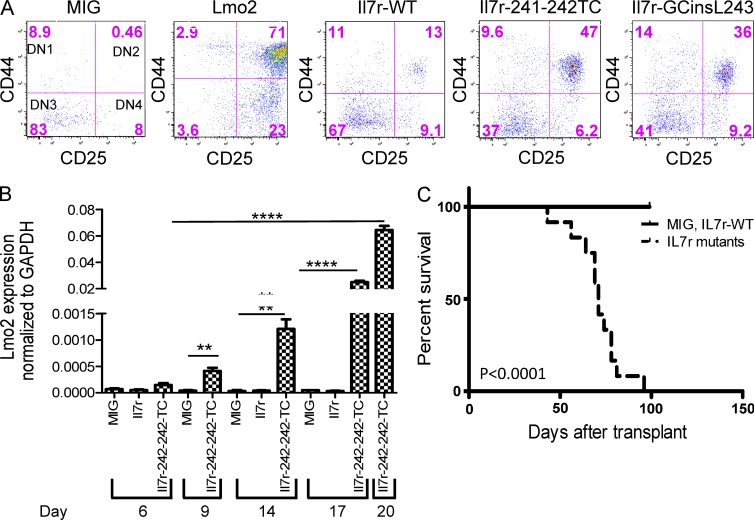
A significant number of human pediatric ETP-ALL cases contain mutations in cell signaling pathways with 20% of cases harboring activating mutations in the IL7R or in the downstream Janus kinases JAK1 and JAK3 (Zhang et al., 2012). These IL7R mutations are most commonly in-frame insertions that introduce a cysteine in the transmembrane domain and result in ligand-independent receptor dimerization and cytokine-independent proliferation in cell lines and primary murine BM cells (Zhang et al., 2012). To determine whether these IL7R mutations are oncogenic drivers in ETP-ALL, DN thymocytes from Arf−/− mice were transduced with MSCV vectors that expressed murine homologues of either of two IL7R mutations identified in human ETP-ALL, IL7R-241-242TC and IL7R-GCinsL243 (Zhang et al., 2012). When these transduced thymocytes were cultured on OP9-DL1 stromal cells, the Il7r mutants caused a partial block at the DN2 stage (Fig. 1 A), similar to the block seen with enforced expression of the Lmo2 transcription factor (Larson et al., 1995; Neale et al., 1995; Treanor et al., 2011). These results show that these mutant Il7r receptors induce a block in differentiation early in thymocyte development and at a stage before irreversible commitment to the T cell lineage. Given that a DN2 block was also seen with enforced Lmo2 expression, we measured Lmo2 mRNA expression in cultured thymocytes transduced with the Il7r-241-241TC vector. Increasing accumulation of endogenous Lmo2 mRNA was observed as the Il7r-241-241TC–transduced cells were cultured for 20 d (Fig. 1 B). These results suggest that signaling from the mutant Il7r receptor specifically induces Lmo2 expression and thereby leads to the block at the DN2 stage of maturation.
Figure 1.
Activating mutations in Il7r induce a DN2 stage block in thymocyte development, result in Lmo2 overexpression, and are highly leukemogenic in vivo. (A) CD4−/CD8− thymocytes from Arf−/− mice were transduced with the MSCV-IRES-GFP, MSCV-Lmo2-IRES-mCherry, MSCV-Il7r-WT-GFP, MSCV-Il7r 241-242TC-GFP, or MSCV-Il7r-GCins243-GFP vector as indicated and cultured on OP9-DL1 stromal cells. The FACS plots show expression patterns for CD44 and CD25 within the transduced CD4−/CD8− population for each of the vectors tested at day 20 in culture. The top right quadrant represents the DN2 stage of development, and the percentage of cells at each stage is indicated. (B) Lmo2 mRNA expression in transduced p19Arf−/− thymocytes is shown relative to GAPDH mRNA levels. DN thymocytes were transduced with the indicated vectors, grown for 6, 9, 14, 17, and 20 d on OP9-DL1 cells and FACS sorted for the GFP+ cells. The histograms represent the Lmo2/GAPDH ratio of each GFP+ FACS sorted sample by qPCR assay. There is a significantly higher amount of Lmo2 in the Il7r-241-241TC-GFP vector transduced thymocytes, **, P < 0.005; ****, P < 0.0001, compared with MSCV-GFP control. (C) Kaplan-Meier curves are shown, indicating the survival of mice transplanted with thymocytes transduced with control vectors (either MSCV-GFP or Il7R-WT, n = 4, solid line) or the Il7r mutant vectors (n = 11; Il7r-241-242TC, n = 6; and Il7r-GCinsL243, n = 5, dashed line). The median survival for mice transplanted with thymocytes transduced with the Il7R mutant vectors was 71 d. In contrast, no deaths were seen in the control group for the first 100 d. This difference in median survival was highly significant (P < 0.0001). This shows the same experiment repeated at multiple time points.
We next tested whether these transduced DN2 thymocytes would induce leukemia in transplanted recipient mice. Thymocytes transduced with the WT or mutant Il7r vectors or empty GFP-expressing vector (GFP) were isolated and transplanted into sublethally irradiated, Rag2−/−, γc−/− recipient mice. Mice transplanted with thymocytes transduced with the mutant Il7r vectors died of leukemia with a median latency of 71 d (Fig. 1 C). By 100 d after transplant, all 11 mice that received cells transduced with either of the two Il7r mutants had died from acute leukemia (Fig. 1 C). In contrast, none of the mice transplanted with Arf−/− thymocytes transduced with the MIG or the Il7r WT vector developed leukemia over this period of time.
Each Il7r mutant receptor induces a distinct ETP-ALL phenotype
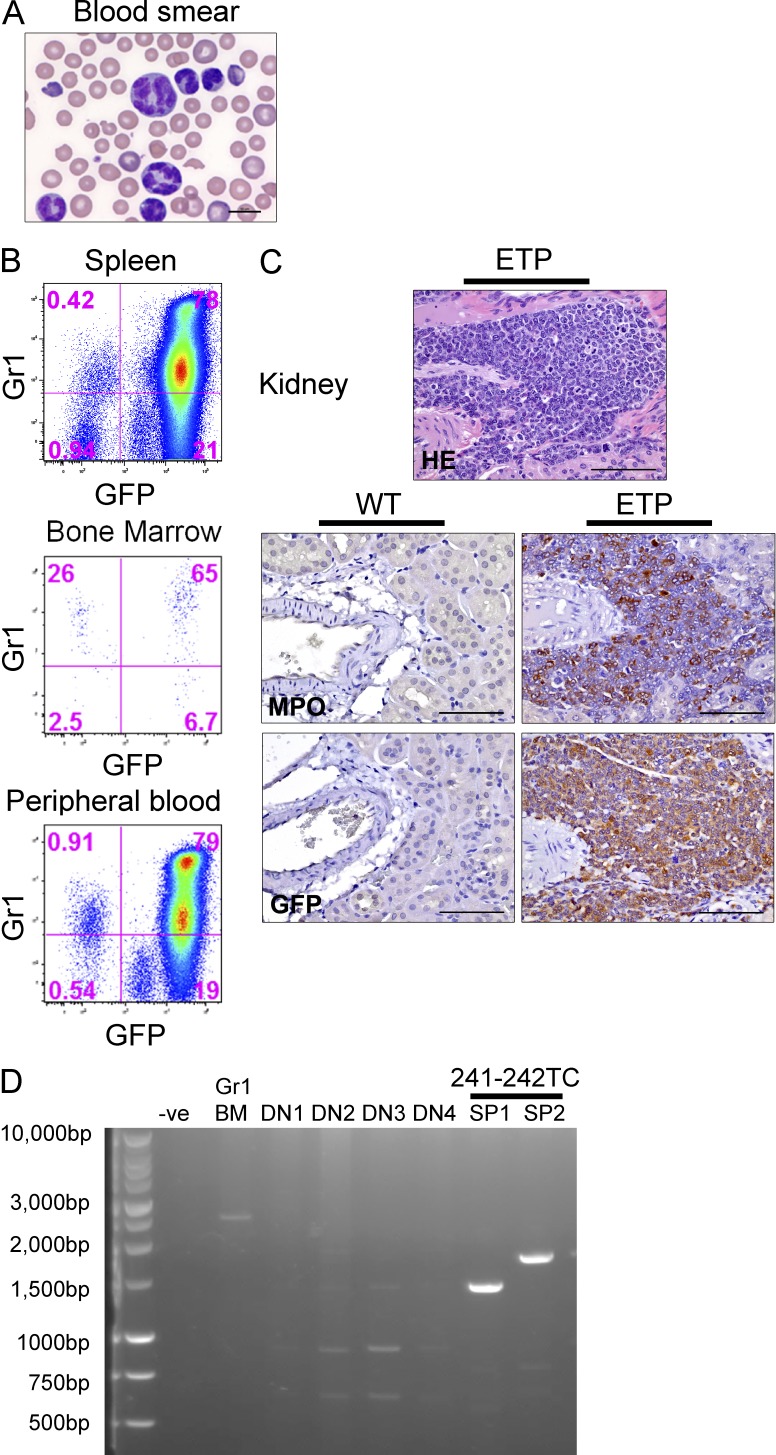
The Il7r-241-242TC Il7r mutant resulted in an aggressive acute leukemia in six mice, with marked elevation of the peripheral leukocyte count, significant splenomegaly, and myeloid morphology of leukemic cells (Fig. 2 A). Leukemic cells expressed the vector-encoded GFP gene, Gr1, Mac1, and myeloperoxidase myeloid markers (Fig. 2, B and C). Leukemic infiltration of the kidney and other solid organs was noted and the tumor cells were uniformly positive for expression of the vector-encoded GFP gene (Fig. 2, B and C). In most of these cases, the myeloid tumor cells also expressed intracellular CD3, a hallmark for the ETP-ALL phenotype (Table 1). Monoclonal rearrangement of the TCRβ loci was demonstrated via PCR (King et al., 2002) in tumor cell DNA derived from the spleen of two independent cases, proving that these tumors originated from an early thymocyte and not a contaminating myeloid cell (Fig. 2 D).
