Abstract
The recent years have seen a significant shift in the expectations for the therapeutic management of disseminated melanoma. The clinical success of BRAF targeted therapy suggest that long-term disease control may one day be a reality for genetically defined sub-groups of melanoma patients. Despite this progress, few advances have been made in developing targeted therapeutic strategies for the 50% of patients whose melanomas are BRAF wild-type. The most significant sub-group of BRAF wild-type tumors is the 15–20% of all melanomas that harbor activating NRAS mutations. Emerging preclinical and clinical evidence suggests that NRAS mutant melanomas have patterns of signal transduction and biological behavior that is distinct from BRAF mutant melanomas. This overview will discuss the unique clinical and prognostic behavior of NRAS mutant melanoma and will summarize the emerging data on how NRAS-driven signaling networks can be translated into novel therapeutic strategies.
Introduction
For many years, the prognosis for patients with disseminated melanoma has been poor. Responses to standard chemotherapy have been historically low and associated with median survival times of 6–10 months (1). While immunotherapy with interleukin-2 and ipilimumab provides more durable responses in approximately 10% of metastatic melanoma patients, there are no established biomarkers to select patients and serious toxicities are common (2). The recent years have seen significant shifts in our understanding of the drivers of malignant transformation. Cancer is now known to be a genetic disease, with defined tumor histologies often being uniquely dependent upon (or “addicted” to) the signaling activity of a restricted number of oncogenes. This knowledge, and the ability to target “druggable” oncogenes using small molecule inhibitors, has led to significant therapeutic breakthroughs in a number of cancers including the targeting of the Bcr-Abl fusion protein in chronic myeloid leukemia (CML), the ALK-EML4 fusion and mutant EGFR in non-small cell lung cancer (NSCLC) and c-KIT in gastrointestinal stromal tumors (GIST) (3–5). A major turning point in our understanding of melanoma biology was the 2002 discovery that the majority (>50%) of all cutaneous melanomas harbored activating mutations in the serine/threonine kinase BRAF (6). The identification of mutant BRAF as a bona fide oncogene in melanoma led to the rapid development of small molecule BRAF inhibitors (such as vemurafenib and dabrafenib), which were subsequently shown to increase the overall survival of patients with metastatic BRAF mutant melanoma (6–8). Following the FDA-approval of vemurafenib in August of 2011, attention has now turned to the 50% of melanoma patients whose tumors are BRAF wild-type, with the hope of making similar therapeutic breakthroughs. Among the cutaneous melanomas that lack BRAF mutations, 15–20% harbor mutations in the small GTPase NRAS (9). At present, little progress has been made in developing targeted therapy strategies for NRAS mutant melanoma. In this review we will discuss the biological and signaling characteristics that are unique to NRAS mutant melanomas and will outline possible future strategies for their therapeutic management.
Ras proteins
Ras genes were first identified as encoding for the proteins that mediated the transforming activity of Rat sarcoma viruses. It is now known that three Ras family members, NRAS, HRAS (Harvey Rat Sarcoma Virus) and KRAS (Kirsten Rat sarcoma virus) are often mutated in human cancers, and that >20% of all tumors harbor activating mutations in one of their RAS genes (10). Ras proteins constitute a major family of low molecular weight plasma membrane-associated GTP-binding proteins. Within this super-family (which also includes Rab, Rho, and Arf), the Ras proteins primarily regulate growth, and function as molecular switches that link signals from receptor tyrosine kinases (RTKs) at the cell surface to transcription factors and cell cycle proteins in the nucleus (11, 12). Under physiological conditions, Ras proteins exist in either an active (GTP-bound) state or an inactive (GDP-bound) state. The exchange of GDP for GTP, and the switching of Ras to its activated state is catalyzed by a family of guanine nucleotide exchange factors (GEFs), such SOS1, SOS2 and RASGRF (10) (Figure 1), and is opposed by the GTPase stimulatory activity of Ras GTPase activating proteins (Ras-GAPs) (13)(Figure 1). In normal cells, Ras activation typically proceeds following the binding of ligand to its cognate RTK. This then leads to RTK autophosphorylation and dimerization, and the recruitment of adaptor molecules, such as GRB2. Signals from the activated RTK are relayed to Ras by the binding of Grb2 to the RTK at its SH2 domain. This then recruits SOS to the plasma membrane through binding of two SH3 domains, and SOS in turn catalyzes the GDP to GTP exchange on Ras (14) (Figure 1). Once activated, Ras recruits and stimulates a number of intracellular signaling pathways including the Raf/MEK/ERK mitogen activated protein kinase (MAPK) pathway, the phosphoinositide 3-kinase (PI3K)/AKT pathway, the Ral guanine nucleotide exchange factors (RAL-GEFs) such as Ral-GDS and phospholipase C epsilon (Figure 2) (15).
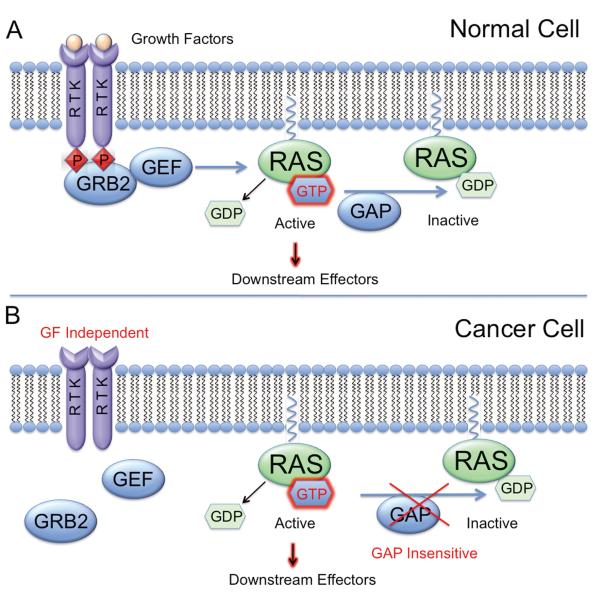
Figure 1. Mechanisms of Ras activation in normal and cancer cells.
A. Upon RTK stimulation, GRB2 adaptor molecules facilitate signal transduction to Ras via guanine nucleotide exchange factors (GEFs). GEFs accelerate GDP release by Ras, allowing GTP binding and activation. The activating effects of GEFs are opposed by GAPs which promote a rapid rate of GTP hydrolysis, returning Ras to its inactive state. B. While Ras activation is dependent on growth factor signaling in normal cells, mutations in oncogenic Ras “lock” the GTPase in an active, GTP-bound conformation and render it insensitive to inactivation by GAPs.
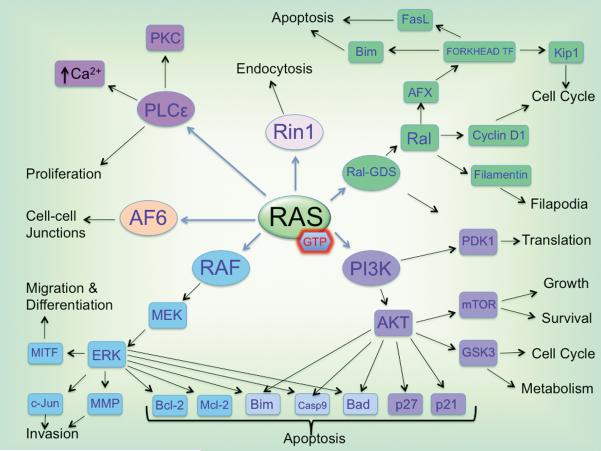
Figure 2. Downstream mediators of Ras signaling.
