Abstract
Eragrostis curvula includes biotypes reproducing through obligate and facultative apomixis or, rarely, full sexuality. We previously generated a “tetraploid-dihaploid-tetraploid” series of plants consisting of a tetraploid apomictic plant (T), a sexual dihaploid plant (D) and a tetraploid artificial colchiploid (C). Initially, plant C was nearly 100% sexual. However, its capacity to form non-reduced embryo sacs dramatically increased over a four year period (2003–2007) to reach levels of 85–90%. Here, we confirmed high rates of apomixis in plant C, and used AFLPs and MSAPs to characterize the genetic and epigenetic variation observed in this plant in 2007 as compared to 2003. Of the polymorphic sequences, some had no coding potential whereas others were homologous to retrotransposons and/or protein-coding-like sequences. Our results suggest that in this particular plant system increased apomixis expression is concurrent with genetic and epigenetic modifications, possibly involving transposable elements.
Apomixis in plants refers to a diverse group of developmental behaviors resulting in asexual reproduction through seeds1. Apomictic individuals bypass both meiotic reduction and egg cell fertilization to produce offspring that are genetic replicas of the maternal plant2. Few genotypes appear to be obligate for the apomictic trait, and those that do not totally exclude sexual processes are termed “facultative apomictic”, because they are able to produce seeds through both sexual and asexual means3. The line between apomixis and sexual reproduction is somewhat blurry, fueling the perception that the trait might have emerged from deregulation of sexuality rather than from the establishment of a new function4. Indeed, previous studies have shown that apomixis and sexual reproduction share key regulatory mechanisms5.
Given the established relationship between sexual and apomictic pathways, different models involving genetic and epigenetic mechanisms have been proposed to explain the occurrence of apomixis at the molecular level6. There is increasing evidence to support epigenetic control of apomixis7, including the observations that in most apomicts chronological and structural reproductive pattern variations occur in individual plants in response to environmental changes3,8. Variations in apomictic reproduction rates in response to environmental influences have been reported for several species and conditions9,10,11.
Weeping lovegrass (Eragrostis curvula [Schrad.] Nees) is a perennial grass native to Southern Africa that displays a type of apomixis called pseudogamous diplospory12. The megasporocyte of E. curvula undergoes two rounds of mitotic division to form a non-reduced tetranucleate embryo sac with an egg, two synergids, and one polar nucleus13. The E. curvula complex includes cytotypes with different ploidy levels (from 2x to 8x) that may undergo sexual reproduction, facultative apomixis or obligate apomixis14. Diploid (2n = 2x = 20) plants are sexual and rare15. Polyploids reproduce mainly by obligate apomixis, although both sexuality and facultative apomixis have also been reported14.
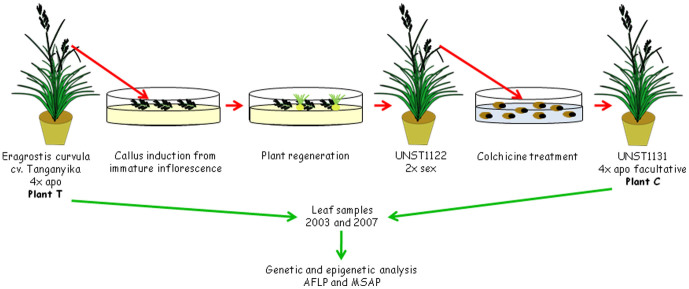
An euploid “back-and-forth” (4x - 2x - 4x) plant series was constructed displaying different ploidy levels and reproductive modes but sharing a common genetic background16. In order to construct the series, a fully sexual diploid plant D was generated from an apomictic tetraploid plant T, through inflorescence in vitro culture16. Seeds from a first generation diploid plant, derived from D after self-pollination, were treated with colchicine, leading to the artificial generation of a tetraploid genotype named C. This colchiploid initially produced variable progeny (as tested with RAPDs) and was subsequently classified as “highly sexual”16. However, the possibility of a low level of apomixis expression was not discarded, because a plant with an increased ploidy level was detected among the progeny, probably originating from fertilization of an unreduced egg cell16. Four years later, the reproductive mode of C was re-analyzed. Surprisingly, the plant exhibited a high proportion of apomictic events (around 85–90%, depending on the technique used to estimate the reproductive mode)13. Based on these results, Meier et al.13 proposed that the temporary expression of high levels of sexual reproduction observed in genotype C might be explained by the genomic stress caused by the in vitro culture and colchicine-induced chromosome duplication procedures.
