Abstract
Lung cancer is the leading cause of tumor-related death. The lack of effective treatments urges the development of new therapeutic approaches able to selectively kill cancer cells. The connection between aberrant microRNA (miRNA – miR) expression and tumor progression suggests a new strategy to fight cancer by interfering with miRNA function. In this regard, LNAs (locked nucleic acids) have proven to be very promising candidates for miRNA neutralization. Here, we employed an LNA-based anti-miR library in a functional screening to identify putative oncogenic miRNAs in non-small-cell lung cancer (NSCLC). By screening NIH-H460 and A549 cells, miR-197 was identified as a new functional oncomiR, whose downregulation induces p53-dependent lung cancer cell apoptosis and impairs the capacity to establish tumor xenografts in immunodeficient mice. We further identified the two BH3-only proteins NOXA and BMF as new miR-197 targets responsible for induction of apoptosis in p53 wild-type cells, delineating miR-197 as a key survival factor in NSCLC. Thus, we propose the inhibition of miR-197 as a novel therapeutic approach against lung cancer.
Keywords: lung cancer, microRNAs, p53, apoptosis, LNA
Recent advances in cancer research candidate microRNAs (miRNAs) as potential tools and targets for the development of antitumoral therapies. miRNAs are short RNAs (∼22 nt) with an extraordinary regulatory activity, fine tuning the expression of several genes and thus modulating cellular homeostasis, proliferation and apoptosis.1 Altered miRNA expression has been observed in tumor samples, signifying a possible role of these molecules in the oncogenic process.2 The capacity of miRNAs to simultaneously repress the expression of many target genes, often functionally linked in a common pathway or biological process, has made them attractive candidates to block or boost the production of several proteins by adding artificial miRNAs or anti-miRNAs, respectively.3 However, it is unclear which of the aberrantly expressed miRNAs may represent therapeutic targets to be exploited for developing more effective treatments.
Lung cancer is a highly heterogeneous disease, classified on histological bases into small cell lung cancer (SCLC) and non-SCLC (NSCLC). The latter can be further divided into subgroups, including the most frequent histotypes such as adenocarcinomas, squamous cell and large cell carcinomas.4 Despite the development of new drugs directed against some genome abnormalities, such as EGF-R mutations or ALK translocations, effective response with targeted therapies is still transitory and confined into a relative small percentage of patients.5 New targets are therefore needed to improve the efficacy of lung cancer therapy, which at present leads to death of the majority of diagnosed patients.4, 6, 7 To satisfy such a request, it is essential the understanding of the molecular networks responsible for tumor growth and the identification of the best therapeutic target candidates.
The uncontrolled proliferation of cancer cells is frequently associated with loss of apoptotic response to genotoxic stress or external stimuli. Several proteins identified as critical checkpoints in the induction of programmed cell death are often altered in cancer. Many miRNAs act as oncogenes by controlling the expression of such proteins and give rise to a complex network of interactions that promote tumor survival and spreading. p53 is a master regulator of cell response to stress,8 governing both cell cycle and cell death, and its function is altered in a high percentage of tumors. A number of publications have described an interplay between p53 and miRNAs,9, 10 some of which are regulated transcriptionally and post-transcriptionally by p53 itself,11, 12, 13, 14, 15 whereas some others are suppressing p53 expression.16 These observations provide a rationale to explore the role of putative oncogenic miRNAs in the regulation of apoptosis and cell cycle progression, with the final goal to identify potential tools or targets for anticancer therapy.
We exploited an unbiased high-content approach to identify miRNAs regulating cell proliferation and tumorigenesis in lung cancer. We have investigated the role of all known miRNAs through the neutralization of their function, taking advantage of a new generation locked nucleic acid (LNA)-based anti-miRNA library (miRCURY LNA microRNA inhibitor library, Exiqon, Vedbaek, Denmark). With the aim to functionally select miRNAs endowed with oncogenic activity, we identified miR-197 as a key repressor of p53-dependent apoptotic cascade. We demonstrated that miR-197 allows the survival of lung cancer cells by controlling the expression of p53, NOXA and Bcl-2-modifying factor (BMF). We propose that targeting of miR-197 could be a promising therapeutic strategy against wild-type p53 lung cancer.
Results
Genome-wide screening of miRNAs' function
To comprehensively investigate how individual miRNAs affect cancer cell viability, we employed a high-content cell screening approach based on anti-miR transfection and subsequent luminescence-based proliferation analysis. We leveraged an LNA library inclusive of 866 mature human miRNAs annotated in miRBase version 12.0 (www.miRBase.org) and other 40 potential, currently not annotated, miRNAs. NIH-H460 and A549 cells were chosen as target systems to identify potential oncogenic miRNAs in NSCLC, whose knockdown would result in cell death and/or growth arrest. Individual LNAs were transfected by a lipid-based transfection protocol that was first optimized to obtain ∼90% efficiency. To monitor the transfection efficiency in every experiment, two control small interfering RNAs (siRNAs) were added to the library plates: one affecting cell viability (PLK-1 siRNA) and another leading to a complete knockdown of the target mRNA (NF1b siRNA). Moreover, the LNA library was integrated with a control LNA as negative control.