Activated Ras can stimulate a wide array of downstream effectors, mediating many cellular processes such as proliferation, survival, invasion, endocytosis, cell-cell signaling and differentiation.
Mutant Ras as an initiating melanoma oncogene
Despite much attention being focused upon BRAF mutant melanoma, NRAS was actually the first melanoma oncogene to be identified. In1984, a screen of melanoma cell lines for genes that possessed transforming properties identified activating mutations in NRAS in 4/30 samples (16). In a further observation, now highly pertinent to the use of BRAF inhibitors in melanoma, it was noted that mutant Ras expression was heterogeneous within metastatic lesions derived from the same patient (found only in 1/5 samples) (16). Currently, mutations in NRAS, KRAS and HRAS are known to be present in 20%, 2% and 1% of all melanomas, respectively (17). With regards to NRAS, the most common oncogenic change (>80% of all NRAS mutations) reported is a point mutation leading to the substitution of leucine to glutamine at position 61, with mutations at positions 12 and 13 occurring less frequently (9). Position 61 mutations also account for the majority of HRAS mutations in melanoma, whereas most KRAS mutations are at position 12 (http://www.sanger.ac.uk/genetics/CGP/cosmic/). From a mechanistic standpoint, Ras mutations at position 61 are associated with impaired GTPase activity and the locking of the Ras protein into its activated (GTP-associated) conformation. Mutations at positions 12 and 13 render Ras insensitive to the physiological mechanisms of inactivation by the Ras-GAPs (9). Once mutated, Ras drives cellular transformation through a network of signal transduction pathways involved in growth, motility and survival (Figure 2). Although it is not clear why NRAS mutations are more frequent in melanoma compared to HRAS or KRAS mutations, there is some suggestion that NRAS may be overexpressed in melanocytes relative to other Ras isoforms. It is also possible that NRAS may activate different signaling pathways to KRAS and HRAS; an idea supported by the observation that NRAS has greater transforming activity than KRAS in experimental models of melanoma (18).
The available evidence suggests that Ras mutations are acquired at an early stage in the oncogenic process. Benign common nevi most commonly (>80%) harbor mutations in BRAF, with NRAS mutation being less frequent (~14%)(19). In contrast, congenital nevi (e.g. those that develop in utero, independently of UV exposure) do not possess BRAF mutations and instead frequently (81%) harbor NRAS mutations (20). Spitz nevi, large benign melanocytic lesions that are histologically indistinguishable from melanoma often (~11% of cases) exhibit concurrent amplification and mutation in HRAS, in the absence of significant proliferative capacity (21). Taken together, these data suggest that although Ras mutations are found in melanoma precursor lesions, these do not appear to initiate oncogenic transformation in isolation and require other co-operating genetic events. These ideas are supported by the observation that most nevi rarely progress to melanoma and typically exhibit signs of replicative senescence (22, 23).
Genetically engineered mouse models of melanoma driven through mutant HRAS or NRAS have suggested a role for concurrent loss or inactivation of the tumor suppressor p16INK4A, deletion of p53 or exposure to ultraviolet light (24, 25). These studies are supported by the finding that NRAS mutant, but not BRAF mutant, melanoma is frequently associated with methylation of the p16INK4A promoter (26). Cell culture studies have confirmed the role of NRAS mutations in the growth of melanoma cell lines, with the siRNA mediated knockdown of NRAS found to inhibit growth associated with decreased expression of cyclins D1 and Cyclin E2 (27). At the same time it was noted that siRNA knockdown of NRAS reversed invasive capacity, an effect associated with the decreased expression of the cytoskeletal proteins paxillin, leupaxin and vinculin (27).
One other sub-group of melanomas with Ras-dependence is those with “low-activity” BRAF mutations, such as those found at positions 465, 463 and 596 (28). Cell lines with low activity BRAF mutations, show an impaired activation of MAPK signaling in isolated kinase assays and often harbor concurrent NRAS mutations at positions 12 and 13 (29). Unlike melanoma cell lines with BRAF V600E/D mutations, low activity mutant BRAF cell lines are sensitive to the growth inhibitory effects of Ras-neutralizing antibodies as well as cell death induced by the CRAF/multi-kinase inhibitor sorafenib (28, 30). It is therefore likely that strategies designed to target Ras mutant melanoma could also be effective in low activity BRAF mutant melanoma.
Clinical and prognostic significance of NRAS mutations in melanoma
It is now accepted that melanoma is a diverse collection of melanocytic neoplasms whose biological behavior can be defined by their pattern of oncogenic mutations (31, 32). A recent histopathological analysis of a large series of melanoma specimens, grouped according to their BRAF/NRAS mutational status showed BRAF-mutated melanomas to possess morphological features that were distinct from tumors that were BRAF wild-type (31). It was found that BRAF-mutated melanomas had an increased tendency to upward migration and nest formation and gave rise to larger, rounded and more pigmented tumor cells. In contrast, NRAS mutated melanomas were not found to exhibit these morphological and phenotypic characteristics (31).
The presence of an NRAS mutation in melanoma is also of prognostic significance. The typical patient with NRAS mutant melanoma tends to be older (>55 years of age) with a more chronic pattern of UV exposure than a patient with a BRAF mutant melanoma (33, 34). Overall, patients with NRAS mutant melanomas have been found to have thicker tumors at presentation. These tumors are typically located at the extremities and have greater rates of mitosis than BRAF mutant melanoma (33, 35). Despite NRAS mutant melanomas having lower rates of ulceration than BRAF mutant melanoma, overall survival for NRAS mutant melanoma patients is worse than their wild-type counterparts (33–35).
Intracellular signaling in NRAS mutant melanoma
Although NRAS and BRAF mutant melanomas share a set of common signaling pathways, significant differences exist in how these are regulated. In normal human melanocytes, MAPK activation occurs following stimulation with growth factors such as stem cell factor (SCF), fibroblast growth factor (FGF) and hepatocyte growth factor (HGF) (36) (Figure 3). Under these normal physiological conditions stimulation of the pathway occurs following the Ras-mediated activation of Raf (37). MAPK signaling in melanocytes proceeds through BRAF rather than CRAF (also called Raf-1), with Raf-isoform selectivity being determined through the concurrent growth factor mediated activation of adenylate cyclase (AC) (37, 38) (Figure 3). Increased AC activity leads to the accumulation of cyclic AMP (cAMP) and the activation of protein kinase A (PKA). This then inactivates CRAF via its direct phosphorylation at two inhibitory sites (S43 and S233) (39, 40) (Figure 3). The high frequency of BRAF mutations found in melanoma likely stems from the lineage-specific reliance of melanocytes upon BRAF signaling for their growth factor mediated MAPK activation.
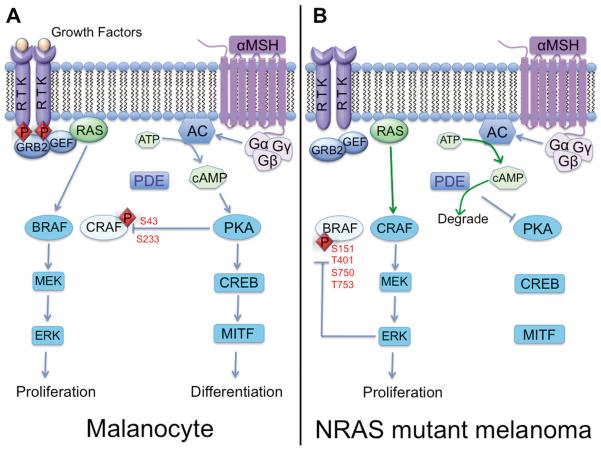
Figure 3. Raf isoform switching in NRAS mutant melanoma.
