Abstract
Receptor tyrosine kinases (RTKs) are co-deregulated in a majority of glioblastoma (GBM), the most common and most deadly brain tumor. We show that the RTKs MET, EGFR, and PDGFR regulate microRNA-134 (miR-134) in GBM. We find that miR-134 is downregulated in human tumors and cancer stem cells and that its expression inversely correlates with the activation of MET, EGFR, and PDGFR. We demonstrate that miR-134 inhibits cancer cell and stem-cell proliferation, survival, and xenograft growth, as well as cancer stem-cell self-renewal and stemness. We identify KRAS and STAT5B as targets of miR-134, and establish molecular and functional links between RTKs, miR-134, KRAS/STAT5B and malignancy in vitro and in vivo. We show that miR-134 induction is required for the anti-tumor effects of RTK inhibitors. We also uncover the molecular pathways through which RTKs regulate miR-134 expression and demonstrate the involvement of MAPK signaling and the KLF4 transcription factor. We therefore identify miR-134 as a novel RTK-regulated tumor-suppressive hub that mediates RTK and RTK-inhibitor effects on GBM malignancy by controlling KRAS and STAT5B.
Keywords: glioblastoma, miR-134, MET, receptor tyrosine kinases, KLF4
microRNAs (miRNAs) regulate a wide variety of physiological and pathological processes.1, 2, 3 miRNAs modulate protein expression by binding to the 3′ untranslated region (3′UTR) of target mRNA and promoting RNA degradation and/or inhibiting translation. Single miRNAs can regulate multiple molecules, highlighting a powerful mechanism for the regulation of redundant and cross-talking signal transduction pathways.4 miRNA dysregulation is a common feature of neoplasia and numerous miRNAs have been characterized as oncogenes or tumor suppressors in many cancers including in glioblastoma (GBM).3, 5, 6, 7, 8, 9, 10, 11
microRNA-134 (miR-134) has been implicated in the regulation of physiological and developmental processes. It was shown to promote mouse embryonic stem-cell differentiation, dendritogenesis, and stage-specific cortical development.12, 13, 14, 15, 16, 17 miR-134 is downregulated by SIRT1 and involved in synaptic plasticity and memory formation.18 miR-134 expression is regulated by MEF2 in dendritogenesis14 and GBM.19
GBM is the most common and most deadly primary malignant brain tumor.20, 21 The Cancer Genome Atlas and other studies have shown that aberrant expression or activation of the receptor tyrosine kinases (RTKs) EGFR, MET, and PDGFR occurs in a majority of GBM and correlates with poor prognosis.22 Importantly, multiple RTKs are co-activated in the same GBM tumors and tumor cells.23, 24 KRAS, STAT3, and STAT5 are activated by RTKs and mediate their oncogenic effects.25, 26, 27, 28 The failure of current treatments for GBM is arguably due to the presence in the tumors of GBM stem cells (GSCs) that are resistant to radio- and chemo-therapy and capable of maintaining and propagating the tumors.29, 30, 31 EGFR, MET, PDGFR, and miRNAs have been implicated in the regulation of GSC functions.32, 33, 34, 35, 36, 37, 38, 39, 40
This study identifies for the first time miR-134 as a critical RTK-regulated tumor suppressive hub that targets KRAS and STAT5B and mediates RTK and RTK inhibitor effects on GBM malignancy.
Results
MET regulates the expression of a subset of miRNAs in GBM
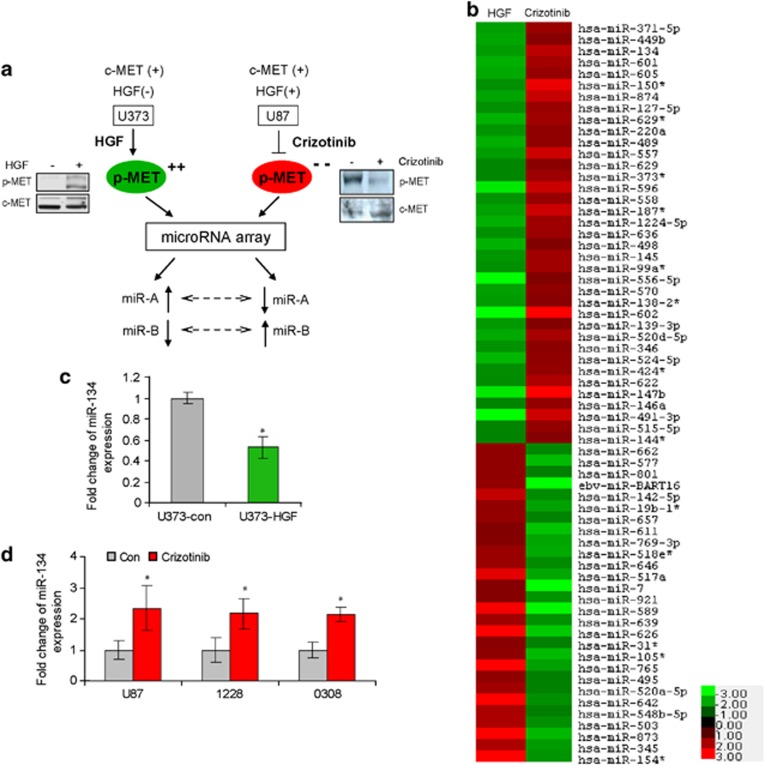
To identify miRNAs that are regulated by MET, we assessed the effects of MET activation and MET inhibition on global miRNA expression using miRNA microarrays. We activated MET in U373 cells that have low MET activation by treatment with the MET ligand HGF and inhibited MET in U87 cells that have high MET activation by treatment with the small molecule kinase inhibitor PF-2341066 (Crizotinib). We then assessed the changes in miRNA expression using miRNA microarrays and searched for common miRNAs that are oppositely regulated in the two cell lines (Figure 1a). This stringent approach was used to minimize false positive changes. We identified 65 miRNAs that were oppositely regulated at >1.8-fold, P<0.05 (Figure 1b). We focused our further studies on miR-134 because it was among the most changed after MET activation/inhibition and because it has been shown to have an important regulatory role in embryonal stem cells and brain development. Using quantitative RT-PCR (qRT-PCR), we confirmed that MET activation inhibits while MET inhibition induces miR-134 in the GBM cells used for the microarrays as well as in GSCs 0308 and 1228 (Figures 1c and d).
Figure 1.
MET regulates the expression of a subset of miRNAs that includes miR-134. (a) Schematic diagram of the strategy used to identify MET-regulated miRNAs using miRNA microarrays. GBM cells U373 (HGF negative) and U87 (HGF positive) were treated with HGF or Crizotinib, respectively. MET activation in U373 and inhibition in U87 were verified by immunoblotting. RNA from the cells was subjected to microarray hybridization and common miRNAs that were oppositely regulated (dotted arrows) were identified. (b) Heatmap of oppositely changed miRNAs. (c) qRT-PCR confirmation of miR-134 downregulation in HGF-treated cells. (d) qRT-PCR confirmation of miR-134 upregulation in Crizotinib-treated cells and stem cells (1228, 0308). *P<0.05
miR-134 is also regulated by the RTKs EGFR and PDGFR
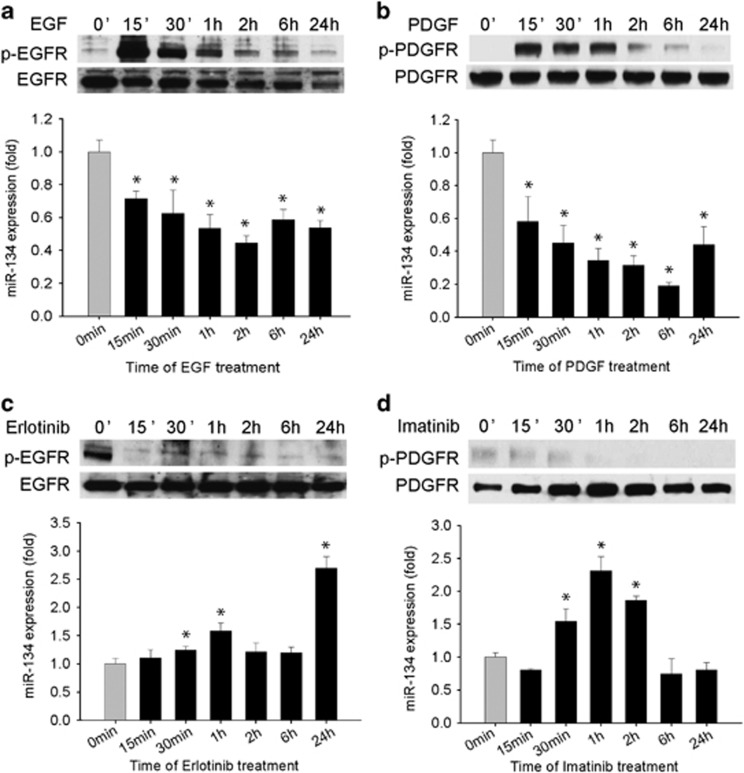
Since MET and other RTKs crosstalk and share downstream signaling pathways, we hypothesized that miR-134 might also be regulated by other RTKs. To test this hypothesis, we assessed the effects of activation or inhibition of EGFR and PDGFR on miR-134 expression levels in GBM cells. EGFR and PDGFR activation by treatment of the cells with EGF and PDGF significantly repressed miR-134 expression by 55 and 81%, respectively (P<0.05) (Figures 2a and b). Conversely, inhibition of EGFR or PDGFR by the small molecule kinase inhibitors Erlotinib or Imatinib significantly upregulated miR-134 by 170 and 131%, respectively (P<0.05) (Figures 2c and d). EGFR and PDGFR activation and inhibition were verified by immunoblotting (Figure 2). The above data indicate that miR-134 is also regulated by EGFR and PDGFR. Interestingly, we found that miR-134 inhibits MET and EGFR phosphorylation (Supplementary Figure 1), but not PDGFR activation (data not shown) suggesting the existence of a feedback loop between miR-134 and select RTKs in GBM.
