Abstract
Adenosine deaminases acting on RNA 1 (ADAR1) catalyzes cellular RNA adenosine-to-inosine editing events on structured RNA molecules. In line with this critical role, ADAR1 exhibits ubiquitous expression and is essential for embryonic development. However, regulation and developmental significance of this RNA editor in a spatiotemporal context are largely elusive. Here we unveil a novel tissue-specific role of ADAR1 in skeletal myogenesis. ADAR1 expression displayed programmed alteration that is coordinated with differentiation cues, and mediated negatively by miRNA-1/206. Coincidently, ADAR1 exerts stage-dependent functions—suppression of apoptosis at the onset of differentiation and preservation of timely myotube formation through later phase. Furthermore, the post-transcriptional aspect of its myogenic role was illustrated by the spectrum of binding RNAs, as revealed by high-throughput approach, as well as by direct regulation of myogenesis-associated targets such as dynamin 1/2 (Dnm1/2) and annexin A4. Consequently, maintenance of target gene expression profiles likely contributes to a state of cytoskeleton and membrane dynamics that is amenable to myoblast morphogenesis. Collectively, these findings uncover a critical link of ADAR1 to myogenesis, and further highlight an epigenetic mechanism by which ADAR1 and miR-1/206 interplay to control scheduled myoblast–myotube transition.
Keywords: ADAR1, dynamins, miRNA-1/206, myogenesis, RIP-Seq
The full complement of RNA molecules in a cell, or the transcriptome, is finely and dynamically regulated to meet the cellular need associated with a particular developmental or physiological state. Among mechanisms that shape the transcriptome, RNA editing of the adenosine-to-inosine (A-to-I) type represents an integral post-transcriptional process. Such conversion is mediated by ADARs (adenosine deaminases acting on RNA), which catalyze the hydrolytic deamination of adenosines in specific double-stranded RNA substrates.1 Through this enzymatic reaction, transcripts are covalently modified in a manner that results in an RNA sequence different from that encoded by the genomic DNA, thereby diversifying the cellular RNA signatures.2, 3, 4 And given that the nucleoside inosine is interpreted as guanosine by cellular machineries, such as the translation and RNA processing apparatuses, the A-to-I editing represents a form of genetic recoding with widespread implications on the expression and functions of coding and non-coding RNAs. Although extensive efforts have been devoted to profile sequence alterations that constitute the RNA editome, regulation and functional consequence of ADAR activity remain largely unresolved.
This metazoa-specific ADAR protein family comprises three members – ADAR1, ADAR2 (or ADARB1), and ADAR3 (or ADARB2) – and is characterized by the catalytic deaminase domain as well as multiple double-stranded RNA-binding domains. The ADAR1 gene encodes two isoforms of the enzymes through alternative promoter usage – an interferon (IFN)-inducible ∼150 kDa protein (p150) and a constitutively expressed N-terminally truncated ∼110 kDa protein (p110).5 The p110 form is exclusively nuclear in the cells, whereas the long form exhibits cytoplasmic localization.6 Among the ADAR family members, ADAR1 is absolutely essential for life in mammals, as inactivation of this gene in mice leads to embryonic lethality attributable to widespread apoptosis.7, 8 Although this is consistent with its pervasive expression and direct link to a critical step of gene expression, demonstration of ADAR1's developmental significance in a spatiotemporal context has been largely elusive. More significantly, how ADAR1's editing and/or transcriptomic roles are manifested in the developmental processes are not fully characterized. In this regard, multiple lines of evidence have provided support to a tissue-specific role of this protein, particularly in the maintenance of hematopoietic stem cells9, 10 as well as in the neuronal differentiation.11, 12 In line with the emerging scenario that ADAR1 may exert context-dependent developmental functions, Meltzer et al have recently demonstrated ADAR1 activation in response to inflammatory stressors, such as TNF-α, IFN-γ, or lipopolysaccharide, in the skeletal myoblast cells, and its possible function in limiting stress response and associated atrophy pathways.13
The formation of skeletal muscle, during development or in response to muscle damages, is accomplished through myogenesis. It is a complex process composed of temporally separate but highly coordinated events – determination, the phase through which multipotent stem cells are first committed to the myogenic lineage, and differentiation, the step through which myoblasts cease proliferation and fuse into multinucleated myotubes.14 At the molecular level, this process entails tight integration of extracellular and intracellular cues at the chromatin of muscle progenitors, which leads to a dynamic and physiologically relevant state of gene expression network.15, 16, 17 The important transcriptional regulators of myogenesis are a family of basic helix–loop–helix (bHLH) transcription factors known as muscle regulatory transcription factors, which has been shown to establish the myogenic lineage during embryogenesis and regulate the myogenic program.18, 19 The status of the muscle-specific genes is also governed by a multi-component epigenetic system comprising a myriad of histone modifiers and chromatin structure modulators.15
In addition, the post-transcriptional regulators of gene expression, principally microRNAs (miRNAs) and RNA-binding proteins, constitute another integral part of myogenesis regulation. In this capacity, a small number of muscle-specific miRNAs, termed myo-miRs, has been identified and shown to impact myogenesis and/or cardiac functions.20, 21, 22, 23, 24 An exemplar member is the miR-1 family, which is composed of six miRNA genes distributed in three paralogous clusters, and has been functionally implicated in the development of human skeletal muscle25 and in a Sprague–Dawley rat model of myocardial infarction.26 Being an important and well-characterized component of this regulatory circuit, miR-1 and miR-206 are closely related in terms of seed sequence, expression, and function, and are known to promote myoblast-to-myotube differentiation.27 Likewise, several RNA-binding proteins, HuR, KSRP, CUGBP1, PABPN1, Lin-28, and TTP, have been implicated in the regulation of alternative splicing, RNA stability, and translation, and ultimately gene expression associated with the myogenic program.28 However, this remains a less understood facet of gene regulation that underlies muscle development.
Results from our present work revealed a novel tissue-specific role of ADAR1 and are further consistent with the scenario that this RNA editor is an integral regulatory constituent of the myogenesis pathway. In response to differentiation cues, expression of ADAR1 underwent stage-specific alterations, which are triggered by both transcriptional and miRNA-mediated mechanisms and lead to near absence of this essential factor in late myogenesis. Consistent with such profiles, ADAR1 provides fundamental, distinct functions at multiple steps – in remedying stress arisen during early differentiation and facilitating timely myoblast–myotube transition. We further demonstrated that its suppressive function in myotube maturation occurs through targeting and modulating the expression of certain myogenesis-associated genes. These findings collectively define a myogenic role of ADAR1 and establish a regulatory circuitry of miR-1/206-ADAR1 that contributes to scheduled gene program transitions and consequently skeletal myogenesis-associated cytoskeleton and membrane dynamics.
Results
Stage-dependent regulation of ADAR1 expression during myogenic differentiation
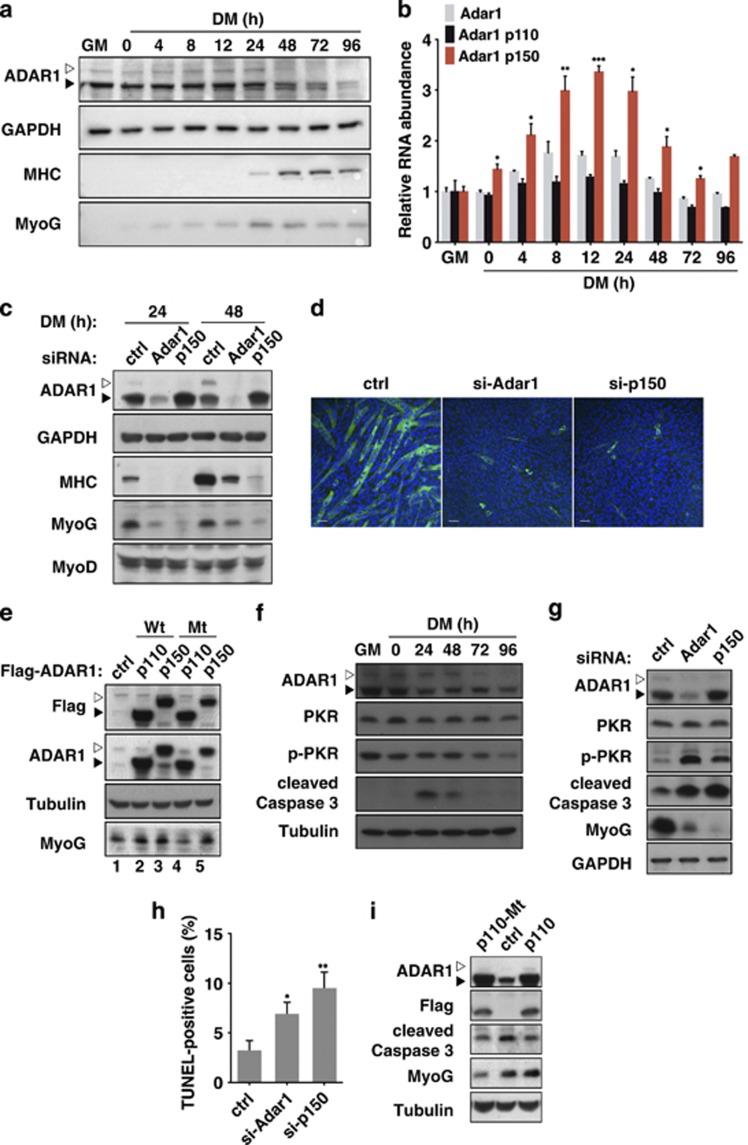
Despite its seemingly ubiquitous expression and functional indispensability, it presently remains unresolved whether or how ADAR1 expression could be regulated in a context-dependent manner. Previous studies have revealed a scarcity of Adar1 in the skeletal and cardiac muscle tissues,29 thus implying a tissue-specific mode of expression regulation. To further explore this possibility, we profiled the expression levels of ADAR1 during myoblast differentiation. Myoblast C2C12 cells cultured under growth condition (growth medium) or undergoing differentiation (DM or differentiation medium) were harvested for mRNA and protein expression analyses (Figures 1a and b). A surge in the expression of IFN-inducible ADAR1 isoform (p150), at both the transcript and protein levels, was observed within the first day of differentiation (<24 h). However, coincidently with the upregulation of skeletal muscle-specific markers myogenin (MyoG) and MHC, such increase in expression subsided. Interestingly, although the mRNA levels of the constitutively expressed isoform of Adar1 (p110) remained invariable throughput the course of differentiation (Figure 1b), protein expression underwent a progressive decline from mid- to late-stage of differentiation (48–96 h; Figure 1a). Such expression alterations thus signify a stage-dependent, isoform-specific regulation of ADAR1 during skeletal myogenesis.