A. In normal melanocytes, cyclic AMP pathway activation promotes PKA-mediated inhibition of CRAF. Therefore MAPK signaling proceeds through BRAF rather than CRAF in these cells (left panel). B. In NRAS mutant melanoma, inhibition of PKA signaling due to enzymatic activity of phosphodiesterase (PDE) and negative feedback inhibition of BRAF due to activation of the MAPK pathway promotes CRAF-mediated MAPK signaling instead (right panel).
Interestingly, BRAF is not required for MAPK activation in every melanoma, with NRAS mutant melanomas being instead reliant upon CRAF signaling (37) (Figure 3). CRAF-mediated MAPK signaling in NRAS mutant melanoma cells is dependent upon two parallel mechanisms; the inactivation of BRAF (permitting Raf isoform switching) and deregulation of PKA signaling (that prevents CRAF inactivation) (37). The inactivation of BRAF is achieved through a negative feedback mechanism in which ERK limits RAF signaling by phosphorylating it at S151, T401, S750 and T753 (41) (Figure 3). The S151 site in BRAF is close to the Ras-binding domain, and its phosphorylation appears to prevent the Ras/BRAF interaction (38). Phosphorylation of BRAF at T401, S750 and T753 disrupts the dimerization of BRAF with CRAF (41). The deregulation of PKA signaling occurs as a result of increased expression of the cAMP degrading enzyme phosphodiesterase IV (PDE4) that limits PKA activity and in turn prevents CRAF from being phosphorylated at its inhibitory sites (38) (Figure 3). The potential therapeutic implications of these findings were demonstrated by the ability of selective PDE4 inhibitors to prevent the growth and increase apoptosis in NRAS mutant melanoma cell lines (38).
Constitutive MAPK activity is known to be essential for melanoma initiation and progression (42). Signals through this pathway are responsible for the uncontrolled growth that characterizes melanoma, and occur via regulation of the G1 checkpoint through the increased expression of cyclin D1 and downregulation of the cell cycle inhibitor p27KIP1 (43, 44). MAPK signaling also plays a critical role in melanoma survival through the phosphorylation (at S69) and proteasomal degradation of the pro-apoptotic protein BIM and via p90RSK-mediated effects upon BAD (45, 46).
In addition to the well-known role of MAPK signaling in melanomagenesis, in vitro and animal modeling studies have suggested that the PI3K/AKT cascade also plays a critical role in melanoma development (47, 48). Once activated at the plasma membrane, PI3K phosphorylates phosphotidylinositol-4,5, biphosphate (PIP2) to PIP3 which in turn activates the downstream kinases PDK1 and AKT. Once active, AKT promotes melanoma cell survival through direct phosphorylation of BAD and through the negative regulation of BIM expression via the phosphorylation and inactivation of FOXO3a (45, 49–51). Signals through the PI3K/AKT pathway are also critical for cell cycle entry, and regulate the G1 cell cycle checkpoint by phosphorylating and inactivating glycogen-3 synthase kinase (GSK3)-β, leading to modulation of cyclin D1 (52). PI3K/AKT signaling also has important downstream effects upon protein turnover and cell glucose metabolism via the regulation of the mTOR/S6K and GSK3β signaling pathways (53). Like the MAPK pathway, PI3K/AKT signaling is differentially regulated between NRAS and BRAF mutant melanoma, with NRAS mutant melanomas rarely harboring either mutations in, or silencing of the negative regulator of the PI3K pathway, phosphatase and tensin homolog (PTEN) (54, 55). Additionally, activating mutations in AKT3, which are sometimes found in BRAF mutant melanoma, have not been observed in NRAS mutant melanomas (56). Despite data implying that mutant NRAS should activate MAPK and PI3K/AKT signaling concurrently, a recent analysis of clinical melanoma specimens and cell lines, showed NRAS mutant melanomas to exhibit lower levels of constitutive AKT signaling than those with BRAF mutations (55). The significance of this observation is still under investigation.
Another family of less well-characterized mediators of mutant NRAS are the Ral-GEFs such as Ral-GDS, RGL1 and RGL2 (15). Ral signaling is known to play a role in cell growth through transcriptional activation of cyclin D1, and (in concert with AKT) in cell survival, through the regulation of BIM and FasL expression (15, 57). Increased Ral activity has been also implicated in cytoskeletal reorganization, associated with increased filopodia formation (57). Although there is little evidence of melanomas harboring Ral-GDS mutations, Ral activity appears to be required for the development of NRAS mutant melanoma (58–60). In animal xenograft models, shRNA knockdown of RalA was noted to delay the growth of human NRAS mutant melanoma cell lines (59). A recent analysis of the role of Raf, PI3K and Ral signaling in immortalized melanocytes revealed distinct requirements for each pathway in oncogenic transformation (60). Specifically, it was shown that whereas Raf and PI3K were important for overcoming oncogene-induced senescence and invasion, signaling through Ral was a prerequisite for anchorage independent growth (60). The requirement for multiple signaling pathways in the development of NRAS mutant melanoma development was demonstrated by the fact that no one individual signaling pathway fully recapitulated the transforming activity of oncogenic Ras (60).
Therapeutic strategies for NRAS mutant melanoma
Farnesyl transferase inhibitors
NRAS has so far proven to be intractable to conventional drug discovery. The main “Ras-directed” therapies to be investigated thus far have been the farnesyl transferase inhibitors (FTIs), a class of drugs designed to prevent the post-translational modification of Ras and its insertion into the plasma membrane (61). Although this strategy showed some preclinical promise, the clinical experience has been disappointing with serious off-target effects reported and few responses observed, even in tumors with high rates of Ras mutations (such as colorectal carcinoma) (61–63). The lack of success with FTIs is attributable to the fact that many critical cellular proteins are farnesylated in addition to Ras and the fact that alternative post-translational modifications exist that allow for membrane targeting of Ras in the absence of FT activity. In melanoma, a small phase II trial of FTIs in genetically unselected patients showed very little clinical activity. Other attempts to directly target Ras have involved the downregulation of Ras protein expression using either antisense oligonucleotides or small interfering RNAs (siRNAs). Despite preclinical evidence that Ras knockdown can be achieved using siRNA, the approach has proven technically challenging in the clinical setting (27). Current approaches are now centered upon improved methods of siRNA delivery in vivo, using nanoparticle-based delivery systems (64).
MEK inhibitors
Given the lack of success thus far in directly targeting NRAS, focus has instead shifted to targeting the critical signal transduction pathways that drive Ras-mediated transformation. Of these, targeting the MAPK pathway, using allosteric MEK inhibitors has received the most attention. In preclinical studies, melanoma cell lines with oncogenic NRAS showed variable response to MEK inhibitors, with some cell lines exhibiting high sensitivity and some demonstrating near-total resistance (43, 65). A recent high throughput genomic analysis, correlating cell line mutational status with drug response (the cancer cell line “encyclopedia”), demonstrated a link between the sensitivity of NRAS mutant melanoma cell lines to MEK inhibition and elevated expression of the aryl hydrocarbon receptor (AHR) (66). The dependence of NRAS mutant melanoma cell lines upon AHR receptor signaling was confirmed through shRNA knockdown and it was noted that MEK inhibition in these cell lines was associated with inhibition of AHR signaling function (66).
