Abstract
A grand challenge impeding optimal treatment outcomes for cancer patients arises from the complex nature of the disease: the cellular heterogeneity, the myriad of dysfunctional molecular and genetic networks as results of genetic (somatic) and environmental perturbations. Systems biology, with its holistic approach to understanding fundamental principles in biology, and the empowering technologies in genomics, proteomics, single-cell analysis, microfluidics, and computational strategies, enables a comprehensive approach to medicine, which strives to unveil the pathogenic mechanisms of diseases, identify disease biomarkers and begin thinking about new strategies for drug target discovery. The integration of multi-dimensional high throughput “omics” measurements from tumor tissues and corresponding blood specimens, together with new systems strategies for diagnostics, enables the identification of cancer biomarkers that will enable presymptomatic diagnosis, stratification of disease, assessment of disease progression, evaluation of patient response to therapy, and the identification of reoccurrences. While some aspects of systems medicine are being adopted in clinical oncology practice through companion molecular diagnostics for personalized therapy, the mounting influx of global quantitative data from both wellness and diseases, is shaping up a transformational paradigm in medicine we termed predictive, preventive, personalized, and participatory (P4) medicine, which requires new strategies, both scientific and organizational, to enable bringing this revolution in medicine to patients and to the healthcare system. P4 medicine will have a profound impact on society—transforming the healthcare system, turning around the ever escalating costs of healthcare, digitizing the practice of medicine and creating enormous economic opportunities for those organizations and nations that embrace this revolution
Keywords: Systems medicine, cancer complexity, quantized cell populations, blood biomarkers, molecular diagnostics, P4 medicine
I. Introduction
A new paradigm in medicine is arising which is termed predictive, preventive, personalized, and participatory, (P4) Medicine [1, 2]. This approach is driven by systems strategies and new technological advancements for disease diagnostics[3], therapeutics and prevention, coupled with ever-increasing digitization of medicine and consumerism. At its heart is the transformation of medicine from a reactive discipline that responds only after symptoms of disease arise to one that is focused deeply on maintaining wellness. The challenge of dealing with disease is its complexity and P4 medicine deals with this complexity by generating an enormous amount of data on the individual patient. Indeed, our prediction is that some 10 years into the future each patient will be surrounded by a virtual cloud of billions of data points and that we will have the computational tools to reduce this enormous data dimensionality to simple hypotheses about health and disease (Figure 1).
Figure 1.
A schematic diagram of the billions of different types of digital data points that will become a typical part of a patient's record in 10 years. Note the very different types of data ranging from molecular and cellular to typical medical records to the environmental influences captured by social networks.
It should be stressed that the fundamental principle of a systems approach to medicine is that disease arises as a consequence of one or more disease-perturbed networks in cells of the relevant organ, which we are able to read now in increasing detail. Because diseases result from perturbed networks there are early signals that can be tracked – even pre-symptomatically – as we come to a deep understanding of their functioning. Thus, a major challenge for P4 medicine pertains to the growing appreciation of disease complexity – reflected in these disease-perturbed molecular networks as unveiled through increasingly sophisticated analysis of ever increasing omics data.
Cancers are perhaps among the most complex diseases, and pose significant challenges for treatment— as the last several decades have readily demonstrated. The challenge of cancer has been dual in nature — both theoretical and practical — namely to gain a deep understanding of the mechanistic underpinnings of cancer and, from this knowledge, to develop strategies for better diagnoses and therapies for patients. As a community, we have been extraordinarily successful at deepening mechanistic understanding of cancer. At least ten hallmarks or phenotypes of cancer contributing to the tumorigenic behavior have been enumerated to date [4]. Translating this deep and increasing wealth of knowledge about cancer biology into clinical practice is of utmost importance and remains a daunting task. Translation of knowledge to practice is due to the complexity challenge of cancer, and new experimental and computational tools are now providing powerful means to deal with these challenges more effectively. Prior to the genomic era, oncologists relied on very limited molecular testing in addition to conventional clinical presentation and histopathological analyses to guide practice. Dramatic advances in genomics and proteomics over the last decade have enabled innovative strategies for interrogating cancers through detailed molecular analyses, spawning scores of composite molecular markers (or marker panels) of putative or demonstrated clinical relevance. Most cancers arising in an organ (e.g. breast cancer) are composed of distinct subtypes, each with unique molecular phenotypes, and each warranting distinct clinical management strategies. One of the grand challenges in P4 medicine is to develop tools and strategies to stratify cancers into their distinct subtypes so that proper impedance matches can be achieved with therapeutic reagents. As demonstrated in human prostate and ovarian cancers, cancer arises from disease-perturbed networks [5]. Different cancers perturb distinct combinations of networks, and the ability to identify these different combinations of disease-perturbed networks allows one to stratify cancers or classify the different subtypes of tumors of particular organs. Disease-perturbed networks change with progression, so there are two key dimensions to decouple when using networks to analyze disease, i.e. one must distinguish between the disease-perturbed networks reflecting different disease stratifications as opposed to those expanding disease-perturbed networks that reflect disease progression. As a vanguard in clinical oncology, personalized cancer medicine is thriving upon the successful development of novel companion molecular diagnostic markers for the purposes of patient disease stratification, clinical outcome predictions, and therapeutic interventions [6-9]. It is also employing the identification of disease-perturbed networks in tumor tissues with genomic sequencing to identify specific molecular lesions that will dictate the choice of appropriate drugs—one of the first powerful examples of personalized medicine [10-16].