Figure 2.
miR-134 is also regulated by EGFR and PDGFR. (a and b) GBM U373 cells were treated with EGF (a) or PDGF (b) for different times and assessed for miR-134 expression by qRT-PCR. EGFR and PDGFR activations were verified by immunoblotting for p-EGFR and p-PDGFR. The results show that miR-134 expression is inhibited by EGFR and PDGFR activation. (c and d) GBM cells were treated with the EGFR inhibitor Erlotinib (c) or the PDGFR inhibitor Imatinib (d) for different times and assessed for miR-134 by qRT-PCR. EGFR and PDGFR inhibitions were verified by immunoblotting for p-EGFR and p-PDGFR. The results show that miR-134 expression is induced by inhibition of EGFR and PDGFR. *P<0.05
miR-134 expression is downregulated in GBM cells, GSCs, and GBM tumors, where expression inversely correlates with MET, EGFR, and PDGFR activation levels
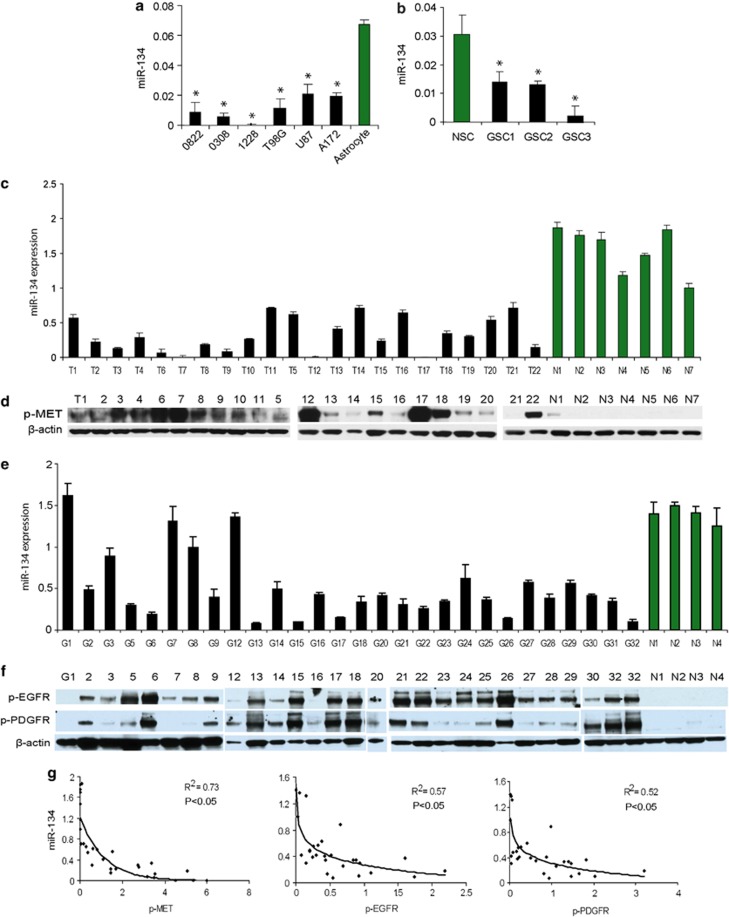
Since MET, EGFR, and PDGFR are frequently overactivated in GBM and since they negatively regulate miR-134, we speculated that miR-134 might be downregulated in these tumors. To determine whether that is true, we measured miR-134 levels in GBM cells, GSCs, and human tumor specimens as well as in normal human astrocytes, neural stem cells (NSCs), and normal brain using qRT-PCR. Furthermore, we analyzed the correlation between miR-134 expression and phospho-MET (p-MET), phospho-EGFR (p-EGFR), and phospho-PDGFR (p-PDGFR) levels in GBM specimens measured by quantitative immunoblotting. We found that miR-134 was significantly lower in GBM cells and GSCs as compared with astrocytes and NSCs (P<0.05) (Figures 3a and b) and significantly lower in GBM tumors as compared with normal brain (P<0.05) (Figures 3c and e). We also found a statistically significant inverse correlation between miR-134 expression and p-MET, p-EGFR, and p-PDGFR in human GBM specimens and normal brain tissues (Figures 3d–g). These results show that miR-134 is downregulated in GBM and GSC and suggest that MET, EGFR, and PDGFR are important regulators of miR-134.
Figure 3.
miR-134 expression is downregulated in GBM cells, GSCs, and human tumors, where it inversely correlates with RTK activation levels. (a) qRT-PCR of miR-134 in GBM cells and GSCs showing lower levels of expression as compared with normal human astrocytes. (b) Quantitative RT-PCR of miR-134 in GSCs showing lower levels of expression as compared with NSCs. (c) Quantitative RT-PCR of miR-134 in GBM surgical specimens (T) showing lower levels of miR-134 than in normal brain (N). (d) Immunoblots showing p-MET levels in the same GBM specimens and normal brains as in (c). (e) Quantitative RT-PCR of miR-134 in GBM surgical specimens (G) showing lower levels of miR-134 than in normal brains (N). (f) Immunoblots showing p-EGFR and p-PDGFR levels in the same GBM specimens and normal brains as in (e). (g) Correlations between miR-134 and p-MET (left panel), miR-134 and p-EGFR (middle panel), and miR-134 and p-PDGFR (right panel) levels in GBM and normal brain tissues from (c)/(d) and (e)/(f), respectively. Correlation coefficient R values between miR-134 and p-MET, p-EGFR, and p-PDGFR were R=0.875 (P<0.05), R=0.754 (P<0.05), and R=0.721 (P<0.05), respectively. *P<0.05
miR-134 directly targets and regulates KRAS and STAT5B in GBM
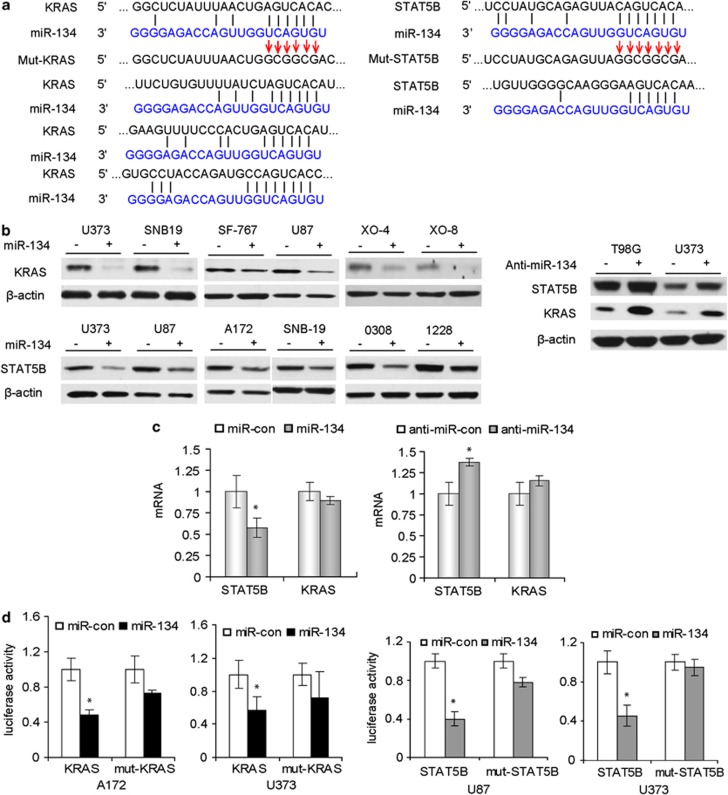
To identify mRNA targets of miR-134 in GBM, we first used bioinformatic databases (Targetscan, Pictar, RNhybrid) and identified several potential targets of miR-134 including KRAS and STAT5B (Figure 4a). To verify whether miR-134 downregulates KRAS and STAT5B proteins, we determined the effects of miR-134 overexpression and inhibition on KRAS and STA5B proteins in GBM cells and GSCs with immunoblotting. miR-134 significantly reduced KRAS and STAT5B proteins in all tested cells (Figure 4b). Conversely, miR-134 inhibition by anti-miR-134 in U373 and T98G cells, which express relatively high levels of endogenous miR-134, increased the protein levels of KRAS and STAT5B (Figure 4b). We also assessed the effects of miR-134 overexpression and inhibition on KRAS and STAT5B mRNA levels with qRT-PCR. We found that miR-134 overexpression slightly downregulates STAT5B mRNA, while anti-miR-134 transfection slightly enhances STAT5B mRNA (Figure 4c). miR-134 did not affect KRAS mRNA levels (Figure 4c). These data suggest that miR-134 reduces the levels of KRAS and STAT5B primarily via inhibition of translation and partly via mRNA degradation. We also found that miR-134 levels inversely correlated with STAT5B expression in human GBM specimens (R=0.52, P=0.016) (Supplementary Figure 2). We also tested other predicted targets of miR-134 including RAB27 and RAN, but could not experimentally verify them (data not shown). To determine whether KRAS and STAT5B are direct targets of miR-134, KRAS and STAT5B 3′UTR and mutant reporter vectors were constructed and transfected into GBM cells. Ectopic expression of miR-134 (confirmed by q-PCR; data not shown) significantly decreased luciferase activity in experimental but not in mutant or control-transfected cells (Figure 4d). Altogether, these data show that miR-134 inhibits KRAS and STAT5B by directly binding to their 3′UTRs.