Figure 2.
The 241-242TC Il7r vector induces ETP-ALL with a myeloid phenotype when introduced into immature thymocytes. (A) A peripheral blood smear from leukemia induced by the Il7r 241-242TC mutant in a recipient mouse 64 d after transplant. Bar, 10 µm. (B) Co-expression of the vector (GFP) and the myeloid lineage marker Gr1 in leukemic cells from the spleen, BM, and peripheral blood as analyzed by flow cytometry. (C) Leukemic infiltrates were present in the kidney and other solid organs and tumor cells expressed myeloperoxidase (MPO) and GFP by immunohistochemistry. Bars, 75 µm. (D) PCR analysis for TCR rearrangements in ETP-ALL tumor cells. Two Il7r 241-242TC mutant ETP-ALL leukemic samples from the spleen (SP1, SP2) have monoclonal TCR rearrangements. Other controls show the unrearranged germline TCR in BM granulocytes and polyclonal TCR rearrangements (multiple bands) in normal DN thymocytes.
Table 1.
Features of the murine ETP-ALLs observed in transplanted recipients
| Mouse ID | Vector | Latency (days) | Peripheral WBC/µl (×103) | Immunophenotype by flow cytometrya | bImmuno-histochemistry | Pathology |
| 647 | Il7r-IL241-242TC-GFP | 69 | ND | ND | GFP+,IC-Cd3+,Mpo+ | Splenomegaly (spleen = 0.73 g). Leukemic infiltrates in brain, kidney, and liver |
| 649 | Il7r-IL241-242TC-GFP | 69 | ND | ND | ND | Splenomegaly (spleen = 0.55 g). Animal was found dead and tissues were partially autolysed. Leukemic infiltrates in BM, brain, liver, and spleen. |
| 650 | Il7r-IL241-242TC-GFP | 64 | 91 | Gr1+Mac1+GFP+ | GFP+, Mpo+ | Splenomegaly (spleen = 0.8 g). Leukemic infiltrates in brain, kidney, liver, and lung. |
| 651 | Il7r-IL241-242TC-GFP | 81 | 29.4 | Gr1+Mac1+GFP+ | IC-Cd3+, Mpo+, Cd117 low | Splenomegaly (spleen = 0.88 g). Leukemic infiltrates in liver and lung. Secondary transplants developed tumors with the same immunophenotype at 24 d. |
| 652 | Il7r-IL241-242TC-GFP | 78 | 64.78 | Gr1+Mac1+GFP+ | IC-Cd3+, Cd117 low | Splenomegaly (spleen = 0.73 g). Leukemic infiltrates in brain, eye, kidney, liver, and lung. |
| 653 | Il7r-IL241-242TC-GFP | 74 | 33.72 | Gr1+Mac1+GFP+ | GFP+, IC-Cd3+, Mpo+ | Splenomegaly (spleen = 1.25 g). Leukemic infiltrates in brain, kidney, liver, and lung. |
| 655 | Il7r-GcinsL243-GFP | 71 | 30.76 | Sca1+Cd71+GFP+ | GFP+, IC-Cd3+, Gata1+, Runx1+, Ly6b+, TdT+ Cd34+ | Splenomegaly (spleen = 0.96 g). Leukemic infiltrates in liver, lung, focally in brain/meninges. |
| 656c | Il7r-GcinsL243-GFP | 78 | 32 | Cd117+Cd71+GFP+ | Pop1: IC-Cd3+, Runx1+, Pop2: IC-Cd3+, Runx1+ and Gata-1+ | Splenomegaly (spleen = 0.77 g). Leukemic infiltrates in liver and lung. |
| 657 | Il7r-GcinsL243-GFP | 43 | 49.46 | Sca1+Cd71+GFP+ | IC-Cd3+, Gata1+, Runx1+, Ly6b+, TdT+, Cd117+, Fe/80+, Mac2+ | Splenomegaly (spleen = 0.88 g). Leukemic infiltrates in liver, lung, focally in brain/meninges. |
| 658c | Il7r-GcinsL243-GFP | 96 | 99.9 | Cd71+GFP+ | Pop1: IC-Cd3+, Runx1+, Pop2: IC-Cd3+, Runx1+ and Gata-1+ | Splenomegaly (spleen = 0.9 g). Leukemic infiltrates in liver and lung. Secondary transplants developed identical tumors at 24 d. |
| 659 | Il7r-GcinsL243-GFP | 71 | 48.89 | Cd4+Cd8+GFP+ | IC-Cd3+Tdt+ | The only T-ALL case. Splenomegaly (spleen = 0.54 g). Enlarged thymus (thymus = 0.132 g). Leukemic infiltrates in liver, lung, brain, and kidney. |
| 242d | Lmo2-mCherry | 89 | 44.3 | mCherry+Gr1+Mac1+ | IC-Cd3+Mpo+ | All animals had splenomegaly (range: spleen = 0.258–1.163 g). Enlarged thymus (range: thymus = 0.064–0.116 g). Leukemic infiltrates in lungs, liver, lymph nodes, and meninges. |
| 243d, 244d, 245d | Lmo2-mCherry | 71 | 195-528 | mCherry+Gr1+Mac1+ | ||
| 611 | Lmo2-mCherry | 154 | 36.18 | Cd117+, Cd71+, B220+, mCherry+ | Cd117 weakly + | Splenomegaly (spleen = 0.385 g). Leukemic infiltrates in lung, spleen, liver, BM, and meninges. |
| 612 | Lmo2-mCherry | 111 | 20.42 | Gr1+, Mac1+, Cd117+, Cd71+, Cd5low, mCherry+ | Cd117+, Cd43+ | Splenomegaly (spleen = 0.825 g). Leukemic infiltrates in liver, spleen, and kidney. |
IC-Cd3− Intracytoplasmic Cd3, Mpo-myeloperoxidase, TdT-terminal deoxynucleotidyl transferase.
Where present, the spleen, thymus, BM, and peripheral blood were immunophenotyped. All tissues were tested for Cd4, Cd8, B220, Cd3, and Cd25. All tumor samples were positive for Cd44.
In the majority of cases where IHC was performed, the samples were tested for Mpo, cKit/Cd117, Cd43, RUNX1, Cd3, Cd5, TdT, Gata-1, B220, Pax5, and Cd34. The markers that had positive reactivity are indicated in the text
These animals contained two distinct populations in their spleen, which could be distinguished by immunohistochemistry as indicated in the text.
Secondary transplant Rag2−/−, γC−/− recipients that received vector-positive thymocytes from a WT C57BL/6J animal that had been transplanted with 2 × 105 Lmo2, Arf−/−, and DN2 thymocytes 21 wk before sacrifice.
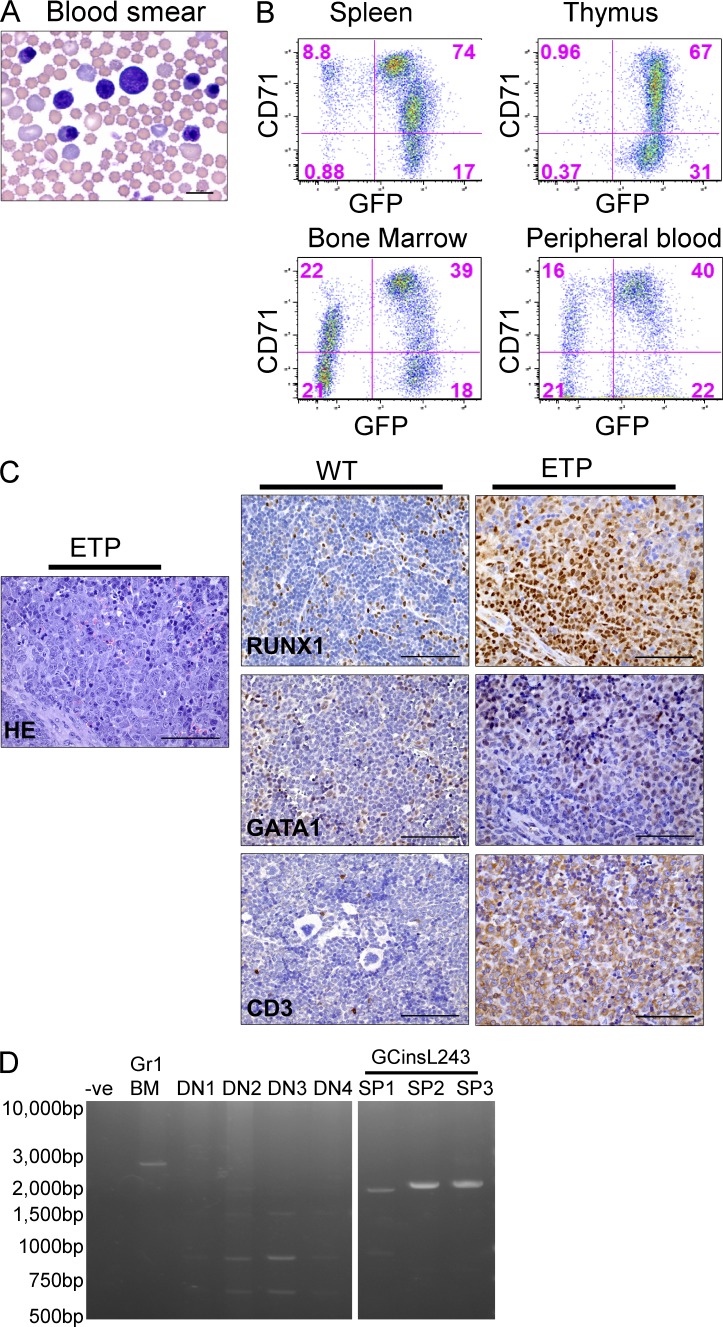
A stem cell leukemia with erythroid features was observed in five of six mice transplanted with Il7r-GCinsL243 expressing thymocytes (Fig. 3 and Table 1). The peripheral blood of leukemic mice contained numerous immature erythroid precursors and blast cells (Fig. 3 A). Tumor cells were present at high levels in the spleen, thymus, BM, and peripheral blood and expressed the vector-encoded GFP gene (Fig. 3 B). Leukemic cells also expressed high levels of the erythroid marker CD71 but did not express the Ter119 erythrocyte marker, as has been described for other murine erythroleukemias (Torchia et al., 2007). Leukemia cells also expressed Gata1 and Runx1 by immunohistochemistry, consistent with an early erythroid/stem cell phenotype, and also expressed intracellular CD3 as would be expected for ETP-ALL (Fig. 3 C). Monoclonal rearrangement of the TCRβ loci was verified in three cases, indicating the thymocyte origin of these tumors (Fig. 3 D). This phenotype suggests a previously unrecognized erythroid potential of early DN thymocytes, perhaps manifest only in the setting of Il7r mutant-induced leukemic transformation.