In this review, we will put forth our view of systems (P4) medicine using cancer as a model. Systems medicine embraces a systems approach to cancer, transformational new technologies (genomic, proteomic, single-cell analyses and high throughput phenotypic assays), and powerful computational methods for delineating relevant biological networks fundamental to the cellular and molecular origins of cancer. Moreover, insights gained from these cancer applications may be adopted to explore powerful new diagnostic and therapeutic strategies for dealing with other human diseases. P4 medicine includes all the elements of systems medicine and also includes the societal challenges that arise from attempting to bring P4 medicine to patients, as will be discussed later.
II. Origins of cancer complexity
Heterogeneity in cancer arises in large part from genetic variation, where the random mutation frequency in human cancer cells is over 100 to 500-fold greater than in adjacent normal cells[17-19]. This information is digital in nature and can be very precisely determined. The histo-pathological heterogeneity of cancer reflects these changes as well as dynamically and environmentally responsive changes to the epigenome, transcriptome, and proteome etc. Taken together, there are extensive alterations in molecular networks that lead to a much-heightened level of molecular heterogeneity in human cancers. These heterogeneous properties contribute to the treatment-refractory nature and differential response arising from individual therapeutic regimes. In this sense the term “cancer” does not represent a disease, but rather a highly diverse set of diseases with highly variable molecular causes that lead to the aforementioned shared phenotypic hallmarks. All these levels of complexity call for global systems analysis of tumor tissues–molecular, cellular and phenotypic—where these data are all organized into models that are predictive and actionable.
A. Quantized cellular heterogeneity
In addition to the histologically defined cell types in a given organ, two types of studies suggest that the metazoan cells in particular organs may be quantized digitally into separate cell types characterized by distinct and stably expressed transcriptomes. First, for example classic studies with sea urchin have demonstrated that cells taken at different stages of development are successively locked into a discrete series of quantized transcriptome patterns [20]. Each of these quantized cell populations has distinct functions. Second, the study of cell types in various organs (brain, liver, kidney) suggests that there are distinct types of cells that carry out interrelated but distinct functions. The emerging picture is that each organ has an unknown number of distinct cell types that presumably are defined by distinct and persistent patterns of gene expression [21]. The same appears to be true of cancers that are often epithelial in nature. For example, several reports have suggested that the true neoplastic potential of tumors lies in cancer stem cells that may constitute less than 1% of the cancer cell population [22-24]. We believe that most cancers may indeed be composed of distinct epithelial cell populations that play important but distinct roles in the neoplastic process. Molecular characterization of these distinct cell populations by cutting edge genomic and proteomic technologies – ultimately with single-cell resolution – will be essential to understanding the true nature of cancer pathogenesis.
B. Excessive genomic mutations
One of the most significant features of cancer is the fact the frequency of random mutations in human cancer cells is more than 100 to 500-fold greater than that in adjacent normal cells [17-19]. This observation has significant consequences for understanding the mechanisms of neoplasia, and for developing effective diagnostic and therapeutic approaches. In particular, the increased rate of mutation and the fact that the genomes of cancer cells within a given tumor are heterogeneous means that cancers have more inherent variability to accelerate their pace of evolution when compared to their cells of origin. Cancer-enabling mutations such as increased growth rate, the ability to invade surrounding tissues or to metastasize to distant sites will experience tremendous positive selective pressures. The challenge of this genomic mutational diversity is that the signal to noise issues are significant. Some mutations actively drive the neoplastic process (driver mutations) whereas other mutations with significant frequencies can be carried along as passengers not requiring selection and not contributing to the disease process (passenger mutations). This genetic diversity will be reflected in transcriptional (and translational) diversity—both in coding mutations and in altered levels of expression. Identifying causal mutations from passenger mutations is highly complex, particularly given that mutations that are passengers can become important in the context of other mutations (and vice versa). Greatly deepening our understanding of tumors as evolving systems is fundamental to addressing the challenge of cancer complexity and to developing therapies that will work for the long term.
This increased mutational level, of course, makes it possible for cancer cells to mutate away from being responsive to drugs—and drug resistance frequently comes as a consequence of treatment with a single drug. This increased mutation level means that different cancers may alter different combinations of networks, thus leading to distinct subtypes of cancers derived from a particular organ. These different subtypes may respond to distinct drugs, may have different prognoses, and will require new approaches for diagnostic stratification. Systems approaches for the development of combination therapies that take into account the underlying evolution of tumors will thus be key. As the tumor of a particular subtype or stratification progresses, mutation continues and the patterns of expressed information (mRNAs, miRNAs and proteins) continue to change. Thus, it is challenging to distinguish the consequences of tumor stratification from the consequences of tumor progression in humans because it is difficult to acquire temporal information (how the tumor changes with time). Overcoming this challenge will require sophisticated new diagnostic techniques. Valid animal models will be critical in studying the dynamics of cancer progression [25].