The objective of the work reported here was to re-analyze the levels of apomixis in colchiploid plant C and characterize the range and nature of the genetic and epigenetic changes occurring during the period from 2003 to 2007, when the change in apomixis expression occurred, and then during the period 2007–2013.
Results
Analysis of reproductive mode
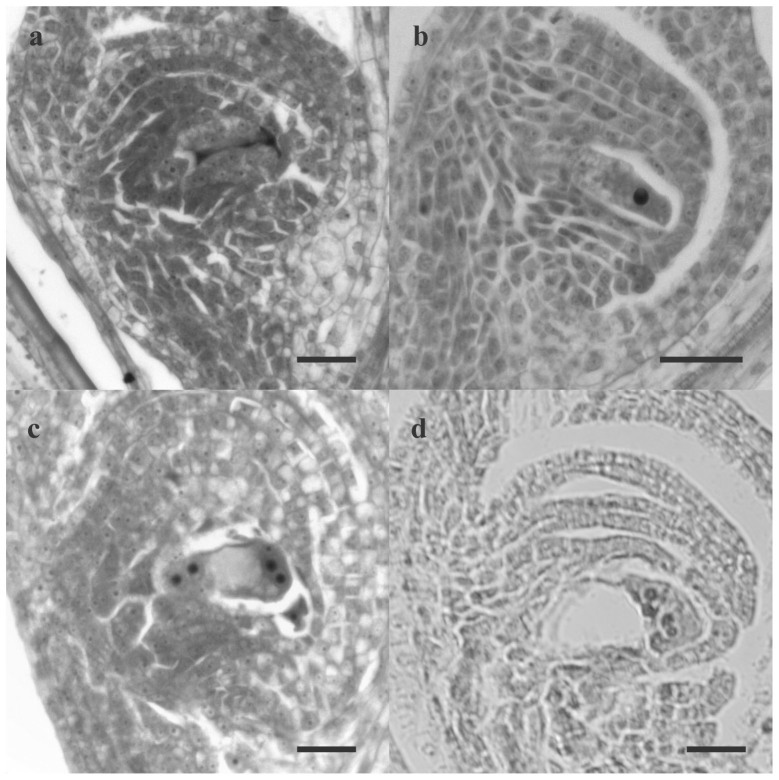
Plants were obtained as is shown in Fig. 1. As was mentioned above, the reproductive mode of plant C was analyzed in 200316 and 200713. An updated cytoembryological analysis of plant C reproductive mode was carried out in year 2009. A total of 233 ovaries at the correct stage were observed. Among these, 24 were found to be meiotic and 209 apomeiotic, corresponding to a potential sexuality level of 10%. Figure 2 shows the embryo sac development in the colchiploid plant C showing sexual (2a and 2c) and apomictic (2b and 2d) processes.
Figure 1. Schematic representation for the generation of the ‘back-and-forth' ploidy series.
This image was created by D.Z.
Figure 2. Cytoembryological analysis.
Sexual (a, c) and diplosporous (b, d) development. Bar: 20 μm. (a) Funtional chalazal megaspore and degenerated micropilar megaspores, (b) elongated megaspore mother cell, (c) tetranucleated stage of sexual embryo sac, (d) tetranucleated stage of apomictic embryo sac.
These results indicated that in 2009 the plant continued to display a level of apomixis as high as that in 200713. Therefore, the rate of apomixis in colchiploid plant C shifted from near 0% in 2003 (21) to 85–90% in 2007 (16), and remained at similar values in 2009 (89,7%).
Genetic and epigenetic structure of plants T and C
We searched for concurrent genetic and/or epigenetic modifications in plant C in 2003 and 2007, when the change in the rate of apomixis was observed. Genomic DNA samples extracted in 2003 and 2007 from plant C and the apomictic tetraploid control plant T were used to produce AFLP and MSAP profiles. Comparisons were performed between plants for the same year and between years for the same plants. A new MSAP analysis of both plants was conducted during the years 2007, 2011 and 2013.
