E4bp4 is required for commitment to the NK lineage and promotes NK development by directly regulating the expression of Eomes and Id2.
Abstract
The transcription factor E4bp4 (Nfil3) is essential for natural killer (NK) cell production. Here, we show that E4bp4 is required at the NK lineage commitment point when NK progenitors develop from common lymphoid progenitors (CLPs) and that E4bp4 must be expressed at the CLP stage for differentiation toward the NK lineage to occur. To elucidate the mechanism by which E4bp4 promotes NK development, we identified a central core of transcription factors that can rescue NK production from E4bp4−/− progenitors, suggesting that they act downstream of E4bp4. Among these were Eomes and Id2, which are expressed later in development than E4bp4. E4bp4 binds directly to the regulatory regions of both Eomes and Id2, promoting their transcription. We propose that E4bp4 is required for commitment to the NK lineage and promotes NK development by directly regulating the expression of the downstream transcription factors Eomes and Id2.
NK cells are innate lymphocytes that kill virally infected and malignant cells. Their importance for the control of disease is highlighted by the susceptibility of NK-deficient patients to viral infections (Orange, 2006). NK cells also play a role preventing the development and progression of cancer: cancer deaths are less frequent in patients who have higher levels of natural cytotoxic activity (Imai et al., 2000), and stem cell transplant–derived NK cells are an important determinant of survival after radiotherapy for acute myeloid leukemia (Velardi et al., 2012).
NK cells develop in bone marrow from common lymphoid progenitors (CLPs; Kondo et al., 1997), which derive from lymphoid-primed multipotent progenitors (LMPPs; Adolfsson et al., 2005). NK progenitors (NKPs) were originally defined as Lin− NK1.1− DX5− CD122+ (Rosmaraki et al., 2001), but the progenitor frequency in this population is only 1 in 12, so the majority of these cells are not NKPs. Recently, the NKP phenotype has been refined, identifying populations that contain NKPs at a frequency of one in two (Carotta et al., 2011; Fathman et al., 2011).
Several transcription factors that affect NK production have been identified, and the phenotypes of their knockouts suggest that these genes are required at different stages in development. Both immature and mature NK (iNK and mNK, respectively) numbers are reduced in the absence of Ets1 (Ramirez et al., 2012), T-bet (Townsend et al., 2004; Gordon et al., 2012), and E4bp4 (Gascoyne et al., 2009; Kamizono et al., 2009), suggesting that they act relatively early in development. Eomes (Gordon et al., 2012), Id2 (Yokota et al., 1999; Boos et al., 2007), IRF2 (Lohoff et al., 2000; Taki et al., 2005), MEF (Lacorazza et al., 2002), and Tox (Aliahmad et al., 2010) act later, whereas NK cells in CEBPγ (Kaisho et al., 1999) and Gata3 (Samson et al., 2003) knockout mice reach maturity but have functional defects. Transcription factor networks controlling the production of some immune lineages have already been described in detail, but although there has been some progress in defining the relationships between various transcription factors required for NK development, the precise hierarchy is still poorly understood.
E4bp4 (Nfil3) is a bZIP transcription factor with diverse roles in immune cell function (Male et al., 2012). Both iNK and mNK numbers are severely reduced in its absence, consistent with a role early in NK development (Gascoyne et al., 2009; Kamizono et al., 2009). Therefore, E4bp4 is a good candidate for a factor required at the NK lineage commitment point, and we might expect to find a network of transcription factors downstream of E4bp4. Here, we show that E4bp4 is essential for the development of NK-committed progenitor cells from CLPs and identify several transcription factors, notably Eomes and Id2, that act downstream of it.
RESULTS AND DISCUSSION
E4bp4 is required for the development of NKPs
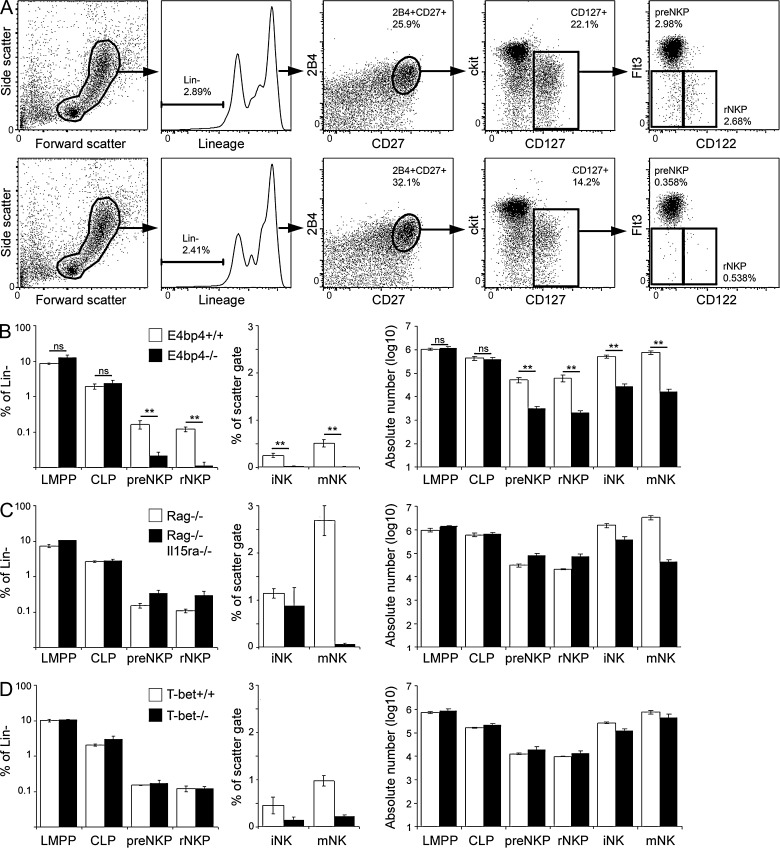
It has previously been reported that NKPs, but not iNK, are present in E4bp4−/− bone marrow, leading to the suggestion that E4bp4 is required at the NKP to iNK transition (Gascoyne et al., 2009; Kamizono et al., 2009), but this work used the original working definition of NKPs (Rosmaraki et al., 2001). We sought to investigate whether the newly defined pre-NKPs (Lin− 2B4+ CD27+ CD127+ Flt3− CD122−) and rNKPs (Lin− 2B4+ CD27+ CD127+ Flt3− CD122+; Fathman et al., 2011) were still present in the absence of E4bp4. Both pre-NKPs and rNKPs were present in E4bp4+/+ bone marrow (Fig. 1 A, top), but their frequency was reduced by 7.8- and 10.6-fold, respectively, in E4bp4−/− bone marrow (Fig. 1 A, bottom). In contrast, the frequency of LMPPs (Lin− Sca1+ ckit+ Flt3+ CD127−) and CLPs (Lin− Sca1int ckitint Flt3+ CD127+) was unchanged (Fig. 1 B). Absolute numbers of pre-NKPs and rNKPs were reduced in E4bp4−/− bone marrow, but LMPPs and CLPs were unchanged (Fig. 1 B).