To better characterize the genomic complexity of cancer, The Cancer Genome Atlas (TCGA) project was launched several years ago (http://cancergenome.nih.gov/). One of the first cancers being studied by TCGA is glioblastoma multiforme (GBM)[10]a deadly brain tumor with a median survival of just over one year. TCGA is generating multi-dimensional genomics data sets including DNA copy number, gene expression, DNA methylation, and sequence aberration from a large number of GBM tumors. This data is tightly integrated and computationally analyzed to identify disease-associated alterations. Among the first discoveries from TCGA was the genomic deregulation of three core GBM biological pathways [RB, p53, and RTK/RAS/PI(3)K], leading to a novel hypothesis for a chemo-resistance mechanism[10, 26]. However, one of the concerns about the TCGA project has been the issue of signal to noise arising from the very large number of measurements of highly heterogeneous tumors with much smaller samples sizes (though the TCGA is the largest such project to date). One key source of variance is due to the fact that the tumors were analyzed using DNA and mRNA from mixtures of heterogeneous tumors cells—and indeed other types of normal cells as well. We believe this signal to noise challenge can be addressed through single-cell analyses to identify the quantized populations of tumor cells. These can then be separated by cell sorting into discrete cell populations based on cell-surface markers identified from molecular characterizations (genomes, transcriptomes, and proteomes) of the quantized cells. As we have demonstrated recently, once the complete genome sequences of a family are determined, one can use the principles of Mendelian genetics to identify (and correct) about 70% of the DNA sequencing errors. This high level of accuracy permits the ready identification of genes that encode simple Mendelian disease traits [27]. In that study, we sequenced the genomes of a family where the parents were normal and the two children each had two genetic diseases. With the accuracy of sequencing made possible by the complete genome sequences of the entire family, we were able to reduce the number of candidate genes down to just four—and from there the disease gene assignments could readily be made. Similar strategies can be applied into the studies of cancer where both Mendelian genetics and sporadic mutations are known contributors to tumorigenesis. The identification of a large fraction of the DNA sequencing errors in cancer genomes (by sequencing their families) will allow one to be certain which are real driver and passenger mutations. We propose two highly informative, signal-enhancing steps that will lay the foundation for an effective genomics strategy for cancer research. First, distinct quantized cancer cell populations from an individual's tumor tissues are determined from single-cell molecular characterization using omics technologies. Second, sequence the normal genomes of that individual as well as the members of his or her family will be determined so that the Mendelian error correction can be applied to significantly improve the quality of the tumor DNA sequence data. Information gained from these studies can be applied to better understand with systems approaches (see below) the disease mechanism, to develop better blood diagnostic biomarkers, and to explore better approaches to therapy.
III. Systems biology and the emerging technologies enables systems medicine
The key to understanding the complexity of cancer as a disease is to effectively utilize systems approaches to deeply and correctly interpret data streams made possible by the emerging technologies of contemporary biology. Biological systems employ both digital information encoded by each individual's genome as well as analog information encoded in epigenetic changes, RNAs, proteins, metabolites and networks that can change in response to environmental perturbations.
A. Principles of systems biology
Systems approaches are predicated on the idea that biology is an information science. 1) There are two fundamental types of biological information—the digital genome and its environmental signals— and these are integrated to mediate phenotype and for cancer initiation and progression. 2) Biological information is captured, transmitted, integrated, and modulated by biological networks before being passed on to biological machines, simple and complex, for execution. One of the keys to understanding cancer-inducing mechanisms is delineating the dynamics, both spatial and temporal, of the underlying perturbed networks. 3) Biological information is hierarchical and multi-scale, spanning DNA, RNA, protein, interactions, networks, cells, tissues, individual, populations, and ecologies. To understand biological systems one must ascertain how the environment modifies the digital information at each of these levels—and this process calls for sophisticated multi-scale models that span these scales. The integration of different types of information is one of the keys to dealing with signal to noise issues.
Systems biology has three central elements. 1) It is hypothesis-driven, where a model (which is a formally structured, precise, and potentially complex hypothesis) is formulated from existing data. Hypotheses from model predictions are then tested with systems perturbations and the high throughput acquisition of data. The data then are reintegrated back into the model with appropriate modifications—and this process is repeated iteratively until new predictions from theory and experimental data are in agreement. 2) It is based on high throughput data that should i) be global (comprehensive), ii) generated for different data types that will be integrated, iii) used to monitor networks dynamically, iv) provide deep insight into biology, and v) be integrated using proper statistics and bioinformatics to handle the enormous signal to noise problems. 3) Models may be descriptive, graphical or mathematical as dictated by the amount of available data, but they must be predictive. For medical use, predictions made must be actionable and useful for treating patients.