Figure 3.
The Gcins243 Il7r vector induces ETP-ALL-like disease with a stem cell/erythroid phenotype in transplanted mice. (A) A representative example of the leukemia cell morphology in the peripheral blood from the Il7r mutant Gcins243 96 d after transplant in a primary transplant recipient. Bar, 10 µm. (B) Flow cytometry analysis of cells in the spleen, thymus, BM, and peripheral blood showed high levels of expression of the vector (GFP) and the CD71 erythroid marker. No expression of Ter119 was noted. (C) Leukemic cells infiltrating the spleen were positive for Runx1, GATA1, and intracellular CD3 by immunohistochemistry. This marker pattern indicated mixed expression of stem cell, erythroid, and T cell markers. Sections from WT tissues are shown as staining controls. The Gata1+ cells in the WT sample represent normal erythroid cells in the spleen. Bars, 75 µm. (D) PCR analysis for T cell receptor rearrangements in ETP-ALL tumor cells. DNA extracted from the spleen of tumor samples of three Il7r Gcins243 mutant ETP-ALL leukemic samples (SP1, SP2, and SP3) show monoclonal TCR rearrangements. Controls are as described in Fig. 2 D.
Tumor cells from both a myeloid and a stem/erythroid ETP-like ALL case were transplanted into secondary recipients and resulted in aggressive leukemias at ∼24 d post-transplant. The tumor-initiating cell frequency in leukemic cells was ∼1/1,500 for the Il7r-241-242TC–induced cases (Table 2). This relatively high tumor-initiating cell frequency is consistent with what has been previously noted in Notch1-induced tumors originating from Arf−/− thymocytes (Volanakis et al., 2009). Altogether, these results demonstrate that overexpression of activating Il7r mutants in early Arf−/− thymocytes are potent oncogenic drivers and function in part by blocking thymocyte development in a manner similar to that seen with Lmo2 overexpression. Interestingly, each mutant displayed a unique and specific phenotype, perhaps reflecting quantitative and/or qualitative differences in constitutive Il7r signaling with each mutant.
Table 2.
Tumor initiating cell frequency
| Cell dose | Number of animals presented with leukemia | Number of animals per group |
| 200,000 | 3 | 3 |
| 100,000 | 5 | 5 |
| 10,000 | 5 | 5 |
| 1,000 | 2 | 5 |
| 100 | 1 | 5 |
LMO2 overexpression is seen in Il7r mutant ETP tumors and is sufficient to generate ETP-ALL
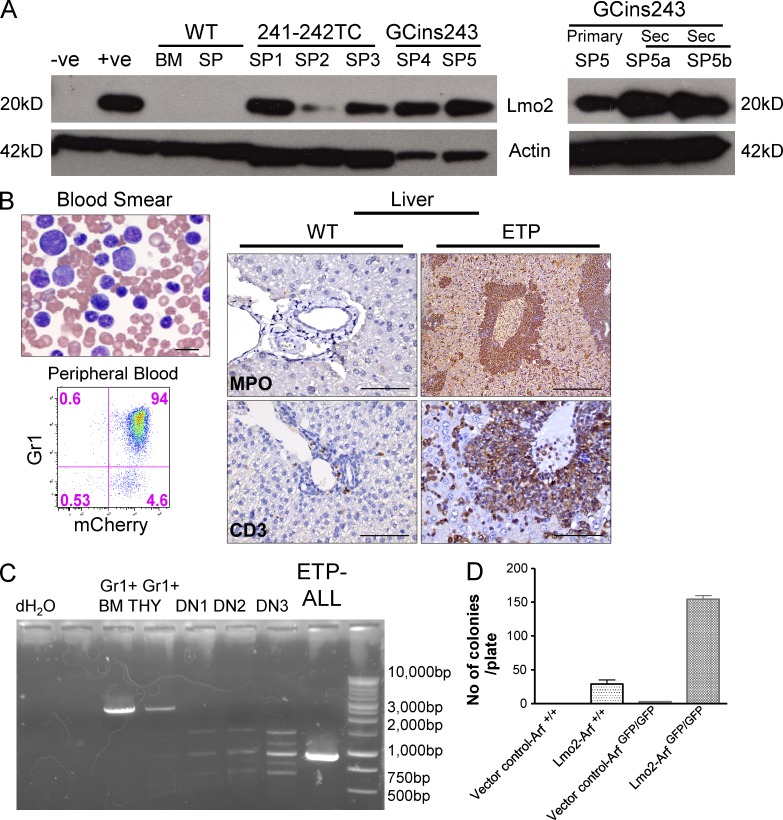
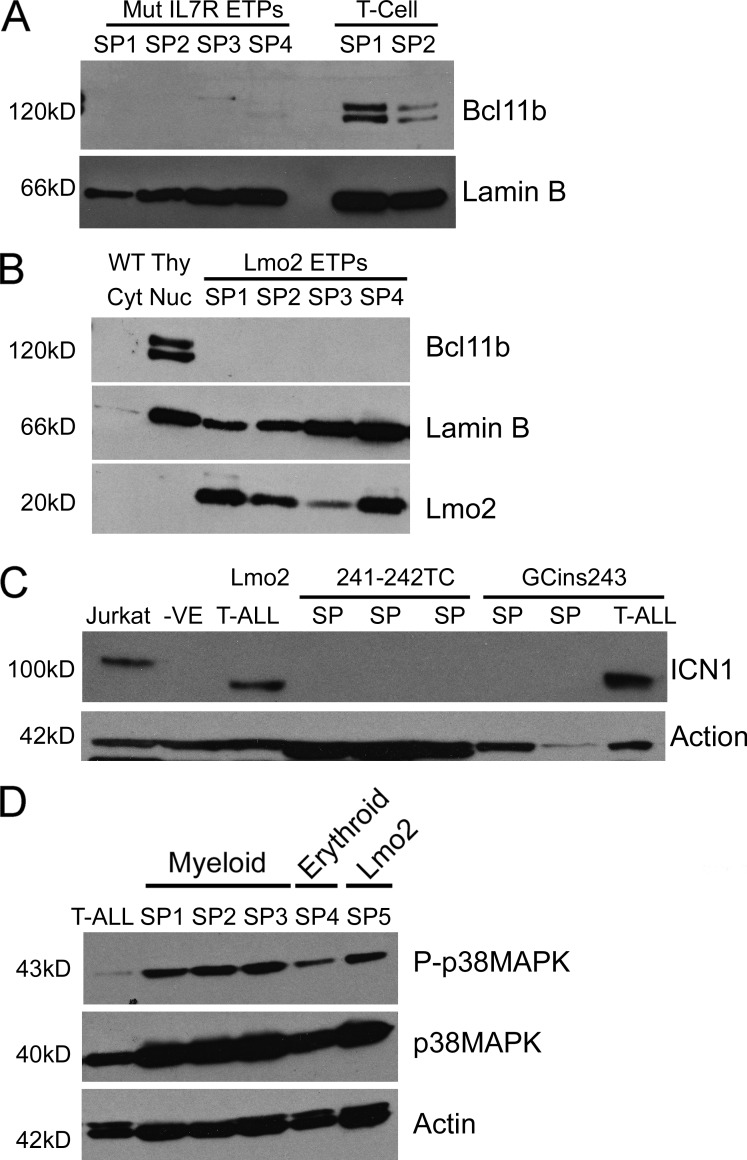
Because the Il7r mutant receptors induce a DN2 block in transduced thymocytes that recapitulates that observed with enforced LMO2 expression, we measured endogenous Lmo2 protein expression in murine ETP-ALL tumor cells by Western blot analysis. Relatively high levels of Lmo2 protein expression were seen in splenic tumor samples arising from both the 241-242TC mutant receptor and from the GCins243 receptor (Fig. 4 A) and approximated that seen in the retroviral producer clone for the MSCV-Lmo2-GFP vector (+ve).