Although MEK inhibitors, such as CI-1040 and AZD6244 have been investigated in melanoma patients, most of the studies were performed prior to the era of routine genetic testing. In most of the published trials to date, patients were unselected and significant intratumoral phospho-ERK inhibition was not observed at the maximum tolerated doses (67). There were some notable exceptions with one NRAS mutant melanoma patient remaining on AZD6244 therapy for 3 cycles, associated with >70% tumor shrinkage and near-total inhibition of intratumoral phospho-ERK (67). Overall the results of clinical trials with early generation MEK inhibitors were disappointing (~10% objective response rate) and retrospective genotyping for the NRAS/BRAF mutations did predict for clinical benefit.
Enthusiasm for targeting MEK in NRAS mutant melanoma patients has received new attention with the clinical development of the third generation MEK inhibitors trametinib (GSK1120212) and MEK162 (ARRY-438162). Both trametinib and MEK162 are potent inhibitors of MEK1/2 with sustained MAPK pathway inhibition at clinically achievable doses (68, 69). Trametinib has now demonstrated strong activity in BRAF mutant melanoma patients treated in the phase III METRIC study (70). The overall response rate (ORR) with trametinib was 24% with a median progression free survival (mPFS) of 4.8 months, both significantly better than standard chemotherapy. Similarly, MEK162 has shown promising clinical activity (ORR 23%) in BRAF mutant melanoma patients in a recent phase II study (71). While these response rates are lower than the reported data for first line treatment with the BRAF inhibitors vemurafenib and dabrafenib, it is important to note that patients in both of the MEKi studies had received multiple prior therapies and perhaps had more aggressive disease biology.
The first prospective clinical data on NRAS mutant melanoma patients treated with targeted therapy was presented at ASCO 2012 (71). As part of a combined study of BRAF and NRAS mutant melanoma patients, 30 metastatic melanoma patients whose tumors harbored an NRAS mutation were enrolled and treated with MEK162. The ORR was 21% and the mPFS was 3.65 months. Further study in this patient population will be necessary to confirm its clinical activity in comparison to other standard therapies. While prospective data with trametinib in NRAS mutant melanoma patients is not available, early retrospective data from ongoing clinical studies suggests that trametinib may have activity in a subset of NRAS mutant melanoma patients (72).
Combination therapy with MEK and PI3K pathway inhibitors
The evolving lesson of using small molecule BRAF inhibitors in patients with BRAF V600E mutant melanoma suggest that resistance will always limit responses to single agent therapy and that multi-drug combinations are necessary (8, 73–76). There is already good evidence from animal models of Ras-driven cancers that the simultaneous inhibition of MEK + PI3K leads to greater extent of tumor regression than either inhibitor alone (77). From a mechanistic standpoint, the MEK and PI3K/AKT pathways seem to converge at the level of cell survival, through the coregulation of the pro-apoptotic protein BAD, as well as the level of cap-dependent protein translation, through the translational repressor 4E-BP1 (78, 79). Similar findings have also been reported in xenograft studies of NRAS mutant human melanoma cell lines, with shRNA knockdown of BRAF + CRAF or BRAF + PI3K expression being found to delay and inhibit tumor formation (80). In melanoma cell lines where BRAF inhibitor resistance is mediated through an acquired NRAS mutation, the combination of a MEK inhibitor (trametinib) with a PI3K/mTOR inhibitor (GSK2126458) was noted to overcome drug resistance and inhibit cell survival (75).
It is not yet clear what the optimal combination of signal transduction inhibitors will be for NRAS mutant melanoma and even whether any one combination will be effective for all melanomas with oncogenic NRAS. Presently, there are 14 active or recently completed phase I clinical trials investigating the combination of PI3K pathway and MEK inhibitors (Table 1). Data presented on the dose escalation of the phase I study of BKM120 and trametinib was promising for tolerability and potential efficacy (81). A total 66 patients were treated at increasing doses of both drugs, reaching the maximum tolerated doses of 70mg daily for BKM120 and 1.5mg daily for trametinib. Of note, the dose of trametinib in phase III study of BRAF mutant patients (METRIC study) was 2mg daily (70). Enrollment focused on RAF and RAS mutant tumors, with 10 melanoma patients treated in total. While the strongest clinical activity was seen in KRAS mutant ovarian carcinoma patients (3 partial responses by RECIST criteria), there was tumor regression that did not meet RECIST criteria seen in 3 melanoma patients. Completed data on this study and others are eagerly awaited.
Table 1.
Active or recently completed phase I studies with combination MEK and PI3K pathway inhibitors.
| PI3K inhibitor | MEK inhibitor | Clinical Trial Number | Status |
| BKM120 | MEK162 | NCT01363232 | Recruiting |
| BKM120 | Trametinib | NCT01155453 | Recruiting |
| BAY86-9766 | BAY90-6946 | NCT01392521 | Recruiting |
| BYL719 | MEK162 | NCT01449058 | Recruiting |
| GDC-0941 | GDC-0973 | NCT00996892 | Recruiting |
| GSK2126458 | Trametinib | NCT01248858 | Recruiting |
| Dual PI3K/mTOR inhibitor | MEK inhibitor | Clinical Trial Number | Status |
| BEZ235 | MEK162 | NCT01337765 | Recruiting |
| SAR245409 | MSC1936369B | NCT01390818 | Recruiting |
| PF-04691502 | PD-0325901 | NCT01347866 | Recruiting |
| AKT inhibitor | MEK inhibitor | Clinical Trial Number | Status |
| GSK2110183 | Trametinib | NCT01476137 | Recruiting |
| GSK2141795 | Trametinib | NCT01138085 | Completed |
| MK-2206 | AZD6244 | NCT01510444 | Recruiting |
| GDC-0068 | GDC-0973 | NCT01562275 | Recruiting |
| mTOR inhibitor | MEK inhibitor | Clinical Trial Number | Status |
| Temsirolimus | AZD6244 | NCT01166126 | Active, not recruiting |
www.clinicaltrials.gov accessed July 7, 2012.
Feedback inhibition and RTK signaling
The experience of targeted therapy in many cancer types has demonstrated that resistance can often occur through the derepression of feedback inhibition and compensatory upregulation of signaling through parallel pathways (82). Understanding and developing strategies that limit these complex adaptive signaling responses is likely to prove critical in the use of targeted therapy regimens for the long-term management of disease. The scale of this problem was recently indicated by an integrated genomic/proteomic study of triple negative breast cancer in which MEK inhibition led to the rapid remodeling the RTK kinome associated with increased expression of multiple RTKs (83). Mechanistically, it was observed that the short-term inhibition of MAPK signaling decreased the expression c-Myc that in turn initiated autocrine signaling through HER2, RON, Axl, VEGFR and PDGFRβ (83). The drug-induced increase in RTK signaling allowed the cells to escape from MEK inhibitor therapy and could be reversed through the combination of a MEK inhibitor with the pan-RTK inhibitor sorafenib (83).