Figure 1.
E4bp4−/− mice, but not other NK-deficient strains, lack NKPs. (A) Flow cytometry gating strategy for identification of pre-NKP and rNKP in bone marrow from E4bp4+/+ (top) and E4bp4−/− (bottom) mice. Frequency of gated population is shown. (B–D) The graphs show the frequency of LMPPs, CLPs, pre-NKPs, and rNKPs as a percentage of the total lineage-negative population in bone marrow, the frequency of iNK and mNK as a percentage of the scatter gate, and absolute numbers of all NK developmental intermediates. (B) E4bp4+/+ and E4bp4−/−; six mice per genotype. (C) Rag−/− and Rag−/−Il15ra−/−; three mice per genotype. (D) T-bet+/+ and T-bet−/−; three mice per genotype. Error bars show SEM. **, P < 0.005.
To determine whether NKPs are reduced in other conditions in which NK production is perturbed, we examined two other strains of mice that have severe NK deficiencies: the Il15ra knockout cannot mediate IL-15 signaling, which is critical for NK production (Lodolce et al., 1998), whereas the T-bet (Tbx21) knockout lacks a transcription factor crucial for NK cell development (Townsend et al., 2004; Gordon et al., 2012). Pre-NKP and rNKP frequencies and absolute numbers were unaffected in the absence of both IL-15 signaling (Fig. 1 C) and T-bet (Fig. 1 D), indicating that a reduction in NKP frequency does not occur in all situations in which NK development is affected. E4bp4, then, is so far the only transcription factor to be identified as essential for the production of NK-committed progenitors.
In the absence of E4bp4, IL-15–responsive CD122+ NKPs fail to develop (Fig. 1 A), suggesting that E4bp4 is required for expression of the IL-15 receptor and thus that E4bp4 acts upstream of IL-15 signaling. Furthermore, the absence of E4bp4 perturbs NK cell development earlier than the absence of IL-15 signaling (Fig. 1 C). These surprising results suggest that E4bp4 acts before IL-15, which has previously been considered the definitive factor required for the production of NK cells. The suggestion that E4bp4 expression is required for detection of IL-15 is not necessarily inconsistent with our previous proposal that E4bp4 acts downstream of IL-15 signaling (Gascoyne et al., 2009) because it is possible that E4bp4 acts both up- and downstream of IL-15, forming a self-reinforcing positive feedback loop that drives commitment to the NK lineage.
E4bp4 is expressed before Eomes, Id2, and T-bet
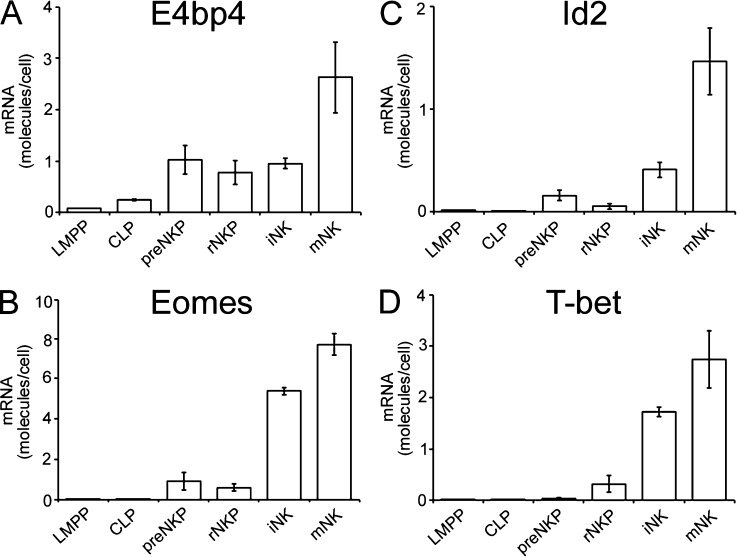
Our finding that E4bp4 is required for NKP production whereas T-bet is not suggests that E4bp4 may act before T-bet in NK development. To investigate this and to compare expression of E4bp4 with that of transcription factors for which no knockout is available on the C57BL/6 background, we sorted cells at various stages of NK cell differentiation from wild-type bone marrow and measured their expression of transcription factor mRNAs. E4bp4 transcript was detectable in both LMPPs and CLPs, increasing at later stages of NK development (Fig. 2 A). Eomes (Fig. 2 B), Id2 (Fig. 2 C), and T-bet (Fig. 2 D) were barely detectable in LMPPs and CLPs. Therefore, E4bp4 is expressed earlier in development than Eomes, Id2, and T-bet.
Figure 2.