Figure 4.
Lmo2 protein expression in Il7r mutant tumors and ETP-ALL directly induced by overexpression of Lmo2. (A) Immunoblot analysis of Lmo2 expression in extracts of spleen cells from Il7r-241-242TC (SP1-3) and Il7r-GCins243-induced (SP4-5) murine ETP-ALLs. The samples on the left are from five independent cases in spleens from primary recipients. As controls, Lmo2 producer cells (+ve) and untransduced packaging cells (-ve) were used as well as normal BM and spleen (SP) cells. (right) Lmo2 expression in spleen cells from both primary and secondary tumors arising from the Il7R-GCins243 vector. (B) ETP-ALL in mice transplanted with thymocytes transduced with a MSCV-Lmo2-IRES-mCherry vector and cultured in vitro on OP9-DL1 cells. At 21 wk after transplant, thymocytes from this primary recipient were sorted for mCherry+ cells and were transplanted into secondary, nonirradiated Rag2−/−, γC−/− mice. At 71 d, 3 out of 4 secondary recipients (Table 1) developed the leukemia. Bar, 10 µm. Flow cytometry showed that these leukemia cells expressed both mCherry and Gr1. The liver was infiltrated with tumor cells that expressed both myeloperoxidase (Bar, 300 µm) and intracellular CD3 (Bar, 75 µm; bottom). WT liver tissue from a normal mouse is shown as a staining control. Bars, 75 µm. (C) PCR analysis for T cell receptor rearrangements in Lmo2-induced ETP-ALL tumor cells. DNA was extracted from the spleen of tumor samples shown in B and analyzed for rearrangements of the β locus of the TCR. The leukemic sample (ETP-ALL) showed a monoclonal TCR rearrangement. The germline TCR configuration was seen in Gr1+ BM and thymic cells (THY) from normal mice. (D)DN (CD4−/CD8−) thymocytes from Arf+/+ (WT) and Arfgfp/gfp animals were transduced with either an MSCV-IRES-mCherry vector control (I) or the MSCV-Lmo2-IRES-mCherry (Lmo2), cultured on OP9-DL1 stromal cells for 21 d, sorted for the DN2 thymocytes, and plated in semisolid cultures specific for myeloid CFU-Cs. The histogram shows the number of myeloid CFU-C obtained for each of the four conditions (n = 3). Lmo2-Arfgfp/gfp have a greater number of colonies when compared with Lmo2-Arf+/+ (WT) thymocytes. **, P < 0.005.
To determine if enforced Lmo2 expression was sufficient to induce murine ETP, we transduced DN thymocytes with an Lmo2 expressing retroviral vector and transplanted the cells into irradiated recipient mice. Thymocytes from reconstituted primary recipients were then transplanted into secondary recipients. ETP-ALL leukemias arose in three independent cases in transplanted secondary recipients that showed a myeloid phenotype and CD3 coexpression (Fig. 4 B) and rearrangement of the TCR-β chain (Fig. 4 C). Note that thymic granulocytes do have faint TCR rearrangements and are known to arise from early thymocyte precursors. Immature thymocyte populations were sorted from normal mice (DN1-DN3) and showed polyclonal TCR rearrangements, as expected. Enforced Lmo2 expression in Arf-null thymocytes blocked development at the DN2a stage (unpublished data) and results in an expanding thymocyte population with significant myeloid differentiation capacity demonstrated in myeloid CFU-C assays (Fig. 4 D). The accumulation of DN2 cells with myeloid potential appears to be caused by a block in differentiation rather than a developmental reversion arising in mature thymocytes (unpublished data).
Alterations in Bcl11b and Notch1 signaling in murine ETP-ALL
BCL11B is a transcription factor that is required for thymocytes to progress beyond the DN2 checkpoint and is required for irreversible commitment to the T cell lineage (Li et al., 2010). Mutations in BCL11B have also been associated with T-ALL and may play a role in leukemic transformation (Gutierrez et al., 2011). BCL11B expression is also known to be negatively regulated by IL-7 signaling during normal thymocyte development (Ikawa et al., 2010), suggesting the possibility that the constitutive signal from Il7r mutants could repress Bcl11b expression in ETP-ALL. To test this possibility, splenic leukemia cells derived from either the Il7r-241-242TC or GCinsL243 vectors were analyzed by Western blot analysis and shown to have little or no Bcl11b expression (Fig. 5 A). Specifically, no Bcl11b was detected in nuclear extracts from three independent splenic tumors initiated with Il7r-241-242TC or in one tumor from the GCins243 mutant. In contrast, classical CD4+ CD8+ murine T-ALLs derived either from LMO2 overexpression in hematopoietic stem cells (HSCs; Treanor et al., 2011) or from the one case arising from Il7r GCinsL243 mutant (Table 1) had significantly elevated levels of Bcl11b expression (Fig. 5 A, T cell). Likewise, no Bcl11b expression was detectable in nuclear extracts from four independent splenic ETP-ALL tumors initiated with the Lmo2 vector (Fig. 5 B). These results show that repression of Bcl11b is characteristic of murine ETP-ALL and may distinguish ETP-ALL from classic T-ALL cases. Furthermore, it is possible that repression of Bcl11b is necessary for the developmental block in DN thymocyte progression seen with either the Il7r mutants or the Lmo2 vector.
Figure 5.
Bcl11b, Notch1, and p38Map kinase protein expression in murine ETP-ALL versus classical T-ALL. (A) Bcl11b protein expression in nuclear extracts from spleens of ETP tumors that arose from the Il7R-241-242TC mutant (SP1-SP3) and GcinsL243 mutant (SP4). The last two lanes show nuclear extracts of spleens from a mature T-ALL tumor that arose from the Il7r-Gcins243 mutant (SP1) and a Lmo2 transduced hematopoietic stem cell induced CD4+CD8+ tumor (SP2). Lamin B serves as a loading control. (B) Bcl11b levels in a nuclear extracts from spleen samples of Lmo2-induced ETP tumors. Four tumors arising directly from the Lmo2 expression vector were analyzed for Bcl11b expression (SP1-4). The positive and negative controls are nuclear and cytoplasmic extract from a WT thymus, respectively. (C) Levels of the Notch1 intracellular domain (ICN1) were assessed in splenic ETP tumor cell extracts arising from the mutant Il7r receptors. Also shown is a tumor cell extract from a CD4+ CD8+ T-ALL case obtained with the GCins243 vector (T-ALL) and a Lmo2-induced classical T-ALL. Jurkat cells were used as positive controls and GPE-86 cells were used as the negative control (-ve). (D) Western blot analysis for Map kinase expression and activation (P-p38MAPK) in various murine ETP-ALL samples as indicated. For comparison, a case of CD4+ CD8+ T-ALL is included in the left hand lane.
NOTCH1 mutations leading to constitutively increased signaling are frequently present in classical human T-ALL cases (Weng et al., 2004), but are significantly less frequent in human ETP-ALL (Zhang et al., 2012). To determine if this was also true in the murine ETP-ALL tumor samples, the level of stabilized intracellular Notch1 (ICN) was evaluated in several murine ETP-ALL samples derived from each of the two Il7r mutants. ICN was not expressed in any of five independent murine ETP-ALL samples derived from either of the two Il7r mutations (Fig. 5 C). Notch activation was detected in a control case of Lmo2-derived classical T-ALL as previously described (Treanor et al., 2011), and in the one case of CD4+, CD8+ T-ALL that was derived from the Il7r-GCinsL243 vector (Fig. 5 C). These studies show that murine ETP-ALL, like human ETP-ALL, lacks Notch 1 activation and this feature distinguishes ETP-ALL from classical T-ALL. We also detected activation of p38 Map kinase in ETP-ALL tumor samples arising from all three vectors in contrast to very low levels of activation in murine CD4+ CD8+ T-ALL (Fig. 5 D).
Gene expression patterns in human pediatric ETP-ALL cases parallel that seen in the murine ETP-ALL model
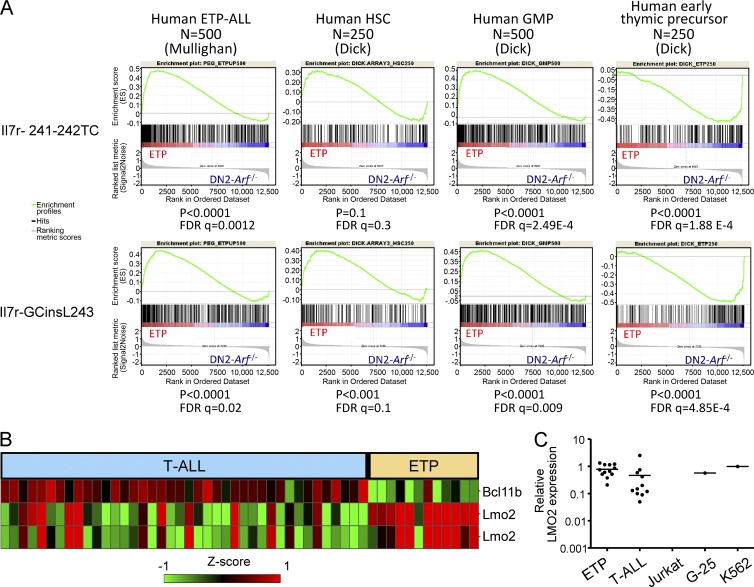
Having established that our mouse model phenotypically resembled human ETP-ALL, we next tested whether the murine model recapitulated the molecular gene expression profile of human ETP-ALL using gene set enrichment analysis (GSEA) from expression array data. Murine ETP-ALL shared statistically significant positive correlations with several gene expression sets that were highly correlated with human ETP-ALL (Zhang et al., 2012), including the profile from human hematopoietic stem cells and from human granulocyte-macrophage precursors and negative correlation with human early thymic precursor cells (Fig. 6 A). Furthermore, the murine ETP-ALL cases highly resembled a GSEA expression profile derived from the top 500 most overexpressed genes in human ETP-ALL (Zhang et al., 2012). These results show that the molecular characteristics of the murine model parallel those seen in human ETP-ALL.