Figure 4.
miR-134 regulates KRAS and STAT5B expressions by directly binding to their mRNA 3′UTRs. (a) Alignment of KRAS, STAT5B 3′UTRs and miR-134 sequences showing the predicted binding sites. The sites of targeted mutagenesis for the generation of mutant controls are indicated by the arrows. (b) Immunoblots (left panel) showing the effects of miR-134 overexpression on the protein levels of KRAS and STAT5B in GBM cells and GSCs. Immunoblots (right panel) showing the effects of miR-134 inhibition with anti-miR-134 on the protein levels of KRAS and STAT5B in GBM cells. (c) Quantitative RT-PCR showing the effects of miR-134 and anti-miR-134 on the mRNA levels of STAT5B and KRAS. (d) 3′UTR luciferase assays for KRAS and STAT5B showing the inhibition of luciferase activity by miR134 in GBM cells. No significant alteration in luciferase activity was observed when 3′UTR binding site was mutated. *P<0.05
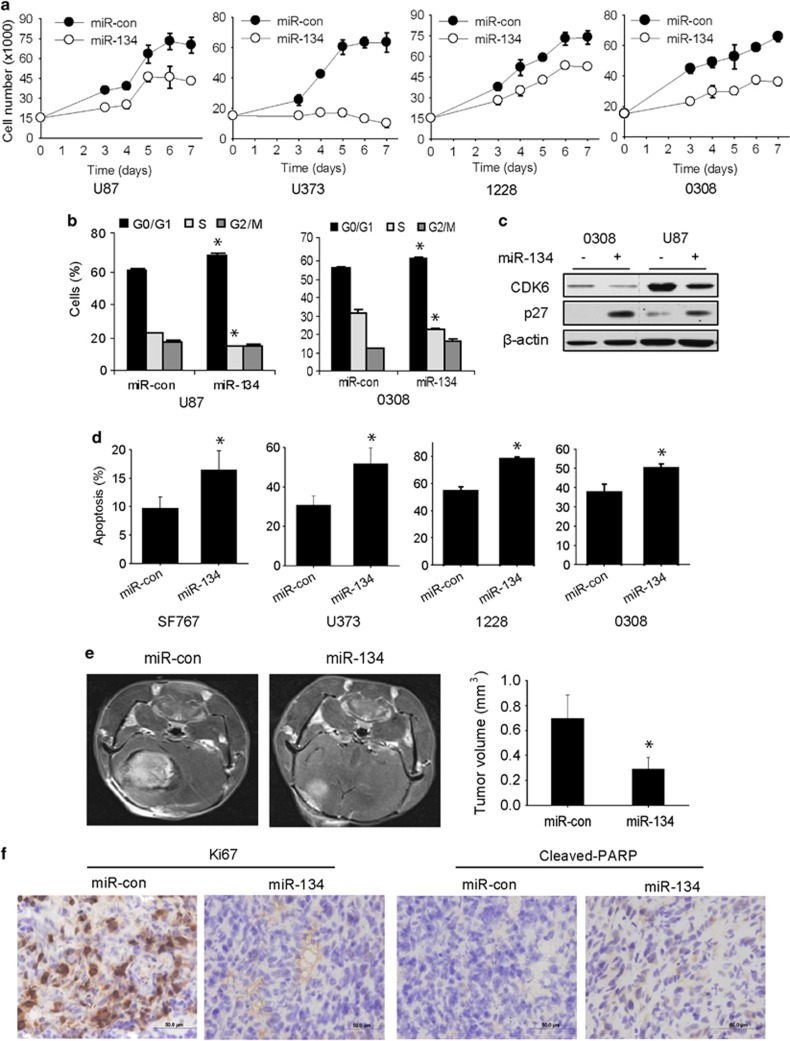
miR-134 inhibits GBM cell proliferation, induces cell-cycle arrest and apoptosis, and inhibits in vivo GSC-derived xenograft growth
We next assessed the effects of miR-134 on cell growth and survival in GBM cells and GSCs. Overexpression of miR-134 significantly inhibited the proliferation of GBM cells and GSCs (P<0.05) (Figure 5a). miR-134 also induced G1/S arrest in GBM cells and GSCs (Figure 5b) and changed the expressions of the cell-cycle regulatory proteins CDK6 and p27, which are known to be regulated by KRAS in GBM (Figure 5c). Moreover, AnnexinV-7AAD flow cytometry assays revealed that miR-134 significantly induced apoptosis and cell death in all cells (P<0.05) (Figure 5d). To determine whether miR-134 suppresses in vivo tumor growth, GSC 1228 was transfected with pre-miR-134 or control miRNA and implanted into the brains of immunodeficient mice (n=6). Tumor sizes were measured using MRI 4 weeks after implantation. GSC-derived xenografts overexpressing miR-134 were significantly smaller than control xenografts (Figure 5e) (P<0.05). Immunohistochemical analysis of the xenografts revealed downregulation of the proliferation marker Ki67 and upregulation of the apoptosis marker cleaved PARP in miR-134-overexpressing xenografts (Figure 5f). These data show that miR-134 exerts tumor suppressive effects in GBM.
Figure 5.