E4bp4 transcript is expressed before those of Eomes, T-bet, and Id2. (A) Real-time PCR analysis of E4bp4 expression by LMPP, CLP, pre-NKP, rNKP, iNK, and mNK cells sorted from wild-type bone marrow. (B–D) Expression of Eomes (B), Id2 (C), and T-bet (D). (A–D) Data are representative of four (A) or three independent experiments (B–D). Error bars show SEM.
NK production requires E4bp4 expression in CLPs
The requirement of E4bp4 for the production of CLP-derived NK developmental intermediates, together with the early expression of E4bp4 in LMPPs and CLPs, led us to speculate that E4bp4 might be a lineage commitment factor controlling the development of NKPs from CLPs. In a seminal experiment, Pax5 was shown to mediate B cell lineage commitment at the pro-B stage by restoring Pax5 expression in Pax5−/− pro-B cells and examining their ability to produce mature B cells (Nutt et al., 1999). To test the hypothesis that E4bp4 is required for NK lineage commitment, we took a similar approach, restoring E4bp4 expression in purified E4bp4−/− CLPs and examining whether this was sufficient to reestablish NK cell development.
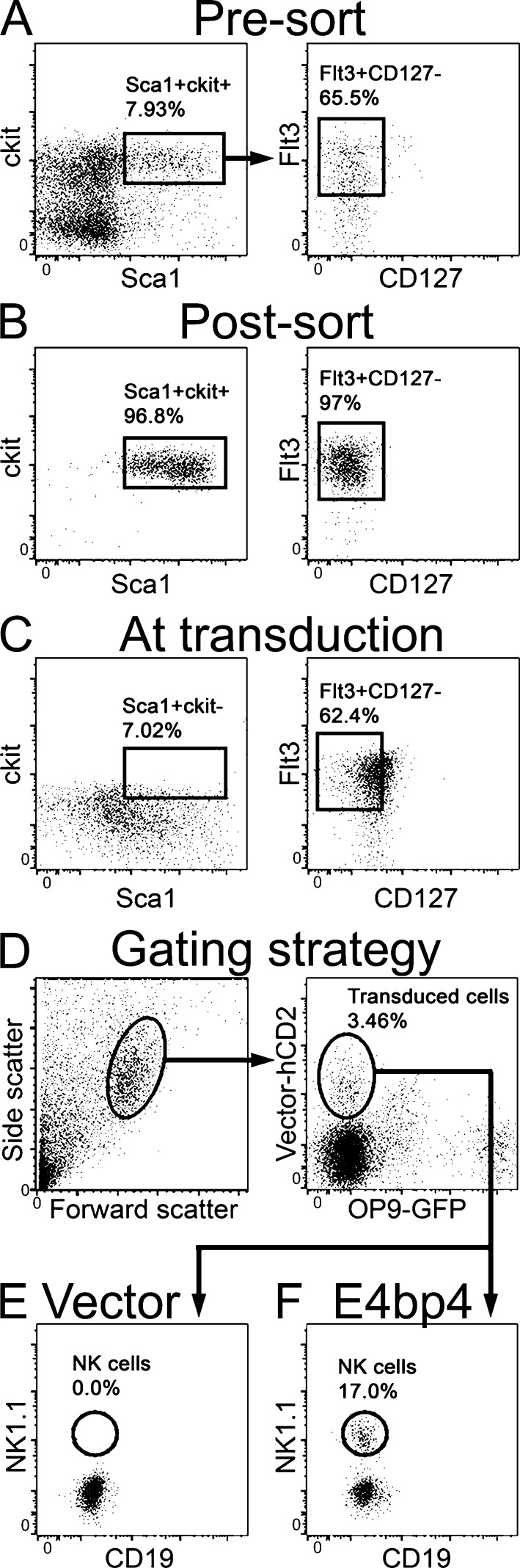
We first sorted CLPs from E4bp4−/− bone marrow and cultured them in lymphocyte-inducing conditions, transducing at 48 h with either E4bp4 or empty vector (negative control) before moving to NK-inducing conditions (Gascoyne et al., 2009). Cells transduced with empty vector did not produce NK cells, but neither did those transduced with E4bp4 (not depicted). Because transduction occurred after 48 h in culture, we reasoned that to transduce CLPs, as opposed to cells merely derived from CLPs, it might be necessary to initiate the culture with an earlier developmental stage and allow the cells to differentiate into CLPs. Indeed, by starting the cultures with sorted LMPPs (Fig. 3, A and B), we found that at the time of transduction, the cells exhibited a CLP phenotype (Fig. 3 C). By taking this approach, the cells that were transduced at 48 h were confirmed CLPs.
Figure 3.
NK development from E4bp4−/− CLPs can be rescued by restoration of E4bp4 expression. (A–C) LMPPs were sorted from E4bp4−/− lineage-depleted bone marrow. The phenotype of the cells presort (A) and postsort (B) is shown. The cells were cultured for 48 h before transduction with either MSCV-hCD2 (vector control) or MSCV-hCD2-E4bp4. The phenotype of the cells at transduction is shown in C. At day 5 the cells were moved to OP9 stromal cells and IL-15 and cultured for a further week. (D–F) Phenotype of cells at the end of culture. Cells were gated by scatter and on transduced (hCD2+) cells, excluding OP9 (D). The presence of NK cells after transduction with the vector control (E) and E4bp4 (F) is shown. Data are representative of three independent experiments.
Transduced cells (Fig. 3 D) were examined for their expression of the NK marker NK1.1. CLPs transduced with E4bp4 had given rise to NK cells, but those transduced with vector control had not (Fig. 3, E and F). The ability of E4bp4 to rescue NK production from E4bp4−/− cells when its expression is restored in a pure CLP population indicates that E4bp4 can act at the CLP stage, just before NK cell lineage commitment. The proposal that E4bp4 is required specifically at the lineage commitment point is supported by the recent report that E4bp4 is not required for the maintenance of cells that are already committed to the NK lineage (Firth et al., 2013). In agreement with our finding that E4bp4 is required for the production of NK-committed progenitors in vivo (Fig. 1 B), this in vitro approach provides further evidence that E4bp4 controls the production of NKPs from CLPs.