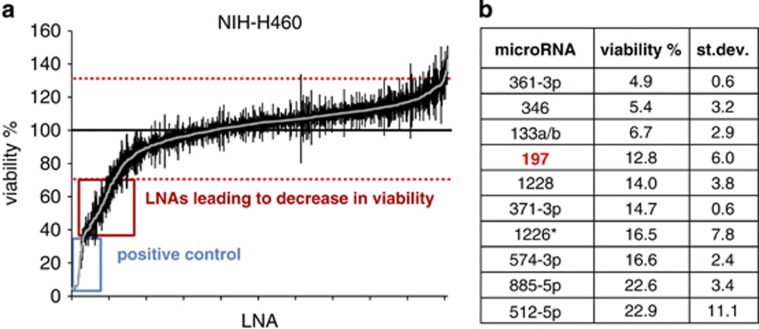
Several LNAs were selected for their ability to impair cell viability (Figures 1a and b and Supplementary Figures S1a and b). Interestingly, numerous of these LNAs resulted effective on both the screened cell lines. Among the top hits, we focused on miR-197, whose inhibition resulted in a strong decrease of cell viability in both cell lines. The choice of miR-197 was also based on recent literature sustaining its potential oncogenic role. MiR-197 was found to be overexpressed in lung, prostate and pancreas cancer tissues compared with normal specimen,17, 18, 19 while levels of circulating miR-197 in combination with other miRNAs were used to generate a signature of biomarkers for early detection of lung cancer.20, 21 Moreover, increased expression of miR-197 contributes to carcinogenesis and it is considered a reliable marker for diagnosis, patient stratification and prognosis assessment in follicular thyroid carcinomas,22, 23, 24 while promoting EMT and metastasis in pancreatic adenocarcinoma.25
Figure 1.
Functional anti-miR screening in NIH-H460 cells. (a) Diagram showing results of LNA library (∼900 LNAs) screening in NIH-H460 cells. The viable cell number was evaluated 72 h after transfection by Cell Titer-Glo luminescent cell viability assay and normalized to the average of all samples, excluding controls. Values above and below two fold standard deviation (red dashed lines) were considered as significant. Reported is the mean±S.E.M. of the screening performed in triplicate; each dot represents an individual LNA. (b) Table showing 10 LNAs that resulted most effective in NIH-H460 cells and relative viability
miR-197 inhibition has oncosuppressive effect on lung cancer cells
We initially confirmed the results obtained during the screening of the LNA library in a series of in vitro experiments. To exclude nonspecific side effects of anti-miR-197 LNA (hereafter LNA-197), a miR inhibitor with different chemical modifications was tested. Also in this case, the depletion of miR-197 in NIH-H460 cells resulted in a marked decrease in cell number (Supplementary Figure S2).
The neutralization of miR-197 in NIH-H460 and A549 cells was able to significantly impair cell growth (Figure 2a and Supplementary Figure S3a) and anchorage-independent colony formation (Figure 2b and Supplementary Figure S3b), thus suggesting a pro-proliferative role of miR-197. In addition, knockdown of miR-197 promoted the induction of the apoptotic pathway, as shown by CASPASE 3–7 activation and positivity for Annexin V staining (Figures 2c and d and Supplementary Figures S3c and d). Furthermore, few hours after LNA-197 transfection, we observed CASPASE 3 activation and cleaved PARP-1 protein by western blotting analysis (Figure 2e and Supplementary Figure S3e), confirming that miR-197-depleted cells undergo apoptosis.
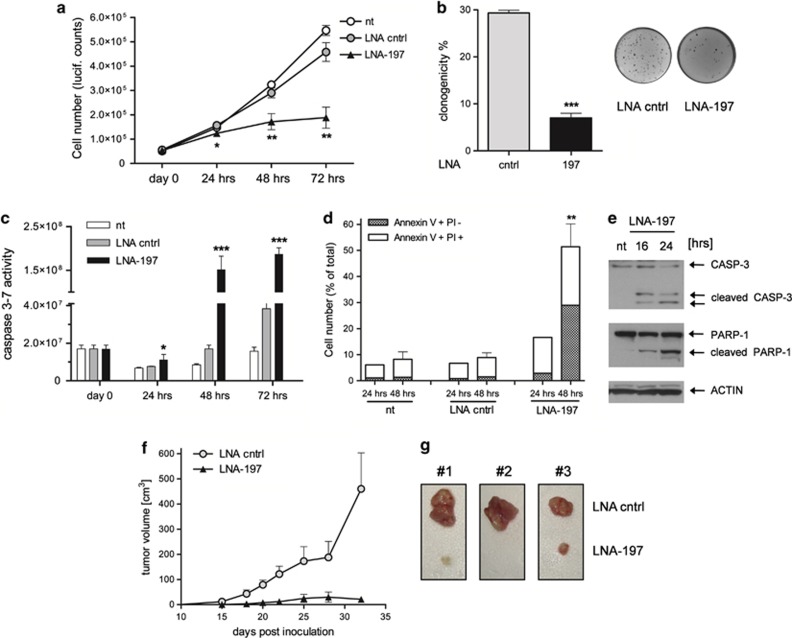
Figure 2.
miR-197 depletion impairs cell proliferation and induces apoptosis. (a) Growth curve of NIH-H460 cells untreated (nt), transfected with control LNA or LNA-197 at 25 nM; cell number was assessed by Cell Titer-Glo assay at the indicated time points after transfection – mean±S.D., *P=0.034, **P<0.001. (b) Clonogenic capacity of NIH-H460 cells after anti-miR-197 treatment; the graph shows the percentage of plated cells that gave rise to colonies – mean±S.D., ***P<0.0001. (c) Activation of the apoptotic pathway after LNA-197 transfection is shown by CASPASE 3–7 activity, Annexin V staining (d) and immunoblot using the specified antibodies at the indicated time points (e) – mean±S.D., *P=0.04, **P=0.0086, ***P<0.0005. (f) NIH-H460 cells were transfected in vitro with control LNA or LNA-197; 16 h after transfection, 105 viable cells were injected into the flank of CD1/nude mice. Shown is the tumor growth of xenografts as defined by mass volume – mean±S.E.M., N=8. In the LNA-197-treated group, the volume of the only three measurable tumors was graphed. (g) Comparison of tumor engraftment sizes of LNA-197-treated versus control LNA-treated NIH-H460 cells. Tumors explanted from three mice 32 days after injection are shown
Supported by these in vitro results, we hypothesized that miR-197 targeting may exert a therapeutic activity by inhibiting tumor growth in vivo. We therefore transfected NIH-H460 cells with control LNA or LNA-197 and subsequently injected the cells subcutis into the flank of nude mice. Depletion of miR-197 strongly inhibited tumor growth, as five out of eight mice did not develop any tumor mass, whereas, the remaining three mice developed tumors later and markedly smaller as compared with the control counterparts (only 15% of control tumors' mean volume) at the end of the experiment (Figures 2f and g). Thus, downmodulation of miR-197 exerts a considerable in vitro and in vivo antitumor activity against NSCLC.