Figure 1.
Stage-dependent expression profile and anti-apoptotic function of ADAR1 during early myogenesis. (a and b) C2C12 cells were cultured in growth medium (GM), and when cells reached confluence, the medium was replaced with differentiation medium (DM). C2C12 cells were harvested under GM and DM (0, 4, 8, 12, 24, 48, 72, 96 h post-differentiation) conditions. (a) Cell extracts were prepared for immunoblot analysis of ADAR1 and differentiation markers, MyoG and MHC. GAPDH serves as the internal control. Filled arrowheads indicate the position of p110 isoform, whereas open arrowheads denote p150. (b) RNA was isolated from these cells and subjected to real-time RT-PCR with primers specific to total Adar1, Adar1 p110, and Adar1 p150. Data presented were normalized to the levels in GM, with mean±S.D. from at least three experiments (*P<0.05; **P<0.01; ***P<0.001). (c) C2C12 cells were transfected with siRNAs targeting Adar1 p150 (p150), both isoforms of Adar1 (Adar1) or control siRNA (ctrl; targeting GFP). After differentiation induction, cells were harvested at 24 or 48 h for western blot analysis of ADAR1 and the indicated myogenic markers. GAPDH serves as the loading control. Filled arrowheads indicate the position of the p110 isoform, whereas open arrowheads denote p150. (d) Morphological characterization of myoblast differentiation in response to Adar1 knockdown (Adar1, total Adar1; p150, p150 isoform). Myotubes (DM-48 h) were visualized by indirect immunofluorescence analysis antibody against MHC (green) and nuclear counterstaining with DAPI (blue). Scale bar, 50 μm. (e) Effect of overexpressing the wild-type (Wt) or catalytic mutant (Mt) variants of the ADAR1 (p110 or p150 isoform) on MyoG. Western blot analysis was done as in c, for cells harvested at 24 h DM. (f) Profiling of stress-associated markers in differentiating C2C12 myoblasts. Cell extracts were prepared at the indicated culture conditions for detecting the expression of total PKR, phosphorylated PKR (p-PKR), activated/cleaved Caspase 3, ADAR1, and loading control Tubulin by immunoblot analysis. (g–i) Role of ADAR1 on the stress response of differentiating C2C12 cells. For loss-of-function experiments (g and h), cells were transfected with siRNAs targeting ADAR p150 (p150), both isoforms of Adar1 (Adar1), or GFP (ctrl), and subsequently harvested at DM-24 h for immunoblot analysis (g) or terminal deoxynucleotidyl transferase dUTP nick end labeling assay for apoptosis detection (h). For ADAR1 p110 overexpression (i), cells were transfected with expression plasmids encoding wild-type (p110) or deaminase-dead variant (p110-Mt). The empty vector was used as a control (ctrl). Cells were collected at DM-48 h and analyzed for expression levels of the indicated proteins. GAPDH and Tubulin were used as loading controls (For statistical analyses shown in this figure: NS, not significant or P>0.05; *P<0.05; **P<0.01; ***P<0.001.)
ADAR1 facilitates initial myogenic progression through its anti-apoptotic function
To next investigate whether alteration in the ADAR1 levels is functionally correlated with the scheduled progression of myogenic differentiation, we initially assessed the effect of ADAR1 knockdown on muscle differentiation. We found a lesser extent of differentiation in the ADAR1 total knockdown culture undergoing differentiation, based on the accumulation of myogenic markers (Figure 1c) and multinucleated myotube formation (Figure 1d). In the specific absence of the p150 isoform, we also observed downregulation of myogenic markers and myotubes (Figures 1c and d). Further in line with a positive regulation, overexpression of both forms of ADAR1 elevated the expression of MyoG (Figure 1e). We also generated a catalytically defective mutant as a means for loss-of-function characterization, as no siRNAs were available for exclusive downregulation of the p110 isoform. Our results showed that cells harboring ADAR1 p110 mutant similarly exhibited abrogated differentiation (Figure 1e). Intriguingly, although overexpression of both wild-type and deaminase-dead forms of p150 promoted the expression of MyoG (Figure 1e), we noted an increased expression of the p110 protein in the p150 overexpressing cells. As transfection of a C-terminally FLAG-tagged construct also gave rise to the expression of FLAG-tagged p110 (Supplementary Figure S1), we presumed that such upregulation was a result of alternative translation initiation from an internal Met codon for the in-frame p110 form. Therefore, as compared with the knockdown experiments, ectopic expression assays may not be suitable for clarifying the role of p150 in myogenesis. Next, to exclude the possibility that the delayed differentiation arose from abnormal cell growth, we performed a BrdU-based cell proliferation assay and subsequently found that the state of cell proliferation was not altered in the knockdown or overexpression cultures (Supplementary Figure S2). Together, these results indicate that both isoforms of ADAR1 are positive regulators of the initial phase of myogenic differentiation program.
We next sought to further delineate functional consequence of ADAR1 expression in early myogenesis. Profiling of the stress-associated cellular state in differentiating myoblasts showed transiently increased levels of phospho-PKR and cleaved form of caspase 3 in the early DM culture (Figure 1f), indicative of elevated stress and apoptotic responses immediately associated with differentiation initiation. Depletion of the total ADAR1 pools or ADAR1 p150 alone augmented these molecular features (Figure 1g) as well as the extent of apoptosis, as assessed by the terminal deoxynucleotidyl transferase dUTP nick end labeling assay (Figure 1h). Conversely, ectopic expression of both the wild-type and deaminase-defective ADAR1 p110 moderated the levels of cleaved caspase 3 in the DM-48 h cells, as compared with the control (Figure 1i). Taken together, these data are consistent with the notion that ADAR1 serves to sustain early commitment window of myogenesis by suppressing exaggerated apoptosis. Intriguingly, ADAR1's involvement in myogenesis-associated stress response was independent of its catalytic activity (Figure 1i), implying that significance of the enzymatic function in myogensis (Figure 1e) may be manifested in other aspects of differentiation.