Compensatory signaling of a similar nature was also observed following the siRNA knockdown of RAS, with recent studies showing that inhibition of KRAS in colorectal carcinoma led to a rebound increase in PI3K signaling (84). Mechanistically this appeared to result from a suppression of MEK-mediated TORC1 activity leading to derepression of IRS-1 and increased AKT signaling mediated through insulin like growth factor receptor (IGFR1) (84). Although not yet studied in the context of NRAS mutant melanoma, functional IGF-1R autocrine signaling loops are known to be present in both melanoma cell lines and tumor specimens, raising the possibility these may contribute to the escape from MEK inhibitor therapy (85). A possible role for rebound IGF-1R signaling in the de novo resistance of a subset of BRAF mutant melanoma cells to MEK inhibition has already been demonstrated (86). In this instance, MEK inhibitor sensitivity was restored through combination with either a TORC1/2 inhibitor or an IGF-1R inhibitor (86). In addition to IGF-1R, comprehensive phospho-proteomic screening of melanoma cell lines revealed a diverse array of RTKs to be expressed including TYRO3, Axl, MERTK, EPHB2, MET, IGFR1, EGFR, KIT, HER3 and HER4 (85, 87). The potential role of these RTKs in NRAS mutant melanoma, and their involvement in adaptive signaling responses following kinase inhibition remains to be determined. Of the RTKs expressed in melanoma, the TAM family (Tyro3, Axl, MER) receptor kinase Axl was noted to be overrepresented in NRAS mutant melanoma (87, 88). In the majority of NRAS mutant cell lines examined, Axl was activated through a Gas6-driven autocrine loop and was linked to activation of AKT signaling (88). Functional studies indicated that Axl signaling was required for the invasive behavior of NRAS mutant melanoma cell lines (88).
While inhibition of the PI3K pathway downstream of RTK signaling may be one approach to disrupt cellular adaptation to RAF and MEK inhibition, preclinical and early clinical data is promising for direct inhibition of RTKs and other tyrosine kinases Recent preclinical work with RAF-265, an inhibitor of BRAF, CRAF, VEGFR2, PDGFR, and c-Kit, has shown induction of tumor regression in BRAF wild-type melanoma xenografts, including one specimen with a Q61 mutation in NRAS (89). RAF-265 is now being studied in combination with MEK162 in patients with BRAF or RAS mutant tumors, including NRAS mutant melanoma (NCT01352273). A similar concept is being explored with the combination of pazopanib (VEGFR, PDGFR, c-Kit and FGFR inhibitor) with trametinib in a phase I study (NCT01438554).
Recent data on broad tyrosine kinase inhibition has also been presented in metastatic melanoma patients treated with the combination of ARQ 197, a specific c-Met inhibitor, and sorafenib, an inhibitor of VEGFR, PDGFR, c-Kit, and RAF (90). In this dose-escalation study, a total of 16 melanoma patients were treated at the recommended phase II dose level. The ORR was 25% with a mPFS of 5.3 months. Interestingly, half of the patients in this cohort had NRAS mutant melanoma. Efficacy data showed one complete response and one partial response in this subgroup, along with an overall mPFS of 9.2 months. These findings suggest that tyrosine kinase inhibition may prove to be valuable therapeutic strategy in NRAS mutant melanoma and further study is warranted.
NRAS mutations as mechanism of therapeutic escape from BRAF inhibitor therapy: mutational heterogeneity in melanoma?
Despite the increases in both progression-free and overall survival seen in BRAF mutant melanoma patients receiving vemurafenib therapy, resistance occurred in nearly every case (8, 73). In a number of instances therapeutic escape was associated with the acquisition of position 61 mutations in NRAS that seemed to be lacking in the original tumor (91). This phenomenon was also reported in cell culture studies, with the emergence of RAS mutations being noted in BRAF mutant melanoma cell lines following chronic vemurafenib/dabrafenib treatment (75, 92, 93). Insights into the potential mechanism underlying this apparent mutational switch came from a recent study into the role of acquired KRAS mutations in the resistance of colorectal carcinoma to EGFR inhibition (94). Using clinical specimens and mathematical modeling, it was demonstrated that low-levels of mutant KRAS pre-existed within ostensibly KRAS wild-type tumors, and that EGFR blockade led to the expansion of these sub-clones (94).
Although BRAF and NRAS mutations are mutually exclusive at the single melanoma cell level there is evidence that a minor fraction of patients have melanomas containing co-existent clones with different oncogenic mutations. At least two independent studies have reported the presence of BRAF mutant and NRAS mutant cells within the same melanoma specimen (95, 96). If confirmed in a larger patient cohort, the issue of mutational polyclonality could have important clinical implications, particularly in light of the overwhelming pre-clinical evidence that BRAF inhibitors confers a growth and survival advantage to NRAS-mutant melanoma cells (97–99).
The paradoxical activation of MAPK signaling observed in Ras mutant cells following BRAF inhibitor treatment occurs following the increased formation of Raf dimers consisting of either CRAF homodimer pairs or dimers between drug-bound BRAF and CRAF (99, 100) (Figure 4). Once formed, the dimers activate downstream MEK signaling. The requirement for dimer formation in the paradoxical activation of MAPK signaling was confirmed by the lack of MAPK activation seen in BRAF inhibitor treated Ras mutant cell lines transfected with dimerization-deficient isoforms of Raf (99, 101). Other studies have provided evidence that the BRAF-inhibitor-mediated paradoxical MAPK activation also proceeds via upstream Ras signaling through a mechanism involving the disruption KRAS and NRAS nanoclustering at the cell membrane (102). Intriguingly, it was noted that although Ras nanoclustering promoted Raf signaling, this unexpectedly led to an inhibition of AKT signaling (102). From a clinical standpoint, the ability of BRAF inhibitors to paradoxically stimulate the growth of Ras mutant cells underlies the emergence of HRAS-mutant squamous cell carcinomas, commonly seen in patients on vemurafenib/trametinib therapy (103, 104). There is evidence that BRAF inhibitor therapy may also lead to the development of new, BRAF wild-type primary melanomas (of which 1/12 was noted to be NRAS mutant) (105). Together these findings underscore the need for the future development of highly sensitive BRAF/NRAS mutational testing protocols that gives a systemic readout of mutational status prior to the initiation of BRAF inhibitor therapy.
Figure 4. Transactivation of Raf signaling by BRAF inhibitors in Ras mutant melanoma.
A. Typically, NRAS mutant melanomas rely on CRAF-mediated MAPK activation, with BRAF locked into an inactive conformation. B. BRAF inhibitors relieve the auto-inhibition of BRAF leading to its recruitment to the plasma membrane. This then leads to the dimerization of BRAF and CRAF leading to enhanced MAPK pathway signaling and proliferation.
NRAS mutations as a predictor of immune therapy response
Despite the current lack of effective targeted therapies for NRAS mutant melanoma, there is some evidence that NRAS mutational status may predict for response to other therapies. A recent retrospective analysis of patients treated with high dose interleukin (IL)-2 demonstrated that the majority of the responders were NRAS mutant and that patients with either BRAF mutant or BRAF/NRAS wild-type melanoma were less likely to respond (106). It is not yet clear whether BRAF or NRAS mutational status predicts for better responses in melanoma patients receiving the anti-CTLA-4 antibody ipilimumab or anti-PD-1 antibodies. In the only published analysis to date, the objective response rate of melanoma patients on ipilimumab therapy was noted to be 30% and 33% for those with and without BRAF mutations, respectively (107). A further breakdown of the BRAF wild-type patients into those with NRAS mutations was not available.
Future strategies
While the current immunotherapeutic agents may offer some hope to NRAS mutant melanoma patients, these therapies are not mutation specific, are associated with modest response rates, and carry the risk of significant toxicities. Many targeted strategies are now being evaluated for NRAS mutant melanoma, although these tumors appear to be more heterogeneous than those with BRAF mutations. It is therefore likely that multiple strategies may have to be developed for NRAS mutant melanoma in concert with more sophisticated mutational screening. So far, the most promising data from clinical investigations are with MEK inhibition. However, the relatively short progression free survival indicates that either combination strategies or other targeted approaches will be necessary to achieve more clinically important disease responses. Further prospective biomarker-driven investigations will be crucial for PI3K pathway, RTK, and other targeted therapeutic approaches.