miR-134 inhibits GBM cell and GSC proliferation, survival, and in vivo tumor growth. (a) Proliferation assay showing the inhibition of GBM cell and GSC proliferation by miR-134 transfection. (b) Flow-cytometric cell-cycle analysis showing cell-cycle arrest induced by miR-134 transfection in GBM cells and GSCs (left panel). (c) Immunoblot analysis of cell-cycle regulatory proteins in response to miR-134 transfection into GBM cells and GSCs (right panel). (d) AnnexinV-PE and 7-AAD flow-cytometric analysis of GBM cells and GSCs showing induction of apoptosis after miR-134 transfection. (e) GSC 1228 was transfected with pre-miR-134 or pre-miR-con and implanted into the brains of immunodeficient mice (n=6). After 4 weeks, the mice were subjected to MRI and tumor volume calculation. The results show that miR-134 inhibits in vivo GBM xenograft growth. (f) Immunohistochemical staining of xenograft sections from (e) for the proliferation marker Ki67 and the apoptotic marker cleaved-PARP showing significantly reduced Ki67 and increased cleaved-PARP in miR-134 overexpressing xenografts (section at × 400 magnification). *P<0.05
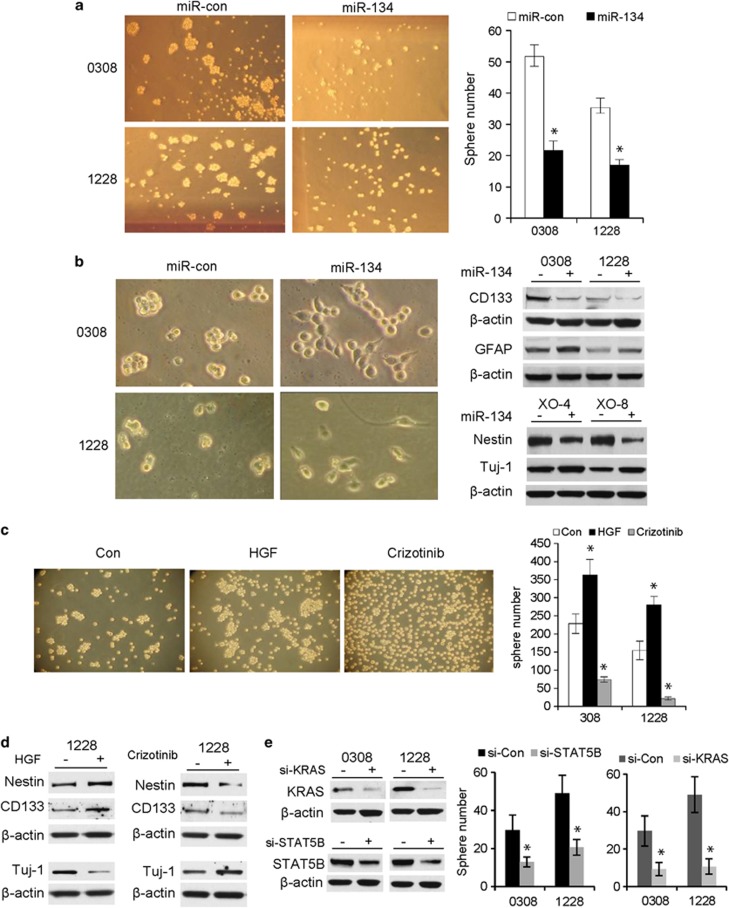
miR-134, MET, KRAS, and STAT5B regulate GSC neurosphere formation and differentiation
Since miR-134 has been associated with mouse embryonal stem-cell biology, we speculated that it might also regulate GSC functions. We therefore studied the effect of miR-134 and one of its RTK regulators (MET) on GSC neurosphere formation and differentiation. Overexpression of miR-134 resulted in a significant reduction in neurosphere number and size in GSCs 1228 and 0308 (Figure 6a). We observed that miR-134 transfection into GSCs 0308, 1228, XO-4, and XO-8 induced the cells to dissociate from the neurospheres, attach to the bottom of cell-culture plates and spread (Figure 6b), suggesting that the stem cells were undergoing differentiation. miR-134 overexpression inhibited the expressions of stem-cell/progenitor markers CD133 and nestin and induced the expressions of the differentiation markers GFAP (astrocytes) and Tuj1 (neurons) (Figure 6b). The above data suggest that miR-134 inhibits GSC self-renewal and induces GSC differentiation. Since MET regulates miR-134, we also assessed the effects of MET activation or inhibition on neurosphere formation and GSC differentiation. We activated MET with HGF or inhibited it with Crizotinib and assessed GSC sphere formation and differentiation as described above. MET activation enhanced while MET inhibition reduced GSC neurosphere formation (Figure 6c). MET activation induced the expressions of stem-cell markers and MET inhibition reduced the expression of differentiation markers (Figure 6d). Conversely, MET inhibition reduced the expressions of stem-cell markers and induced the expression of differentiation markers (Figure 6d). Since miR-134 regulates GSC sphere formation and directly targets KRAS and STAT5B, we also determined the role of KRAS and STAT5B in GSC sphere formation. Knockdown of KRAS and STAT5B expressions with siRNA significantly inhibited GSC neurosphere formation (Figure 6e). The above data show that miR-134, its regulator MET, and its targets KRAS and STAT5B regulate GSC self-renewal and differentiation.
Figure 6.
miR-134 overexpression and MET inhibition repress neurosphere formation and induce stem-cell differentiation. (a) Neurosphere formation assay in response to miR-134 transfection in GSCs. The data show that miR-134 reduces the number and size of GSC neurospheres (left panel shows photomicrographs of neurospheres; right panel shows quantification of the previous). (b) Immunoblots showing the effects of miR-134 on GSC stem-cell markers CD133 and nestin and differentiation markers GFAP and Tuj1. (c) Neurosphere formation assay in response to MET activation with HGF or inhibition with Crizotinib in GSCs. The data show that HGF induces and Crizotinib inhibits neurosphere formation (left panel shows photomicrographs of neurospheres; right panel shows quantification of the previous). (d) Immunoblots showing the effects of MET activation with HGF or inhibition with Crizotinib on GSC stem-cell and differentiation markers. (e) Neurosphere formation assay showing that siRNA-mediated knockdown of KRAS and STAT5B reduces GSC neurosphere formation (left panel shows siRNA transfection efficiency by immunoblots and right panel shows quantification of the neurosphere numbers). *P<0.05
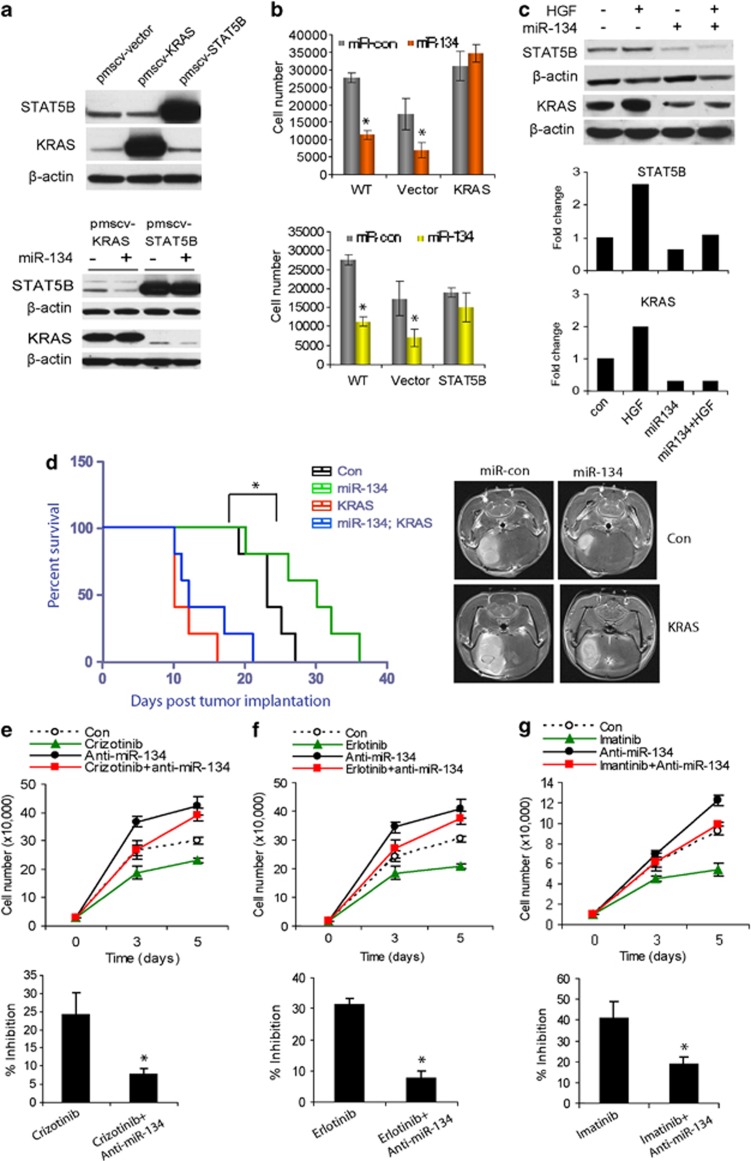
KRAS and STAT5B mediate the effects of miR-134 on GBM cell proliferation and in vivo xenograft growth
To determine whether the tumor suppressive effects of miR-134 are mediated by KRAS or STAT5B, we constructed KRAS and STAT5B cDNA plasmids that lack the 3′UTRs and thus cannot be inhibited by miR-134 and used them to generate GBM clones that constitutively express KRAS or STAT5B (U373-KRAS and U373-STAT5B). KRAS and STAT5B expressions were confirmed by immunoblotting (Figure 7a). miR-134 overexpression had no effect on KRAS and STAT5B in these cells as confirmed by immunoblotting (Figure 7a). The effects of miR-134 on proliferation were subsequently determined in the cells. Overexpression of miR-134 significantly reduced cell numbers in wild-type and control-transfected cells, but not in KRAS or STAT5B expressing cells (Figure 7b, P<0.05). To determine whether the tumor suppressive effects of miR-134 are mediated by KRAS in vivo, we generated the GBM U87 lines that constitutively express KRAS as described above (U373 cells do not form tumors in vivo). The cells were transfected with pre-miR-134 or control and implanted into the brains of immunodeficient mice (n=5) and animal survival was assessed. The tumors were also visualized by MRI 2 weeks post tumor implantation. miR-134 induced a significant prolongation of animal survival in animals implanted with control U87 xenografts (P<0.05) but not in animals implanted with U87-KRAS (Figure 7d). These data suggest that the tumor suppressive effects of miR-134 in GBM are mediated by KRAS and STAT5B inhibition.
Figure 7.