Identification of transcription factors that complement E4bp4 function
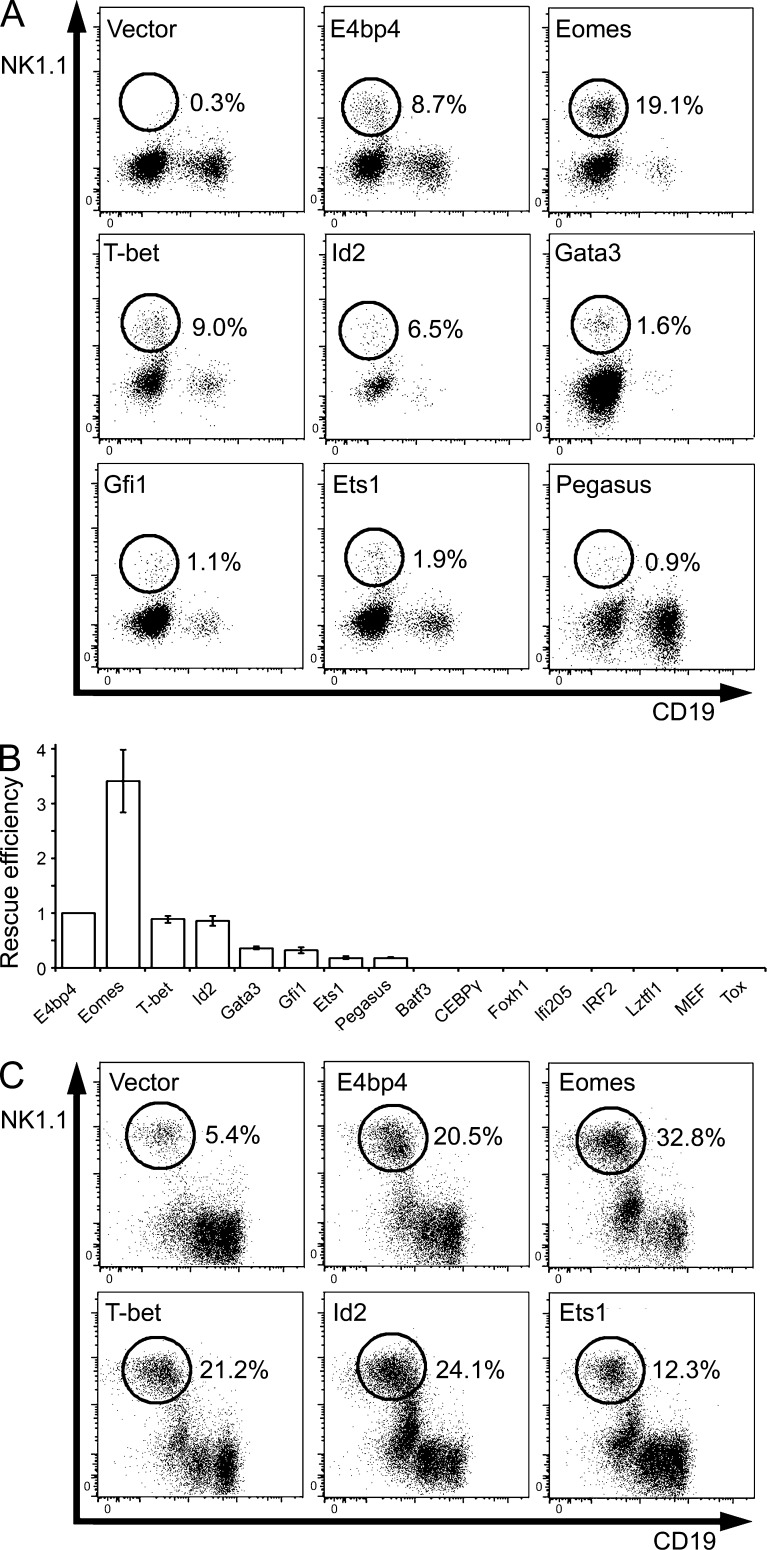
In light of these indications that E4bp4 acts at the earliest possible point in NK development, we sought to define its relationship with other transcription factors that are likely to act downstream. We generated a panel of candidates to be regulated by E4bp4. Eomes, T-bet, Id2, Gata3, Gfi1, Ets1, CEBPγ, IRF2, MEF, and Tox were included because of the phenotypes of their knockouts. Batf3 acts downstream of E4bp4 in CD8α+ cDC development (Kashiwada et al., 2011), so we wished to see whether it acts similarly in NK development. We included Ifi205, Foxh1, Lztfl1, and Pegasus (Ikzf5) because they were expressed at reduced levels in E4bp4−/− lineage-depleted bone marrow in a microarray analysis (Table S1). To determine whether any of these genes act downstream of E4bp4, we set up a screen using a complementation approach. E4bp4−/− lineage-depleted bone marrow was cultured in lymphocyte-inducing conditions and transduced at 48 h with empty vector (negative control), E4bp4 (positive control), or a transcription factor of interest before being moved into NK-inducing conditions. E4bp4−/− lineage-depleted bone marrow cells did not give rise to NK cells under these conditions, but restoring E4bp4 expression rescued NK cell production (Fig. 4 A; Gascoyne et al., 2009; Kamizono et al., 2009). If a transcription factor other than E4bp4 is able to rescue NK cell production in the absence of E4bp4, this could indicate that it acts downstream of E4bp4.
Figure 4.
Identification of transcription factors that complement the function of E4bp4. (A) E4bp4−/− lineage-depleted bone marrow cells were cultured in NK-producing conditions and transduced with the indicated MSCV-hCD2 vectors. Cells were gated as in Fig. 3 D. The percentage of transduced cells that are NK1.1+ is shown. (B) Efficiency of the rescues effected by the 15 cDNAs examined, normalized to that of the positive control (E4bp4). Data are representative of five (Eomes and T-bet), four (Id2), three (Gata3, Gfi1, Ets1, and Pegasus), or two (Batf3, CEBPγ, Foxh1, Ifi205, IRF2, Lztfl1, MEF, and Tox) independent experiments. Error bars show SEM. (C) E4bp4+/+ lineage-depleted bone marrow cells were cultured as in A. Data are representative of two independent experiments.