ADAR1 expression in differentiated myotubes is downregulated by myo-miRNAs miR-1 and miR-206
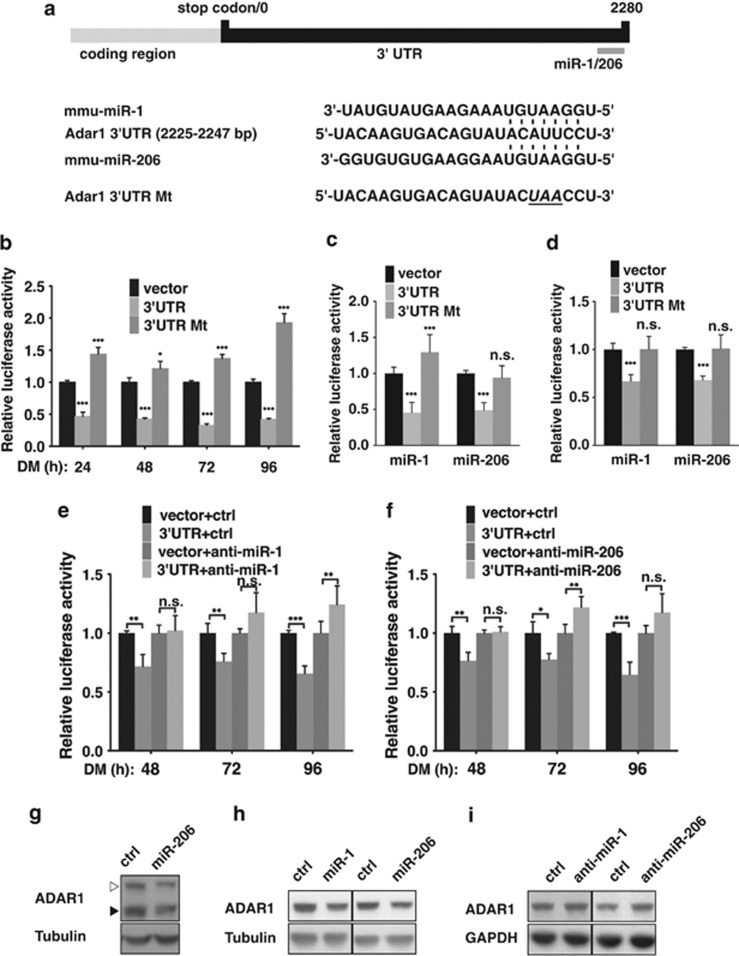
Based on our expression profiling data, ADAR1 p110 protein underwent downregulation in differentiated myoblasts (48–96 h post-DM), whereas the corresponding mRNA levels exhibited no significant changes (Figures 1a and b). To further interrogate the mechanism underlying such control, we turned our attention to miRNA-mediated post-transcriptional regulation owing to its established link to myogenesis.30 To this end, sequence analysis of mouse Adar1 3′ untranslated region (3′UTR) using TargetScan uncovered the unique presence of putative target site for two miRNAs – miR-1 and miR-206 – with high degree of conservation and complete complementarity in seed region (Figure 2a). Consistent with previous studies and their roles in promoting myogenic differentiation, expression profiling revealed that miR-1 and miR-206 were clearly induced when C2C12 myoblasts underwent differentiation (Supplementary Figure S3). Such accumulation in differentiating cells coincided with ADAR1 protein reduction in later stage of myogenic program. To further assess the importance of Adar1 3′UTR in the downregulation of ADAR1 protein in late differentiation, we generated an Adar1 3′UTR reporter by grafting the entire 3′UTR to the 3′ end of a luciferase reporter (Figure 2a). We subsequently observed in the differentiating C2C12 marked reduction in the luciferase activity that closely correlated with miR-1/206 induction (Figure 2b). On the contrary, reporter construct with modified miR-1/206 presumptive site in the 3′UTR (Mt) lost its responsiveness to differentiation signals (Figure 2b).
Figure 2.
miR-1/206 target the Adar1 3′UTR in differentiating myoblast. (a) Putative miRNA target site within the Adar1 3′UTR (top). The alignment of the indicated Adar1 3′UTR/miRNA complement (bottom). Numbers denote sequences relative to the stop codon (0). (b) Luciferase reporter constructs that contain wild-type 3′UTR sequence of Adar1 (3′UTR) or 3′UTR with mutated miR-1/206 target sites (3′UTR Mt) was transfected into C2C12 myoblasts, which were subsequently induced to differentiate. Luciferase activity was determined 0, 24, 48, 72, and 96 h after DM replacement, and is shown with respective vector controls being represented as 1. (c and d) Synthetic miR-1 or miR-206 was co-transfected with the above reporter constructs into C2C12 myoblasts (c) or HeLa cells (d), as indicated. Luciferase activity was determined 24 h after transfection, with vector controls being represented as 1. (e and f) In C2C12 myoblasts, antisense oligonucleotides against miR-1 (e) or miR-206 (f) were co-transfected with control or wild-type 3′UTR reporter constructs, as shown. Upon differentiation induction, luciferase activity was determined at 48, 72, and 96 h post-DM, and displayed with controls being represented as 1. (g and h) Effect of ectopic miR-1/206 on the expression of endogenous ADAR1. C2C12 (g) or HeLa (h) cells were transfected with synthetic miR-1 or miR-206, as indicated, and subsequently subjected to immunoblot analysis of ADAR1 (p110). Tubulin serves as an internal control. (i) Immunoblots of ADAR1 (p110) in C2C12 myoblasts transfected with synthetic control, anti-miR-1, or anti-miR-206 oligomers, as indicated. Expression of GAPDH represents equal loading (For statistical analyses shown in this figure, which were based on three independent experiments: NS, not significant or P>0.05; *P<0.05; **P<0.01; ***P<0.001.)
In further support to the miR-1/206-ADAR1 link, overexpression of synthetic miR-1 and miR-206 in either the C2C12 (Figure 2c) or HeLa (Figure 2d) cell lines inhibited the ectopic 3′UTR reporter in a target sequence-dependent manner, whereas the wild-type reporter was negatively regulated by the co-transfected miRNAs and the mutant reporter ceased to respond to such treatment. In parallel, delivery of anti-miR-1 (Figure 2e) and anti-miR-206 (Figure 2f) oligomers to the differentiating C2C12 cells reversed the differentiation-associated suppression of the 3′UTR. In addition, the significance of miR-1/206 regulation was corroborated by the effects of altering these myo-miRs on the expression of endogenous ADAR1 proteins – the levels of both isoforms reduced under the overexpression of miR-1/206 in C2C12 (Figure 2g) or HeLa (Figure 2h) cells, yet higher levels of ADAR1 p110 remained in differentiated myoblasts (DM-72 h) that harbored miR-1/206 antagomers (Figure 2i). Overall, these results are in full agreement with the scenario that miR-1/206 directly targets Adar1 3′UTR in C2C12 cells and mediates its expression repression in response to myogenic differentiation.
The downregulation of ADAR1 is important for scheduled transition from nascent to mature myotubes and for muscle development
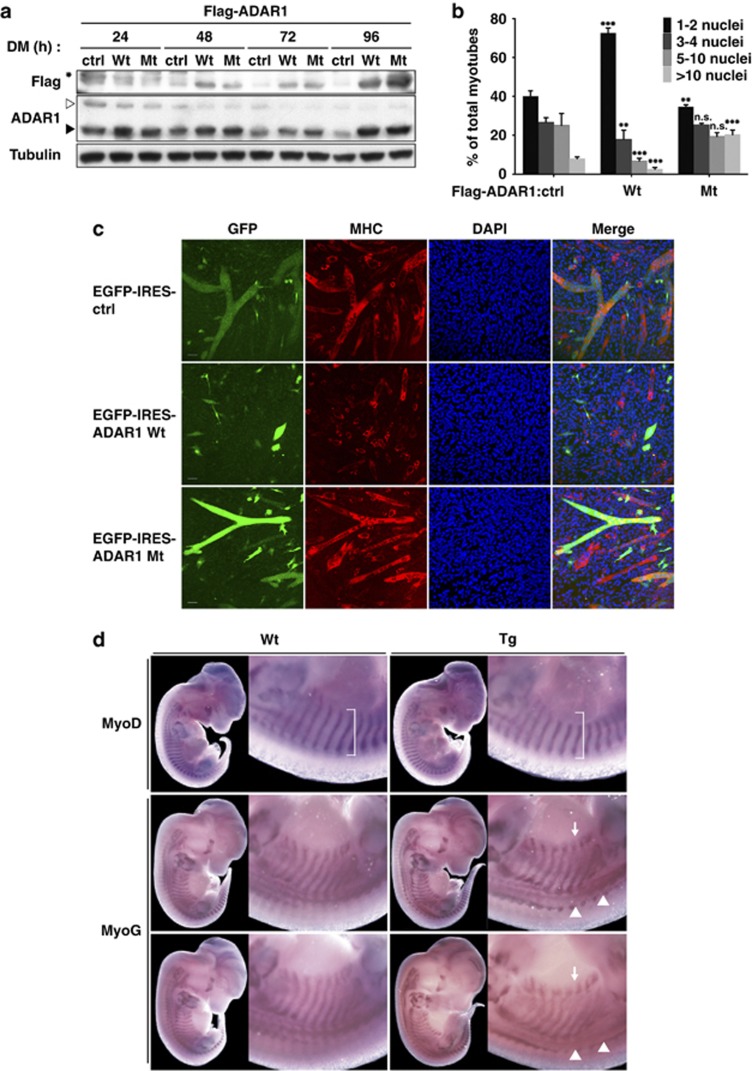
Having established the stage-specific, miR-1/206-mediated downregulation of ADAR1, we next aimed to deduce the biological relevance of such regulation in myogenic maturation. To address this issue, we first set out to reconstitute the expression of ADAR1 p110 in late phase of differentiation and observe its effect on multinucleated myotube formation, a hallmark of terminal differentiation. A expression construct was generated that induces ectopic expression of ADAR1 p110 by a muscle-specific muscle creatine kinase (MCK) promoter/enhancer, which is transcriptionally restricted to differentiated muscle cells.31 Such exogenous construct resulted in higher expression of ADAR1 p110 in the late phase of differentiation course (48–96 h; Figure 3a), complementing the depleted pools of this protein at this stage. Quantitative examination of the extent of differentiation was done by measuring the extent of myotube fusion, defined as the percentage of total nuclei in myotubes that comprise multiple nuclei. As shown in Figures 3b and c, ectopic expression of ADAR1 p110 in late-differentiation culture significantly moderated the myotube formation and fusion extent as compared with the vector control, thus implying severely impaired myotube formation. In contrast, extent of myotube maturation was promoted upon overexpression of the deaminase-deficient ADAR1 p110 (Figures 3b and c), suggestive of a dominant-negative effect. Collectively, these results attest to the notion that downregulation of ADAR1 activity may be an important determinant in the scheduled transition from nascent to mature myotube.