Acknowledgments
Grant support: Work in the Smalley lab is supported by U54 CA143970-01 and R01 CA161107-01 from the National Institutes of Health, The Harry Lloyd Trust and the State of Florida (09BN-14).
References
- 1.Atkins MB. In: The role of cytotoxic chemotherapeutic agents either alone or in combination with biological response modifiers. Kirkwood JK, editor. Marel Dekker; NY: 1997. p. 219. [Google Scholar]
- 2.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010 Aug 19;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001 Apr 5;344(14):1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 4.Duensing S, Duensing A. Targeted therapies of gastrointestinal stromal tumors (GIST)--the next frontiers. Biochem Pharmacol. 2010 Sep 1;80(5):575–583. doi: 10.1016/j.bcp.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007 Aug 2;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 6.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002 Jun 27;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 7.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010 Aug 26;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011 Jun 30;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bos JL. ras oncogenes in human cancer: a review. Cancer Research. 1989 Sep 1;49(17):4682–4689. [PubMed] [Google Scholar]
- 10.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nature reviews Cancer. 2003 Jun;3(6):459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 11.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004 Sep 14;2004(250):RE13. doi: 10.1126/stke.2502004re13. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCormick F. Ras-related proteins in signal transduction and growth control. Mol Reprod Dev. 1995 Dec;42(4):500–506. doi: 10.1002/mrd.1080420419. [DOI] [PubMed] [Google Scholar]
- 13.Lowy DR, Willumsen BM. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 14.Buday L, Downward J. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell. 1993 May 7;73(3):611–620. doi: 10.1016/0092-8674(93)90146-h. [DOI] [PubMed] [Google Scholar]
- 15.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003 Jan;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 16.Albino A, LeStrange R. Transforming ras genes from human melanoma: a manifestation of tumor heterogeneity? Nature. 1984;308(5954):69–72. doi: 10.1038/308069a0. [DOI] [PubMed] [Google Scholar]
- 17.Milagre C, Dhomen N, Geyer FC, Hayward R, Lambros M, Reis-Filho JS, et al. A mouse model of melanoma driven by oncogenic KRAS. Cancer Res. 2010 Jul 1;70(13):5549–5557. doi: 10.1158/0008-5472.CAN-09-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitwam T, Vanbrocklin MW, Russo ME, Haak PT, Bilgili D, Resau JH, et al. Differential oncogenic potential of activated RAS isoforms in melanocytes. Oncogene. 2007 Jul 5;26(31):4563–4570. doi: 10.1038/sj.onc.1210239. [DOI] [PubMed] [Google Scholar]
- 19.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003 Jan;33(1):19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 20.Bauer J, Curtin JA, Pinkel D, Bastian BC. Congenital Melanocytic Nevi Frequently Harbor NRAS Mutations but no BRAF Mutations. J Invest Dermatol. 2006 Aug;:3. doi: 10.1038/sj.jid.5700490. [DOI] [PubMed] [Google Scholar]
- 21.Bastian BC, LeBoit PE, Pinkel D. Mutations and copy number increase of HRAS in Spitz nevi with distinctive histopathological features. Am J Pathol. 2000 Sep;157(3):967–972. doi: 10.1016/S0002-9440(10)64609-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mooi WJ, Peeper DS. Oncogene-induced cell senescence--halting on the road to cancer. N Engl J Med. 2006 Sep 7;355(10):1037–1046. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 23.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005 Aug 4;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 24.Bardeesy N, Bastian BC, Hezel A, Pinkel D, DePinho RA, Chin L. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol Cell Biol. 2001 Mar;21(6):2144–2153. doi: 10.1128/MCB.21.6.2144-2153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997 Nov 1;11(21):2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonsson A, Tuominen R, Grafstrom E, Hansson J, Egyhazi S. High Frequency of p16 (INK4A) Promoter Methylation in NRAS-Mutated Cutaneous Melanoma. Journal of Investigative Dermatology. 2010 Dec;130(12):2809–2817. doi: 10.1038/jid.2010.216. [DOI] [PubMed] [Google Scholar]
- 27.Eskandarpour M, Huang F, Reeves KA, Clark E, Hansson J. Oncogenic NRAS has multiple effects on the malignant phenotype of human melanoma cells cultured in vitro. Int J Cancer. 2009 Jan 1;124(1):16–26. doi: 10.1002/ijc.23876. [DOI] [PubMed] [Google Scholar]
- 28.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004 Mar 19;116(6):855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 29.Lin WM, Baker AC, Beroukhim R, Winckler W, Feng W, Marmion JM, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008 Feb 1;68(3):664–673. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smalley KS, Xiao M, Villanueva J, Nguyen TK, Flaherty KT, Letrero R, et al. CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009 Jan 8;28(1):85–94. doi: 10.1038/onc.2008.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Viros A, Fridlyand J, Bauer J, Lasithiotakis K, Garbe C, Pinkel D, et al. Improving melanoma classification by integrating genetic and morphologic features. PLoS medicine. 2008 Jun 3;5(6):e120. doi: 10.1371/journal.pmed.0050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005 Nov 17;353(20):2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 33.Devitt B, Liu W, Salemi R, Wolfe R, Kelly J, Tzen CY, et al. Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. Pigm Cell Melanoma R. 2011 Aug;24(4):666–672. doi: 10.1111/j.1755-148X.2011.00873.x. [DOI] [PubMed] [Google Scholar]
- 34.Jakob JA, Bassett RL, Jr., Ng CS, Curry JL, Joseph RW, Alvarado GC, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2011 Dec;:16. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellerhorst JA, Greene VR, Ekmekcioglu S, Warneke CL, Johnson MM, Cooke CP, et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin Cancer Res. 2011 Jan 15;17(2):229–235. doi: 10.1158/1078-0432.CCR-10-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu M, Hemesath TJ, Takemoto CM, Horstmann MA, Wells AG, Price ER, et al. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 2000 Feb 1;14(3):301–312. [PMC free article] [PubMed] [Google Scholar]
- 37.Dumaz N, Hayward R, Martin J, Ogilvie L, Hedley D, Curtin JA, et al. In Melanoma, RAS Mutations Are Accompanied by Switching Signaling from BRAF to CRAF and Disrupted Cyclic AMP Signaling. Cancer Res. 2006 Oct 1;66(19):9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 38.Marquette A, Andre J, Bagot M, Bensussan A, Dumaz N. ERK and PDE4 cooperate to induce RAF isoform switching in melanoma. Nat Struct Mol Biol. 2011 May;18(5):584–591. doi: 10.1038/nsmb.2022. [DOI] [PubMed] [Google Scholar]