Figure 6.
mRNA expression analysis in murine and human pediatric ETP-ALL cases. (A) Gene set enrichment analysis comparing both the Il7r-241-242TC and Il7r-GCinsL243 induced murine ETP-ALLs to characteristic gene expression sets from human ETP-ALL, human hematopoietic stem cells (HSC), and human granulocyte-macrophage precursor (GMP) cells. False discovery rate (FDR) and p-values are indicated at the bottom of each analysis. (B) Transcription factor microarray analysis of human pediatric leukemia samples from the St. Jude cohort of 40 T-ALL and 12 ETP cases. The top row shows expression of BCL11B mRNA and the two bottom rows show two different LMO2 oligo spots. Red indicates relatively high levels of expression and green indicates lower levels. (C) Quantitative real-time RT-PCR assay for LMO2 RNA expression in 11 ETP and 11 T-ALL samples. Jurkat cells do not express LMO2 and were used as a negative control. K562 and G25, two cell lines known to express high levels of LMO2 protein, were used as the positive control. There is a trend toward higher Lmo2 expression in ETP-ALL (P = 0.1); however, some classical T-ALL cases were associated with high Lmo2 expression.
Also paralleling murine ETP-ALL, human pediatric ETP-ALL specifically show high levels of LMO2 mRNA expression and low levels of BCL11B expression. Analysis of microarray (Fig. 6 B) and quantitative RT-PCR (Fig. 6 C) data of 12 ETP cases versus 40 non-ETP T-ALL cases showed higher LMO2 expression in ETP versus non-ETP T-ALL and lower levels of Bcl11b expression. All ETP cases had LMO2 mRNA levels that were equivalent to a Jurkat T cell clone (G-25) with engineered retroviral vector insertions in the LMO2 locus (Ryu et al., 2008) and K562 cells, both of which express high levels of LMO2 protein on Western blot analysis. In contrast, the classical T-ALL cases showed variable and, on average, lower levels of LMO2 mRNA. A highly significant decrease in BCL11B expression was noted in the 12 pediatric ETP cases versus classical T-ALL in the expression array studies (Fig. 6 B). Together, these results demonstrate that high levels of LMO2 expression, repression of Bcl11b, and lack of Notch1 mutations are hallmarks of ETP-ALL in both human pediatric cases and in our mouse model and suggest that these alterations may be functionally important for the development of ETP-ALL.
Targeting JAK–STAT signaling in murine ETP-ALL cells with ruxolitinib
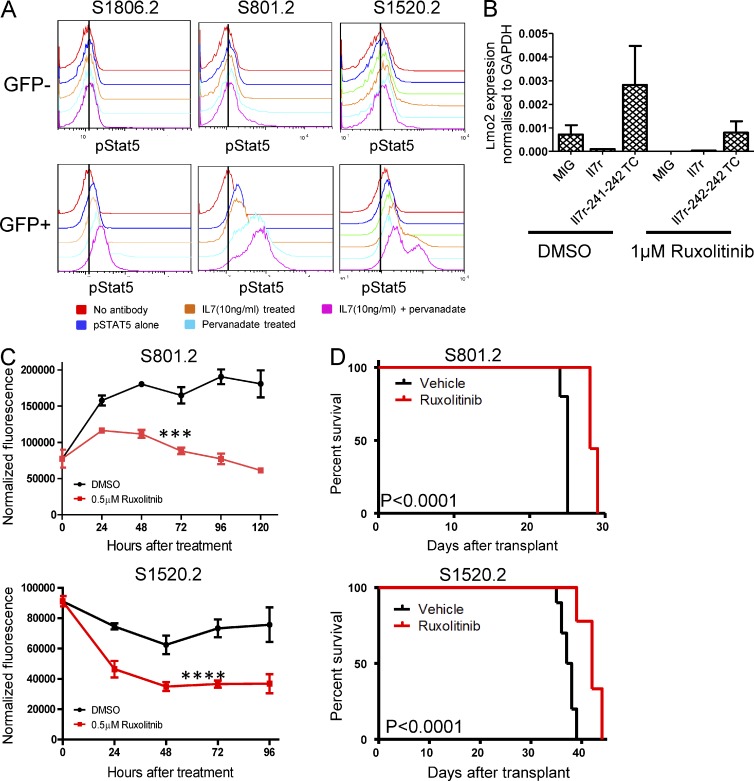
Mutations in the α chain of the IL7R receptor that lead to ligand independent activation function via activation of JAK1 kinase and the STAT5 transcription factor but do not require common gamma chain or JAK3 signaling (Zenatti et al., 2011), suggesting that pharmacologic inhibition of this pathway could inhibit growth of ETP-ALL tumor cells. Phospho-flow analysis for activated Stat5 was performed on ETP-ALL cells derived from the Il7r-241-241TC vector and showed Stat5 phosphorylation from three independent ETP-ALL lines (Fig. 7 A). Stat5 phosphorylation was detected only in cells containing the Il7r mutant vectors (GFP+) but not in GFP− nontransduced control cells (Fig. 7 A). The pStat5 signaling was augmented by the phosphatase inhibitor pervanadate and treatment with exogenous IL7.
Figure 7.
Ruxolitinib inhibition of Jak/Stat signaling and tumor cell growth in Il7r mutant-induced ETP-ALL. (A) Phospho-flow analysis for activated Stat5 in three cases of murine ETP-ALL induced by the Il7R-241-242TC mutant vector. The red histogram shows the background fluorescence without phospho-Stat5 antibody. The blue, green, orange, and cyan curves show phospho-Stat5 staining in cells cultured in media alone, treated with Il7 ligand, treated with pervanadate, and treated with both pervanadate and Il7, respectively. Note that analyses were done on gated GFP+ cells that contained and expressed the mutant Il7r vector and on GFP− cells in the same sample that were either untransduced or did not express the vector. This was repeated on four independent ETP-ALL lines. (B) Lmo2 expression by qPCR in thymocytes transduced with various vectors and cultured in ruxolitinib. Cells were transduced with the WT Il7R vector, with a MIG control vector, or with the Il7r-241-241TC mutant vector and then cultured as in B. RNA was extracted and analyzed for Lmo2 expression after 4 d of drug exposure. There is a trend toward lower Lmo2 expression in the Il7r241-242TC mutant vector transduced thymocytes treated with Ruxolitinib compared with DMSO but this did not reach statistical significance (P = 0.25). (C) Murine ETP-ALL tumor lines tested for sensitivity to ruxolitinib in bulk liquid culture. Tumor cells arising from the Il7r-241-242TC mutant vector were cultured for 96–120 h in the presence of 0.5 µM ruxolitinib or carrier and the number of viable cells was measured every 24 h using a fluorometric assay. Experiments for two independent cell lines are shown and viability assays were repeated in triplicate. A two-factor analysis of variance showed a significant effect of ruxolitinib on the ETP-ALL cells. ***, P < 0.0005 for S801; ****, P < 0.0001 for S1520. (D) Animals transplanted with ETP-ALL line S801.2 or S1520.2 were treated with 60 mg/kg ruxolitinib or vehicle twice daily by oral gavage. Kaplan-Meier curves are shown indicating the survival of mice treated with vehicle alone or ruxolitinib (P < 0.0001).
Ruxolitinib is a selective drug inhibitor of Jak1 and 2 that is approved for the treatment of myelofibrosis due to activating JAK2 mutations (Harrison et al., 2012). To determine whether Lmo2 expression was linked to Jak–Stat activation, thymocytes transduced with the Il7r-241-241TC vector were cultured in the presence of ruxolitinib and Lmo2 mRNA expression was measured at 96 h of exposure. Endogenous Lmo2 mRNA expression was decreased by ruxolitinib treatment (Fig. 7 B) relative to DMSO controls demonstrating that Lmo2 expression is at least in part controlled by the mutant Il7r signal.