Out of the 14 AFLP primer combinations used in samples from 2003 and 2007, five (5) showed no amplification, four (4) showed absence of amplification in at least one lane, and five (5) showed amplification in all lanes (E36-M37; E42-M36; E33-M38; E33-M37; E33-M33). The initial number of markers counted for each primer combination was 174, as follows: E36-M37: 44 markers; E42-M36: 32 markers; E33-M38: 35 markers; E33-M37: 31 markers; E33-M33: 32 markers. However, 39 individual bands showed inconsistencies in one of the technical replicates. Therefore, the corresponding markers were eliminated from analyses, rendering a total of 135 final markers with 74% (99) being monomorphic and 26% (36) polymorphic. Polymorphic bands were distributed into three categories: 1) patterns that were variable between plants but conserved over time; 2) patterns that showed chronological variation but were conserved between plants; and 3) patterns that were conserved neither between plants nor over time (Table 1). A significant number of variations (5% of the total bands and 20% of the polymorphic ones) occurred equally in both plants, suggesting that they do not represent random mutations. These “conservative” changes were based mainly on the emergence of new bands (0/0/1/1 patterns) (see Supplementary Table S1: the values 0 and 1 represent marker absence or presence, respectively and data from the samples were taken in the following order: T (2003), C (2003), T (2007) and C(2007). Thus, a 0/0/1/1 pattern indicates a band that was absent in both plants in 2003 and is present in both plants in 2007). Among the polymorphic patterns, 72% were not conserved between plants or between years. Notably, four bands were identified only in the colchiploid plant in 2007 (0/0/0/1 patterns) and three bands were associated with apomictic expression (1/0/1/1 patterns).
Table 1. AFLP profiles produced from Tanganyika (T) and UNST1131 (C) from 2003–2007.
| Profiles | Number of bands | Percentage (total) | Percentage (polymorphic) |
|---|---|---|---|
| Monomorphic patterns | 99 | 74% | |
| Polymorphic patterns that were variable between plants and conserved over time | 3 | 2% | 8% |
| Polymorphic patterns that were conserved between plants and variable over time (conservative variations) | 7 | 5% | 20% |
| Polymorphic patterns that were conserved neither between plants nor over time | 26 | 19% | 72% |
| Subtotal polymorphic patterns | 36 | 26% | 100% |
| Total | 135 | 100% | 100% |
Markers with missing data in at least one of the entries were eliminated from the matrix.
To examine genome evolution in each particular plant during the period considered, band profiles were analyzed individually for each plant (Table 2). The total amount of variation over time was similar between plants (15–17%) (Table 2), with the appearance of new bands more frequent than the disappearance of former ones (Supplementary Table S2). This suggests the occurrence of de novo sequence insertion in the genome, because random point mutations would have led to both appearance and disappearance of bands at the same rate, while deletions would have led to most bands vanishing.
Table 2. Variation in the AFLP profiles in each individual plant (T and C) from 2003–2007.
| Profile | T Plant | C Plant | ||
|---|---|---|---|---|
| Absence of changes over time | 104 | 83% | 107 | 85% |
| Presence of changes over time | 22 | 17% | 19 | 15% |
| Total | 126 | 100% | 126 | 100% |
Markers with missing data in at least one of the entries were eliminated from the matrix.
Cluster analysis was performed to estimate genomic similarity of the two plants (T and C) in 2003 and 2007. The results showed higher similarity in 2003 (90%) compared with 2007 (83%) (Fig. 3a). This observation indicates that genome sequences diverged during this four-year period, since both plants have genetically changed mostly in an independent fashion. Whereas some of the loci changed in a conserved manner (Table 1), many of them experienced independent mutations, leading to genomes that were more variable in 2007 than in 2003. Cluster analysis also indicated that T and C similarly diverged from their original genomic structures (81% vs. 83% similarity).
Figure 3. Dendrograms of T and C genotypes for genetic and epigenetic markers.