Figure 3.
Ectopic expression of ADAR1 retards myotube fusion of C2C12 myoblasts and muscle development. (a–c) MCK promoter-driven constructs encoding Adar1 p110 (Wt) and its catalytic mutant (Mt) were generated in either the Flag-tag fusion (a and b) or the EGFP-IRES chimeric expression (c) system. C2C12 cells were transiently transfected with these constructs for further analyses. (a) Upon DM culture (24, 48, 72, and 96 h), the expression of indicated proteins (Flag and ADAR1) was examined by western blot analysis, in comparison to cells with the vector control (ctrl). Tubulin was used as an internal control. Asterisk denotes non-specific signals. Filled and open arrowheads, respectively, indicate the positions of ADAR1 p110 and p150 isoforms. (b) The number of nuclei per myotube in C2C12 culture harboring the expression constructs as shown in a was calculated at DM-72 h. Extent of myotube fusion, based on the percentages of myotubes containing the indicated numbers of nuclei, was determined and shown. Data shown are means±S.D. from three experiments (n=1500 MHC+ cells per experiment). Statistical significance of comparisons versus control (ctrl): NS, not significant or P>0.05; *P<0.05; **P<0.01; ***P<0.001. (c) Cells (DM-72 h) with the indicated constructs were subjected to morphological characterization of myotube fusion extent, which was visualized by immunostaining (MHC, red; GFP, green; DAPI, nuclei counterstain). Individual and merged (Merge) images were captured by laser scanning confocal microscope, and single sections are shown. Scale bar, 50 μm. (d) WISH on wild-type (Wt) and transgenic (Tg; with MCK-Adar1 p110) embryos of identical somite numbers at embryonic stage E13.5. Probes corresponding to MyoD and MyoG were used to monitor marker expression and somite structure, as indicated. Lateral view of whole embryo is shown on the left of each panel, and magnification of the thoracic regions on the right. The ventral expression domain of MyoD in interlimb somites is denoted by brackets, and exhibited reduction in Tg embryos. Arrows point to altered distribution of myogenin in the ventral regions of interlimb somites in Tg embryos, and arrowheads indicate posterior extension of myogenin expression in dorsomedial dermomyotome
To further assess the role of ADAR1 in skeletal muscle development in vivo, we studied mice harboring a muscle-specific transgenic (Tg) construct – an Adar1 transgene driven by a MCK upstream regulatory sequence. Whole-mount RNA in situ hybridization (WISH) was used to characterize the spatiotemporal expression of the myogenic bHLH factors myognic differentiation 1 (Myod) and Myogenin. To this end, wild-type and Tg embryos of equivalent somite numbers at embryonic day 13.5 (E13.5) were collected and analyzed (Supplementary Figures S4a and b). Although MCK is active in differentiating muscle cells, there was a rather restricted, but distinct, pattern of ectopic Adar1 expression, most discernably in the interlimb regions (Supplementary Figure S4b, box). Tg animals were viable and without visible anatomical malformation (data not shown), possibly as a result of such limited expression. Interestingly, however, distribution of the myogenic factors was altered in the Tg ectopic sites – the ventral expression domain of MyoD in interlimb somites exhibited reduction in Tg embryos (Figure 3d, brackets; Supplementary Figure S4c), and the ventral expansion of Myogenin expression in interlimb somites was altered in E13.5 embryos, giving rise to reduced size or even truncation of the expression domain (Figure 3d, arrows). In addition, there was a notable posterior extension of Myogenin expression in the dorsomedial dermomyotome of Tg embryos (Figure 3d, arrowheads). These phenotypes of defective myogenic bHLH factors expression, which correlated with expression patterns of the transgene, indicate that ADAR1 may have an effect on muscle development.
Identification of targets that underlie ADAR1's contribution to myogenesis
Given the putative role of ADAR1 activity in contrasting myogenic maturation, we hypothesized that regulation of myogenesis-associated genes may constitute an underlying mechanism of this function. To consider this possibility, we first examined whether known editing target set was enriched in genes related to myogenic differentiation. By analyzing with the bioinformatics tool MetaCore (GeneGo) the functional and cellular attributes of the editing target genes uniquely archived in the DARNED database32 and a previous deep sequencing study,33 we found myogenesis regulation as one of the most significantly enriched GeneGo map folders (Supplementary Figure S5a).
To complement the pathway analysis, we next aimed to systematically pinpoint ADAR1 targets in the differentiating mouse myoblasts. By virtue of ADAR1's direct binding capacity as a mode of regulating their targets, we performed native RNA immunoprecipitation (RIP) combined with RNA-Seq (RIP-Seq) to capture at the transcriptome-wide level RNAs bound by ADAR1 (Figure 4a). RNAs isolated from C2C12 cells at DM-0 h (early differentiation) or DM-72 h (late) stages were immunoprecipitated by anti-ADAR1 antibody. Bound transcripts as well as total cellular RNA from respective cells (input) were prepared for strand-specific RNA-Seq using the SOLiD technology (see Materials and Methods, and Supplementary Tables S1 and 2 for sequencing results). We did not uncover sufficient amount of RNA in the control IgG immune-complexes for sequencing, and hence used total cellular RNA as a background control for subsequent analyses.
Figure 4.
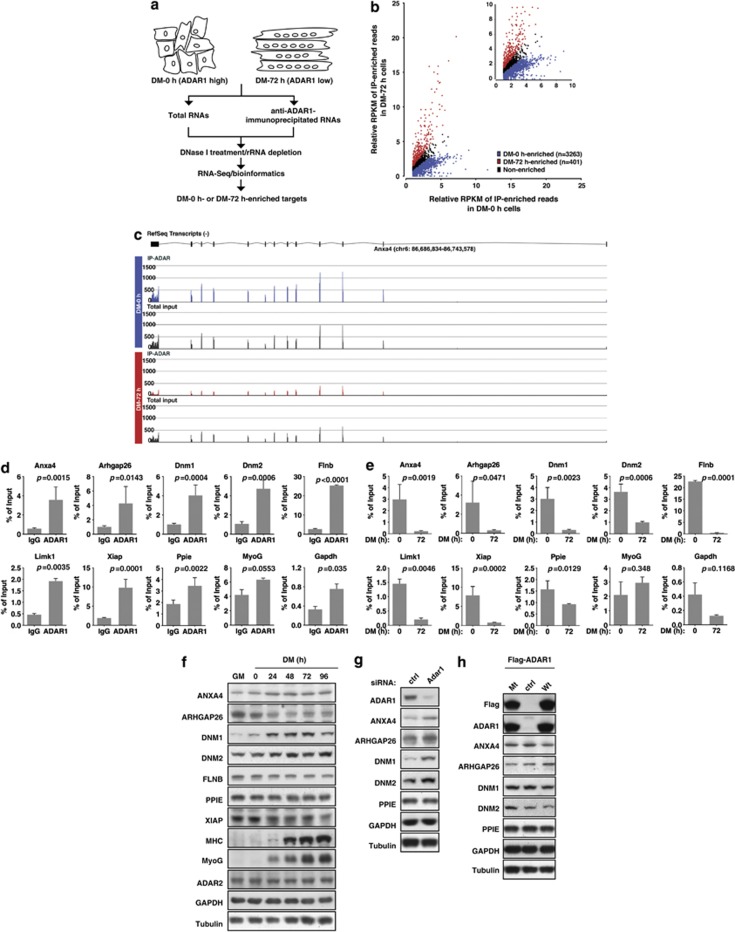
RIP-Seq profiles ADAR1-target RNAs interactions during myogenesis. (a) Schematic depiction of the experimental design of the RIP-Seq study (see Materials and Methods for details). Total RNAs and anti-ADAR1-immunoprecipitated RNAs from C2C12 myoblasts under DM-0 h or DM-72 h culture were subjected to high-throughput whole transcriptome sequencing. (b) The scatter-plot profiles the transcripts by their IP/input values (in RPKM) in the DM-0 h libraries (x axis) and the DM-72 h libraries (y axis). Transcripts that are enriched in DM-0 h, enriched in DM-72 h, or non-enriched are marked, respectively, in blue, red, and black. (c) Read density plot for Anxa4 representation in the four sequencing libraries. x Axis denotes relative position of the gene, whereas y axis corresponds to number of distinct reads. Corresponding gene structure is depicted on top of each plot. (d) Cells cultured under confluence (DM-0 h) were subjected to RIP for validating transcript binding by ADAR1. qRT-PCR was preformed to compare anti-ADAR1 and control (IgG) immunoprecipitations of selected candidate genes uncovered by RIP-Seq experiments. MyoG and Gapdh were used as negative controls. The experiments were done at least three times in triplicate, and shown with mean±S.D. as well as P values. (e) Stage-dependent enrichment of ADAR1 transcript binding. Anti-ADAR1 RIP of DM-0 h and DM-72 h C2C12 cells was assessed by qRT-PCR. Data presented were normalized to IgG RIP values, with mean±S.D. from at least three experiments and P values also shown. (f) Differentiation-associated protein expression profiling of selected ADAR1-targeted candidates. C2C12 cells were cultured in GM or DM for the indicated time lengths before western blot analysis of targets, as noted. Expression of MHC and MyoG serves as differentiation marker, whereas levels of ADAR2, Tubulin, and GAPDH were used as loading control. (g and h) Effect of ADAR1 mis-expression on the protein levels of selected candidate targets. C2C12 cells were transiently transfected with siRNAs targeting Adar1 or control siRNA (ctrl; targeting GFP) (g), or plasmids encoding Flag-tagged wild-type (Wt) or catalytically defective (Mt) ADAR1 (h). Cells were harvested at DM-48 h for immunoblot analysis using antibodies against the indicated proteins. Tubulin and GAPDH serve as internal controls
To explicitly call transcripts an ADAR1-binding target, we defined several criteria that account for transcript expression levels and representation enrichment in IP versus input (see Materials and Methods). We subsequently identified transcripts that were preferentially bound by ADAR1 at one stage or the other (Figures 4b and c, and Supplementary Table S3) – DM-0 h targets were defined as those with a 1.32-fold binding enrichment as compared with DM-72 h (n=3263), whereas DM-72 h targets, with the converse threshold, amounted to 401. Such changes toward a less complex ADAR1-associated transcriptome were in line with the downregulation of ADAR1 expression in late myoblast differentiation. Figure 4c and Supplementary Figure S5b illustrate sequencing results for selected candidate targets with enriched binding of ADAR1 in DM-0 h cells. Notably, we found a preponderance of DM-0 h-enriched target genes implicated in cytoskeleton-associated processes (5 of the top 10 enriched GeneGo processes/networks; Supplementary Figure S5c).