miR-197 controls BMF expression in NSCLC
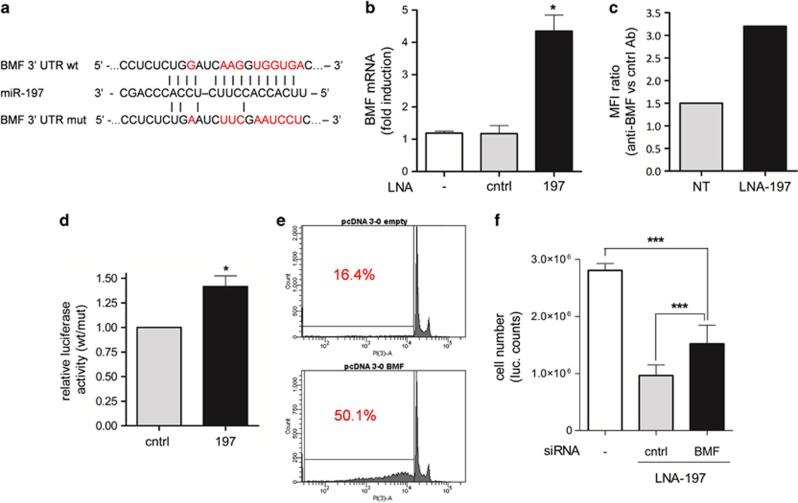
To identify the miR-197 target proteins that are involved in apoptosis induction, bioinformatic analyses were conducted. All miR-197 putative targets (listed by Targetscan) were analyzed by DAVID (The Database for Annotation, Visualization and Integrated Discovery).26, 27 Among the genes belonging to the apoptotic pathway, we found the proapoptotic protein BMF (Figure 3a). BMF is a BH3-only protein that localizes on the light chain of dynein when inactive. On activation by intra or extracellular stimuli, BMF binds to and neutralizes antiapoptotic Bcl2 family members on the mitochondrial membrane. As a direct consequence, proapoptotic proteins BAX and BAK are able to dimerize and promote the cytochrome C release inducing cell death.28 Interestingly, loss of 15q14/15, which includes the BMF gene, has been reported in lung and breast cancer.29 A marked increase of BMF at mRNA and protein level was found when treating the cells with LNA-197 (Figures 3b and c). The direct interaction of miR-197 with the BMF 3′UTR was demonstrated by luciferase reporter assay. To this aim, the 3′UTR of BMF was cloned into pGL3-Control vector downstream of the luciferase coding sequence (pGL3-BMF UTR-wt). The putative miR recognition site was then mutated to generate the pGL3-BMF UTR-mut derivative. Downmodulation of miR-197 by specific LNA transfection determined an increased luciferase activity only in the presence of the wild-type miR-binding site, indicating that BMF was indeed a bona fide target of miR-197 (Figure 3d).
Figure 3.
BMF is a direct target of miR-197. (a) Predicted BMF 3′UTR-binding site for miR-197. The alignment of the seed region of miR-197 with BMF 3′UTR is shown. The sites of target mutagenesis are indicated in red. (b) qRT-PCR showing inverse correlation of BMF mRNA and miR-197 expression – mean±S.D., *P=0.015; (c) cytofluorimetric analysis of BMF expression in NIH-H460 on transfection of LNA-197. The graph shows the ratio of MFI normalized to control Ab – one representative experiment out of three is shown. (d) Luciferase activity in NIH-H460 cells transfected with BMF UTR wt or mut in combination with a control or LNA-197. The ratio of normalized luciferase activity in cells transfected with BMF UTR wt versus mut is indicated – mean±S.E.M., *P=0.0118. (e) Overexpression of BMF-induced apoptosis: the percentage of apoptotic cells is shown as sub-G0 fraction – one representative experiment out of three is shown. (f) Downmodulation of BMF by RNA interference can rescue LNA-197 apoptotic effect in NIH-H460 cells as shown by Cell Titer-Glo assay. Mean±S.D., ***P<0.0001
We next investigated whether BMF was able to induce apoptosis in NIH-H460 cells. Ectopic expression of BMF promoted a strong proapoptotic activity (Figure 3e), thus confirming the pivotal role of this protein in lung cancer. However, consistently with a cooperative model of action of miR-197 targets, the knockdown of BMF by siRNA (Supplementary Figure S6a) only partially restores the cell viability on miR-197 depletion (Figure 3f).
miR-197 regulates p53-dependent apoptosis and cell proliferation
To better characterize the molecular events engendered by the downmodulation of miR-197, we performed gene arrays (Whole Transcript Expression Arrays, HuGene 1.0 St, Affymetrix, Santa Clara, CA, USA) on NIH-H460 cells treated with specific LNA-197, control LNA or not treated. Coherently with the observed phenotype of treated cells, in silico analyses of microarray results by DAVID26, 27 showed a strong induction of genes involved in the apoptotic process and altered expression of cell cycle regulators on miR-197 downmodulation (Figure 4a). In particular, we found that the p53 pathway was among the most upregulated pathways (P=0.00012), suggesting the involvement of p53 in the antitumor activity triggered by miR-197 targeting. The modulation of some p53-related genes and cell cycle regulators was further confirmed by real-time PCR (RT-PCR) and western blotting (Supplementary Figure S4a).