Boosting signal-to-noise in complex biology is essential for deciphering complexity. Two fundamentals have been leveraged by biologists to reduce noise and to enhance statistical power: filters and integrators. Filters are used to winnow down the number of candidates based on the biological assumptions about complexity (e.g. modularity, hierarchical organization, complexity arising from evolution, and inheritance); another is the availability of complementary data of genome, transcriptome, miRNAome, proteome, metabolome, and interactome. [28] Successful application of these strategies in disease will lead to transformational understanding of disease and therapeutics.
B. Systems approaches to medicine
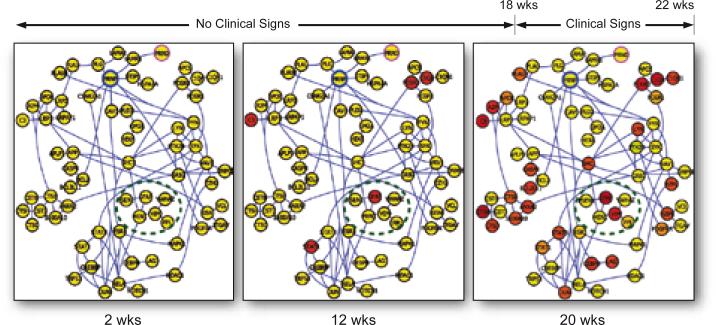
The framework for approaching these studies is a systems approach to disease—the idea that disease arises as a consequence of the disease-perturbation (genetic and/or environmental) of one or more biological networks in the relevant organ. This disease-perturbation alters the envelope of information the network encodes in a dynamic manner that changes during the progression of the disease (e.g., changing levels of mRNAs, miRNAs, or indirectly proteins)—and these altered levels explain the pathophysiology of the disease and provide new insights into diagnosis and therapy. We will illustrate this approach with a recently published systems approach to prion infection in mice (a neurodegenerative disease)[29]. We analyzed the transcriptomes of the infected animals at 10 time points across the approximately 22 weeks of disease progression—and at each time point subtracted these transcriptomes from diseased animals from those from normal liter mates—to identify 7400 differentially expressed genes (DEGs)—which represented a staggering signal to noise problem. We carried this study out in 6 different inbred strain combinations of mice infected with two different strains of prions—and then used a deep biological understanding of the disease process to subtract away noise (e.g. in the double knock out for the prion gene—the animals after injection with infectious prions never get the disease—so any changes in the transcriptomes of these animals are irrelevant to the core prion disease response and can be subtracted away). With seven additional subtractions, we identified a core of about 333 differentially expressed genes that encoded the basic prion-disease process. We mapped these DEGs on to four major biological networks of the prion disease process that had been defined by serial histopathology of diseased brains—and then integrated the transcriptome data with 1) serial brain histopathological analyses of these animals, 2) serial saggital brain sections stained for infectious prions, 3) clinical signs of the disease and 4) blood biomarker analyses. Figure 2 illustrates one of these major dynamically changing networks. We drew the following conclusions. 1) Two thirds of the DEGs mapped into the four known networks—and their dynamics explained virtually every aspect of known prion disease. 2) These four networks were disease-perturbed in a serial manner—first prion replication and accumulation, second, glial activation, third, the degeneration of neuronal axons and dendrites and finally neuron apoptosis. 3) The remaining one third of the DEGs identified six new networks that were not here-to-fore unknown to prion disease—the so-called “dark genes and networks of prion disease” as identified by the global analyses of normal and diseased transcriptomes. These insights emphasize the importance of global analyses of the transcriptomes. 4) These studies suggested new approaches to blood diagnostics that are discussed below. 5) For therapy it is obvious the first and most proximal prion-specific network should be re-engineered with drugs to make it behave in a more normal manner and hopefully abrogate the downstream consequences of this pathological progression. It is clear that multiple drugs will be required to re-engineer biological networks. This systems view of disease will be applied to human cancers—which present special challenges since the disease cannot easily be followed serially in individuals.
Figure 2.
A schematic of the network perturbations of one neural degenerative network over the 20 weeks of the progression of this disease in a mouse model. The red nodes indicate mRNAs that have become disease perturbed as compared with the brain transcripts of normal mice. The spreading of the disease-perturbed networks at the 3 different times points is striking--indicating the progressive disease-perturbation of this neurodegenerative network.