Eomes effected the most striking rescue, consistently producing more NK cells than E4bp4 (mean 3.4-fold change). T-bet and Id2 were able to produce NK cells with roughly the same efficiency as E4bp4 (mean 0.9-fold change). Gata3, Gfi1, Ets1, and Pegasus produced a more modest rescue. Cells transduced with Batf3, CEBPγ, IRF2, MEF, Tox, Foxh1, Ifi205, and Lztfl1 did not produce NK cells. The efficiency of the rescue effected by each of the transcription factors in the screen is shown in Fig. 4 B. As NK production varied between assays, the frequency of NK cells produced by the transcription factor of interest is normalized to the frequency of NK cells in the positive control (E4bp4) condition.
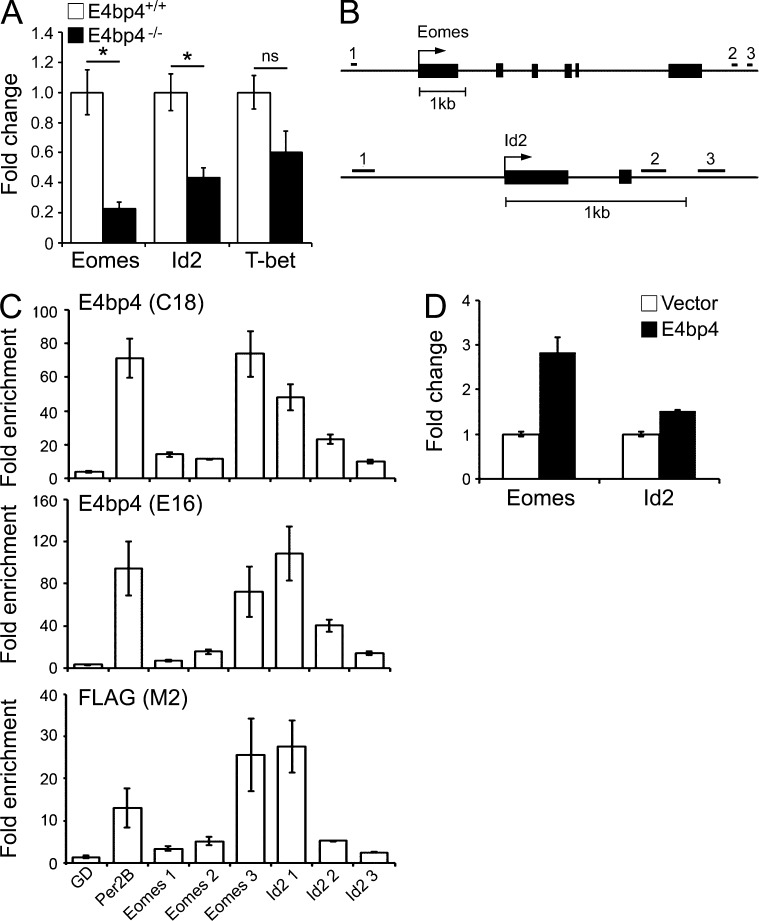
The ability of Eomes, Id2, and T-bet to complement E4bp4 suggests that they act downstream in the hierarchy of transcription factors controlling NK development. Overexpression of E4bp4 in wild-type cells is known to increase NK production (Gascoyne et al., 2009), and we found that Eomes, Id2, and T-bet had a similar effect, consistent with a position downstream of E4bp4 and the law of mass action (Fig. 4 C). Additionally, we observed that Eomes and Id2, but not T-bet, were expressed at lower levels in E4bp4−/− lineage-depleted bone marrow cells compared with E4bp4+/+ cells (Fig. 5 A). Therefore, E4bp4 seems to promote the expression of Eomes and Id2 but not T-bet.
Figure 5.
E4bp4 protein directly binds the Eomes and Id2 loci. (A) Lineage-depleted bone marrow cells from either E4bp4+/+ or E4bp4−/− mice were cultured for 48 h in lymphocyte-inducing conditions, and Eomes, Id2, and T-bet transcripts were measured (six mice per genotype). *, P < 0.05. (B) Eomes and Id2 loci showing the location of the predicted E4bp4-binding sites. (C) ChIP analysis of the enrichment of E4bp4 at the indicated loci in chromatin from MNK-1 cells transduced with 5′ FLAG–E4bp4. C18 and E16 are polyclonal antibodies to E4bp4, and M2 is a monoclonal antibody to FLAG. GD (gene desert) was used as a negative control; Per2B was a positive control. (D) MNK-1 cells were transduced with either vector control or FLAG-E4bp4, and expression of Eomes and Id2 transcripts was determined by real-time PCR. (C and D) Data are representative of three independent biological replicates. Error bars show SEM.
E4bp4 directly regulates the expression of Eomes and Id2
We used chromatin immunoprecipitation (ChIP) to determine whether E4bp4 promotes the transcription of Eomes and Id2 directly by binding in vivo to the regulatory regions of their genes. To do this, we took advantage of the new mouse NK-like cell line MNK-1, which allowed us to use a known E4bp4-binding site in the regulatory region of the mouse Per2 gene, Per2B, as a positive control (Ohno et al., 2007). MNK-1 cells were transduced with FLAG-E4bp4, and protein–chromatin complexes were immunoprecipitated using either IgG, anti-FLAG, or one of two polyclonal antibodies to E4bp4.