KRAS and STAT5B mediate the effects of miR-134 and miR-134 mediates the effects of RTKs on KRAS, STAT5B, cell proliferation, and in vivo xenograft growth. (a) Immunoblots showing expressions of KRAS and STAT5B in GBM cells stably transfected with respective cDNAs lacking the 3′UTR sequences (upper panel), and lack of inhibition of KRAS and STAT5B expressions by miR-134 in the same cells (lower panel). (b) Proliferation assay showing the effects of miR-134 in the clones overexpressing KRAS and STAT5B described in (a). The results show rescue of the effects of miR-134 in the cells. (c) Immunoblots and immunoblot quantifications of KRAS or STAT5B in response to HGF treatment with or without miR-134 overexpression. The data show that miR-134 partially rescues the effects of HGF on KRAS and STAT5B. (d) KRAS expressing stable cells (U87-KRAS) or U87-Control were transfected with pre-miR-134 or pre-miR-con and implanted into the brains of immunodeficient mice (n=5) and animal survival was assessed. The results show that miR-134 significantly enhances animal survival in control but not in KRAS-overexpressing tumor-bearing animals (left panel). *P<0.05. The right panel shows representative MRIs with tumors at 2 weeks post implantation. (e–g) Proliferation assay of GBM cells in response to RTK inhibitors, Crizotinib (e), Erlotinib (f), and Imatinib (g), with or without anti-miR-134 pre-treatment. The data show that anit-miR-134 partially rescues the effects of the inhibitors on cell proliferation. *P<0.05
miR-134 mediates the effects of MET on KRAS and STAT5B
To determine whether miR-134 mediates the effects of MET on KRAS and STAT5B, we assessed the effects of MET activation on KRAS and STAT5B in the setting of overexpressed miR-134. We transfected GBM cells with pre-miR-134 or pre-miR-control before treating them with HGF and measuring the expressions of KRAS and STAT5B by immunoblotting (Figure 7c). KRAS and STAT5B expressions were induced by HGF stimulation in pre-miR-con transfected cells. Induction of KRAS and STAT5B by MET was reduced in miR-134 transfected cells (Figure 7c). These data show for the first time that MET activation induces KRAS and STAT5B protein expressions in GBM and indicate that this induction is partly mediated by miR-134 downregulation.
The anti-tumor effects of RTK inhibitors require miR-134 upregulation
Since RTK inhibitors induce the expression of the tumor suppressive miR-134 (Figure 2), we speculated that miR-134 upregulation might mediate the effects of the inhibitors. We performed rescue experiments to test whether anti-miR-134 pre-treatment is able to reduce the effects of the small molecule kinase inhibitors Crizotinib (MET), Erlotinib (EGFR), and Imatinib (PDGFR) on GBM cell growth. We found that Criziotinb-induced inhibition of GBM cell proliferation was reduced by 67.8% (P<0.05) in cells that were pre-treated with anti-miR-134 (Figure 7e). Similarly, pre-treatment with anti-miR-134 reduced the anti-proliferative effects of Erlotinib and Imatinib by 75.6% (P<0.05) (Figure 7f) and 53.9% (P<0.05) (Figure 7g), respectively. These data indicate that miR-134 mediates the effects of RTKs in GBM and that its upregulation is required for the anti-tumor effects of RTK inhibitors.
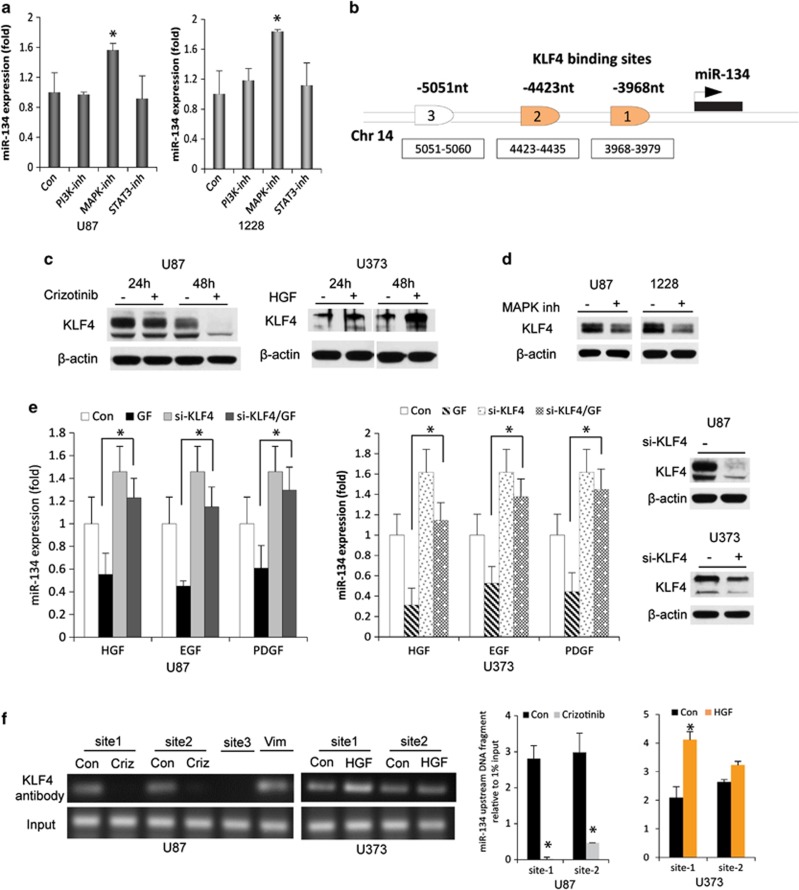
RTKs regulate miR-134 via MAPK and the transcription factor KLF4
To determine the mechanism of miR-134 regulation by RTKs, we first investigated the involvement of the three main pathways that mediate RTK effects (MAPK, PI3K, and JAK/STAT). We treated GBM cells and GSCs with the PI3K inhibitor LY294002, the MAPK inhibitor PD98059, or the STAT3 inhibitor S3I-201 and measured the expression of miR-134. MAPK inhibition led to a significant upregulation of miR-134 levels but PI3K and STAT inhibitions did not affect miR-134 expression (Figure 8a). These data suggest that RTKs regulate miR-134 mainly through MAPK signaling.
Figure 8.
RTKs regulate miR-134 expression via MAPK and KLF4. (a) GBM U87 and GSC 1228 cells were treated with inhibitors of PI3K, MAPK, and STAT3 and assessed for miR-134 expression by qRT-PCR. Only MAPK inhibition led to an upregulation of miR-134 expression in both cell lines. (b) KLF4 predicted binding sites in the putative miR-134 promoter. (c) Immunoblots showing the effect of MET inhibition and activation on KLF4 expression in GBM cells. (d) Immunoblots showing the downregulation of KLF4 expression after MAPK inhibition in GBM cells and GSCs. (e) Rescue experiments showing that siRNA-based KLF4 knockdown reverses the inhibitory effects on miR-134 expression of MET, EGFR, and PDGFR activations with their respective growth factor (GF) ligands in GBM cells (two left panels). Right panel shows immunoblots with KLF4 knockdown. (f) ChIP/qPCR showing the binding of KLF4 to two KLF4 binding sites (site 1 and site 2) in the putative miR-134 promoter in response to MET inhibition or activation with Crizotinib (criz) or HGF, respectively. Vimentin (Vim) was used as a positive control. Left panel shows ChIP/qPCR and right panel shows its quantification. *P<0.05
To identify potential transcription factors that might regulate miR-134 expression, we used the bioinformatics TESS database (http://www.cbil.upenn.edu/cgi-bin/tess/tess) to analyze the putative miR-134 promoter region for transcription factor consensus binding sites. Among several predicted transcription factors, only two (NF-kB and KLF4) are known to exert repressive activities and one (MEF2) has been previously shown to regulate miR-134 expression (Figure 8b). We therefore assessed the involvement of the above three transcription factors in mediating the repressive effects of the RTK MET on miR-134. We first determined whether these transcription factors are regulated by MET. We inhibited MET with Crizotinib or activated it with HGF treatment and determined the effects on NF-kB nuclear and cytoplasmic fractions, MEF2, KLF4 with immunoblotting. MET inhibition reduced KLF4 expression and MET activation increased KLF4 expression in GBM cells and GSCs (Figure 8c). Similar effects were observed for NF-kB nuclear expression (Supplementary Figure 3A and B). MET inhibition/activation did not affect MEF2 expression (not shown). These data suggest that KLF4 and NF-kB, but not MEF2, are regulated by MET activation. Additionally, we demonstrated that MAPK inhibition leads to inhibition of KLF4 in GBM cells and GSCs (Figure 8d).
We next sought to determine whether KLF4 and/or NF-kB regulate miR-134 expression and mediate the effects of RTKs. Knockdown of KLF4 with siRNA significantly enhanced miR-134 expression (P<0.05) and partially reversed the inhibitory effects of HGF, EGF and PDGF on miR-134 (P<0.05) (Figure 8e). Knockdown of NF-kB did not alter miR-134 expression (Supplementary Figure 3C). These data suggest that KLF4 but not NF-kB mediates the inhibitory effects of RTKs on miR-134.