- 39.Dumaz N, Light Y, Marais R. Cyclic AMP blocks cell growth through Raf-1-dependent and Raf-1-independent mechanisms. Molecular and Cellular Biology. 2002 Jun;22(11):3717–3728. doi: 10.1128/MCB.22.11.3717-3728.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dumaz N. Mechanism of RAF isoform switching induced by oncogenic RAS in melanoma. Small Gtpases. 2011 Sep;2(5):289–292. doi: 10.4161/sgtp.2.5.17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritt DA, Monson DM, Specht SI, Morrison DK. Impact of feedback phosphorylation and Raf heterodimerization on normal and mutant B-Raf signaling. Molecular and Cellular Biology. 2010 Feb;30(3):806–819. doi: 10.1128/MCB.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smalley KSM. A pivotal role for ERK in the oncogenic behaviour of malignant melanoma? International Journal of Cancer. 2003 May 1;104(5):527–532. doi: 10.1002/ijc.10978. [DOI] [PubMed] [Google Scholar]
- 43.Haass NK, Sproesser K, Nguyen TK, Contractor R, Medina CA, Nathanson KL, et al. The mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitor AZD6244 (ARRY-142886) induces growth arrest in melanoma cells and tumor regression when combined with docetaxel. Clin Cancer Res. 2008 Jan 1;14(1):230–239. doi: 10.1158/1078-0432.CCR-07-1440. [DOI] [PubMed] [Google Scholar]
- 44.Bhatt KV, Spofford LS, Aram G, McMullen M, Pumiglia K, Aplin AE. Adhesion control of cyclin D1 and p27Kip1 levels is deregulated in melanoma cells through BRAF-MEK-ERK signaling. Oncogene. 2005 May 12;24(21):3459–3471. doi: 10.1038/sj.onc.1208544. [DOI] [PubMed] [Google Scholar]
- 45.Shao Y, Aplin AE. Akt3-mediated resistance to apoptosis in B-RAF-targeted melanoma cells. Cancer Res. 2010 Aug 15;70(16):6670–6681. doi: 10.1158/0008-5472.CAN-09-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boisvert-Adamo K, Aplin AE. Mutant B-RAF mediates resistance to anoikis via Bad and Bim. Oncogene. 2008 May 22;27(23):3301–3312. doi: 10.1038/sj.onc.1211003. [DOI] [PubMed] [Google Scholar]
- 47.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr., et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009 May;41(5):544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheung M, Sharma A, Madhunapantula SV, Robertson GP. Akt3 and mutant V600E B-Raf cooperate to promote early melanoma development. Cancer Res. 2008 May 1;68(9):3429–3439. doi: 10.1158/0008-5472.CAN-07-5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sunters A, Fernandez de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA, et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003 Dec 12;278(50):49795–49805. doi: 10.1074/jbc.M309523200. [DOI] [PubMed] [Google Scholar]
- 50.Madhunapantula SV, Robertson GP. The PTEN-AKT3 signaling cascade as a therapeutic target in melanoma. Pigment Cell Melanoma Res. 2009 Aug;22(4):400–419. doi: 10.1111/j.1755-148X.2009.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer Research. 2011 Apr 1;71(7):2750–2760. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998 Nov 15;12(22):3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem J. 2001 Oct 1;359(Pt 1):1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004 Feb;122(2):337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davies MA, Stemke-Hale K, Lin E, Tellez C, Deng W, Gopal YN, et al. Integrated Molecular and Clinical Analysis of AKT Activation in Metastatic Melanoma. Clin Cancer Res. 2009 Dec 15;15(24):7538–7546. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davies MA, Stemke-Hale K, Tellez C, Calderone TL, Deng W, Prieto VG, et al. A novel AKT3 mutation in melanoma tumours and cell lines. Br J Cancer. 2008 Oct 21;99(8):1265–1268. doi: 10.1038/sj.bjc.6604637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Ruiter ND, Burgering BM, Bos JL. Regulation of the Forkhead transcription factor AFX by Ral-dependent phosphorylation of threonines 447 and 451. Molecular and Cellular Biology. 2001 Dec;21(23):8225–8235. doi: 10.1128/MCB.21.23.8225-8235.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Omholt K, Hansson J. No evidence of RALGDS mutations in cutaneous melanoma. Melanoma Res. 2007 Dec;17(6):410–412. doi: 10.1097/CMR.0b013e3282ef4178. [DOI] [PubMed] [Google Scholar]
- 59.Zipfel PA, Brady DC, Kashatus DF, Ancrile BD, Tyler DS, Counter CM. Ral activation promotes melanomagenesis. Oncogene. 2010 Aug 26;29(34):4859–4864. doi: 10.1038/onc.2010.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mishra PJ, Ha L, Rieker J, Sviderskaya EV, Bennett DC, Oberst MD, et al. Dissection of RAS downstream pathways in melanomagenesis: a role for Ral in transformation. Oncogene. 2010 Apr 22;29(16):2449–2456. doi: 10.1038/onc.2009.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Discov. 2007 Jul;6(7):541–555. doi: 10.1038/nrd2221. [DOI] [PubMed] [Google Scholar]
- 62.Smalley KSM, Eisen TG. Farnesyl transferase inhibitor SCH66336 is cytostatic, pro-apoptotic and enhances chemosensitivity to cisplatin in melanoma cells. Int. J Cancer. 2003 Jun 10;105(2):165–175. doi: 10.1002/ijc.11064. [DOI] [PubMed] [Google Scholar]
- 63.Niessner H, Beck D, Sinnberg T, Lasithiotakis K, Maczey E, Gogel J, et al. The farnesyl transferase inhibitor lonafarnib inhibits mTOR signaling and enforces sorafenib-induced apoptosis in melanoma cells. J Invest Derm. 2011 Feb;131(2):468–479. doi: 10.1038/jid.2010.297. [DOI] [PubMed] [Google Scholar]
- 64.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010 Apr 15;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006 Jan 19;439(7074):358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012 Mar 29;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008 May 1;26(13):2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved Survival with MEK Inhibition in BRAF-Mutated Melanoma. New Engl. J. Med. 2012 Jun;:4. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 69.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res. 2011 Mar 1;17(5):989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 70.Robert C, Flaherty KT, Hersey P, Nathan PD, Garbe C, Milhem MM, et al. METRIC phase III study: Efficacy of trametinib (T), a potent and selective MEK inhibitor (MEKi), in progression-free survival (PFS) and overall survival (OS), compared with chemotherapy (C) in patients (pts) with BRAFV600E/K mutant advanced or metastatic melanoma (MM) J Clin Onc. 2012 Jun 21;30(18_suppl):LBA8509. 2012. [Google Scholar]
- 71.Ascierto PA, Berking C, Agarwala SS, Schadendorf D, Van Herpen C, Queirolo P, et al. Efficacy and safety of oral MEK162 in patients with locally advanced and unresectable or metastatic cutaneous melanoma harboring BRAFV600 or NRAS mutations. J Clin Onc. 2012 May 30;30(15_suppl):8511. 2012. [Google Scholar]
- 72.Smalley KS, Aplin AE, Flaherty KT, Hoeller C, Bosserhoff AK, Haass NK, et al. Meeting report from the 2011 International Melanoma Congress, Tampa, Florida. Pigm Cell Melanoma R. 2012 Jan;25(1):E1–11. doi: 10.1111/j.1755-148X.2011.00943.x. [DOI] [PubMed] [Google Scholar]
- 73.Fedorenko IV, Paraiso KH, Smalley KS. Acquired and intrinsic BRAF inhibitor resistance in BRAF V600E mutant melanoma. Biochem Pharmacol. 2011 May;:25. doi: 10.1016/j.bcp.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Paraiso KH, Haarberg HE, Wood E, Rebecca VW, Chen YA, Xiang Y, et al. The HSP90 Inhibitor XL888 Overcomes BRAF Inhibitor Resistance Mediated through Diverse Mechanisms. Clin. Cancer Res. 2012 May 1;18(9):2502–2514. doi: 10.1158/1078-0432.CCR-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, et al. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol. Cancer Therap. 2012 Apr;11(4):909–920. doi: 10.1158/1535-7163.MCT-11-0989. [DOI] [PubMed] [Google Scholar]