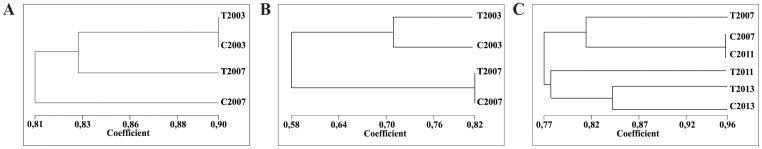
Dendrograms obtained from AFLPs (A) and MSAPs (B) of genotypes Tanganyika (T) and colchiploid (C) from samples collected in 2003 and 2007 and MSAPs (C) of genotypes Tanganyika (T) and colchiploid (C) from samples collected in 2007, 2011 and 2013.
The same genotypes were subjected to MSAP analysis in which the DNA was digested with the isoschizomers HpaII and MspI, which show different sensitivity to cytosine methylation at 5′-CCGG-3′ sites. HpaII will not cleave the DNA if either of the cytosines is fully (double-strand) methylated, but will cleave it if the external C is hemi-methylated (single strand). On the contrary, MspI will not cleave only if the external cytosine is fully- or hemi-methylated. Full methylation of the external or both cytosines prevents digestion by both enzymes.
Out of the total number of MSAP primer combinations used (14) in the first comparison (2003–2007), two (2) did not amplify at all. Three (3) primer combinations did not amplified one or more lanes. Nine primer combinations produced a total of 273 markers, as follows: HM5-E40: 32 markers; HM5-E36: 41 markers; HM6-E32: 17 markers; HM6-E37: 29 markers; HM5-E35: 48 markers; HM5-E37: 32 markers; HM6-E35: 27 markers; HM7-E35: 29 markers; HM4-E35: 18 markers. However, 7 bands showed inconsistencies in one of the technical replicates, so the corresponding markers were eliminated. MSAP experiments produced 266 epigenetic markers (Table 3). Of these, 201 (76%) were invariable (monomorphic) and 65 (24%) were variable (polymorphic). Out of the polymorphic bands, 9% were polymorphic between plants and did not undergo any epigenetic modification in the four-year period; an additional 32% were affected by the same type of epigenetic modifications in both plants, with methylation predominating over demethylation (14 out of 21 vs. 7 out of 21). Finally, some loci underwent unique modifications in each plant (Table 3). A more detailed characterization of the methylation patterns detected is presented in Supplementary Table S3.
Table 3. MSAP profiles produced from Tanganyika (T) and UNST1131 (C) from 2003–2007.
| Profiles | Number of bands | Percentage (total) | Percentage (polymorphic) |
|---|---|---|---|
| Monomorphic patterns | 201 | 76% | |
| Polymorphic patterns that were variable between plants and conserved over time | |||
| Demethylations (T vs C) | 5 | ||
| Methylations (T vs C) | 1 | ||
| Subtotal | 6 | 2% | 9% |
| Polymorphic patterns that were conserved between plants but varied over time (conservative variations) | |||
| Demethylations (2003 vs 2007) | 7 | ||
| Methylations (2003 vs 2007) | 14 | ||
| Subtotal | 21 | 8% | 32% |
| Polymorphic patterns that were conserved neither between plants nor over time | |||
| Demethylations | 12 | ||
| Methylations | 17 | ||
| Demethylations/Methylations | 9 | ||
| Subtotal | 38 | 14% | 59% |
| Subtotal polymorphic | 65 | 24% | 100% |
| Total bands | 266 | 100% |
Polymorphisms arising between 2003 and 2007 were individually analyzed in T and C plants (Table 4). T polymorphic bands corresponding to demethylations were as frequent as those corresponding to methylations (8% demethylations vs. 7% methylations). Interestingly, demethylations mainly involved loci methylated at internal cytosines (12 out of 21) (see Supplementary Table S4). In C, by contrast, the time-variable polymorphic bands corresponded mostly to methylations (15% methylations vs. 5% demethylations). Transformations from internal-cytosine methylation to full methylation were the most frequent ones (See Supplementary Table S4). The rate of methylation occurring in C was higher than that observed in T (15% vs. 7%). In genotype T, internal methylation to full methylation switches (I/M switches) represented 0.7% of the total analyzed loci (2 bands out of 266), whereas in genotype C they reached 6% (16 bands out of 266) (Supplementary Table S4). These results reveal that the I/M switch was 8.5 times more frequent in C than in T. It is important to note that I/M transitions could represent both genetic and/or epigenetic alterations, as pattern M reveals absence of bands after digestion with either enzyme (full methylation) or, alternatively, absence of the site due to sequence variation. However, since similar rates of sequence mutation were detected in T and C by AFLP (15–17%, see Table 2), the variation observed should be of epigenetic origin.