IV. Blood as a window for monitoring health (wellness) and disease
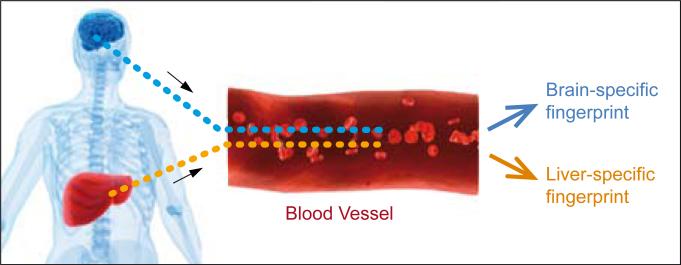
A systems approach to blood diagnostics emerged from two ideas arising from the prion studies. First, some transcripts are expressed in their disease-perturbed networks 8 weeks or more before the first clinical signs (e.g., 10 weeks and 18 weeks, respectively). We were able to demonstrate that several of these DEG transcripts encoded proteins expressed in the blood and we could see the altered protein levels in the blood—hence this was an example of presymptomatic diagnosis, a long-sought keystone of cancer research. However, these DEGs were expressed in several different organs—and hence we could not be certain of the location of the disease-perturbed process directly from observing protein concentration changes in the blood. Second, in order to obtain blood markers with an organ-specific addresses, we identified transcripts that were organ-specific by deep comparative transcriptome analyses across 40 or more different organs in humans and mice. From these analyses—and through an examination of the human and mouse blood protein data bases and experimental mass spectrometry analyses—we were able to identify about 100 brain-specific proteins in humans and mouse (Figure. 3). Of these, about 95% were orthologous between the two species (the presumptions is that they will reflect similar activities in the two species), and these collectively constituted a brain-specific blood fingerprint. We were able to show that some of these brain-specific proteins could also be used for presymptomatic diagnosis of prion disease in mice—and that brain-specific blood proteins encoded by each of the four distinct networks may exhibit concentration changes in the blood in a serial manner consistent with the order of disease-perturbation of these transcriptional networks. These data demonstrate that we will be able to assess both early disease detection and disease progression from the blood. Hence in the organ-specific blood protein fingerprints each individual protein assesses the behavior of its cognate biological network—distinguishing normal functioning from disease-perturbed functioning. Because each disease perturbs different combinations of networks, the brain-specific blood fingerprints will be able to distinguish normal from disease and, if diseased, identify the disease. This will enable the five holy grails of blood disease diagnoses: 1) presymptomatic diagnosis; 2) stratification of disease; 3) assessment of the progression of the disease; 4) following patient response to therapy; and 5) identifying reoccurrences. We are now applying this strategy to identify human organ-specific blood biomarker for several cancer types.
Figure 3.
A schematic drawing indicating brain-specific and liver-specific blood proteins that come to constitute an organ-specific fingerprint in the blood. These organ-specific proteins serve as reporters for their specific cognate networks to differentiate a normal organ from its specific disease counterpart. When a network becomes disease-perturbed--its cognate proteins will change their concentration levels in the blood. Since different diseases perturb different combinations of networks--the organ-specific blood fingerprints can distinguish health from disease--and if a disease which disease--for each organ whose blood fingerprints are measured quantitatively.
In addition to blood proteins as tumor biomarkers, circulating DNAs, mRNAs, and microRNAs, as well as circulating tumor cells have also been studied which can serve as surrogate disease biomarkers and for monitoring cancer recurrence [30-32].
V. Application of emerging technologies in cancer research.
Emerging technologies in genomics, proteomics, microfluidics, and single-cell analysis are transforming cancer research and in the past several years have started to make an impact in the practice of oncology.
A. Genomic sequencing technologies
High throughput sequencing technologies have been widely adopted to identify both known and novel mutations in cancers. For instance, TCGA consortium employed targeted re-sequencing of a few hundred genes in a large cohort of GBM patient samples to delineate their mutation spectrum. Rapidly evolving next generation sequencing techniques (NGS) have been applied broadly in genome-wide exon sequencing, as well as for whole genome sequencing in a variety of cancers including breast cancer, prostate cancer, and leukemia [13, 33-35]. These studies analyzed normal patient DNA (from blood cells) and their tumor DNA to assess the levels of tumor mutation. Successful application of mate-pair NGS (sequencing both ends of the DNA fragment to enable assembly of the NGS short reads into large contigs) has been applied to colon cancer where genomic rearrangements have been used as personalized biomarkers for predicting disease progression [36]. We are the first to employ family whole genome sequencing to identify causal mutations in inheritable diseases [27]. We anticipate more family sequencing strategies being applied in the near future for the identification of germline mutations in cancer patients. We will also be able to generally distinguish more effectively between somatic mutations and DNA sequencing errors in the tumor tissues. NGS has also been used for single-cell genomic sequencing studies in breast cancer cell lines [37]. Although current sequencing throughput and sensitivity is still not sufficiently effective to cover the whole genome of a single cell in a single sequencing run, it did allow the distinction of disparate cell populations at the copy number level. With the exponential improvement of data output and quality, and drastic reduction in sequencing cost, we will see a deluge of genomic sequencing data from an ever-increasing number of cancer patients. The grand challenge that most biologists and clinicians will have to face is to sift through these enormous amounts of data to extract information that will optimally benefit patients. Genomic data delineating which signal transduction networks have been disease-perturbed are already being employed to select complementary therapies for personalized analyses of tumors [38-41]
B. Transcriptomic profiling and disease stratification for personalized medicine
One of the most mature genomic technologies in cancer research is the use of gene expression profiling to molecularly stratify similar cancers for guiding clinical patient management. The prevailing technologies are DNA microarrays and quantitative PCR. For instance, Genomic Health Inc. has developed a 21-gene q-PCR assay which predicts the likelihood of chemotherapy benefit for low grade breast cancer patients and quantifies the likelihood of recurrence[42]. More recently a 12-gene qPCR assay has been developed that provides an individualized score reflective of the risk of colon cancer recurrence for individual patients with stage II colon cancer (Genomic Health Inc, CA). This assay helps provide individualized treatment decisions. Our own research also looked into relative gene expression levels and identified a highly accurate two-gene classifier that separates gastrointestinal stromal tumor (GIST) and leiomyosarcoma (LMS) – two clinically indistinguishable tumors – with very high accuracy [43]. In addition, we have also developed a cancer stem cell-specific (CD133+ subpopulation) transcriptomic signature for molecular staging and subtyping glioblastoma (GBM) based on gene expression profiling of highly purified CD133+/– GBM cells [44]. Significant enrichment of CD133-up gene set in stem cells and higher-grade human cancers, provides molecular support for the stem-cell-like nature of CD133+ cells and enabled identification of a novel aggressive subtype of GBM (younger patients with shorter survival) who accumulated excessive genomic mutations. The CD133 related gene panel provides the potential for an objective means to evaluate cancer aggressiveness, and provides an approach for further developing molecular tests to stratify cancer patients through designing clinical trials for both old and new drugs. This study also established, for the first time, a genetic link between a cancer stem cell signature and a hypermutated genotype.