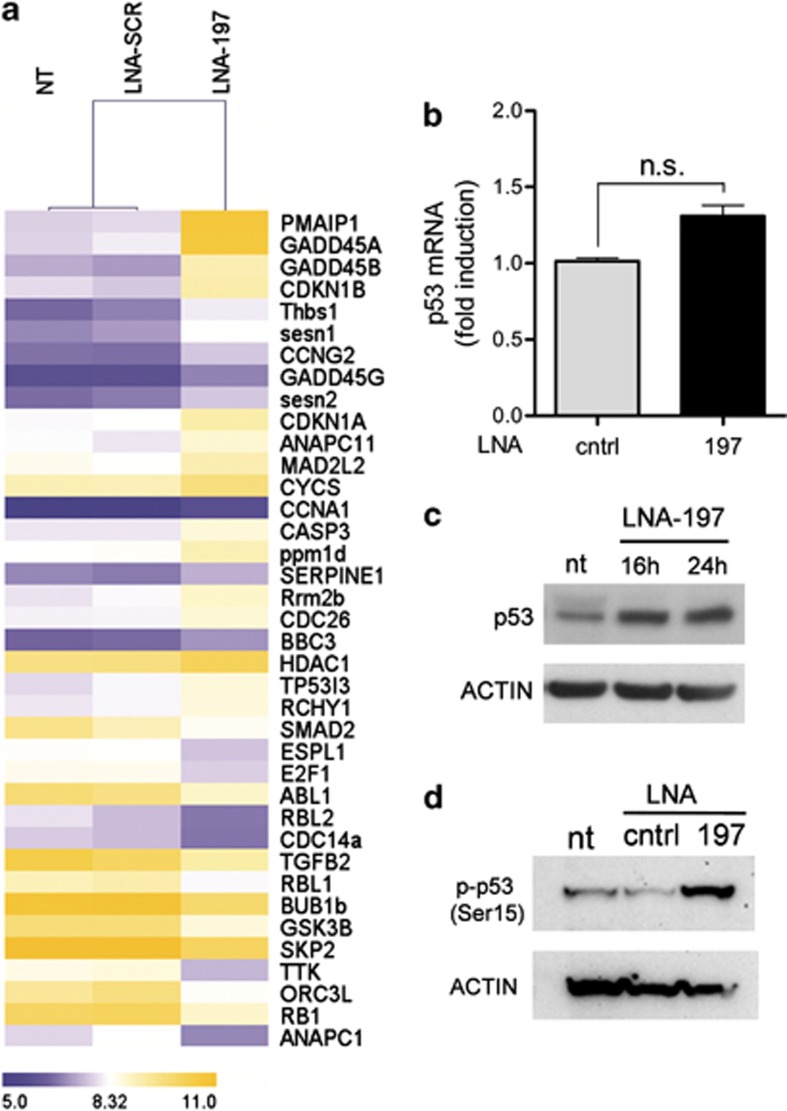
Figure 4.
p53 is activated on miR-197 downmodulation. (a) Unsupervised hierarchical clustering of genes belonging to ‘cell cycle' and ‘p53' pathways (listed by KEGG pathways) that are significantly modulated on miR-197 depletion; genes in the heatmap are shown starting from the most upregulated to the most downmodulated one. (b and c) p53 mRNA and protein levels on LNA-197 treatment in NIH-H460 cells. (d) Western blotting showing p53 phosphorylation at Ser15, 24 h after LNA-197 transfection
To investigate the role of p53 in the induction of apoptosis mediated by LNA-197, the levels of p53 protein were analyzed. Although the mRNA levels of p53 were not significantly affected by miR-197 depletion (Figure 4b), we observed a strong upregulation of the protein already after 16–24 h of LNA treatment (Figure 4c). To elucidate whether miR-197 directly or indirectly regulates the p53 pathway, in silico analyses of miR-197 putative targets were performed (used algorithms being TargetScan, Pictar, miRANDA, MICRORNA.ORG etc.). Although p53 is listed among miR-197 putative targets, neither direct regulation was observed by luciferase assays nor p53 protein downmodulation occurred after miR-197 overexpression (Supplementary Figure S5). The increase in p53 protein levels is likely due to protein stabilization and reduced turnover rate, as it correlates with the increment of the active form, phosphorylated at Ser15 (Figure 4d). This modification is known to cause the release of p53 from the E3 ubiquitin-protein ligase MDM2, thus determining a delayed proteolytic turnover and increased p53 stability.
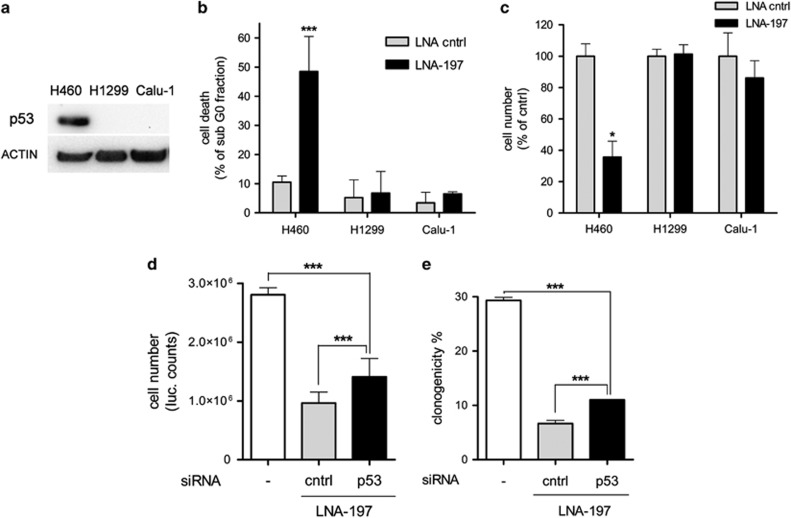
Strikingly, in lung cancer cell lines lacking functional p53 (H1299 and Calu-1), the effect of LNA-197 on cell growth and apoptosis was absent (Figures 5a–c). We next performed p53 silencing in NIH-H460 cells. Notably, the concomitant transfection of anti-p53 siRNA and LNA-197 does not completely knock down p53 protein, but restores p53 levels of untreated cells (Supplementary Figures S6b and c). Such inhibition of p53 induction relieves the effect of LNA-197 on cell viability and clonogenic capacity (Figures 5d and e), confirming that p53 activity is required for the antitumoral effect of miR-197 targeting in NSCLC cells.