Table 4. Variation in the MSAP profiles in each individual plant (T and C) from 2003–2007.
| Profile | T | C | ||
|---|---|---|---|---|
| Absence of changes over time | 227 | 85% | 213 | 80% |
| Methylations occurring over time | 18 | 7% | 39 | 15% |
| Demethylations occurring over time | 21 | 8% | 14 | 5% |
| Total bands | 266 | 100% | 266 | 100% |
A similarity matrix was constructed from the MSAP data. Cluster analysis showed that after the four-year period, the plants underwent cytosine methylation modifications that made them more similar epigenetically (Fig. 3b). Both plants displayed methylation patterns that were only 58% similar to those they had in 2003. However, the similarity between the plants increased from 71% to 82%. These observations indicate that cytosine methylation patterns in both plants changed over time, with a tendency to acquire the same epigenetic structure at least in some loci. Examples of AFLP and MSAP gels are shown in Fig. 4.
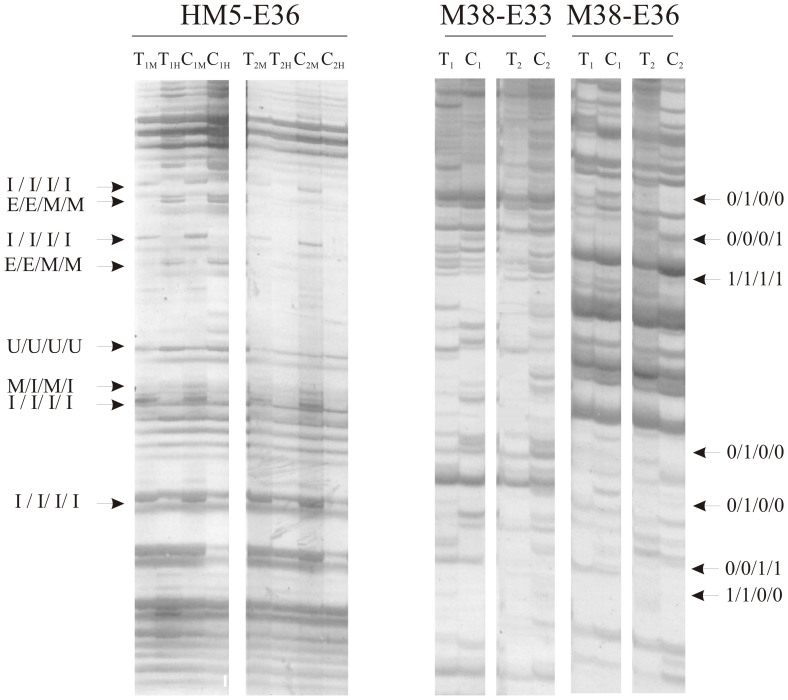
Figure 4. Examples of MSAP and AFLP profiles.
Left panel: MSAP amplicon produced with the HM5-E36 primer combination. 1 indicates samples obtained in 2003. 2 indicates samples obtained in 2007. M indicates digestion with MspI. H indicates digestion with HpaII (e.g., sample T1M corresponds to genotype Tanganyika, year 2003, MspI digestion). Different cytosine methylation patterns are indicated by arrows on the left. Right panels: AFLP amplicons produced with the M38-E33 and M38-E36 primer combinations. 1 indicates samples obtained in 2003. 2 indicates samples obtained in 2007 (e.g., sample T1 corresponds to genotype Tanganyika, year 2003). Different genetic patterns produced by primer combination M38-E36 are indicated by arrows on the left. Full length blots are presented in the Supplementary Information.