- 76.Atefi M, von Euw E, Attar N, Ng C, Chu C, Guo D, et al. Reversing melanoma cross-resistance to BRAF and MEK inhibitors by co-targeting the AKT/mTOR pathway. PLoS ONE. 2011;6(12):e28973. doi: 10.1371/journal.pone.0028973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008 Dec;14(12):1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.She QB, Solit DB, Ye Q, O'Reilly KE, Lobo J, Rosen N. The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell. 2005 Oct;8(4):287–297. doi: 10.1016/j.ccr.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010 Jul 13;18(1):39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jaiswal BS, Janakiraman V, Kljavin NM, Eastham-Anderson J, Cupp JE, Liang Y, et al. Combined targeting of BRAF and CRAF or BRAF and PI3K effector pathways is required for efficacy in NRAS mutant tumors. PLoS One. 2009;4(5):e5717. doi: 10.1371/journal.pone.0005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bedard P, Tabernero J, Kurzrock R, Britten CD, Stathis A, Perez-Garcia JM, et al. A phase lb, open-label, multicenter, dose-escalation study of the oral pan-PI3K inhibitor BKM120 in combination with the oral MEK1/2 inhibitor GSK1120212 in patients (pts) with selected advanced solid tumors. J Clin Onc. 2012 May 30;30(15_suppl):3003. 2012. [Google Scholar]
- 82.Chandarlapaty S. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discov. 2012 Apr;2(4):311–319. doi: 10.1158/2159-8290.CD-12-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012 Apr 13;149(2):307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J. Clin Invest. 2011 Nov;121(11):4311–4321. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Molhoek KR, Shada AL, Smolkin M, Chowbina S, Papin J, Brautigan DL, et al. Comprehensive analysis of receptor tyrosine kinase activation in human melanomas reveals autocrine signaling through IGF-1R. Melanoma Res. 2011 Aug;21(4):274–284. doi: 10.1097/CMR.0b013e328343a1d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gopal YN, Deng W, Woodman SE, Komurov K, Ram P, Smith PD, et al. Basal and treatment-induced activation of AKT mediates resistance to cell death by AZD6244 (ARRY-142886) in Braf-mutant human cutaneous melanoma cells. Cancer Res. 2010 Nov 1;70(21):8736–8747. doi: 10.1158/0008-5472.CAN-10-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tworkoski K, Singhal G, Szpakowski S, Zito CI, Bacchiocchi A, Muthusamy V, et al. Phosphoproteomic Screen Identifies Potential Therapeutic Targets in Melanoma. Mol. Cancer Res. 2011 Jun;9(6):801–812. doi: 10.1158/1541-7786.MCR-10-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sensi M, Catani M, Castellano G, Nicolini G, Alciato F, Tragni G, et al. Human Cutaneous Melanomas Lacking MITF and Melanocyte Differentiation Antigens Express a Functional Axl Receptor Kinase. J. Invest. Derm. 2011 Dec;131(12):2448–2457. doi: 10.1038/jid.2011.218. [DOI] [PubMed] [Google Scholar]
- 89.Su Y, Vilgelm AE, Kelley MC, Hawkins OE, Liu Y, Boyd KL, et al. RAF265 inhibits the growth of advanced human melanoma tumors. Clin. Cancer Res. 2012 Apr 15;18(8):2184–2198. doi: 10.1158/1078-0432.CCR-11-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Means-Powell JA, Adjei AA, Puzanov I, Dy GK, Goff LW, Ma WW, et al. Safety and efficacy of MET inhibitor tivantinib (ARQ 197) combined with sorafenib in patients (pts) with NRAS wild-type or mutant melanoma from a phase I study. J Clin Onc. 2012 May 30;30(15_suppl):8519. 2012. [Google Scholar]
- 91.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010 Nov 24;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Su F, Bradley WD, Wang QQ, Yang H, Xu LZ, Higgins B, et al. Resistance to Selective BRAF Inhibition Can Be Mediated by Modest Upstream Pathway Activation. Cancer Res. 2012 Feb 15;72(4):969–978. doi: 10.1158/0008-5472.CAN-11-1875. [DOI] [PubMed] [Google Scholar]
- 93.Gowrishankar K, Snoyman S, Pupo GM, Becker TM, Kefford RF, Rizos H. Acquired Resistance to BRAF Inhibition Can Confer Cross-Resistance to Combined BRAF/MEK Inhibition. J. Invest Derm. 2012 Mar;:22. doi: 10.1038/jid.2012.63. [DOI] [PubMed] [Google Scholar]
- 94.Diaz LA, Jr., Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012 Jun 28;486(7404):537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sensi M, Nicolini G, Petti C, Bersani I, Lozupone F, Molla A, et al. Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene. 2006 Jun 8;25(24):3357–3364. doi: 10.1038/sj.onc.1209379. [DOI] [PubMed] [Google Scholar]
- 96.Jovanovic B, Egyhazi S, Eskandarpour M, Ghiorzo P, Palmer JM, Bianchi Scarra G, et al. Coexisting NRAS and BRAF mutations in primary familial melanomas with specific CDKN2A germline alterations. J Invest Dermatol. 2010 Feb;130(2):618–620. doi: 10.1038/jid.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kaplan FM, Shao Y, Mayberry MM, Aplin AE. Hyperactivation of MEK-ERK1/2 signaling and resistance to apoptosis induced by the ongenic B-RAF inhibitor, PLX4720, in mutant N-Ras melanoma cell lines. Oncogene. 2010;30:366–371. doi: 10.1038/onc.2010.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Halaban R, Zhang W, Bacchiocchi A, Cheng E, Parisi F, Ariyan S, et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res. 2010 Apr;23(2):190–200. doi: 10.1111/j.1755-148X.2010.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010 Mar 18;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010 Jan 22;140(2):209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010 Mar 18;464(7287):431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 102.Cho KJ, Kasai RS, Park JH, Chigurupati S, Heidorn SJ, van der Hoeven D, et al. Raf inhibitors target ras spatiotemporal dynamics. Current Biol. 2012 Jun 5;22(11):945–955. doi: 10.1016/j.cub.2012.03.067. [DOI] [PubMed] [Google Scholar]
- 103.Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. The New England journal of medicine. 2012 Jan 19;366(3):207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oberholzer PA, Kee D, Dziunycz P, Sucker A, Kamsukom N, Jones R, et al. RAS mutations are associated with the development of cutaneous squamous cell tumors in patients treated with RAF inhibitors. J. Clin Onc. 2012 Jan 20;30(3):316–321. doi: 10.1200/JCO.2011.36.7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zimmer L, Hillen U, Livingstone E, Lacouture ME, Busam K, Carvajal RD, et al. Atypical Melanocytic Proliferations and New Primary Melanomas in Patients With Advanced Melanoma Undergoing Selective BRAF Inhibition. J. Clin Onc. 2012 May;:21. doi: 10.1200/JCO.2011.41.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Joseph RW, Sullivan RJ, Harrell R, Stemke-Hale K, Panka D, Manoukian G, et al. Correlation of NRAS mutations with clinical response to high-dose IL-2 in patients with advanced melanoma. J. Immunother. 2012 Jan;35(1):66–72. doi: 10.1097/CJI.0b013e3182372636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shahabi V, Whitney G, Hamid O, Schmidt H, Chasalow SD, Alaparthy S, et al. Assessment of association between BRAF-V600E mutation status in melanomas and clinical response to ipilimumab. Cancer Immunol Immunother. 2012 May;61(5):733–737. doi: 10.1007/s00262-012-1227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]