Figure 5.
LNA-197 apoptotic effect is p53 dependent. (a) Western blotting showing p53 status of NIH-H460, H1299 and Calu-1 lung cancer cell lines. H1299 and Calu-1 cells, mutated in p53 locus, are not sensitive to miR-197 inhibition, as shown by FACS analysis of PI-stained cells (b) and Cell Titer-Glo assay (c). Downmodulation of p53 by RNA interference can partially rescue LNA-197 effect in NIH-H460 cells; (d) Cell Titer-Glo assay; (e) colony formation assay. Mean±S.D., *P<0.05, ***P<0.0001
NOXA is a direct target of miR-197 in NSCLC
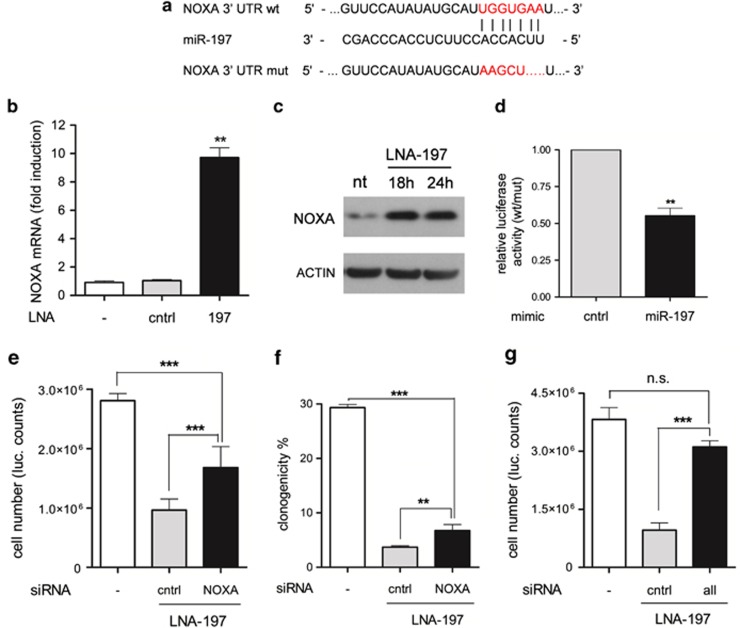
NOXA is a BH3-only proapoptotic protein transcriptionally activated by p53 that contains a putative binding site for miR-197 in the 3′UTR (Figure 6a). Gene arrays results showed that NOXA was the top upregulated gene on miR-197 depletion (Figure 4; PMAIP1: 7.3 mean fold induction, P<0.05). Consistently, knockdown of miR-197 by LNA increased NOXA expression at both mRNA and protein level in NIH-H460 cells (Figures 6b and c). The observed increment of NOXA expression was by far higher than the sole transcriptional activation of the p53-regulated gene p21 (Supplementary Figure S4a; CDKN1A: 1.85-fold increase from gene array), thus suggesting that NOXA overexpression is not only due to p53 activation. To demonstrate the direct interaction with miR-197, the NOXA 3′UTR was cloned into the pGL3-Control vector, downstream to the luciferase gene (pGL3-NOXA UTR-wt). As a control, the miR-197 putative binding site was mutated (pGL3-NOXA UTR-mut). Co-transfection of wt UTR together with a miR-197 mimic caused the decrease of luciferase activity in NIH-H460 cells. Conversely, mutation of the miR-197-binding site abolished the ability of miR-197 to regulate NOXA expression (Figure 6d). Notably, NOXA knockdown by siRNA (Supplementary Figures S6d and e) partially restores cell viability and clonogenic capacity of NIH-H460 cells depleted for miR-197 (Figures 6e and f). Finally, the concomitant depletion of p53, NOXA and BMF by siRNAs completely rescued the LNA-197 effect on cell viability (Figure 6g), indicating that the described mechanisms of apoptosis regulation recapitulate the activity of miR-197 in lung cancer cells.
Figure 6.
NOXA is a direct target of miR-197. (a) Predicted NOXA 3′UTR-binding site for miR-197. The alignment of the seed region of miR-197 with NOXA 3′UTR is shown. The sites of target mutagenesis are indicated in red. (b) qRT-PCR showing inverse correlation of NOXA mRNA and miR-197 expression – mean±S.D., **P=0.0032; (c) western blotting showing the upmodulation of NOXA protein on miR-197 neutralization. (d) Luciferase activity in NIH-H460 cells transfected with NOXA UTR wt or mut in combination with a control or miR-197 mimic. The ratio of normalized luciferase activity in cells transfected with NOXA UTR wt versus mut is indicated – mean±S.E.M., **P=0.0031. Downmodulation of NOXA by RNA interference can rescue LNA-197 apoptotic effect in NIH-H460 cells; shown is Cell Titer-Glo assay (e) and colony formation assay (f) – mean±S.D. **P=0.0086, ***P<0.0001. (g) Downmodulation of p53, NOXA and BMF by RNA interference completely rescues LNA-197 apoptotic effect in NIH-H460 cells as shown by Cell Titer-Glo assay – mean±S.D., ***P<0.0001
Discussion
Lung cancer is one of the leading causes of cancer-associated death worldwide, with NSCLC accounting for 85% of all lung cancers.4, 7 Despite advances in early detection and refining of therapeutic regimens, NSCLC is still characterized by a poor clinical outcome that can profit from a deep comprehension of the molecular bases of lung cancer biology.