We searched regulatory regions within 2 kb 5′ and 3′ of the Eomes and Id2 genes and identified six putative E4bp4-binding sites (Fig. 5 B and Table S2). We examined their enrichment in chromatin immunoprecipitated with anti-FLAG or anti-E4bp4 compared with that immunoprecipitated with IgG. To rule out the possibility of spurious enrichment, we used primers that amplify a region in a gene desert on chromosome 11 as a negative control. We did not observe enrichment of the predicted binding site 5′ of Eomes, but we did observe enrichment in one of the predicted binding sites 3′ of the gene. For Id2, we observed enrichment at the predicted binding site 5′ of the transcriptional start site but not at either of the binding sites 3′ of the gene (Fig. 5 C). These results indicate that E4bp4 binds the regulatory regions controlling Eomes and Id2 transcription in vivo, suggesting that their transcription is regulated by E4bp4. To confirm that E4bp4 promotes transcription at these loci, we transduced MNK-1 cells with FLAG-E4bp4 and examined expression of Eomes and Id2 transcript. Both Eomes and Id2 increased when E4bp4 was overexpressed (Fig. 5 D), consistent with E4bp4 acting as a transactivator at these loci.
Eomes knockout mice die in utero, but experiments using conditional knockouts point to its role in mediating the iNK to mNK transition (Gordon et al., 2012). Its requirement at this point is consistent with a position for Eomes downstream of E4bp4. T-bet seems to be required earlier than Eomes, promoting the survival of iNK cells (Townsend et al., 2004; Gordon et al., 2012). Both Eomes and T-bet can complement E4bp4. This is consistent with both acting downstream of it, but the finding that E4bp4 promotes expression of Eomes and not T-bet is perhaps at odds with this proposal. One possible explanation is that T-bet, although able to rescue NK cell production from E4bp4−/− cells, acts in a different pathway from Eomes and E4bp4. In support of this, those NK cells that develop in the absence of T-bet express Eomes at normal levels (Townsend et al., 2004), and there is mounting evidence that Eomes is primarily required for NK development in the bone marrow, whereas T-bet mediates extramedullary development of NK cells (Gordon et al., 2012; Sciumé et al., 2012; Rankin et al., 2013). The Id2 knockout displays a reduction in mNK but not iNK, suggesting that Id2, like Eomes, acts at the iNK to mNK transition (Boos et al., 2007). The relatively late point at which Id2 is required, together with our findings that Id2 is expressed later than E4bp4 and that Id2 is a direct transcriptional target of E4bp4, places Id2 downstream of E4bp4 in the hierarchy of transcription factors controlling NK development. Id2 is expressed later than Ets1, is underexpressed in its absence, and is a direct target, suggesting that it is also downstream of Ets1 (Ramirez et al., 2012). The Ets1−/− mouse displays a small reduction in NKPs (Ramirez et al., 2012), and, although the phenotype is less striking than that observed in the E4bp4 knockout, the point at which the defect is first observed, together with the findings that both E4bp4 and Ets1 directly regulate Id2 transcription, could be consistent with E4bp4 and Ets1 working at a similar point upstream of Id2. Therefore, there is mounting evidence that Id2 is not as high in the hierarchy of transcription factors controlling NK development as previously thought.
Here we have shown that NK-committed progenitors are not produced in the absence of E4bp4 and that E4bp4 expression in CLPs promotes production of NK cells. These findings suggest that E4bp4 is required at the NK lineage commitment point and hold out the possibility that E4bp4 may even be the NK lineage–defining transcription factor. Consistent with a position for E4bp4 at the top of a hierarchy of transcription factors, we have also shown that Eomes and Id2 both act downstream of E4bp4 and that their transcription is directly regulated by E4bp4. Improving our understanding of the molecular basis of NK cell development is likely to have practical consequences because expression of these core transcription factors could have applications in the development of NK-based immunotherapies for viral and malignant disease.
MATERIALS AND METHODS
Mice.
E4bp4−/− mice (Gascoyne et al., 2009) and T-bet−/− mice (Townsend et al., 2004) were generated as described previously. Rag−/− IL15ra−/− and Rag−/− control mice were obtained from the Jackson Laboratory. All animals used were on a C57BL/6 background, between 6 and 12 wk old, and were matched for age and gender. Where possible, littermates were used. Animal husbandry and experimental procedures were performed according to UK Home Office regulations and institute guidelines.
Flow cytometry.
Cells were stained with the following antibodies, all of which were anti–mouse and from eBioscience unless otherwise specified: 2B4 (clone m2B4(B6)458.1; BioLegend), CD2 (RM2-5), CD3 (17A2), CD11b (M1/70), CD19 (1D3), CD27 (LG.7F9), CD122 (TM-b1), CD127 (A7R34), B220 (RA3-6B2), ckit (ACK2), Flt3 (A2F10), Gr1 (RB6-8C5), NK1.1 (PK136), Sca1 (D7), Ter119 (TER119), and anti–human CD2 (RPA-2.10). The lineage cocktail contained B220, CD2, CD11b, Gr1, NK1.1, and Ter119. Cells were analyzed on a Fortessa system (BD) or sorted using FACSAria (BD).
Microarray analysis.
Lin− bone marrow cells were obtained from 6 × E4bp4+/+ and 6 × E4bp4−/− mice, and RNA was extracted using RNeasy Micro columns (QIAGEN). RNA was converted to cDNA using the Ambion WT Expression kit (Affymetrix), and cDNA was labeled using GeneChip WT Terminal Labeling and Controls kit (Affymetrix). The labeled cDNA was hybridized to a GeneChip Mouse Gene 1.0 ST Array (Affymetrix). The data generated was imported into GeneSpring GXv11 (Agilent Technologies) and normalized using the Robust Multi-Array (RMA16) algorithm. Genes were filtered on expression value between the 20th and 100th percentile to remove those with very low or no expression across all the samples. A moderated Student’s t test was applied to the data, and p-values were adjusted for multiple testing using a Benjamini–Hochberg false discovery rate correction. The microarray data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession no. GSE55014.
Real-time PCR.