C. Targeted proteomics approach – Selected Reaction Monitoring (SRM)
Targeted proteomics techniques such as the recently-developed selected reaction monitoring (SRM) approach [45] are enabling efficient and specific detection and quantification of potential protein markers in patient tumor tissues and blood samples. SRM analysis is performed by triple quadruple mass spectrometry. Typically, the first mass analyzer allows one or more ideally proteotypic peptides (unique to the protein) to be selected for further fragmentation in the second mass analyzer (collision cell). The third analyzer monitors for multiple user-defined fragment ions (transitions) produced by collision-induced dissociation of proteotypic peptides in the second mass analyzer. These techniques require one to have a predetermined set of protein biomarker candidates or transitions. This SRM assay can analyze several hundred proteins at the mid-atomole level in an hour. The benefit of SRM assays is that detection of multiple targets in blood no longer requires the complexities associated with ELISA development, so that marker validation time is no longer such a significant issue. We have initiated a human proteome atlas project in which we have developed SRM mass spectrometry assays for virtually all human proteins—thus further reducing the assay development time (http://www.srmatlas.org/).
Using a genetically engineered mouse model of prostate cancer and targeted blood proteomics via SRM-MS coupled with glycoprotein capturing, a novel panel of protein markers was identified from mouse tumors and validated in mouse and then human bloods derived from patients with prostate cancer. This protein panel outperformed the current gold standard blood protein test for prostate cancer, PSA [46]. Our own efforts using a tissue-specific protein panel and SRM assays also established blood protein essays capable of stratifying and detecting early acetophenamine toxicity (Hu and Lausted, et. al. submitted).
D. Microfluidics and single cell analysis
Microfluidics devices have been developed for genomics assays analyzing hundreds to a thousand transcriptomes simultaneously [47], after appropriate linear mRNA amplification and bar-coding, in a single run of a next generation sequencer. We are collaborating with a microfluidics company, Fluidigm, to develop a 1000 plate single-cell analyzer for highly multiplexed transcriptome analyses on single cells. We believe that single-cell analyses will be one of the transformational technologies in cancer biology, as well as in biology and medicine in general.
E. Information technology for healthcare poses many challenges
The world of P4 medicine poses many challenges for generating sufficient data to deal with the enormous signal to noise problems. 1) How do we identify sufficient patient populations to deal with the extensive disease stratification that will, for example, divide human breast cancer into at least five different subtypes of disease [1]? 2) As we suggested earlier, the average patient in 10 years will be surrounded by a virtual cloud of billions of data points (Figure. 1). How will we reduce this enormous dimensionality into simple hypotheses about health and disease? 3) In 10 years we suggest that the human genome will be a routine portion of each individual patient's medical record. If so, how will we generate the computational tools to be able to mine comparatively the 340 million genomes in the US, for example, for the predictive medicine of the future? 4) How will we deal with the enormous amounts of data that will be generated with the extensive in vivo imaging possibilities of the future as well as single-cell analyses? The opportunities are staggering; the informational technology challenges are striking.
VI. Concluding remarks
Rapidly advancing genomics, proteomics, metabolomics, single-cell analysis, phenotyping, microfluidics and imaging technologies, as applied to various human organs as well as tumor tissues and blood, will change the way cancer is diagnosed (early detection, stratification into different subtypes, assessment of stage of progression and response to therapy) and treated, will enable using old drugs more effectively through an impedance match with the stratified subtypes and, of course, will facilitate the creation of drug combinations that can re-engineer disease-perturbed networks to behave in a normal fashion.