A new comparison between plants was performed between the years 2007–2011 and 2011–2013. Using six (6) primer combinations (HM4-E37, HM4-E40, HM6-E32, HM6-E40, HM7-E32 and HM7-E40) 26 out of 240 MSAP markers were polymorphic between years (Table 5). A new similarity matrix was constructed using these markers in the same way than before (2003–2007) (Fig. 3c). This analysis showed that the level of variation between plants along the years was lower than before (2003–2007) showing closer similarity between plants. It seems like with time both plants tend to stabilize their epigenomes and reach a similar methylation status. Plant C in the periods 2007–2011 and 2011–2013 showed one (1) methylation and one (1) de-methylation and three (3) methylations and six (6) de-methylations, respectively. In the same periods T showed nine (9) and six (6) methylations and two (2) and eight (8) de-methylations, respectively. Comparisons between plants showed that five (5) loci were methylated in T in 2011, but were not affected in C. The same five (5) loci were de-methylated in T in 2013. The rest of the polymorphic loci were affected in both plants in the following way: one (1) locus was de-methylated in 2011 and two (2) loci in 2013 in both plants T and C. Related to methylations, one (1) locus was methylated in T and C (2013), one (1) locus in T (2011) and the same locus in C two years later C (2013). Another locus experiences the same situation but in C (2011) and later in T (2013). Other methylations and de-methylations also occurred in independent loci.
Table 5. Variation in the MSAP profiles in each individual plant (T and C) between years 2007–2011 and 2011–2013.
| T | C | |||
|---|---|---|---|---|
| Profile | 2007–2011 | 2011–2013 | 2007–2011 | 2011–2013 |
| Methylations | 9 | 6 | 1 | 3 |
| Demethylations | 2 | 8 | 1 | 6 |
MSAP analysis of three (3) individuals of the cv. Tanganyika of the same age growing in the same environmental conditions using three primer combinations (HM4-E32, HM5-E32 and HM5-E34) showed no detectable changes in their epigenetic landscapes.
Cloning and sequencing of genetic and epigenetic polymorphic bands
Some of the polymorphic bands obtained with AFLP or MSAP were cloned, sequenced, and compared with sequences in the NCBI non-redundant protein sequence (nr) and MIPS databases. The cloned bands were selected to represent different patterns of variation. Most of the AFLP sequences showed similarity to protein-coding genes in short segments (4 out of 5) but no significant similarities were found for the AFLP sequences when searching against the MIPS database (Supplementary Table S5). By contrast, nearly half of the MSAP sequences (3 out of 8) showed similarity to Gypsy or Copia retrotransposons (Supplementary Table S6). The presence of sequences with similarity to proteins was further analyzed (Supplementary Table S6). We isolated 27 additional polymorphic bands originated from AFLP from the same series. All these 27 sequences were also similar to retrotransposons, pseudogenes or retrotransposons + pseudogenes (unpublished).
Based on these results, it can be concluded that some of the variations detected using MSAP occurred in genomic regions showing similarity to mobile elements.
Survey of retrotransposons in cDNA libraries generated from E. curvula genotypes T and C
To analyse the presence of transcripts related to retrotransposons in the plants under study, we screened EST databases corresponding to inflorescence cDNA libraries derived from T and C in 200319, constructed shortly after the colchicine-induced formation of plant C.
The percentage of sequences showing similarity to retrotransposons (E value ≤ 1 e-08) in the different EST libraries showed that such transcripts were more abundant in plant C at that time (Fig. 5). The Gypsy subfamily comprised the most common class of retrotransposons found, and it was especially abundant in plant C.
Figure 5. Contribution of retrotransposon sequences to cDNA libraries of E. curvula.