Real-time PCR was performed using TaqMan (Applied Biosystems) primer/probe sets recognizing Hprt1 (Mm00446968_m1), E4bp4 (Nfil3; Mm00600292_s1), Eomes (Mm01351985_m1), Id2 (Mm00711781_m1), and T-bet (Tbx21; Mm00450960_m1). Ct values from samples were compared with a standard curve made from a known number of molecules of plasmid (E4bp4, Eomes, Id2, and T-bet) or a known number of cells (housekeeping control gene Hprt1) to define mRNA expression in molecules/cell.
In vitro NK cell development assay.
pMSCV constructs were cloned as described previously (Gascoyne et al., 2009), and retroviral vectors were generated by transfection into LinXE cells. The Gfi1 cDNA was a gift from B. Scheijen (Nijmegen Medical Centre, Nijmegen, Netherlands). Total Lin− bone marrow cells, sorted LMPPs, or CLPs were cultured for 48 h in DMEM supplemented with 10% FCS (STEMCELL Technologies), 50 µM β-mercaptoethanol (Gibco), 10 ng/ml Flt3L (PeproTech), 10 ng/ml IL-7 (PeproTech), and 100 ng/ml SCF (PeproTech). Cells were transduced either by spinfection at 700 g and 20°C for 45 min with 8 µg/ml Polybrene (lineage-depleted bone marrow) or by addition of the virus with 4 µg/ml Polybrene (sorted progenitors) and incubated for a further 72 h. Cells were washed and replated at 3 × 104 cells/ml on OP9 stromal cells in α-MEM supplemented with 20% FCS, β-mercaptoethanol, and 30 ng/ml IL-15 (PeproTech). Cells were cultured for a further 7 d, with a complete medium change at day 4.
ChIP.
Regulatory regions of Eomes and Id2 were searched for putative E4bp4-binding sites using MatInspector (Genomatix) matrix V$E4BP4.01 nrttayGTAAyu. MNK-1 cells (originating from the laboratory of D.S.J. Allan and J.R. Carlyle) were maintained in DMEM supplemented with 10% FCS (Gibco), 50 µM β-mercaptoethanol, 1 mM sodium pyruvate, and 10 ng/ml IL-2 (PeproTech). Lentivirus was produced by transfecting 293T cells with pCSGW or pCSGW–5′ FLAG–E4bp4 and the packaging plasmids psPAX2 and pMD2.G (deposited at Addgene as plasmids p12259 and p12260, respectively, by D. Trono, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland). MNK-1 cells were transduced by spinfection at 700 g and 20°C for 45 min with 8 µg/ml Polybrene and cultured for a further 48 h before cross-linking with 1% formaldehyde and shearing on a Diagenode sonicator (5 × 30-s pulses). Protein–DNA complexes were precleared on protein G–coupled agarose beads (EMD Millipore) before immunoprecipitation with IgG (EMD Millipore), M2 antibody to FLAG (Sigma-Aldrich), or polyclonal antibodies E4bp4 C18 or E16 (Santa Cruz Biotechnology, Inc.). For each sample, 106 cell equivalents of chromatin were incubated with 10 µg antibody. Protein G–coupled agarose beads were used to isolate immune complexes, and cross-links were reversed by heating (65°C, 4 h), followed by RNase A and proteinase K treatment. DNA was purified on spin columns (EMD Millipore) and amplified using ABsolute Blue qPCR SYBR Green master mix (Thermo Fisher Scientific) and primers designed to recognize putative E4bp4-binding regions (Table S2). The identity of the products was confirmed by running on an agarose gel.
Statistical testing.
Differences between populations were detected using the Mann–Whitney U Test. Where the number of data points was insufficient for comparison using nonparametric statistics, no indication of significance is shown.
Online supplemental material.
Tables S1 and S2 show candidate targets of E4bp4 identified by microarray and primers used to amplify putative E4bp4-binding sites in Eomes and Id2 regulatory regions. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20132398/DC1.
Supplementary Material
Acknowledgments
We thank J. Srivastava, G. Barton, P. Mealyer, A. Williams, S. Sesay, M. Caulfield, R. Subramanian, B. Seddon, A. Badamchi, and B. Pennycook for technical assistance.