The current evidence-based medicine is largely a reactive response to disease rather than the proactive response of P4 medicine. Evidence-based medicine has been important in advancing the state of healthcare—but it may well have reached it limits—and pouring large sums of support into its advancement may yield increasingly marginal returns in the future. The contrasts between evidence-based medicine and P4 medicine are really striking—proactive vs. proactive, population based vs. individual based, clinical trials with large undifferentiated populations vs. clinical trials on small stratified populations, etc. (see Table 1).
Table 1.
P4 Medicine is Revolution in How to Practice Medicine
| Reactive Medicine—Evidence-based Medicine | Proactive P4 Medicine |
|---|---|
| Reactive--respond after a patient is sick (symptoms based) | Proactive--responds before a patient is sick (based on pre-symptomatic markers) |
| Disease-treatment system | Wellness-maintenance system |
| Few measurements | Many measurements, including complete genome sequencing, high-parameter blood diagnostics, many longitudinal omics measurements |
| Disease-centric, with standard of care associated with a disease diagnosis | Individual-centric, with standard of care tailored more fully to multiple measurements |
| Records not highly linked | Deeply integrated data that can be mined for continued improvement of healthcare strategies |
| Large-scale diffusion of medical information mediated mostly through physicians alone | Social networking of patients to enhanced shared experiences and diffusion of knowledge in consultation with their physicians |
| Drugs tested against large populations—10s of thousands to develop statistics for FDA | Stratification of disease populations into small groups, 50 or so, that can be effectively treated to achieve FDA approval |
One important question is how the average cancer biologist is going to be provided access to all of these emerging systems strategies and technologies. Another challenging question is how physicians will be informed (educated) as to the power of the new systems (P4) medicine. A third question is how will medical researchers be given access to these powerful new techniques. The Institute for Systems Biology has created a cross-disciplinary culture where many different types of scientists (biologists, computer scientists, chemists, engineers, mathematicians, physicists and physicians) learn one another's languages and work together on teams to develop the new technologies and analytical tools that are required by the frontier problems of contemporary medicine. ISB has data generation facilities (genomics, proteomic, single-cell, phenotype, imaging, etc) and data analyses facilities that are available to any ISB scientist—to attack big or small scientific problems. This systems-driven, cross-disciplinary, and integrative environment is ideal for attacking challenging problems in science [48].
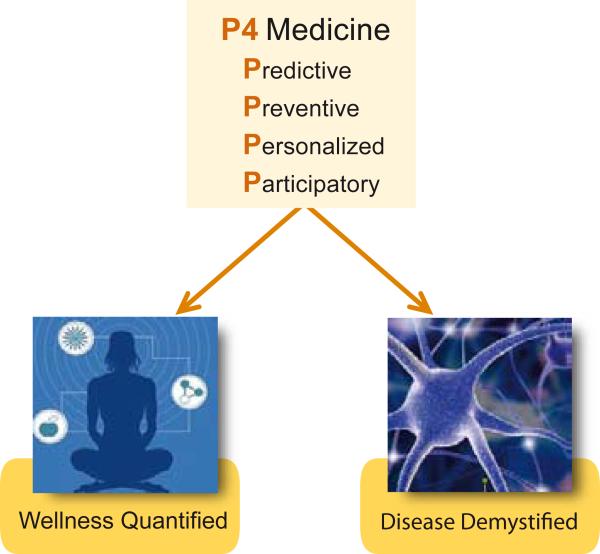
It is clear that P4 medicine will pioneer two revolutions—quantifying wellness and demystifying disease (Figure. 4). A fascinating question is how to bring systems (P4) medicine to the medical world and to patients. There are two challenges in doing so. First, the technical challenges that have been discussed above. Second, the societal challenges that include how do you educate patients, physicians and the medical community as to the challenges of systems medicine, how you convince a well entrenched and conservative medical community to accept the P4 revolution—as well as many ethical, social and legal issues including privacy, confidentiality, security, policy, etc. In our view the societal issues are by far the most challenging. ISB has decided to attack the challenge of bringing P4 medicine to society by strategic partnerships. We have several different types of partnerships. 1) We have a partnership with the Grand Duchy of Luxembourg to attack two fundamental problems of P4 medicine--how to decipher the genome and how to understand the phenome and convert these data into an understanding of how biological networks mediate health and disease [1]. 2) In the human proteome atlas project, a collaboration with Ruedi Aebersold at the ETH and several companies including Agilent, Origene and ABsciex—we have developed SRM mass spectrometry assays for all human proteins. This will lead to a democratization of all proteins just as the human genome project democratized all genes (e.g. all scientists were given access to all genes and now to all proteins). 3) ISB created the P4 Medicine Institute (a non-profit organization) to help create a network of medical centers that would employ a series of clinical assays developed at ISB, together with conventional medical tests, in the context of pilot projects to demonstrate the power of P4 medicine. Ohio State was our first partner and together we are formulating pilot projects on wellness and heart failure. More recently PeaceHealth, a community hospital in the Northwest, has joined this network. We are looking for an additional 3-4 members for this medical network. The P4 Medicine Institute is also actively involved in considering other of the societal challenges of P4 medicine including its economics.