RNA–ADAR1 interactions derived from the deep sequencing studies were independently validated using RIP-qPCR. As shown in Figure 4d and Supplementary Figure S5d, candidate transcripts showed significant enrichment in the anti-ADAR1 versus IgG immunoprecipitates. In addition, we also compared anti-ADAR1 pull-down of RNAs in DM-0 h relative to DM-72 h C2C12 cells and found stronger enrichment for all selected candidate RNAs (Figure 4e and Supplementary Figure S5d). This differentiation-responsive decrease in RNA binding was thus consistent with the downregulation of ADAR1. On the contrary, sequences of the negative controls Gapdh, MyoG, and Acta1 were not enriched in either ADAR1-specific or differentiation-associated RIPs (Figures 4d and e, and data not shown). Interestingly, we also examined and confirmed ADAR1's occupancy of the Arhgap26 transcript, a known myogenesis regulator34 that initially was not identified in the RIP-Seq analysis because of transcript abundance.
Next, to gain further insight into how ADAR1 operates, we characterized whether ADAR1 regulates the expression of candidate genes. For a more definite evaluation, we first sought to identify targets whose expression patterns correlate with the myoblast differentiation process. Expression profiling revealed that some of these selected candidates underwent expression alteration during the course of differentiation, albeit with variable profiles (Figure 4f and Supplementary Figure S5e). In view of the post-transcriptional mechanism of ADAR1's action, we then focused on three transcripts – Dnm1, Dnm2, and annexin A4 (Anxa4) – which displayed progressively elevated protein expression upon myoblast differentiation. Incidentally, these genes share an ‘ultra-editing' feature, characterized by substantial RNA editing events (respectively, 84, 33, and 229, according to the DARNED database), and presumably should be most prone to transcript alteration in response to ADAR1 downregulation. At the mRNA level, ADAR1 seemed to have marginal effects on their expression in the differentiating myoblast (Supplementary Figure S5f). However, knockdown of ADAR1 markedly augmented their protein levels (Figure 4g). In contrast, overexpression of ADAR1 had a negative effect on the expression of these proteins (Figure 4h). Moreover, ectopic expression of the deaminase-defective ADAR1 p110 did not result in expression alteration as compared with the control, indicating that its enzymatic activity may be indispensable for its repressive role. Therefore, these results collectively demonstrate ADAR1's direct involvement in the negative regulation of certain myogenesis-associated genes and, together with the above findings, serve as strong evidence that maturation of muscle tissue hinges on the temporal regulation of this activity.
Regulation and myogenic roles of Anxa4 and dynamins
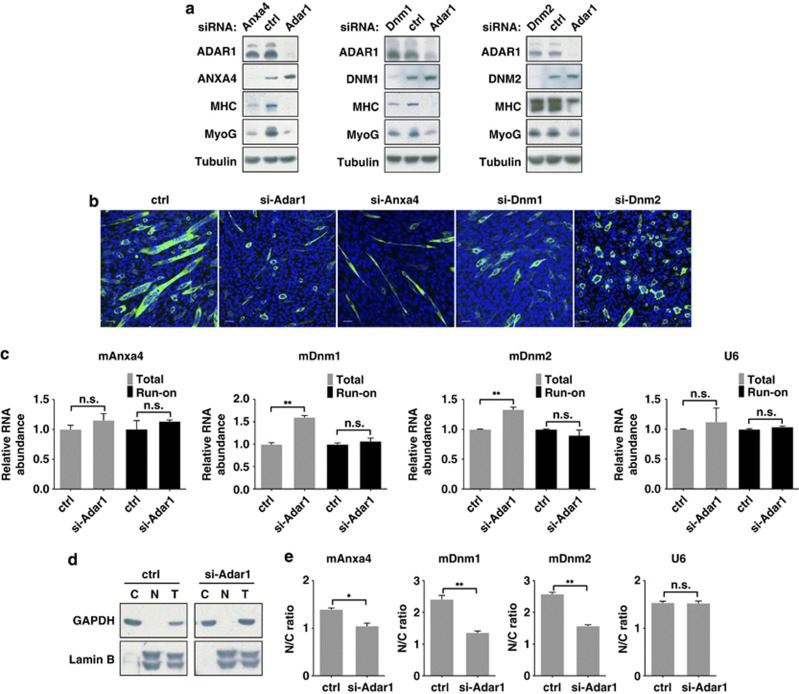
To further link ADAR1's regulatory role to myogenesis, we next investigated whether alteration in the target gene levels is functionally correlated with the scheduled progression of differentiation. To this end, we examined whether knockdown of target gene (Anxa4, Dnm1, and Dnm2) might impact C2C12 differentiation, and found a lesser extent of multinucleated myotubes formation in the knockdown culture undergoing differentiation (Figure 5a). Such outcome phenocopies the effects of ADAR1 abrogation in this respect. Interestingly, we observed gene-specific effects on the extent of myotube formation and morphology – each culture seemed to exhibit distinct characteristics of arrested myotube fusion (Figure 5b). Furthermore, although the presence of Anxa4 was critical for the upregulation of myogenic markers associated with differentiation, Dnm1 and Dnm2 were dispensable for this attribute. Viewed together, these findings strongly suggest differential requirements for these ADAR1 targets at distinct steps of the initial myogenic differentiation, and thus a possibly broad involvement of ADAR1 in this process.
Figure 5.