As two of putative KLF4 binding sites were identified (3968–3979 nt and 4423–4435 nt) upstream of the miR-134 start codon, we next investigated MET-induced KLF4 binding to the putative miR-134 promoter using chromatin immunoprecipitation and qPCR (ChIP)/qPCR. Taking into account the predicted KLF4 binding sites, three chromatin regions were analyzed: two spanning the KLF4 binding sites and the third 5051 nt upstream of the miR-134 5′ end, as a negative control that does not contain a binding site for KLF4. The ChIP showed remarkable KLF4 binding at the putative KLF4 biding sites (Figure 8f). Importantly, this binding was significantly reduced by MET inhibition with Crizotinib and enhanced by MET activation with HGF (Figure 8f). No binding was observed in the control region. These data demonstrate that KLF4 binds to the putative miR-134 promoter in an MET-dependent manner.
Altogether, the data from this section show that RTKs regulate miR-134 via MAPK and KLF4.
Discussion
Our study uncovers for the first time miR-134 as a tumor suppressive miRNA that is downregulated by MET, EGFR and PDGFR via upregulation of the transcriptional repressor KLF4, and that mediates RTK effects and the anti-tumor effects of their inhibitors by regulating the expressions of KRAS and STAT5B (Supplementary Figure 4).
We find that miR-134 is downregulated in GBM cells as compared with normal astrocytes, in GSCs as compared with NSCs, and in tumor specimens as compared with normal brain. RTKs are activated in a majority of GBM, providing a mechanism for miR-134 downregulation in these tumors. We demonstrate an inverse correlation between miR-134 and p-MET, p-EGFR, and p-PDGFR in GBM specimens, suggesting that these RTKs are critical regulators of miR-134. Our data also show tumor suppressive effects of miR-134 in GBM cells and GSCs. We therefore propose miR-134 as a novel tumor suppressor that regulates GBM malignancy.
An important finding of our study is the regulation of miR-134 expression by the three most frequently activated RTKs in GBM. According to TCGA data, 88% of GBM harbor one or more functional mutations in RTK pathways. Transcriptional and autocrine activation of RTKs is even more frequent. EGFR amplification associates with worse prognosis and decreases overall GBM patient survival.41 Activation of MET in GBM enhances tumorigenicity, tumor growth, and tumor-associated angiogenesis.42 PDGF is a critical regulator of gliomagenesis and neurogenesis and regulates tumor cell proliferation, migration, and angiogenesis.43, 44 Our data identify miR-134 as a mediator of the effects of multiple RTKs by controlling KRAS and STAT5B. Interestingly, miR-134 also regulates MET and EGFR activation suggesting the existence of a feedback loop between RTKs and miR-134. Importantly, we also find that RTK inhibitor-induced miR-134 is required for the anti-tumor effects of these clinically applicable drugs. New and daunting findings indicate that even within the same GBM tumor, individual cells may have amplifications of different RTKs including EGFR, PDGFR, and MET.23, 24 This poses a major hurdle for a personalized medicine approach that aims at inhibiting RTK activation in GBM. Our data suggest that this hurdle might be potentially overcome by the use of the common RTK mediator miR-134.
The study uncovers MAPK and KLF4 as mediators of miR-134 regulation by RTKs. KLF4 is a zinc-finger transcription factor critical for maintaining self-renewal, survival, and multi-lineage differentiation potential of embryonic stem cell45, 46 and cancer stem cells, including GSCs.34, 47 Our experiments demonstrate that KLF4 mediates RTK-regulated miR-134 expression by binding to the miR-134 promoter region. These data highlight a new RTK-MAPK-KLF4-miR-134 signaling pathway that regulates GBM and GSC malignancy parameters. Another study previously implicated MEF2 in the regulation of miR-134 expression19 but RTK-dependent inhibition of miR-134 was not mediated by MEF2 in our system. However, miR-134 regulation by RTKs and other factors is likely to be complex and involve additional mechanisms than the ones described by us and others.
Oncogenic KRAS activates the RAS/MAPK pathway, a major cell survival pathway in cancers and GBM.48 KRAS mutations are relatively rare in GBM, but activation of the pathway is a common occurrence.49 Our study uncovers a new mechanism of KRAS regulation by RTKs. We show that RTKs not only activate KRAS but also regulate KRAS protein levels via inhibition of miR-134. Importantly, we also show that KRAS mediates the effects of miR-134 on GBM malignancy. RTKs signal through several effector arms, including JAK-STAT, which regulate cellular proliferation, cell scattering and migration, and cytokine stimulation in GBMs.21, 50 STAT5B is a transcription factor that has been reported to regulate cancer and GBM cell proliferation and apoptosis.27, 51 It has been shown that nuclear EGFRvIII-STAT5B complexes contribute to GBM cell survival by direct activation of the Bcl-XL promoter.52 However, a link between MET and STAT5B in GBM has not been established before. We show for the first time that STAT5B protein is regulated by MET via miR-134 and that STAT5B mediates the effects of MET and possibly also other RTKs in GBM.
Accumulating evidence implicates GSCs in GBM recurrence and resistance to therapy.29, 30 We show for the first time that miR-134 (similar to MET) regulates GSC self-renewal and differentiation. Differentiated GSCs lose long-term repopulation potential and fail to propagate tumors, suggesting that induction of GSC differentiation might be a practical strategy to eliminate the GSC population. This is consistent with our in vivo results showing that miR-134 inhibits GSC-derived xenograft growth. Together with the data obtained from GBM cells, these data indicate that miR-134 inhibits both GSCs and differentiated tumor cells.
Altogether, our study shows for the first time that RTKs converge on miR-134 via MAPK and KLF4 to regulate GBM cell and stem pathobiology by controlling KRAS and STAT5B. The data also establish and characterize miR-134 as a novel tumor suppressor miRNA.
Materials and Methods
Reagents
Human recombinant EGF and PDGF were from R&D Systems (Minneapolis, MN, USA). HGF was a kind gift from Genentech (South San Francisco, CA, USA). The MET inhibitor Crizotinib was from Pfizer (New York, NY, USA), the EGFR inhibitor Erlotinib was from Sigma (St. Louis, MO, USA), the PDGFR inhibitor Imatinib was from LC Laboratories (Woburn, MA, USA), the PI3K inhibitor LY294002 and MAPK inhibitor PD98059 were from LC Laboratories and STAT3 inhibitor S3I-201 was from Santa Cruz (Santa Cruz, CA, USA). Pre-miR-134, anti-miR-134, and the corresponding controls (pre-miR-con, anti-miR-con) were from Ambion (Grand Island, NY, USA). siRNA for KRAS, STAT5B, KLF4, NF-kB and control siRNA are from Life Technologies (Grand Island, NY, USA). Oligofectamine transfection reagent and Lipofectamine RNAimax were from Invitrogen (Grand Island, NY, USA). The Dual Luciferase kit was from Promega (Fitchburg, WI, USA). The Fugene 6 transfection reagent was from Roche (Pleasanton, CA, USA). The miScriptRNA Extraction kit, miScript Reverse Transcriptase kit, and miR-134 Primer Assay were from Qiagen (Chatsworth, CA, USA). All primers for realtime PCR were from Integrated DNA Technology (San Jose, CA, USA). Propidium iodide, Annexin V-PE, and 7 AAD were from BD Pharmingen (San Diego, CA, USA).
GBM cells, GSCs, and tumor specimens
GBM cells lines U87, U373, A172, T98G, SNB-19, and SF-767 were from ATCC repository. GSCs 1228 and 0308 (a kind gift from Dr. Jeongwu Lee, Cleveland Clinic) were isolated from patient surgical specimens and characterized for in vivo tumorigenesis, pluripotency, self-renewal, stem-cell markers, and neurosphere formation.53 GSCs XO-4 and XO-8 were isolated from GBM tumor specimens obtained after surgery from patients undergoing treatment at the University of Pittsburgh. Expression of stem-cell markers CD133 and Nestin was confirmed by immunoblotting and in vivo intracranial tumor formation was verified in our laboratory (data not shown). CD133-positive GSCs (GSC1, 2, and 3) isolated from GBM surgical specimens and CD133-positive NSCs isolated from brain subventricular zone specimens were kindly provided by Dr. Rainer Glass (Neurosurgical Research, University Clinics Munich, Germany). GBM surgical specimens were obtained from the University of Virginia Brain Tumor Bank according to procedures that were reviewed and approved by the Review Board of the University of Virginia Health System.
Vectors
The 3′UTR reporter plasmids were constructed via insertion of 3′UTR of STAT5B or KRAS downstream of the Firefly or the Renilla luciferase stop codon in respective pGL3 or pRL vectors (Promega, Madison, WI, USA). Site-directed mutagenesis of predicted miR-134 target sites was performed to generate mutant-control vectors. Human STAT5B and KRAS cDNAs were purchased from Open Biosystem (Huntsville, AL, USA), and cloned into the pMSCV retroviral vector. STAT5B primer pairs, FW: 5′- ggcggcgaattcatggctgtgtggatacaagctc-3′ and RW: 5′-attattgcggccgctcacgattgtgcgtgcgggatcc-3′, and KRAS primer pairs, FW: 5′-ggcg gcggatccatgactgaatataaacttgt-3′ and RW: 5′-ggaggcgcggccgcttacataattacacactttgtctttgac-3′, were used for PCR amplification. The production of retroviral particles and stable KRAS and STAT5B expressing cell lines were generated following standard protocols.