Figure 4.
Two central conceptual themes of P4 medicine--the quantification of wellness and the demystification of disease.
In closing, it is clear that the grand challenge for all scientific and engineering disciplines in the 21st century is complexity. What is unique about biology is that the various elements of the systems approaches described above (e.g. biology as an information science, holistic or systems experimental approaches, emerging technologies and transformational analytical tools) afford a powerful series of strategies for biology to attack the various forms of its complexity. These same approaches allow biology to approach many of society's most fundamental problems—healthcare, global health, agriculture, energy, environment, nutrition, animal health and the like. Those institutions that have provided their scientists with a cross-disciplinary, systems-driven and integrative infrastructure will be in a uniquely powerful position to attack these problems [48]. It is a wonderful time to be in biology!
Acknowledgements
We gratefully acknowledge funding from the Grand Duchy of Luxembourg, NIH/NCI NanoSystems Biology Cancer Center (U54 CA151819A), NIH/NIGMS Center for Systems Biology (P50 GM076547), an NIH Howard Temin Pathway to Independence Award in Cancer Research (R00CA126184), and a Roy J. Carver Young Investigator Grant.
Footnotes
Conflict of interest statement
No conflicts of interest were declared
References
- 1.Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011;8(3):184–7. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- 2.Price ND, et al. In: Systems biology and systems medicine, in Genomic and Personalized Medicine: From Principles to Practice. Ginsburg G, Willard H, editors. Elsevier; 2009. pp. 131–141. [Google Scholar]
- 3.Hood L, et al. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306(5696):640–3. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Lin B, et al. Evidence for the presence of disease-perturbed networks in prostate cancer cells by genomic and proteomic analyses: a systems approach to disease. Cancer Res. 2005;65(8):3081–91. doi: 10.1158/0008-5472.CAN-04-3218. [DOI] [PubMed] [Google Scholar]
- 6.Schiffer CA. BCR-ABL tyrosine kinase inhibitors for chronic myelogenous leukemia. N Engl J Med. 2007;357(3):258–65. doi: 10.1056/NEJMct071828. [DOI] [PubMed] [Google Scholar]
- 7.Mok TS, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Karapetis CS, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 9.Tiacci E, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364(24):2305–15. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361(11):1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465(7297):473–7. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 17.Bielas JH, et al. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103(48):18238–42. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salk JJ, Fox EJ, Loeb LA. Mutational heterogeneity in human cancers: origin and consequences. Annu Rev Pathol. 2010;5:51–75. doi: 10.1146/annurev-pathol-121808-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer. 2011;11(6):450–7. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proc Natl Acad Sci U S A. 2008;105(51):20063–6. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–78. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapidot T, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–8. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 23.Al-Hajj M, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh SK, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–8. [PubMed] [Google Scholar]
- 25.Chow LM, et al. Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain. Cancer Cell. 2011;19(3):305–16. doi: 10.1016/j.ccr.2011.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328(5978):636–9. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ideker T, Dutkowski J, Hood L. Boosting signal-to-noise in complex biology: prior knowledge is power. Cell. 2011;144(6):860–3. doi: 10.1016/j.cell.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang D, et al. A systems approach to prion disease. Mol Syst Biol. 2009;5:252. doi: 10.1038/msb.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106(11):4402–7. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin EH, et al. Elevated circulating endothelial progenitor marker CD133 messenger RNA levels predict colon cancer recurrence. Cancer. 2007;110(3):534–42. doi: 10.1002/cncr.22774. [DOI] [PubMed] [Google Scholar]
- 32.Iinuma H, et al. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J Clin Oncol. 2011;29(12):1547–55. doi: 10.1200/JCO.2010.30.5151. [DOI] [PubMed] [Google Scholar]
- 33.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40(6):722–9. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maher CA, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc Natl Acad Sci U S A. 2009;106(30):12353–8. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470(7333):214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leary RJ, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2(20):20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verstovsek S, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med. 2010;363(12):1117–27. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paik S, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 43.Price ND, et al. Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leiomyosarcomas. Proc Natl Acad Sci U S A. 2007;104(9):3414–9. doi: 10.1073/pnas.0611373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan X, et al. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc Natl Acad Sci U S A. 2011;108(4):1591–6. doi: 10.1073/pnas.1018696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Picotti P, et al. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138(4):795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cima I, et al. Cancer genetics-guided discovery of serum biomarker signatures for diagnosis and prognosis of prostate cancer. Proc Natl Acad Sci U S A. 2011;108(8):3342–7. doi: 10.1073/pnas.1013699108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petriv OI, et al. Comprehensive microRNA expression profiling of the hematopoietic hierarchy. Proc Natl Acad Sci U S A. 2010;107(35):15443–8. doi: 10.1073/pnas.1009320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hood L. Biological Complexity Under Attack, A Personal View of Systems Biology and the Coming of “Big” Science. Genetic Engineering & Biotechnology News. 2011 [Google Scholar]