To determine whether ruxolitinib would have an antiproliferative effect on ETP-ALL cells, we cultured two independent Il7R-241-241TC ETP-ALL cell lines in physiologically achievable concentrations of drug and measured cell proliferation relative to control cultures. Both cell lines showed a significant decrease in cell growth after 96 h of culture in 0.5 µM ruxolitinib relative to DMSO control cultures (Fig. 7 C). Finally, we tested whether ruxolitinib treatment of mice inoculated with these two ETP-ALL cell lines would have antileukemic effects in vivo. Mice were treated with 60 mg/kg of ruxolitinib via oral gavage twice daily beginning 2 d after transplant of ETP-ALL cells. In the S801.2 group, all of the animals treated with vehicle succumbed to leukemia by 25 d after transplant (Fig. 7 D), whereas animals receiving ruxolitinib survived until days 28 and 29 (P < 0.0001). In the second ETP-ALL line, S1520.2, all of the animals treated with vehicle died of leukemia by 39 d after transplant, whereas the ruxolitinib-treated group showed a significantly prolonged survival, with some mice living until day 44 (P < 0.0001; Fig. 7 D). These results demonstrate that ruxolitinib has significant antileukemic activity in vivo as a single agent. Further studies will be required to understand the mechanisms of leukemic breakthrough in this model and whether other agents can be added to achieve longer disease-free intervals.
DISCUSSION
Several recent studies have suggested that constitutively active IL7R mutants may be oncogenic drivers in ETP-ALL (Shochat et al., 2011; Zenatti et al., 2011), but did not test transformation capacity in primary hematopoietic cells or define how these mutant receptors contribute to the ETP-ALL phenotype. We describe the first de novo mouse model of ETP-ALL showing that two different Il7r mutant alleles, homologous to those identified in human ETP-ALL, are sufficient to induce murine ETP-ALL when expressed in primitive, Arf-null thymocytes, proving that mutant IL7R receptors can be leukemia drivers in vivo and cause an ETP-ALL like disease in mice. This murine disease highly resembles human ETP-ALL pathologically, immunophenotypically, and molecularly with regard to gene expression profiles. These activating mutations in Il7r result in activation of Stat5 in primary tumor cells, suggesting that inhibition of Jak activation could be an effective therapy. We indeed show that ruxolitinib has significant antileukemic activity in vivo, indicated by a significant prolongation of survival in treated mice with ETP-ALL. This result indicates that the ETP-ALL may require continual Il7r signaling for maintenance and growth. This mouse model should also be useful for studying the activity of other therapeutics and for studying mechanisms of treatment resistance.
In addition to providing information about therapy for ETP-ALL, this work illustrates several biological insights into the mechanism of ETP-ALL. All oncogenes that cause ETP-ALL in our model lead to a block in thymocyte development at the DN2 stage, resulting in transformation at a stage in T cell development at which significant stem cell and myeloid potential is retained. At the DN2 stage, thymocytes retain the ability to generate myeloid, NK, and dendritic cells (Bell and Bhandoola, 2008; Wada et al., 2008); however, the physiological significance of this multipotentiality has been questioned (Schlenner et al., 2010; Schlenner and Rodewald, 2010; Richie Ehrlich et al., 2011). Recent work has shown that thymic granulocytes do arise from early thymocyte progenitor cells (De Obaldia et al., 2013). Our results further establish the significance of this developmental plasticity by showing that the cellular mechanism of ETP-ALL results from transformation of an early thymocyte that has retained myeloid differentiation potential.
The expression patterns of LMO2, BCL11B, and NOTCH1/ICN1 transcription factors clearly distinguished ETP-ALL cases from non-ETP T-lineage ALL in both human and murine tumors and suggest that these alterations are characteristic of this leukemia subtype. The cellular stage at which LMO2 overexpression and/or IL7R mutations are acquired could determine whether ETP versus non-ETP-ALL arises; e.g., if the mutations are acquired after DN2 progression, classical T-ALL would likely be favored. In this regard, it is notable that activating IL7R mutants have been identified in classical T-ALL cases as well as in ETP-ALL (Zenatti et al., 2011; Zhang et al., 2012). The absolute expression level of LMO2 may also determine whether ETP T-ALL versus non-ETP-ALL will arise. In previous studies in transgenic mice using nonviral promoters to express LMO2, a partial DN3 block was noted before development of classical T-ALL (Larson et al., 1995; Neale et al., 1995, 1997). Our experiments use the stronger MSCV retroviral promoter to express Lmo2, resulting in an earlier, partial DN2 block. We suggest that the critical DN2-DN3 checkpoint may be sensitive to the absolute level of Lmo2 expression in transitioning thymocytes and that differentiation block before the DN2 stage is required for ETP-ALL. Consistent with this idea, high levels of Lmo2 are strongly selected against in mature CD4+/CD8+ T-ALLs (Treanor et al., 2011). Another possibility, that is not mutually exclusive, is that additional cooperating mutations may play a dominant role in determining the leukemia phenotype.
We propose a model for ETP-ALL in which transformation is initiated at a cell stage at or before DN2 because of altered expression of key transcription factors such as LMO2 and/or BCL11b. During normal T cell development, LMO2 is expressed at high levels in hematopoietic stem cells, in the earliest thymic immigrants from the BM, and in DN1 cells. Expression of LMO2 is then sharply down-regulated as cells pass from DN2a to DN2b (Herblot et al., 2000). At this critical transition, IL7R signaling decreases and Bcl11b is induced, permitting commitment to the T cell lineage (Ikawa et al., 2010). For instance, deletion of Bcl11b results in a DN2 differentiation block and loss of development of mature T cells (Wakabayashi et al., 2003). Thymocytes that are blocked at this stage retain both myeloid and T cell differentiation potential and the phenotype and biological behavior of these leukemic blasts reflect this bipotentiality. The exact mechanisms by which Lmo2, Bcl11b, and Il7r signaling are related are not fully understood, however we do show that pharmacologic inhibition of Il7r signaling results in decreased expression of Lmo2, suggesting that the pathways are linked.
ETP-ALL tumors in mice do not express activated Notch1, and Notch1 mutations are relatively infrequent in human cases. Notch 1 signaling normally increases after the DN2 checkpoint and is necessary for development of double- and single-positive thymocytes (Tanigaki and Honjo, 2007). Activating Notch1 mutations are also commonly seen in classical T-ALL and presumably reflect a requirement for this signal in the transformation process. The lack of Notch1 signaling in our murine ETP-ALLs most likely reflects the stage-specific independence from Notch1 signals at this early stage of thymocyte development.
Currently, only ∼25% of pediatric patients with ETP-ALL are cured of their disease and new therapeutic approaches need to be identified. Drug sensitivity studies on cultured human leukemic blasts can be used to screen for new therapeutic approaches (Holleman et al., 2004); however, primary leukemia cells often require complex culture conditions to grow and proliferate (Nishigaki et al., 1997) and this assay may not represent drug sensitivity in the actual leukemia-initiating/stem cells. Another approach is to establish xenografts in immunodeficient mice using human leukemia cells, as has been done for acute myeloid leukemia (Eppert et al., 2011); however, primary ETP-ALL samples are not widely available for this purpose. Therefore, our mouse model for ETP-ALL has value in providing an approach to screen and test candidate drugs for antileukemic activity, such as we have done with ruxolitinib. Although we do detect significant antileukemic activity, it is not surprising that this single drug therapy does not result in curative eradication of leukemia, at least not in our initial studies. Newer drugs and combination therapy will likely be required for more durable responses. Other possible targets are the characteristic alterations in transcription factor expression in ETP-ALL that we describe. Prior studies have shown that tumor growth can depend upon deregulated expression of oncogenic transcription factors (Felsher and Bishop, 1999; Felsher, 2003; Zuber et al., 2011), but drugs for targeting transcription factors have been more difficult to identify than those associated with oncogene addiction based on signaling proteins, such as RAS (Singh et al., 2009) and BCR-ABL1 kinases (Shah et al., 2008). However, there has been recent success in targeting specific oncogenic transcription factors in the Myc family, so that targeting certain oncogenic transcription factors may be possible for ETP-ALL.
MATERIALS AND METHODS
Viral vectors and production.
A cDNA encoding mouse Lmo2 (National Center for Biotechnology Information reference sequence BC057880.1) was inserted into a mouse stem cell vector containing an internal ribosome entry site–GFP gene reporter (MSCV-IRES-GFP) as previously described (Shou et al., 2006). To facilitate experiments in which p19Arfgfp/gfp cells were used (Sherr, 2004), analogous mCherry vectors were constructed by removing the GFP cassette and replacing it with a cDNA encoding mCherry. A polyclonal population of ecotropic retroviral producer GPE-86 cells was generated for each vector and freshly collected supernatants containing high-titer vector were generated.
The Il7R IL241-242TC and Il7R GCinsL243 vectors have been described previously (Zhang et al., 2012). For production of ecotropic retroviral supernatants, vectors were prepared by transient transfection of vector plasmids and the pMD gagpol and pCAG4-Eco packaging plasmids.
Mouse strains.
Arf−/− (Kamijo et al., 1997)and Arfgf/gfp mice (Sherr, 2004) are both p19Arf null strains, were all on a backcrossed C57BL/6J background and were provided by C. Sherr (St. Jude Children’s Research Hospital, Memphis, TN). They were used between 8 and 12 wk of age. The experiments in Figs. 1 and 2 used the Arfgf/gfp mice, all the other experiments were done using Arf−/− mice. Female C57BL/6J and Rag2−/− γc−/− mice (Taconic) were used as transplantation recipients. Transgenic mice on a B6 background carrying an enhanced GFP gene whose expression is controlled by the proximal lck promoter (plck-GFP mice) were a gift from T. Nakayama (Chiba University, Chiba, Japan).
All experimental procedures were reviewed and approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
Thymocyte culture.