Regulation and myogenic roles of Anxa4 and dynamins. (a and b) C2C12 cells were transfected with siRNAs targeting the indicated genes. After differentiation induction, cells were harvested at 48 h for (a) western blot analysis of the indicated genes and myogenic markers (Tubulin serves as the loading control), or (b) morphological characterization of myoblast differentiation. Myotubes (DM-48 h) were visualized by indirect immunofluorescence analysis antibody against MHC (green) and nuclear counterstaining with DAPI (blue). Scale bar, 50 μm. (c) Effect of ADAR1 knockdown on the transcription rates of the indicated target genes. Nuclear run-on assays were performed to monitor newly transcribed RNA from nuclei of control (ctrl) and ADAR1 (si-Adar1) knockdown C2C12 cells. U6 snRNA, which remained unchanged in both cell types, was used to demonstrate uniformity of input RNA. Quantitative results are shown by bar graph, and represent the means±S.D. of three independent experiments (**P<0.01; NS, P>0.05). (d and e) Control (ctrl) and ADAR1 (si-Adar1) knockdown C2C12 cells were separated into nuclei (N) and cytoplasm (C) fractions (d). Proteins from both fractions and whole-cell extracts (T) were probed with antibodies against GAPDH and Lamin B, respectively, as the cytoplasmic and nuclear markers, to show extent of fractionation. (e) Bar graphs show relative distribution of the indicated target transcripts between the nuclear and cytoplasmic compartments, as assessed by real-time RT-PCR. U6 serves as a control. Means±S.D. were calculated from three independent experiments (*P<0.05; **P<0.01; NS, P>0.05)
Next, we set out to elucidate the mechanism whereby ADAR1 modulates the expression of these targets. To this end, we first interrogated whether these selected candidate genes are subjected to RNA editing, by performing Sanger sequencing to check for A-to-I(G) changes at sites previously reported in the mouse Dnm1 and Dnm2 transcripts.35 However, we failed to detect any editing events at known sites (Supplementary Figure S6) as well as in the analogous intronic regions reportedly harboring RNA editing in the human counterparts (data not shown). We then sought to characterize the involvement of ADAR1 in regulating the transcription efficiency of these genes. We conducted nuclear run-on assay on control and knockdown cells and quantitatively measured changes in the transcription activity by real-time RT-PCR analysis. As shown in Figure 5c, we did not detect significant changes in the levels of newly synthesized transcripts of Dnm1, Dnm2, and Anxa4, thus excluding the possibility that ADAR1 exerts its regulation at the transcriptional level. Finally, as transcript binding by the nucleus-resident ADAR1 implies a spatial regulation of target RNAs' localization, we assessed this possibility by using subcellular fractionation of RNA combined with real-time RT-PCR analysis (Figure 5d). Our results revealed that, in the absence of ADAR1, abundance of target transcripts in the nucleus relative to cytoplasm underwent notable decrease (Figure 5e). Such reduced nuclear accumulation of target transcripts therefore indicates that their subcellular localization depends on ADAR1, and that altered gene expression during differentiation may arise from transcript re-distribution.
Discussion
During cell lineage commitment and differentiation, transcription and epigenetic factors constitute a significant regulatory mechanism contributing to the timely expression of lineage-specific genes. Compared with the extensive knowledge of various chromatin regulatory complexes involved in the muscle differentiation program, much less is understood about the post-transcriptional regulation of myogenic transcriptome changes. In this study, we identified a novel myogenesis regulator, ADAR1, which exhibited transient expression fluctuation consistent with a dynamic regulation and its stage-dependent functions. Consequently, the binding and regulation of certain myogenesis-associated gene transcripts by ADAR1 were implicated in conferring a proper state of gene networks that maintains progression of myoblast-to-myotube maturation. To our knowledge, our study is the first report of a hitherto unrecognized link of the ADAR1 protein to muscle development. Our findings also uncovered a new component of the post-transcriptional regulatory network that underlies myogenesis.
One of the important findings of this study is the programmed downregulation of the seemingly ubiquitous and essential RNA editor ADAR1 by the myo-miRs, miR-1/206. Although early microarray-based screening has revealed ADAR1 as a candidate target of miR-1,36 we provided experimental evidence that confirms miR-1/206's negative role in expression, firmly indicating that ADAR1 is subject to a distinct mode of regulation employed by the myogenic program. Our studies thus uncovered and characterized a new target of the myo-miRs, expanding their substrate spectrum of the gene regulatory molecules. More importantly, this muscle-specific miR-1/206-ADAR1 axis highlights a regulatory cross-talk between different post-transcriptional mechanisms in the context of myogenesis, which may serve to increase the robustness of the transcriptomic transition during muscle differentiation process. Intriguingly, this scenario also suggests that ADAR1 and RNA editing are dispensable in maturing muscle tissue, which may be refractory to a less heterogeneous state of transcriptome.
Our current findings implicated ADAR1 in mitigating apoptosis of differentiating myoblasts (Figure 1). Interestingly, Meltzer et al have recently demonstrated ADAR1 activation in response to inflammatory stressors such as TNF-α, IFN-γ, or lipopolysaccharide, and a possible function of this protein in limiting stress response and associated muscle atrophy pathways.13 This regulatory role is reminiscent of the previously reported involvement of ADAR1 in hematopoiesis, where it acts to suppress IFN signaling and to block premature apoptosis in hematopoietic stem cells undergoing differentiation.10 This parallelism in the upregulation of stress and apoptotic signaling (i.e., increased expression of IFN and activated caspase 3) between the hematopoietic and myogenic programs lends support to the paradigm that apoptosis facilitates differentiation during maturative progression,37, 38 and further signifies a more widespread role of ADAR1 in safeguarding cell differentiation programs. In addition, this anti-apoptotic aspect of ADAR1's myogenic roles lies partly in the association with and inhibition of the dsRNA-dependent protein kinase PKR. Given the known link of PKR to myogenesis,39, 40 our studies thus extended the ADAR1-PKR functional antagonism in stress response, which has been prevalently observed in the context of virus infection,41, 42, 43 to muscle development.
Although a regulatory function on myogenic differentiation as well as specific muscle genes has been ascribed to ADAR1 by our results, how such a role is manifested remains unresolved. Owing to the extensive substrate spectrum of ADAR1, it is conceivable that its activity, or lack thereof upon differentiation, may impinge on a myriad of genes – an expression network intimately linked to myogenic program. Such notion is supported by our in silico and deep sequencing analyses, which revealed a substantial extent of overlap between the ADAR1-targeted transcripts and those associated with myogenesis (Figure 4 and Supplementary Figure S5). Therefore, although we have successfully pinpointed a few bona fide targets of ADAR1, regulation of which presumably contributes to myogenesis progression, developmental relevance of this enzyme should certainly be considered at the systems level. Nevertheless, given the known functions of the myogenesis-associated editing targets, such as cytoskeleton organization and membrane dynamics, ADAR1 may act, rather than stochastically, via targeting specific cellular/biological pathways that underlie skeletal muscle development.
An intriguing outcome of our mechanistic characterization of ADAR's myogenic roles was the failure to detect RNA editing in the candidate target transcripts, despite analysis of previously annotated sites as well as human-analogous regions. It remains a formal possibility that ADAR1 might target as yet unreported sites. A more comprehensive sequence analysis of the candidate targets may thus be needed to discern whether ADAR1 contributes to gene regulation in a catalysis-dependent manner. As previous studies reported underrepresentation of RNA editing in non-primate species, and particularly in the non-brain tissues,44, 45 another likely explanation for the dearth of editing sites is the reduced enzymatic activity of ADAR1 in murine myoblasts and/or cell lines. In this regard, we also failed to detect any RNA editing in the selected candidate transcripts in another mouse neuronal cell line, N2A (data not shown). Without excluding the possibility of RNA editing, our results nonetheless revealed an alternative scenario through which ADAR1 affects target gene expression (i.e., transcript localization).
An important feature of skeletal myogenesis that is unique from other developmental pathways is the formation of myotube, which entails progressive membrane fusion of myocytes. Incidentally, several functional attributes of ADAR1 are highly suggestive of its involvement in this process: (1) breakdown of the known binding targets revealed a preponderance of genes associated with cytoskeleton functions (Supplementary Figure S5c). (2) ADAR1 was further found associated with transcripts of certain candidate genes directly implicated in membrane fusion or myogenesis, and requisite for their timely differentiation-associated expression profiles (Figures 4g and h). (3) Differentiation-associated downregulation of ADAR1 coincided temporally with the maturation of myotube structure; ectopic expression of this protein at this stage stalled the progression of myotube formation, giving rise to immature, altered myotube organization. Therefore, myo-miRs-mediated downregulation of ADAR1 may underpin a state of membrane dynamics and/or cytoskeleton network that is amenable to the coalescence of single cells into multi-nucleated myotubes. Alternatively, it remains a formal possibility that expression and activity of the RNA editor serve as a critical determinant in the maintenance of single-cell identity.
Our current study uncovered in skeletal myogenesis the hitherto uncharacterized upregulation of DNM1, which is otherwise known to express mainly in brain. DNM1 is one of the three dynamin GTPases, whose roles in intracellular membrane-related events such as endocytosis/exocytosis and mitochondrial network dynamics have been extensively established.46 Presumably, catalysis of membrane severing or fusion may also contribute to membrane changes that underlie myoblast fusion. Although no myogenic role has yet to be ascribed to DNM1, mutation in DNM2 is associated with an autosomal dominant centronuclear myopathy.47 Considering that differentiating myoblasts undergo extensive membrane remodeling en route to multinucleated, fused myotube, increased expression of DNM1 and DNM2 may represent a mechanical measure to the rising demand for membrane-modulating activities. In this capacity, DNM1 might cooperate with the myotube fusion facilitator ARHGAP26,34 previously shown to interact strongly with DNM1.48 Intriguingly, expression of an miRNA miR-199b, for which Dnm1 is a host gene, are highly correlated with dystrophic muscle fibers of the mdx mice,49 further strengthening DNM1's myogenic function.