Using an unbiased approach, here we have identified miR-197 as one of the top oncogenic miRNA candidates as therapeutic target in NSCLC. Neutralization of miR-197 promotes the upregulation of p53, BMF and NOXA, with consequent caspase activation and p53-dependent apoptosis.
Our study aimed at disclosing new molecular mechanisms underlying NSCLC, to lay the foundation for the development of anticancer-targeted therapies. The choice of miRNAs as targets for new therapies gets strength from the great regulatory potential of these molecules and their ability to simultaneously repress the expression of several proteins.3 Modulating the expression of a single miRNA can hence modify whole pathways and allows a kind of reprogramming of the cells toward a noncancerous phenotype, for example, by reducing proliferation rate or restoring the apoptotic response to stress.
On this exciting background, the identification and functional characterization of new oncogenic miRNAs will reasonably result in pre-clinical studies with mouse models and finally flow into clinics. The high throughput approach presented here, coupled with the unbiased functional assay, permits the identification of unraveled miRNAs involved in lung cancer biology and provides a strong basis for further clinical translation of the obtained results.
Our findings about miR-197 oncogenic activity candidate this miRNA as a new target to fight lung cancer. The induction of programmed cell death represents remarkable promising approach to develop antitumoral drugs. p53 is considered the ‘great gatekeeper', sensing internal and external conditions, and directing the cell toward the best suited answer to a variety of stresses, namely, inducing cell cycle arrest or cell death. Here we describe a novel molecular network regulating p53 activity, where miR-197 acts at different levels of p53 pathway to counteract apoptosis induction, thus allowing cells to proliferate in an uncontrolled manner. We can speculate that lung cancer cells suffer miR-197 inhibition as a stress stimulus and react by activating the p53 signaling pathway that results in reduced cell growth and apoptosis induction. Our gene array data show that on LNA-197 transfection the ribosome synthesis machinery is strongly upregulated (P=8.1 × 10−21), with up to 39 ribosomal proteins that are overexpressed (Supplementary Figure S7). This unbalance in ribosome components could cause the so-called nucleolar stress, characterized by the release in the cytoplasm of ribosomal proteins.30 It has been shown that some of the ribosomal proteins (mainly, L5, L11 and L23) can bind to MDM2 once translocated to the cytoplasm, thus inhibiting the E3-ligase activity and leading to p53 stabilization and activation.31, 32
We showed that miR-197 is also controlling the expression of NOXA, a downstream target of p53 and another BH3-only protein, BMF. This multistep control of programmed cell death represents the paradigm of miR-dependent regulation and explains the fast and irreversible effect that we obtain by downmodulating miR-197.
Finally, we performed in vivo experiments to establish the therapeutic potential of miR-197 targeting. We showed that LNA-197-treated cells loose the capacity to engraft in immunodeficient mice giving encouraging bases for further analyses.
According to the results presented, we propose inhibition of miR-197 as a possible new therapeutic strategy to fight NSCLC. The pharmacological targeting of miRNAs is particularly interesting, as the current technology allows for RNA-based approaches in vivo.33 Recent studies demonstrated a functional downmodulation of miRNA and mRNA levels using antagomiR, LNA and siRNA treatments in mice, non-human primates34, 35, 36 and even in humans.37 In all of these studies, a functional delivery and a low overall toxicity of the treatment were described. Moreover, LNAs already passed a phase II clinical trial in chronic hepatitis C patients.38 The successful targeting of liver, although in non-oncological patients, will guide the effort to improve the delivery of LNAs to lung and other tissues to establish the power of this novel therapeutic approach in the fight against cancer.
Materials and Methods
Cell culture
Human NSCLC A549 and Calu-1 (American Type Culture Collection – ATCC, Manassas, VA, USA) were grown in DMEM, whereas, NIH-H460 and H1299 (ATCC) in RPMI 1640 (Lonza, Basel, Switzerland). All cell media were supplemented with 10% heat-inactivated fetal bovine serum (PAA, Pasching, Austria).
LNA library screening
For the screening of the LNA library, 2 × 103 cells per well were seeded in 96-well plates in the presence of 1% fetal bovine serum. HiPerfect reagent (Qiagen, Hilden, Germany) was added on LNA and incubated for 15–20 min at room temperature. The transfection mixture was then added to the cells at the final LNA concentration of 25 nM. These conditions were formerly set up to guarantee ∼90% transfection efficiency. The percentage of transfected cells was monitored with a control siRNA against NF1b mRNA (40 nM), and verified by RT-PCR. As a positive control, the siRNA against PLK-1 was added into empty wells of the library plates. Cancer cells were assayed 72 h post transfection with the Cell Titer-Glo luminescent cell viability assay (Promega, Fitchburg, WI, USA) on a Beckman DTX 880 plate luminometer (Beckman Coulter, Fullerton, CA, USA). The relative viability was calculated and normalized to the average of all samples, excluding positive controls. The experiment was performed in triplicate. Values below and above two fold standard deviation were considered significant and were used in further analyses.
Anchorage-independent assay
Cells were seeded in 24-well plates and transfected with indicated LNAs at the final concentration of 25 nM. Sixteen hours post transfection, the cells were tripsinized and counted. A total of 500 cells were resuspended in 1.5 ml of medium with 10% fetal bovine serum and 0.3% Agar Noble (Difco, Kansas City, MO, USA), and plated on 1.5 ml of medium with 10% fetal bovine serum and 0.6% Agar Noble. Colony formation was determined after 2 weeks by staining with crystal violet (Fluka, St. Gallen, Switzerland) and colonies were counted visually.