Thymi were explanted from 4–8-wk-old animals. Whole thymocyte populations were stained with CD4-phycoerythrin (PE) and CD8-PE antibodies (BD), incubated at 4°C, washed with MACS buffer (PBS, 0.5% BSA [Sigma-Aldrich], and 2 mM EDTA), and incubated with anti-PE microbeads (Miltenyi Biotec) for 15 min. Cells washed with MACS buffer were centrifuged and placed on a LS column (Miltenyi Biotec), and unattached CD4−/CD8− cells were collected. An aliquot was analyzed by flow cytometry to ascertain the purity of the “double-negative” thymocyte population.
For transduction with the Lmo2 vector the CD4/CD8-depleted thymocytes were co-cultured for 48 h at a density of 0.5–0.7 × 106 cells/well with irradiated (30 Gy) vector producer cells in α-MEM (Invitrogen) containing 20% FBS, penicillin/streptomycin, glutamine, sodium pyruvate (Invitrogen), and polybrene. After 48 h, thymocytes were removed and plated onto confluent OP9-DL1 stromal cells (gift from J.C. Zuniga-Pflucker, University of Toronto, Toronto, Canada) in 12-well plates in complete medium containing 5 ng/ml each of FLT-3 (R&D Systems) and IL-7 (PeproTech). For the MSCV-IRES-GFP, Il7R-WT, and mutant vectors the cells were plated on OP9-DL1 for 24 h. After 24 h, the cells were transduced with 1 ml of vector supernatant collected from 293T cells, spun at 2,000 RPM for 1 h at room temperature. This was repeated 24 h later. Every 3–5 d, thymocytes were replated onto fresh OP9-DL1 cells. Cells harvested at indicated times were immunophenotyped by flow cytometric analysis.
Thymocyte transplantation and colony forming unit assay.
Vector-transduced thymocytes were removed from OP9-DL1 cultures on day 21 and stained for expression of CD4, CD8, CD44, and CD25 (antibodies from BD). Fluorescence-activated cell sorting was used to isolate vector-positive (mCherry+) CD4/CD8 double-negative, CD44/CD25 DN cells, 105 to 3 × 105 of which were transplanted by tail vein injection into sublethally irradiated (6Gy) female γc−/−Rag2−/− mice.
Lmo2, Arf−/− DN2-sorted thymocytes were plated at 33,000 cells/plate in MethoCult GF M3434 (StemCell Technologies) for myeloid progenitor CFU-C assays. 7 d after plating, primary colonies were counted, pooled, and either DNA was extracted or they were analyzed by flow cytometry.
Immunoblotting and immunohistochemistry.
The M-PER and NE-PER mammalian protein extraction reagents (Thermo Fisher Scientific) were used to prepare lysates from tissues for LMO2, Notch1, and Bcl11b immunoblots together with the Halt protease inhibitor cocktail (Thermo Fisher Scientific). Protein concentration was quantified by the Bradford method (Bio-Rad Laboratories). Samples (30–40 µg protein per lane) were electrophoretically separated on 4–12% Bis-Tris NuPAGE gels (Invitrogen), transferred to polyvinylidene fluoride membranes (Invitrogen), and probed using a goat polyclonal antiserum to LMO2 (R&D Systems) at a 1/500 dilution, a rabbit monoclonal antibody to Notch1 cleaved at val1744 (D3B8; Cell Signaling Technology) at a 1/250 dilution, and a rabbit polyclonal antiserum to Bcl11b at ½,000. The secondary antibodies were either a HRP-conjugated rabbit anti–goat (EMD Millipore) or donkey anti–rabbit (GE Healthcare) used at a dilution of 1/1,000. Antibodies to β-actin (Abcam) were used to control for protein loading.
Immunohistochemistry was done using standard protocols. In brief, slides of 4–6-µm sections were cut from formalin-fixed paraffin-embedded tissues, deparaffinized, and rehydrated, and then epitope retrieval was performed. Slides were probed with a rabbit polyclonal antiserum to GFP (Invitrogen) at a 1/200 dilution, a goat polyclonal antiserum to CD3 (Santa Cruz Biotechnology, Inc.) at a 1/350 dilution, a rabbit monoclonal to RUNX1 (Abcam) at a 1/600 dilution, a goat polyclonal antiserum to GATA-1 at a 1/500 dilution, and a rabbit polyclonal antiserum to Myeloperoxidase (MPO; DAKO) at a 1/1,500 dilution.
TCR-β receptor analysis.
The method was done as previously described (King et al., 2002). In brief, genomic DNA was isolated and amplified by a PCR touchdown method (10 s at 94°C, 30 s at 68–63°C, 2 min at 72°C), followed by a 15-cycle PCR (10 s at 94°C, 30 s at 63°C, 2 min at 72°C) with primers Dβ1.1ext and Jβ1.7ext. For analysis of TCR-β rearrangement in DN1-DN4 populations, colonies and tumor samples the PCR products were separated on a 1.2% gel and 0.2–2 kb was excised to exclude germline bands. The DNA was purified by spin column (QIAquick gel extraction kit; QIAGEN). 0.5–1-µl aliquot from the first amplification was subjected to the second PCR with nested primers Dβ1.1int and Jβ1.7 for 24–30 cycles.
Patient samples.
Tumor samples were obtained from diagnostic BM aspirates or peripheral blood, and comprised at least 90% tumor cells from 12 children with ETP-ALL and 40 children with non-ETP-T-ALL treated at St. Jude Children’s Research Hospital. All cases fulfilled pathological and immunophenotypic criteria for ETP-ALL. The study was approved by the Institutional Review Boards of St. Jude Children‘s Research Hospital.
Gene expression profiling and real-time quantitative RT-PCR.
We examined LMO2 expression in the published U133 Plus 2 data from 12 ETP and 40 non-ETP-T-lineage ALL samples that have been previously reported (Coustan-Smith et al., 2009). Expression signals were normalized in log2 scale by the RMA algorithm.
For qRT-PCR assays, reverse transcription was done with SuperScript VILO cDNA Synthesis kit (Invitrogen). qPCR was done for human with the LMO2 assay (Hs00153473_m1) or the mouse Lmo2 assay (Mm01281680; Applied Biosystems), and HPRT1 (Applied Biosystems) was used for the human or GAPDH (Applied Biosystems) for the mouse endogenous control in a step one plus instrument. Relative LMO2 expression level was calculated as the ratio of LMO2/HPRT1 or Lmo2/GAPDH using standard curves for each of the two transcripts.
Drug treatment.
50,000 sorted GFP+ Il7r-GCinsL243 and Il7r241-242TC ETP leukemic cells were plated in triplicate in a 96-well plate. The cells were cultured in DMEM with 15% FBS with Il-3, IL-6, and stem cell factor containing either 0.5 µM ruxolitinib (LC laboratories) or DMSO (vehicle control; Thermo Fisher Scientific). Cell viability was measured by incubating the cells with Cell Titer-Blue (Promega) for 4 h and reading the plate on a FlUOstar Omega plate reader (BMG Labtech).
Thymocytes were cultured as above on OP9-DL1 and either 1 µM ruxolitinib (LC Laboratories) or DMSO (vehicle control; Thermo Fisher Scientific) were added and the cells were analyzed for cell surface markers by flow cytometry as indicated. For RNA analysis, the cells were sorted for GFP− and GFP+ populations and RNA was extracted using the RNAqueous-micro kit (Ambion). Lmo2 RNA levels were measured using the same qRT-PCR assay as above.
For the in vivo studies, animals were sublethally irradiated and transplanted with 5,000 cells from 2 independent ETP-ALL cell lines, S801.2 and S1520.2. 2 d after transplant, treatment was initiated with ruxolitinib (Chemitek) at a dose of 60 mg/kg twice a day by oral gavage or vehicle (5% methylcellulose; Sigma-Aldrich) twice daily for 27 d. Animals were monitored for signs of illness or hematological abnormalities and the endpoint was survival.
Phospho-Stat5 analysis.
ETP leukemic cells were placed in serum and cytokine-free RPMI for 4 h at 37°C. After 4 h, 8 × 105 ETP leukemic cells were treated with pervanadate as a positive control. Upon removal from culture the ETP leukemic cells were fixed and permeabilized (eBioscience). Nonspecific binding was eliminated using 50 µg/ml rabbit IgG and, after blocking, the cells were incubated with a rabbit monoclonal antibody to pStat5 conjugated with Alexa Fluor 647 (Cell Signaling Technology). The cells were resuspended in PBS and their fluorescence was analyzed on a LSR flow cytometer (BD).
Acknowledgments
The authors thank J.C. Zuniga-Pflucker for providing the OP9-DL1 cells. We thank the St. Jude Children’s Research Hospital flow cytometry core facility for assistance, Nicole Lantz for animal husbandry, David Finkelstein for computational support, and members of the Sorrentino laboratory for constructive suggestions and criticisms throughout the course of these studies.