Materials and Methods
Cell culture and differentiation
Mouse C2C12 myoblast cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen, Grand Island, NY, USA) supplemented with 20% heat-inactivated fetal bovine serum (Invitrogen) and 100 U/ml penicillin and streptomycin solution (Invitrogen; growth medium), at subconfluent densities. Human HeLa cervical cancer cell line was cultured similarly, except with 10% fetal bovine serum. Cells were maintained in a 5% CO2 humidified incubator at 37 °C. To induce C2C12 myotube differentiation, the medium was replaced by Dulbecco's modified Eagle's medium supplemented with 2% heat-inactivated horse serum (DM) at subconfluent culture and maintained for the time indicated in the corresponding experiments.
Tg mice
The MCK-ADAR1 transgene construct consists of a 3-kb MCK 5′ regulatory region, a 2.8-kb mice ADAR1 p110 cDNA sequence, and a 0.5-kb polyadenylation sequence. The MCK upstream region inserted into the muscle-specific expression cassette contains both promoter and enhancer and was based on a previous report.50 MCK-Adar1 Tg FVB/NjNar1 founder mice were generated by injecting linearized transgene into the nuclei of fertilized embryos. Semiquantitative PCR-based genotyping was used to detect a 216-bp fragment of the MCK-ADAR1 transgene. Mice were fed ad libitum with standard diet and housed in a pathogen-free environment on a 12-h light, 12-h dark cycle. Procedures for animal studies were approved by the Chang Gung University Institutional Animal Care and Use Committee.
WISH
WISH was performed basically as described in a previous report.51 Embryos were dissected in PBS, fixed in 4% PFA for overnight at 4 °C, washed twice in PBS, dehydrated in sequentially increasing concentrations of methanol in PBT (25, 50, 75%, 2 × 100%), and stored at –20 °C in 100% methanol. They were subsequently rehydrated following the reverse procedure up to the PBT stage. Embryos were then bleached in 6% H2O2 in PBT for 1 h, washed twice in PBT, treated with Proteinase K solution (10 μg Proteinase K per ml of PBT) for 30 min at room temperature, washed twice in PBT, refixed in 4% PFA+0.2% glutaraldehyde for 20 min. After two washes in PBT, embryos were placed for at least 2 h in hybridization buffer (50% formamide, × 5 SSC, 0.5% Chaps, 0.2% Tween 20, 50 μg/ml yeast tRNA, heparin, pH adjusted to 4.5 with citric acid), before overnight hybridization with 0.5 μg/ml digoxigenin-labeled antisense RNA probe at 70 °C. Embryos were then washed twice in hybridization buffer and twice in washing solution (50% formamide/ × 1 SSC/0.1% Tween 20). Following hybridization/wash, they were incubated sequentially for 1 h in MABT supplemented with 2% blocking powder (MABTB) at room temperature, 2 h in MABTB containing 20% goat serum, and overnight at 4 °C in MABTB containing 20% goat serum and 1/2000 alkaline phosphatase anti-digoxigenin Fab fragments (Roche, Indianapolis, IN, USA). Embryos were washed in MABT, before incubation for 30 min in NTMT (100 mM Tris pH 9.5, 50 mM MgCl2, and 0.1% Tween 20) and staining in NTMT solution containing NBT/BCIP substrates (Roche). Stained embryos were refixed in 4% PFA, and transferred into 80% glycerol.
RIP
Native RIP was carried out largely as described previously.35, 52 Briefly, cells were washed twice with ice-cold × 1 PBS and removed from the culture plate using a cell scraper in 1 ml ice-cold polysomal lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES (pH 7.0), 0.5% NP40, 1 mM DTT, 50 U RNase inhibitor (SUPERase-in; Ambion, Austin, TX, USA), protease inhibitor cocktail (Roche)) per 10-cm dish. Subsequently, suspension was passed through a 27.5-G needle eight times to promote cell lysis. Whole-cell extracts were collected by centrifugation (16 000 g, 15 min) and pre-cleared with magnetic protein-G beads (Invitrogen) at 4 °C for 1 h. Immunoprecipitation was performed by adding the ADAR1 antibody to the precleared extracts and incubating at 4 °C overnight. Magnetic protein-G beads were then added to each IP sample and rotated for 1 h at 4 °C. The beads were pelleted and washed with polysomal lysis buffer. After several washes, 20 U of DNase I (Roche) and 10 × reaction buffer was added and incubated at 37 °C for 15 min to remove all contaminating DNA. Then, 1 ml Trizol reagent was added to the beads and the RNA was extracted according to the manufacturer's protocol. Reverse transcription and quantitative PCR were performed as described in Supplementary Methods.
RIP-Seq
RIP was performed using C2C12 cells under confluence (DM0) or 3 days after differentiation induction (DM3). RNA-Seq was performed by the Genomics Core Laboratory of the Molecular Medicine Research Center, Chang Gung University. SOLiD sequencing libraries were prepared using the SOLiD Total RNA-Sequencing kit (ABI PN4445374; Life Technologies, Grand Island, NY, USA) according to the manufacturer's instructions. About 5 μg of total input or immunoprecipitated RNAs was used as starting materials, except for IP of DM3, of which 1.6 μg was used because of lower recovery of bound RNAs. Samples were then subjected to ribosomal RNA removal using the Ribo-Zero Gold Kits (Human/Mouse/Rat; Epicentre, Madison, WI, USA; Cat. #RZG1224), and then 200–500 ng of the rRNA-depleted RNAs were fragmented by RNAase III. After purification, 100 ng of each fragmented sample were ligated with the RNA adaptors. Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) was used for profiling the distribution of fragmented RNA (with the median size between 125 and 140 nt). After reverse transcription and size selection, each cDNA library was amplified with distinct barcoded 3′ PCR primer from the SOLiD RNA Barcoding kits (PN 4427046). The distribution of insert size and concentration of each library were measured by Agilent 2100 Bioanalyzer. From each library, equal concentration (0.8 pM) was pooled together and sequenced strand-specifically in an ABI SOLiD5500 platform (Life Technologies, Foster City, CA, USA) generating 75-bp tags. Approximately 50 million reads were generated for each library (Supplementary Table S1).
Bioinformatic analysis of RIP reads
The Raw RNA-Seq reads were analyzed by the Whole Transcriptome Analysis workflow, a built-in module in the LifeScope Genomic Analysis Solution version 2.5.1 (Life Technologies, Grand Island, NY, USA). In brief, the adaptor/barcode sequences and homopolymer reads were first detected and filtered, and reads matching the transfer and ribosomal RNAs were excluded (Supplementary Table S1). Remaining reads were mapped to the complete mouse genome (mm9, NCBI Build 37). The initially unmapped reads are sequentially aligned to a virtual exon and junction sequence library (Supplementary Tables S1 and 2). In the final step, the aligned reads were merged into a single BAM file.
To properly calculate the expression level of mapped regions, we first examined the read distribution of several known transcripts (examples are shown in Figure 4c and Supplementary Figure S5b). On the basis that the read distributions of IP-enriched regions on transcripts displayed similar pattern as those of the control input samples, we thus used the RNA-seq workflow in Partek Genome Suite 6.6 (Partek, St. Louis, MO, USA) to calculate the read counts per exon and per gene, and summarize the read counts for all exons within a gene. Sequence hits to the transcripts were converted to RPKM units,53 a normalized representation that allows for comparisons within transcriptomes and between libraries of different sequencing depths.
Identification of ADAR1 targets
For each gene, we first determined its respective RPKM values in the four RNA-Seq samples – DM0/input, DM0/IP, DM3/input, and DM3/IP. To call transcripts an ADAR1-binding target in a more unequivocal manner, we defined several criteria to help exclude potential false positives: (1) the abundance of the transcripts, based on sequence coverage (RPKM), was accounted on the principle that lower-than-background expression might bias binding enrichment analysis. We set the RPKM minimum at 1.5 for genes represented in the input/DM0 library, which is equivalent to ∼5 × coverage of transcript by distinct reads per million. (2) Owing to the lack of sequence data for control IP (by IgG), we considered the relative transcript representation in the IP versus input libraries of DM0 cells, in which ADAR1 was considerably expressed, as the extent of binding enrichment. To establish a threshold for this parameter, we examined negative control transcripts Gapdh, Acta1, and Myog, which are expressed at varying levels and have not been reported as editing targets – these genes did not show significant ADAR1 association in RIP-qPCR experiments, and had IP/input ratios of 9.24:10.18 (=0.91), 41.02:58.95 (=0.70), and 1.93:2.30 (=0.84), respectively. On this basis, the enrichment cutoff was set at 1.1-fold. (3) Finally, we compared the IP/input ratios between DM0 and DM3 (median=1.32), and identified transcripts that were preferentially bound by ADAR1 at one stage or the other. DM0 targets were defined as those with a 1.32-fold difference in the DM0 versus DM3 (n=3263), whereas DM3 targets, with the converse threshold, amounted to 401. Lists of all enriched transcripts can be found in Supplementary Table S3.
Statistical analysis
Data are presented as mean with error bars indicating the standard deviation (S.D.). Student's t-test was used to determine the statistical significance of quantitative comparisons. Degrees of statistical significance (NS, not significant; P>0.05; *P<0.05; **P<0.01; ***P<0.001) are indicated in the respective figure legends.