Treatment with growth factors and inhibitors
GBM cells and GSCs grown in 0.1% FBS or no FBS (GSCs) medium and treated with HGF (20 ng/ml), EGF (25 ng/ml), or PDGF (50 ng/ml) for different times. The cells were also treated with Crizotinib (300 nM), Erlotinib (5 μM), or Imatinib (15 μM). p-MET, p-EGFR, or p-PDGFR and miR-134 expressions in response to the treatments were verified by immunoblotting and q-PCR, respectively. To determine the signaling pathways that mediate the regulation of miR-134 expression by RTKs, the cells were treated with the PI3K inhibitor LY294002 (20 μM), the MAPK inhibitor PD98059 (60 μM), and the STAT3 inhibitor S3I-201(100 μM), and miR-134 expression was determined by q-PCR.
miRNA microarrays
miRNA microarrays were used to identify miRNAs that are regulated by MET. U373 cells that do not express HGF and have low levels of p-MET were treated with HGF (20 ng/μl) and U87 cells that express HGF and display high p-MET levels were treated with the MET inhibitor Crizotinib (300 nM) for 24 h and total RNA was extracted. The RNA was hybridized to Agilent Human miRNA Microarrays containing 723 human and 76 human viral miRNAs in triplicates. Microarray hybridization, analysis, and statistical calculations were performed by Beckmann Genomics (Durham, NC, USA). The experiments were repeated two times. miRNAs that were oppositely changed by at least 1.8-fold (P<0.05) in cells treated with HGF and Crizotinib were considered as differentially expressed (Figure 1b).
Quantitative RT-PCR
Total RNA was extracted from GBM cells, GSCs, and tissues using the miScript RNA extraction kit (Qiagen), according to the instructions of the manufacturer. Each RNA sample was reverse transcribed. cDNA was synthesized using the miScript Reverse Transcriptase kit (Qiagen) and quantitative PCR analysis was performed using the 7500 Real-time PCR System (Applied Biosystems, Carlsbad, CA, USA). A human U6B miRNA from Qiagen was used as an endogenous control. KRAS- and STAT5B-specific primers were KRAS-FW: 5′-gcaatgagggaccagtacatgag-3′, KRAS-RW: 5′-gtattgtcggatctccctcacca-3′ STAT5B-FW: 5′-cgtgctacagtcctggtgtgag-3′, STAT5B-RW: 5′-tactccatgacgcagcagttgt-3′. Human 18s rRNA was used as an endogenous control using specific primers (18sr-FW: 5′-cggctaccacatccaaggaa-3′, 18sr-RW: 5′-gctggaattaccgcggct-3′).
Chromatin immunoprecipitation
To determine whether KLF4 binds to the promoter region of miR-134, ChIP assay was performed according to the protocol of Magna ChIP G (Millipore, Billerica, MA, USA). GBM cells treated with HGF or Crizotinib were used. The cells were crosslinked with 1% formaldehyde for 10 min at 37°C, washed, and collected by centrifugation. Cell pellets were resuspended and sonicated to achieve an average nucleic acid fragment size of 200–500 bp. The DNA–protein complexes were immunoprecipitated using 5 μg of the anti-KLF4 antibody or with rabbit polyclonal IgG control overnight. Crosslinks in the immunoprecipitated chromatin were reversed by heating with proteinase K at 65°C overnight, and DNA was purified by the MinElute Reaction Cleanup column (Qiagen) and resuspended in water. The purified chromatin was subjected to PCR to amplify the genomic region of the putative KLF4-binding sites. Vimentin has been reported as a KLF4 target gene54 and was used as a positive control and GAPDH as a negative control. The PCR primer sequences were for site 1 (3916–4040 nt upstream of the miR-134 start codon) 5′-ccttgctttccaccatgact-3′ and 5′-agtccattttctggctgctg-3′ site 2 (4419–4518 nt upstream of the start codon) 5′-tccattgcttggtgctcata-3′ and 5′-gcatcacagagccgaagatt-3′ Vimentin (Vim) 5′-cttcaagactcggtggacttct-3′ and 5′-cttgtcgatgtagttggcaaag-3′ and GAPDH-F: 5′-cccactcttccaccttcg-3′, GAPDH-R: 5′-tccttggaccccatgtaggccat-3′. Two microliters ChIP DNA was used as a template and the results were normalized by 1% input for each sample.
Cell transfections
Cells were cultured to 50–80% confluence and transiently transfected with 20 nM pre-miRNA or anti-miRNA or siRNA using Oligofectamine or Lipofectamine RNAimax according to the manufacturer's instructions. Plasmid transfections were performed with Fugene 6 reagent according to the instructions of the manufacturer.
3′UTR reporter assays
To determine whether miR-134 directly binds to the STAT5B and KRAS 3′UTR, cells were transfected with pre-miR-134 or pre-miR-con for 24 h. The cells were then transfected with 3′UTR control, 3′UTR-STAT5B, mutant 3′UTR-STAT5B, 3′UTR-KRAS, or mutant 3′UTR-KRAS in addition to control cytomegalovirus-β-galactosidase reporter plasmids for 24 h. Luciferase assays were performed using the Luciferase System Kit (Promega, Madison, WI, USA) and luminescence was measured. Firefly luciferase activity was double normalized by dividing each well first by β-galactosidase activity and then by average luciferase/β-galactosidase value in a parallel set done with constitutive luciferase plasmid.
Immunoblotting
Immunoblotting was performed as previously described55 using antibodies specific for STAT5B (a kind gift from Dr. Barbara Merk, University of Virginia), KRAS, p-MET (T1349), MET, p-EGFR, EGFR, MEF2, GKLF (KLF4), NF-kB, LaminB1 (Santa Cruz Biotechnologies, Santa Cruz, CA, USA), p-PDGFRα, PDGFRα, p27 or CDK6 (Cell Signaling, Danvers, MA, USA). All blots were stripped and re-probed with β-actin (Santa Cruz Biotechnologies) as a loading control.
Cell proliferation assays
Cells (15 000–20 000 cells/well) were transfected with pre-miR-134 or pre-miR-con as described above. After 72 h, the cells were collected every day for 5 days, counted with a hemocytometer, and growth curves were established.
Propidium iodide flow cytometry
The cell-cycle status was assessed using propidium iodide flow cytometry as previously described.55 Briefly, the cells were transfected with pre-miR-134 or pre-miR-con for 72 h. The cells were washed with PBS, harvested, and fixed in 70% (v/v) ethanol. The cells were then treated with 20 μg/ml of DNase-free RNase and stained with propidium iodide. Cell samples were analyzed on a FACscan (Becton Dickinson, Franklin Lakes, NJ, USA) and G0–G1, S, and G2–M fractions were determined.
Annexin V-PE and 7AAD flow cytometry
Cell death and apoptosis were assessed by Annexin V-PE and 7AAD flow cytometry as previously described. Briefly, cells were transfected with pre-miR-134 or pre-miR-con for 96 h. The cells were harvested and stained with Annexin V-PE and 7AAD according to the instructions of the manufacturer. Cell samples were analyzed on a FACscan and apoptotic and dead cell fractions were determined.
Animal experiments
The effects of miR-134 on in vivo tumor growth and survival were tested in an intracranial xenograft model. GSCs (1228) or GMB (U87) cells were transfected with pre-miR-134 or pre-miR-con for 24 h. The cells (3 × 105) were stereotactically implanted into the striata of immunodeficient mice (n=6). Two to four weeks after tumor implantation, the animals were subjected to MRI scan and tumor volumes were quantified according to established and validated protocols. Alternatively, animal survival was determined.
Glioma stem cell sphere formation assay
GSCs were grown in low EGF and FGF medium (20 ng/ml) and treated with HGF, Crizotinib, or vehicle for 24 h. GSCs were also transfected with pre-miR-134/pre-miR-con or KRAS siRNA (si-KRAS), STAT5b siRNA (si-STAT5B), or scrambled control (si-con) for 72 h. The cells were dissociated into single cells with dissociation buffer (EDTA 1 mM, BSA 0.5% in PBS) and 1000 single cells were transferred to 24-well plates and incubated at 37°C for 7 days. Secondary neurospheres containing>30 cells were quantified.
Functional and molecular rescue experiments
Rescue experiments were performed to determine whether KRAS and STAT5B mediate the effects of miR-134 on cell proliferation. The effects of miR-134 on cell proliferation were tested in cells engineered to express either KRAS or STAT5B transcripts lacking the respective 3′UTRs. KRAS and STAT5B could not be inhibited by miR-134 in these cells as verified by immunoblotting. Pre-miR-134 was transfected in U373-KRAS or U373-STAT5B for 96 h. The cells were analyzed for proliferation by cell counting as described above.