This work was supported by the National Heart, Lung, and Blood Institute (grant P01 HL 53749), the National Cancer Center (support grant P30 CA 21765), the Assisi Foundation of Memphis, and the American Lebanese Syrian Associated Charities.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- Bcl11b
- B cell lymphoma/leukemia 11B
- CFU-C
- colony forming unit in culture
- DN
- double negative
- ETP-ALL
- early thymic precursor leukemia acute lymphoblastic leukemia
- GSEA
- gene set enrichment analysis
- ICN1
- intracellular notch 1
- JAK
- Janus kinase
- MPO
- myeloperoxidase
- p38MAPK
- mitogen activated protein kinase 38 kD
- Stat5
- signal transducer and activator of transcription 5
- T-ALL
- T cell acute lymphoblastic leukemia
References
- Bell J.J., Bhandoola A. 2008. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 452:764–767 10.1038/nature06840 [DOI] [PubMed] [Google Scholar]
- Coustan-Smith E., Mullighan C.G., Onciu M., Behm F.G., Raimondi S.C., Pei D., Cheng C., Su X., Rubnitz J.E., Basso G., et al. 2009. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 10:147–156 10.1016/S1470-2045(08)70314-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Obaldia M.E., Bell J.J., Bhandoola A. 2013. Early T-cell progenitors are the major granulocyte precursors in the adult mouse thymus. Blood. 121:64–71 10.1182/blood-2012-08-451773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K., Takenaka K., Lechman E.R., Waldron L., Nilsson B., van Galen P., Metzeler K.H., Poeppl A., Ling V., Beyene J., et al. 2011. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 17:1086–1093 10.1038/nm.2415 [DOI] [PubMed] [Google Scholar]
- Felsher D.W. 2003. Cancer revoked: oncogenes as therapeutic targets. Nat. Rev. Cancer. 3:375–380 10.1038/nrc1070 [DOI] [PubMed] [Google Scholar]
- Felsher D.W., Bishop J.M. 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell. 4:199–207 10.1016/S1097-2765(00)80367-6 [DOI] [PubMed] [Google Scholar]
- Gutierrez A., Kentsis A., Sanda T., Holmfeldt L., Chen S.C., Zhang J., Protopopov A., Chin L., Dahlberg S.E., Neuberg D.S., et al. 2011. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood. 118:4169–4173 10.1182/blood-2010-11-318873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C., Kiladjian J.J., Al-Ali H.K., Gisslinger H., Waltzman R., Stalbovskaya V., McQuitty M., Hunter D.S., Levy R., Knoops L., et al. 2012. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N. Engl. J. Med. 366:787–798 10.1056/NEJMoa1110556 [DOI] [PubMed] [Google Scholar]
- Herblot S., Steff A.M., Hugo P., Aplan P.D., Hoang T. 2000. SCL and LMO1 alter thymocyte differentiation: inhibition of E2A-HEB function and pre-T alpha chain expression. Nat. Immunol. 1:138–144 10.1038/77819 [DOI] [PubMed] [Google Scholar]
- Holleman A., Cheok M.H., den Boer M.L., Yang W., Veerman A.J., Kazemier K.M., Pei D., Cheng C., Pui C.H., Relling M.V., et al. 2004. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N. Engl. J. Med. 351:533–542 10.1056/NEJMoa033513 [DOI] [PubMed] [Google Scholar]
- Ikawa T., Hirose S., Masuda K., Kakugawa K., Satoh R., Shibano-Satoh A., Kominami R., Katsura Y., Kawamoto H. 2010. An essential developmental checkpoint for production of the T cell lineage. Science. 329:93–96 10.1126/science.1188995 [DOI] [PubMed] [Google Scholar]
- Kamijo T., Zindy F., Roussel M.F., Quelle D.E., Downing J.R., Ashmun R.A., Grosveld G., Sherr C.J. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 91:649–659 10.1016/S0092-8674(00)80452-3 [DOI] [PubMed] [Google Scholar]
- King A.G., Kondo M., Scherer D.C., Weissman I.L. 2002. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc. Natl. Acad. Sci. USA. 99:4508–4513 10.1073/pnas.072087899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson R.C., Osada H., Larson T.A., Lavenir I., Rabbitts T.H. 1995. The oncogenic LIM protein Rbtn2 causes thymic developmental aberrations that precede malignancy in transgenic mice. Oncogene. 11:853–862 [PubMed] [Google Scholar]
- Li L., Leid M., Rothenberg E.V. 2010. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 329:89–93 10.1126/science.1188989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale G.A., Rehg J.E., Goorha R.M. 1995. Ectopic expression of rhombotin-2 causes selective expansion of CD4-CD8- lymphocytes in the thymus and T-cell tumors in transgenic mice. Blood. 86:3060–3071 [PubMed] [Google Scholar]
- Neale G.A., Rehg J.E., Goorha R.M. 1997. Disruption of T-cell differentiation precedes T-cell tumor formation in LMO-2 (rhombotin-2) transgenic mice. Leukemia. 11(Suppl 3):289–290 [PubMed] [Google Scholar]
- Nishigaki H., Ito C., Manabe A., Kumagai M., Coustan-Smith E., Yanishevski Y., Behm F.G., Raimondi S.C., Pui C.H., Campana D. 1997. Prevalence and growth characteristics of malignant stem cells in B-lineage acute lymphoblastic leukemia. Blood. 89:3735–3744 [PubMed] [Google Scholar]
- Richie Ehrlich L.I., Serwold T., Weissman I.L. 2011. In vitro assays misrepresent in vivo lineage potentials of murine lymphoid progenitors. Blood. 117:2618–2624 10.1182/blood-2010-05-287102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E.V., Zhang J., Li L. 2010. Multilayered specification of the T-cell lineage fate. Immunol. Rev. 238:150–168 10.1111/j.1600-065X.2010.00964.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu B.Y., Evans-Galea M.V., Gray J.T., Bodine D.M., Persons D.A., Nienhuis A.W. 2008. An experimental system for the evaluation of retroviral vector design to diminish the risk for proto-oncogene activation. Blood. 111:1866–1875 10.1182/blood-2007-04-085506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenner S.M., Rodewald H.R. 2010. Early T cell development and the pitfalls of potential. Trends Immunol. 31:303–310 10.1016/j.it.2010.06.002 [DOI] [PubMed] [Google Scholar]
- Schlenner S.M., Madan V., Busch K., Tietz A., Läufle C., Costa C., Blum C., Fehling H.J., Rodewald H.R. 2010. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 32:426–436 10.1016/j.immuni.2010.03.005 [DOI] [PubMed] [Google Scholar]
- Shah N.P., Kasap C., Weier C., Balbas M., Nicoll J.M., Bleickardt E., Nicaise C., Sawyers C.L. 2008. Transient potent BCR-ABL inhibition is sufficient to commit chronic myeloid leukemia cells irreversibly to apoptosis. Cancer Cell. 14:485–493 10.1016/j.ccr.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Sherr C.J. 2004. An Arf(GFP/GFP) reporter mouse reveals that the Arf tumor suppressor monitors latent oncogenic signals in vivo. Cell Cycle. 3:239–240 10.4161/cc.3.3.744 [DOI] [PubMed] [Google Scholar]
- Shochat C., Tal N., Bandapalli O.R., Palmi C., Ganmore I., te Kronnie G., Cario G., Cazzaniga G., Kulozik A.E., Stanulla M., et al. 2011. Gain-of-function mutations in interleukin-7 receptor-α (IL7R) in childhood acute lymphoblastic leukemias. J. Exp. Med. 208:901–908 10.1084/jem.20110580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou Y., Ma Z., Lu T., Sorrentino B.P. 2006. Unique risk factors for insertional mutagenesis in a mouse model of XSCID gene therapy. Proc. Natl. Acad. Sci. USA. 103:11730–11735 10.1073/pnas.0603635103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Greninger P., Rhodes D., Koopman L., Violette S., Bardeesy N., Settleman J. 2009. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 15:489–500 10.1016/j.ccr.2009.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanigaki K., Honjo T. 2007. Regulation of lymphocyte development by Notch signaling. Nat. Immunol. 8:451–456 10.1038/ni1453 [DOI] [PubMed] [Google Scholar]
- Torchia E.C., Boyd K., Rehg J.E., Qu C., Baker S.J. 2007. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol. Cell. Biol. 27:7918–7934 10.1128/MCB.00099-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treanor L.M., Volanakis E.J., Zhou S., Lu T., Sherr C.J., Sorrentino B.P. 2011. Functional interactions between Lmo2, the Arf tumor suppressor, and Notch1 in murine T-cell malignancies. Blood. 117:5453–5462 10.1182/blood-2010-09-309831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volanakis E.J., Williams R.T., Sherr C.J. 2009. Stage-specific Arf tumor suppression in Notch1-induced T-cell acute lymphoblastic leukemia. Blood. 114:4451–4459 10.1182/blood-2009-07-233346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H., Masuda K., Satoh R., Kakugawa K., Ikawa T., Katsura Y., Kawamoto H. 2008. Adult T-cell progenitors retain myeloid potential. Nature. 452:768–772 10.1038/nature06839 [DOI] [PubMed] [Google Scholar]
- Wakabayashi Y., Watanabe H., Inoue J., Takeda N., Sakata J., Mishima Y., Hitomi J., Yamamoto T., Utsuyama M., Niwa O., et al. 2003. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat. Immunol. 4:533–539 10.1038/ni927 [DOI] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Yui M.A., Feng N., Rothenberg E.V. 2010. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J. Immunol. 185:284–293 10.4049/jimmunol.1000679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenatti P.P., Ribeiro D., Li W., Zuurbier L., Silva M.C., Paganin M., Tritapoe J., Hixon J.A., Silveira A.B., Cardoso B.A., et al. 2011. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat. Genet. 43:932–939 10.1038/ng.924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S.L., Payne-Turner D., Easton J., Chen X., Wang J., Rusch M., et al. 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 481:157–163 10.1038/nature10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J., Rappaport A.R., Luo W., Wang E., Chen C., Vaseva A.V., Shi J., Weissmueller S., Fellmann C., Taylor M.J., et al. 2011. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 25:1628–1640 10.1101/gad.17269211 [DOI] [PMC free article] [PubMed] [Google Scholar]