Cell proliferation and apoptosis detection
For proliferation assay and caspase activity evaluation, NIH-H460 and A549 cells were seeded at the same density (2.5 × 103 cells per well) in a 96-well plate, transfection was performed using HiPerFect reagent (Qiagen) and LNAs (Exiqon) at 25 nM. At the indicated time points, three wells for each condition were analyzed. Growth curve were generated evaluating cell number with Cell Titer-Glo (Promega) luminescent cell viability assay; CASPASE 3–7 activity was assessed by a fluorescence-based assay (Apo-ONE Caspase 3/7 assay, Promega) following manufacturer's instructions.
Apoptosis was measured with the Apoptosis Detection Kit (MBL International, Woburn, MA, USA). Briefly, 4 × 104 cells were seeded and transfected with control LNA and LNA-197; at the indicated time points, cells were collected, counted and stained with Annexin V-FITC and PI, and analyzed using a BD FACSCanto Cytometer (Becton Dickinson, San Jose, CA, USA). On BMF overexpression, cells were collected, incubated with Nicoletti buffer (0.1% sodium citrate, pH 7.4/0.1% NP40/9.65 mM NaCl/200 μg/ml RNAse A/50 μg/ml PI) and FACS analyzed.
Immunoblotting and FACS staining
Whole-cell protein extracts (lysis buffer: 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP40, and 1x Protease Inhibitor Cocktail – Sigma-Aldrich, St Louis, MO, USA) were quantified by BCA assay (Pierce, Rockford, IL, USA), separated onto NuPAGE 4–12% polyacrylamide gels (Invitrogen, Carlsbad, CA, USA) and blotted on nitrocellulose membranes (Amersham, England). The filters were hybridized with polyclonal anti-PARP-1 (#9542 Cell Signaling Technology, Danvers, MA, USA), anti-CASPASE 3 (#9665 Cell Signaling Technology), anti-p53 (DO-1, sc-126 Santa Cruz Biotechnology, Dallas, TX, USA), anti-NOXA (Calbiochem, Darmstadt, Germany), anti-SMAD2 (#3122 Cell Signaling Technology), anti-p21 (#2947 Cell Signaling Technology) and p27 (BD Biosciences, Franklin Lakes, NJ, USA; #610241). Monoclonal antibody anti-ACTIN (CP01, Calbiochem) was used as loading control. Bands were visualized and quantified with FluorChem E System (Protein Simple, Santa Clara, CA, USA). For BMF protein FACS evaluation, NIH-H460 cells were transfected with 50 nM ctrl-LNA and LNA-197; 24 h later, 5 × 105 cells were fixed in 2% paraformaldehyde, permeabilized by PBS-Triton X-100 0.1% and incubated for 1 h with anti-BMF polyclonal antibody (#5889 G81, Cell Signaling Technology). Cells were then washed twice with PBS, incubated for 1 h with Alexa Flour 647-conjugated goat anti-rabbit IgG (Invitrogen A-21244) and FACS analyzed. We observed different basal levels of BMF protein that can probably be ascribed to cell culture conditions (that is, cell density, medium availability and so on); nevertheless, in all the experiments performed, LNA-197 transfection determined an increment in BMF protein.
DNA constructs and transfection experiments
The target genes' 3′UTRs were PCR amplified from human genomic DNA by using the following primers: BMF-3′UTRfor 5′-ATAGGTACCGCTCCGATAGCAGGCACAGG-3′, BMF-3′UTRrev 5′-ATAGCTAGCAAGCCTCCACCGTCTTCAGC-3′ Noxa-3′UTRfor 5′-ATAGGTACCGAAGGTGCATTCATGGGTGC-3′, Noxa-3′UTRrev 5′-ATAGCTAGCGAGGACTGTATATAGCCCAG-3′ p53-3′UTRfor 5′-ATAGGTACCACCCAGGACTTCCATTTGCTTTGTC-3′, p53-3′UTRrev 5′-ATAGCTAGCGATCGCCTGAGCCCAGGAGTTT-3′. The PCR fragments were cloned downstream of the luciferase gene in the pGL3-Control PLK+ vector39 by digestion with KpnI and NheI. The mutant derivatives, lacking the putative miR-binding sites, were generated from these constructs by inverse PCR with the following primers: BMF-Δ-for 5′-ATCCTCAGGCAGCCTGCTGCCGTATGC-3′, BMF-Δ-rev 5′-TCGAAGATTCAGAGAGGGGAGAACAACG-3′ Noxa-Δ-for 5′-AGCTTATACACATACTTAC-3′, Noxa-Δ-rev 5′-TATATGGAACTTTCATTC-3′ p53-Δ-for 5′-AGCTTAGTACCTAAAAGGAAATC-3′, p53-Δ-rev 5′-TAGAGGTTGTCAGACAGGG-3′.
The BMF cDNA was PCR amplified from pBluescript-BMF construct (Open Biosystems, Lafayette, CO, USA) with the following primers: BMF atg 5′-ATGGTACCATGGAGCCATCTCAGTGTG-3′ and BMF stop 5′-GATCTCGAGTCACCTAGGGCCTGCCCCGTTCC-3′. On KpnI and XhoI digestion, the cDNA was cloned into pcDNA 3.0 vector under the control of CMV promoter. Transfection of 0.5 μg of DNA was performed with Fugene HD (Promega) in a 3 : 1 ratio.
All siRNAs (Dharmacon smart pool: PLK1, NF1b; Qiagen: BMF, p53, Noxa) were used at the final concentration of 25 or 50 nM. The miR mimics (Qiagen) and the LNAs (Exiqon) were used at the final concentration of 30 and 25 nM, respectively. The transfection experiments were performed using HiPerFect reagent following manufacturer's instructions.