This work was supported by Medical Research Council Grant G0901737 (to H.J.M. Brady) and U117597139 (to A. Wack). Production of MNK-1 cells was supported by Canadian Institutes of Health Research grant 106491 (to J.R. Carlyle).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- CLP
- common lymphoid progenitor
- iNK
- immature NK
- LMPP
- lymphoid-primed multipotent progenitor
- mNK
- mature NK
- NKP
- NK progenitor
References
- Adolfsson J., Månsson R., Buza-Vidas N., Hultquist A., Liuba K., Jensen C.T., Bryder D., Yang L., Borge O.-J., Thoren L.A.M., et al. 2005. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: A revised road map for adult blood lineage commitment. Cell. 121:295–306 10.1016/j.cell.2005.02.013 [DOI] [PubMed] [Google Scholar]
- Aliahmad P., de la Torre B., Kaye J. 2010. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue–inducer cell and NK cell lineages. Nat. Immunol. 11:945–952 10.1038/ni.1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos M.D., Yokota Y., Eberl G., Kee B.L. 2007. Mature natural killer cell and lymphoid tissue–inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med. 204:1119–1130 10.1084/jem.20061959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotta S., Pang S.H., Nutt S.L., Belz G.T. 2011. Identification of the earliest NK-cell precursor in the mouse BM. Blood. 117:5449–5452 10.1182/blood-2010-11-318956 [DOI] [PubMed] [Google Scholar]
- Fathman J.W., Bhattacharya D., Inlay M.A., Seita J., Karsunky H., Weissman I.L. 2011. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood. 118:5439–5447 10.1182/blood-2011-04-348912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth M.A., Madera S., Beaulieu A.M., Gasteiger G., Castillo E.F., Schluns K.S., Kubo M., Rothman P.B., Vivier E., Sun J.C. 2013. Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J. Exp. Med. 210:2981–2990 10.1084/jem.20130417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne D.M., Long E., Veiga-Fernandes H., de Boer J., Williams O., Seddon B., Coles M., Kioussis D., Brady H.J. 2009. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10:1118–1124 10.1038/ni.1787 [DOI] [PubMed] [Google Scholar]
- Gordon S.M., Chaix J., Rupp L.J., Wu J., Madera S., Sun J.C., Lindsten T., Reiner S.L. 2012. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 36:55–67 10.1016/j.immuni.2011.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K., Matsuyama S., Miyake S., Suga K., Nakachi K. 2000. Natural cytotoxic activity of peripheral-blood lymphocytes and cancer incidence: An 11-year follow-up study of a general population. Lancet. 356:1795–1799 10.1016/S0140-6736(00)03231-1 [DOI] [PubMed] [Google Scholar]
- Kaisho T., Tsutsui H., Tanaka T., Tsujimura T., Takeda K., Kawai T., Yoshida N., Nakanishi K., Akira S. 1999. Impairment of natural killer cytotoxic activity and interferon γ production in CCAAT/enhancer binding protein γ–deficient mice. J. Exp. Med. 190:1573–1582 10.1084/jem.190.11.1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizono S., Duncan G.S., Seidel M.G., Morimoto A., Hamada K., Grosveld G., Akashi K., Lind E.F., Haight J.P., Ohashi P.S., et al. 2009. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 206:2977–2986 10.1084/jem.20092176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada M., Pham N.L., Pewe L.L., Harty J.T., Rothman P.B. 2011. NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood. 117:6193–6197 10.1182/blood-2010-07-295873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672 10.1016/S0092-8674(00)80453-5 [DOI] [PubMed] [Google Scholar]
- Lacorazza H.D., Miyazaki Y., Di Cristofano A., Deblasio A., Hedvat C., Zhang J., Cordon-Cardo C., Mao S., Pandolfi P.P., Nimer S.D. 2002. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 17:437–449 10.1016/S1074-7613(02)00422-3 [DOI] [PubMed] [Google Scholar]
- Lodolce J.P.D.L., Boone D.L., Chai S., Swain R.E., Dassopoulos T., Trettin S., Ma A. 1998. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 9:669–676 10.1016/S1074-7613(00)80664-0 [DOI] [PubMed] [Google Scholar]
- Lohoff M., Duncan G.S., Ferrick D., Mittrücker H.W., Bischof S., Prechtl S., Röllinghoff M., Schmitt E., Pahl A., Mak T.W. 2000. Deficiency in the transcription factor interferon regulatory factor (IRF)-2 leads to severely compromised development of natural killer and T helper type 1 cells. J. Exp. Med. 192:325–336 10.1084/jem.192.3.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Male V., Nisoli I., Gascoyne D.M., Brady H.J. 2012. E4BP4: An unexpected player in the immune response. Trends Immunol. 33:98–102 10.1016/j.it.2011.10.002 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Heavey B., Rolink A.G., Busslinger M. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 401:556–562 10.1038/44076 [DOI] [PubMed] [Google Scholar]
- Ohno T., Onishi Y., Ishida N. 2007. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 35:648–655 10.1093/nar/gkl868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orange J.S. 2006. Human natural killer cell deficiencies. Curr. Opin. Allergy Clin. Immunol. 6:399–409 10.1097/ACI.0b013e3280106b65 [DOI] [PubMed] [Google Scholar]
- Ramirez K., Chandler K.J., Spaulding C., Zandi S., Sigvardsson M., Graves B.J., Kee B.L. 2012. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 36:921–932 10.1016/j.immuni.2012.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin L.C., Groom J.R., Chopin M., Herold M.J., Walker J.A., Mielke L.A., McKenzie A.N., Carotta S., Nutt S.L., Belz G.T. 2013. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14:389–395 10.1038/ni.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosmaraki E.E., Douagi I., Roth C., Colucci F., Cumano A., Di Santo J.P. 2001. Identification of committed NK cell progenitors in adult murine bone marrow. Eur. J. Immunol. 31:1900–1909 [DOI] [PubMed] [Google Scholar]
- Samson S.I., Richard O., Tavian M., Ranson T., Vosshenrich C.A., Colucci F., Buer J., Grosveld F., Godin I., Di Santo J.P. 2003. GATA-3 promotes maturation, IFN-γ production, and liver-specific homing of NK cells. Immunity. 19:701–711 10.1016/S1074-7613(03)00294-2 [DOI] [PubMed] [Google Scholar]
- Sciumé G., Hirahara K., Takahashi H., Laurence A., Villarino A.V., Singleton K.L., Spencer S.P., Wilhelm C., Poholek A.C., Vahedi G., et al. 2012. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 209:2331–2338 10.1084/jem.20122097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki S., Nakajima S., Ichikawa E., Saito T., Hida S. 2005. IFN regulatory factor-2 deficiency revealed a novel checkpoint critical for the generation of peripheral NK cells. J. Immunol. 174:6005–6012 [DOI] [PubMed] [Google Scholar]
- Townsend M.J., Weinmann A.S., Matsuda J.L., Salomon R., Farnham P.J., Biron C.A., Gapin L., Glimcher L.H. 2004. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity. 20:477–494 10.1016/S1074-7613(04)00076-7 [DOI] [PubMed] [Google Scholar]
- Velardi A., Ruggeri L., Mancusi A. 2012. Killer-cell immunoglobulin-like receptors reactivity and outcome of stem cell transplant. Curr. Opin. Hematol. 19:319–323 10.1097/MOH.0b013e32835423c3 [DOI] [PubMed] [Google Scholar]
- Yokota Y., Mansouri A., Mori S., Sugawara S., Adachi S., Nishikawa S., Gruss P. 1999. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 397:702–706 10.1038/17812 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.