Rescue experiments were also performed to determine whether miR-134 mediates the effects of MET on KRAS and STAT5B. Pre-miR-134 or pre-miR-con was transfected in U373 cells for 48 h and cells were then treated with HGF or control. After 24 h, the cells were lysed and immunoblotted for KRAS and STAT5B.
To determine whether miR-134 upregulation is required for RTK inhibitor effects, U373 cells were transfected with anit-miR-134 (20 nM) for 24 h and cells were then treated with RTK inhibitors, Crizotinib (150 nM), Erlotinib (2.5 μM), or Imatinib (15 μM). After 96 h, proliferation was assessed by cell counting.
Statistics
All experiments were repeated at least three times (except microarrays that were conducted twice in triplicates). When appropriate, two group comparisons were analyzed with a t-test, and P-values were calculated. The statistical associations between miR-134 and protein expression were evaluated with regression correlation analyses. The correlation coefficient ‘R' was calculated. P<0.05 was considered as significant and symbolized by an asterisk in the figures.
Acknowledgments
This study was supported by NIH grants RO1 NS045209 and RO1 CA134843 (R Abounader).
Glossary
- GBM
Glioblastoma
- miR-134
microRNA-134
- KLF4
Krueppel-like factor 4
Dr. Christensen was an employee and shareholder of Pfizer, Inc. and patent holder on PF2341066.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by R De Maria
Supplementary Material
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, King KS, Donahue CP, Khrapko K, Kosik KS. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dutta A, Abounader R. The role of microRNAs in glioma initiation and progression. Front Biosci. 2012;17:700–712. doi: 10.2741/3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez MA, Nicoloso MS, Shimizu M, Rossi S, Gopisetty G, Molina JR, et al. miR-29b and miR-125a regulate podoplanin and suppress invasion in glioblastoma. Genes Chromosomes Cancer. 2010;49:981–990. doi: 10.1002/gcc.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563–3572. doi: 10.1158/0008-5472.CAN-10-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guessous F, Alvarado-Velez M, Marcinkiewicz L, Zhang Y, Kim J, Heister S, et al. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112:153–163. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski J, Nowicki MO, Bronisz A, Nuovo G, Palatini J, De Lay M, et al. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. 2010;37:620–632. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fareh M, Turchi L, Virolle V, Debruyne D, Almairac F, de-la-Forest Divonne S, et al. The miR 302-367 cluster drastically affects self-renewal and infiltration properties of glioma-initiating cells through CXCR4 repression and consequent disruption of the SHH-GLI-NANOG network. Cell Death Differ. 2012;19:232–244. doi: 10.1038/cdd.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang H, Wang W, et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells. 2008;26:17–29. doi: 10.1634/stemcells.2007-0295. [DOI] [PubMed] [Google Scholar]
- Fiore R, Khudayberdiev S, Christensen M, Siegel G, Flavell SW, Kim TK, et al. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Christensen M, Larsen LA, Kauppinen S, Schratt G. Recombinant Adeno-Associated Virus-Mediated microRNA Delivery into the Postnatal Mouse Brain Reveals a Role for miR-134 in Dendritogenesis in Vivo. Front Neural Circuits. 2010;3:16. doi: 10.3389/neuro.04.016.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughwin P, Ciesla M, Yang H, Lim B, Brundin P. Stage-specific modulation of cortical neuronal development by Mmu-miR-134. Cereb Cortex. 2011;21:1857–1869. doi: 10.1093/cercor/bhq262. [DOI] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laddha SV, Nayak S, Paul D, Reddy R, Sharma C, Jha P, et al. Genome-wide analysis reveals downregulation of miR-379/miR-656 cluster in human cancers. Biol Direct. 2013;8:10. doi: 10.1186/1745-6150-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CBTRUS. Central Brain Tumor Registry of the United States. CBTRUS: Chicago, IL; 1998. [Google Scholar]
- Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- Network CGAR Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci USA. 2012;109:3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, Ciardiello F. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6:519–527. doi: 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- Liang QC, Xiong H, Zhao ZW, Jia D, Li WX, Qin HZ, et al. Inhibition of transcription factor STAT5b suppresses proliferation, induces G1 cell cycle arrest and reduces tumor cell invasion in human glioblastoma multiforme cells. Cancer Lett. 2009;273:164–171. doi: 10.1016/j.canlet.2008.08.011. [DOI] [PubMed] [Google Scholar]
- Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Kelly JJ, Stechishin O, Chojnacki A, Lun X, Sun B, Senger DL, et al. Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells. 2009;27:1722–1733. doi: 10.1002/stem.98. [DOI] [PubMed] [Google Scholar]
- Mazzoleni S, Politi LS, Pala M, Cominelli M, Franzin A, Sergi Sergi L, et al. Epidermal growth factor receptor expression identifies functionally and molecularly distinct tumor-initiating cells in human glioblastoma multiforme and is required for gliomagenesis. Cancer Res. 2010;70:7500–7513. doi: 10.1158/0008-5472.CAN-10-2353. [DOI] [PubMed] [Google Scholar]
- Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci USA. 2011;108:9951–9956. doi: 10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo KM, Jin J, Kim E, Ho Kim K, Kim Y, Gu Kang B, et al. MET signaling regulates glioblastoma stem cells. Cancer Res. 2012;72:3828–3838. doi: 10.1158/0008-5472.CAN-11-3760. [DOI] [PubMed] [Google Scholar]
- Piccirillo SG, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- Guessous F, Zhang Y, Kofman A, Catania A, Li Y, Schiff D, et al. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9:1031–1036. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst A, Campos B, Meier J, Devens F, Liesenberg F, Wolter M, et al. De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene. 2010;29:3411–3422. doi: 10.1038/onc.2010.83. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, et al. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- Shinojima N, Tada K, Shiraishi S, Kamiryo T, Kochi M, Nakamura H, et al. Prognostic value of epidermal growth factor receptor in patients with glioblastoma multiforme. Cancer Res. 2003;63:6962–6970. [PubMed] [Google Scholar]
- Abounader R, Laterra J. Scatter factor/hepatocyte growth factor in brain tumor growth and angiogenesis. Neuro Oncol. 2005;7:436–451. doi: 10.1215/S1152851705000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Holland EC. Platelet-derived growth factor (PDGF) and glial tumorigenesis. Cancer Lett. 2006;232:139–147. doi: 10.1016/j.canlet.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Yeh HJ, Silos-Santiago I, Wang YX, George RJ, Snider WD, Deuel TF. Developmental expression of the platelet-derived growth factor alpha-receptor gene in mammalian central nervous system. Proc Natl Acad Sci USA. 1993;90:1952–1956. doi: 10.1073/pnas.90.5.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Jun HJ, Bronson RT, Charest A.Inhibition of EGFR induces a c-MET driven stem cell population in glioblastoma Stem Cells 2013. e-pub ahead of print 1 October 2013; doi: 10.1002/stem.1554 [DOI] [PMC free article] [PubMed]
- Holmen SL, Williams BO. Essential role for Ras signaling in glioblastoma maintenance. Cancer Res. 2005;65:8250–8255. doi: 10.1158/0008-5472.CAN-05-1173. [DOI] [PubMed] [Google Scholar]
- Guha A, Feldkamp MM, Lau N, Boss G, Pawson A. Proliferation of human malignant astrocytomas is dependent on Ras activation. Oncogene. 1997;15:2755–2765. doi: 10.1038/sj.onc.1201455. [DOI] [PubMed] [Google Scholar]
- Heldin CH. Protein tyrosine kinase receptors. Cancer Surv. 1996;27:7–24. [PubMed] [Google Scholar]
- Li H, Ahonen TJ, Alanen K, Xie J, LeBaron MJ, Pretlow TG, et al. Activation of signal transducer and activator of transcription 5 in human prostate cancer is associated with high histological grade. Cancer Res. 2004;64:4774–4782. doi: 10.1158/0008-5472.CAN-03-3499. [DOI] [PubMed] [Google Scholar]
- Latha K, Li M, Chumbalkar V, Gururaj A, Hwang Y, Dakeng S, et al. Nuclear EGFRvIII-STAT5b complex contributes to glioblastoma cell survival by direct activation of the Bcl-XL promoter. Int J Cancer. 2013;132:509–520. doi: 10.1002/ijc.27690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Tiwari N, Meyer-Schaller N, Arnold P, Antoniadis H, Pachkov M, van Nimwegen E, et al. Klf4 is a transcriptional regulator of genes critical for EMT, including Jnk1 (Mapk8) PLoS One. 2013;8:e57329. doi: 10.1371/journal.pone.0057329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Guessous F, Kwon S, Kumar M, Ibidapo O, Fuller L, et al. PTEN has tumor-promoting properties in the setting of gain-of-function p53 mutations. Cancer Res. 2008;68:1723–1731. doi: 10.1158/0008-5472.CAN-07-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.