Luciferase reporter assay
In luciferase experiments, NIH-H460 cells were seeded in a 96-well plate (5 × 103 cells per well) and transfected with 40 ng of firefly luciferase vectors (empty pGL3-Control, pGL3-BMF UTR wt or mutant, pGL3-noxa UTR wt or mutant) and 4 ng of Renilla luciferase vector (pRL-TK, Promega), together with 50 nM LNAs (Exiqon) or 30 nM miR mimics (Qiagen), where indicated. 0.15 μl per well of Fugene HD (Roche, Basel, Switzerland) were used for transfection. Experiments were measured 48 or 72 h after transfection by using Victor-X3 (Perkin Elmer, Waltham, MA, USA). Firefly and renilla luciferase activities were measured using the Dual Luciferase Assay kit (Promega); transfection efficiencies were normalized by calculating the ratio firefly/renilla. The experiment was performed three times in quadruplicate.
Quantitative RT-PCR analyses
For mRNA analysis, total RNA was purified with TRIZOL Reagent (Invitrogen) and reverse transcribed with random primers (N6, Roche) and MMLV RT (Invitrogen) after DNase-I treatment (RQ1 DNase, Promega). RT-PCR was performed with SensiMiX (Quantace, London, UK) using the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, USA) according to standard procedures. Human GAPDH was used as endogenous control. Expression of mature miRNAs was determined using miRNA-specific quantitative real-time PCR (qRT-PCR; Applied Biosystems). U6 was used for normalization.
In vivo experiments
CD/1 female athymic nude mice were purchased from the Jackson Laboratory and housed in groups of six in isolated ventilated cages; food and water were provided ad libitum. All animal procedures were performed according to the protocol approved by the Istituto Superiore di Sanità Animal Care Committee.
A total of 106 NIH-H460 cells were transfected with 25 nM LNA; 16 h after transfection, cells were counted using trypan blue staining and 105 viable cells were subcutaneously injected into the flank of 6–8-week-old mice. Tumor size was assessed every 2 days by caliper measurement. Tumor volume was calculated as follows: D × d2 × π/6, where D and d are the longer and the shorter diameters, respectively. N=8.
Cells derived from the same transfection were used to perform anchorage-independent assay and annexin V staining.
Acknowledgments
We thank S. Soddu for critically reading the manuscript. This study was supported by Italian Association for Cancer Research (AIRC) Investigator Grant to RDM.
Glossary
- miRNA, miR-
microRNA
- SCLC
small cell lung cancer
- NSCLC
non small cell lung cancer
- LNA
locked nucleic acids
- BMF
Bcl-2 modifying factor
- PLK
polo like kinase
- siRNA
small interfering RNA
- UTR
untranslated region
- EGF-R
epidermal growth factor receptor
- ALK
anaplastic lymphoma kinase
- EMT
epithelial-mesenchymal transition
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Oren
Supplementary Material
References
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang S, Alexander R, Yao Y, MacLennan GT, Pan CX, et al. The landscape of EGFR pathways and personalized management of non-small-cell lung cancer. Future Oncol. 2011;7:519–541. doi: 10.2217/fon.11.25. [DOI] [PubMed] [Google Scholar]
- Jemal A, Ma J, Rosenberg PS, Siegel R, Anderson WF. Increasing lung cancer death rates among young women in southern and midwestern States. J Clin Oncol. 2012;30:2739–2744. doi: 10.1200/JCO.2012.42.6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant M, Allemani C, Santaquilani M, Knijn A, Marchesi F, Capocaccia R. EUROCARE-4. Survival of cancer patients diagnosed in 1995-1999. Results and commentary. Eur J Cancer. 2009;45:931–991. doi: 10.1016/j.ejca.2008.11.018. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network—another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Xiao J, Lin H, Luo X, Wang Z. miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 2011;30:524–532. doi: 10.1038/emboj.2010.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Schageman JJ, Subauste MC, Saber B, Hammond SM, Prudkin L, et al. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7:1234–1243. doi: 10.1158/1541-7786.MCR-08-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE, et al. Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol. 2011;4:575–586. [PMC free article] [PubMed] [Google Scholar]
- Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–3718. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F, Teresi RE, Broelsch CE, Frilling A, Eng C. A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab. 2006;91:3584–3591. doi: 10.1210/jc.2006-0693. [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keutgen XM, Filicori F, Crowley MJ, Wang Y, Scognamiglio T, Hoda R, et al. A panel of four miRNAs accurately differentiates malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin Cancer Res. 2012;18:2032–2038. doi: 10.1158/1078-0432.CCR-11-2487. [DOI] [PubMed] [Google Scholar]
- Hamada S, Satoh K, Miura S, Hirota M, Kanno A, Masamune A, et al. MiR-197 induces epithelial-mesenchymal transition in pancreatic cancer cells by targeting p120 catenin. J Cell Physiol. 2012;228:1255–1263. doi: 10.1002/jcp.24280. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Villunger A, O'Reilly LA, Beaumont JG, Coultas L, Cheney RE, et al. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293:1829–1832. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- Pinon JD, Labi V, Egle A, Villunger A. Bim and Bmf in tissue homeostasis and malignant disease. Oncogene. 2008;27 (Suppl 1:S41–S52. doi: 10.1038/onc.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Chen LH, Chen JD. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25:5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Liao JM, Liao WJ, Lu H. Scission of the p53-MDM2 loop by ribosomal proteins. Genes Cancer. 2012;3:298–310. doi: 10.1177/1947601912455200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias E, Jin AW, Deisenroth C, Bhat K, Mao H, Lindstrom MS, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell. 2010;18:231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.