Descriptions for gene expression analyses, immunochemical analyses, expression constructs, miRNA manipulation, and reporter assays are provided in Supplementary Methods.
Acknowledgments
We thank Chang Gung University Laboratory Animal Center for generating and maintaining the Tg mice, and are indebted to Yi-Chuan Cheng and Yu-Ting Yan for expert help on WISH. We are grateful to Dr. Scott Schuyler and Simon D Silver and members of the BC-MT laboratory for critical reading of the article and important discussions. This work was supported by grants from the National Science Council of Taiwan (NSC101-2321-B-182-009 and NSC101-2320-B-182-036-MY3), Chang Gung Memorial Hospital (CMRPD1A0322), National Health Research Institute of Taiwan (NHRI-EX102-9923SC), and the Ministry of Education, Taiwan.
Glossary
- ADAR
adenosine deaminases acting on RNA
- Anxa4
annexin A4
- Dnm1
dynamin 1
- Dnm2
dynamin 2
- MCK
muscle creatine kinase
- MyoD
myognic differentiation 1
- MyoG
myogenin
- A-to-I
adenosine-to-inosine
- IFN
interferon
- bHLH
basic helix–loop–helix
- miRNA
microRNA
- DM
differentiation medium
- 3′UTR
3′ untranslated region
- WISH
whole-mount RNA in situ hybridization
- Tg
transgenic
- RIP
RNA immunoprecipitation
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by J Cidlowski
Supplementary Material
References
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci. 2010;35:377–383. doi: 10.1016/j.tibs.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26:221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc Natl Acad Sci USA. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Keegan LP, Lafarga M, Berciano MT, O'Connell M, Carmo-Fonseca M. Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci. 2003;116 (Pt 9:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizasa H, Nishikura K. A new function for the RNA-editing enzyme ADAR1. Nat Immunol. 2009;10:16–18. doi: 10.1038/ni0109-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenberg S, Paz Yaacov N, Safran M, Moshkovitz S, Shtrichman R, Sherf O, et al. Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PLoS One. 2010;5:e11173. doi: 10.1371/journal.pone.0011173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukma M, Tohda M, Watanabe H, Matsumoto K. The mRNA expression differences of RNA editing enzymes in differentiated and undifferentiated NG108-15 cells. J Pharmacol Sci. 2005;98:467–470. doi: 10.1254/jphs.sc0050074. [DOI] [PubMed] [Google Scholar]
- Meltzer M, Long K, Nie Y, Gupta M, Yang J, Montano M. The RNA editor gene ADAR1 is induced in myoblasts by inflammatory ligands and buffers stress response. Clin Transl Sci. 2010;3:73–80. doi: 10.1111/j.1752-8062.2010.00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Serna IL, Roy K, Carlson KA, Imbalzano AN. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J Biol Chem. 2001;276:41486–41491. doi: 10.1074/jbc.M107281200. [DOI] [PubMed] [Google Scholar]
- Saccone V, Puri PL. Epigenetic regulation of skeletal myogenesis. Organogenesis. 2010;6:48–53. doi: 10.4161/org.6.1.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E, Sousa-Victor P, Ballestar E, Munoz-Canoves P. Epigenetic regulation of myogenesis. Epigenetics. 2009;4:541–550. doi: 10.4161/epi.4.8.10258. [DOI] [PubMed] [Google Scholar]
- Guasconi V, Puri PL. Chromatin: the interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 2009;19:286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, et al. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203–214. doi: 10.1128/MCB.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Dey BK, Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell. 2010;21:2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol. 2011;11:34. doi: 10.1186/1471-213X-11-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan ZX, Lin QX, Fu YH, Deng CY, Zhou ZL, Zhu JN, et al. Upregulated expression of miR-1/miR-206 in a rat model of myocardial infarction. Biochem Biophys Res Commun. 2009;381:597–601. doi: 10.1016/j.bbrc.2009.02.097. [DOI] [PubMed] [Google Scholar]
- Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42:1252–1255. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apponi LH, Corbett AH, Pavlath GK. RNA-binding proteins and gene regulation in myogenesis. Trends Pharmacol Sci. 2011;32:652–658. doi: 10.1016/j.tips.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, et al. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Chen J. MicroRNAs in skeletal myogenesis. Cell Cycle. 2011;10:441–448. doi: 10.4161/cc.10.3.14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita F, Takase K, Tozuka M, Saha S, Okuda K, Ishii N, et al. Muscle creatine kinase/SV40 hybrid promoter for muscle-targeted long-term transgene expression. Int J Mol Med. 2007;19:309–315. [PubMed] [Google Scholar]
- Kiran A, Baranov PV. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010;26:1772–1776. doi: 10.1093/bioinformatics/btq285. [DOI] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- Doherty JT, Lenhart KC, Cameron MV, Mack CP, Conlon FL, Taylor JM. Skeletal muscle differentiation and fusion are regulated by the BAR-containing Rho-GTPase-activating protein (Rho-GAP), GRAF1. J Biol Chem. 2011;286:25903–25921. doi: 10.1074/jbc.M111.243030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Nellaker C, McIntyre RE, Buendia-Buendia JE, Bumpstead S, Ponting CP, et al. High levels of RNA-editing site conservation amongst 15 laboratory mouse strains. Genome Biol. 2012;13:26. doi: 10.1186/gb-2012-13-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Luchetti F, Burattini S, Ferri P, Papa S, Falcieri E. Actin involvement in apoptotic chromatin changes of hemopoietic cells undergoing hyperthermia. Apoptosis. 2002;7:143–152. doi: 10.1023/a:1014362415047. [DOI] [PubMed] [Google Scholar]
- Zamai L, Burattini S, Luchetti F, Canonico B, Ferri P, Melloni E, et al. In vitro apoptotic cell death during erythroid differentiation. Apoptosis. 2004;9:235–246. doi: 10.1023/B:APPT.0000018805.63663.a5. [DOI] [PubMed] [Google Scholar]
- Alisi A, Spaziani A, Anticoli S, Ghidinelli M, Balsano C. PKR is a novel functional direct player that coordinates skeletal muscle differentiation via p38MAPK/AKT pathways. Cell Signal. 2008;20:534–542. doi: 10.1016/j.cellsig.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Kronfeld-Kinar Y, Vilchik S, Hyman T, Leibkowicz F, Salzberg S. Involvement of PKR in the regulation of myogenesis. Cell Growth Differ. 1999;10:201–212. [PubMed] [Google Scholar]
- Clerzius G, Gelinas JF, Daher A, Bonnet M, Meurs EF, Gatignol A. ADAR1 interacts with PKR during human immunodeficiency virus infection of lymphocytes and contributes to viral replication. J Virol. 2009;83:10119–10128. doi: 10.1128/JVI.02457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AM, Li Z, Cattaneo R, Samuel CE. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J Biol Chem. 2009;284:29350–29356. doi: 10.1074/jbc.M109.045146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George CX, Li Z, Okonski KM, Toth AM, Wang Y, Samuel CE. Tipping the balance: antagonism of PKR kinase and ADAR1 deaminase functions by virus gene products. J Interferon Cytokine Res. 2009;29:477–487. doi: 10.1089/jir.2009.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg E, Nemzer S, Kinar Y, Sorek R, Rechavi G, Levanon EY. Is abundant A-to-I RNA editing primate-specific. Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, et al. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci USA. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules. Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Durieux AC, Vignaud A, Prudhon B, Viou MT, Beuvin M, Vassilopoulos S, et al. A centronuclear myopathy-dynamin 2 mutation impairs skeletal muscle structure and function in mice. Hum Mol Genet. 2010;19:4820–4836. doi: 10.1093/hmg/ddq413. [DOI] [PubMed] [Google Scholar]
- Lundmark R, Doherty GJ, Howes MT, Cortese K, Vallis Y, Parton RG, et al. The GTPase-activating protein GRAF1 regulates the CLIC/GEEC endocytic pathway. Curr Biol. 2008;18:1802–1808. doi: 10.1016/j.cub.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciotta S, Meregalli M, Cassinelli L, Parolini D, Farini A, Fraro GD, et al. Hmgb3 Is Regulated by MicroRNA-206 during Muscle Regeneration. PLoS One. 2012;7:e43464. doi: 10.1371/journal.pone.0043464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst R, Avetisova E, Burden SJ. Restoration of synapse formation in Musk mutant mice expressing a Musk/Trk chimeric receptor. Development. 2002;129:5449–5460. doi: 10.1242/dev.00112. [DOI] [PubMed] [Google Scholar]
- Correia KM, Conlon RA. Whole-mount in situ hybridization to mouse embryos. Methods. 2001;23:335–338. doi: 10.1006/meth.2000.1145. [DOI] [PubMed] [Google Scholar]
- Hsieh CL, Lin CL, Liu H, Chang YJ, Shih CJ, Zhong CZ, et al. WDHD1 modulates the post-transcriptional step of the centromeric silencing pathway. Nucleic Acids Res. 2011;39:4048–4062. doi: 10.1093/nar/gkq1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.