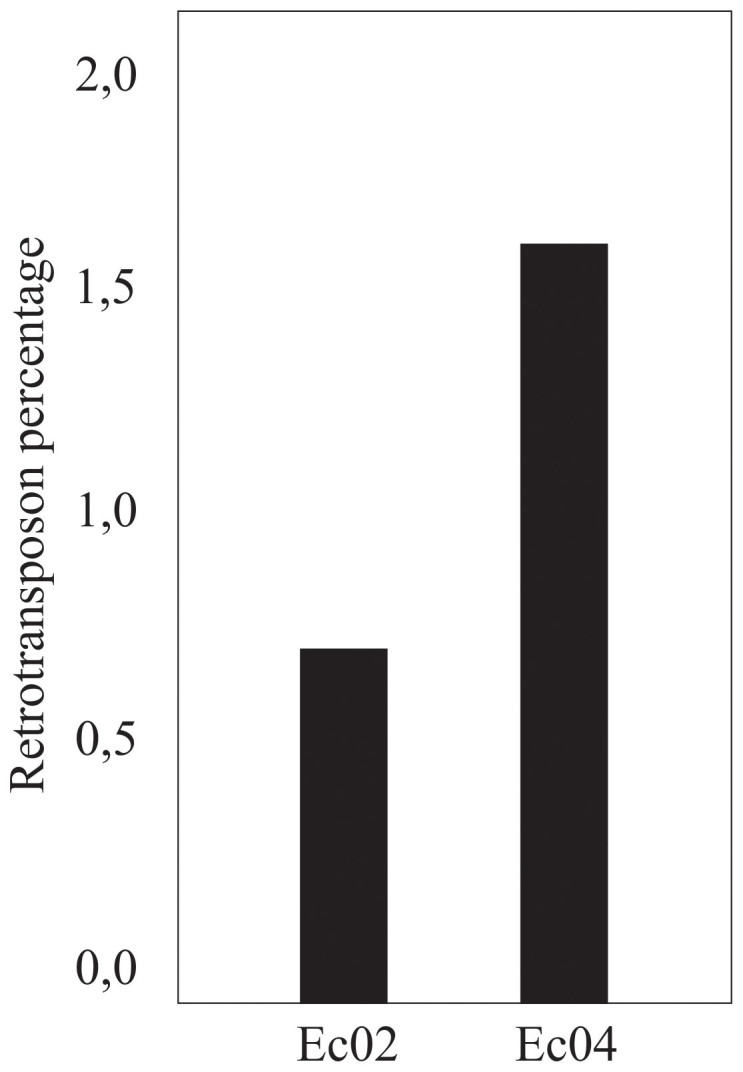
Each bar indicates the percentage of the retrotransposons in the library. Ec02: Library produced from Tanganyika (T) inflorescences, Ec04: Library produced from UNST1131 (C) inflorescences.
Discussion
In this work we confirmed the occurrence of significant variation over time in the rate of asexual reproduction in a newly-synthesized tetraploid E. curvula plant. During the period from 2003 to 2007 the rate of asexual reproduction changed from nearly 0% to almost 90% in the colchiploid plant C, as revealed by cytoembryological studies. This modification of the apomixis level was accompanied by the occurrence of genetic and epigenetic modifications. Genomic polymorphisms were also detected over the same period in the natural tetraploid apomictic plant T, which did not exhibit any variation in the apomixis rate. However, since apomixis is proposed to be governed by one or few loci, any genetic modifications, even when slight, could be relevant. On the other hand, cytosine methylation levels were higher in the colchiploid plant C with respect to the control during the analyzed period. Epigenetic modifications occurring during the same interval appeared to be mostly specific, making the plants more similar. It is interesting to note that even when the level of epigenetic variation between plants C and T was comparable, a higher rate of methylation occurred in the colchiploid plant C. Thus, our results suggest that an increase in apomixis rate occurred along with an increase in global genome methylation. Further experiments should be performed in order to test whether apomixis expression is functionally associated with the establishment of methylation in particular genomic areas.
In previous work, genetic characterization of the “back and forth” ploidy series used herein revealed high levels of genetic rearrangement derived from the chromosome number reduction followed by a duplication event20. The transition from plant T (4x) to plant D (2x) involved the expected occurrence of band loss (due to allele elimination during dihaploidization) but also the unforeseen emergence of novel bands. Surprisingly, the restoration of the original ploidy level by colchiploidization (transition from D to C), revealed recovery of many of the alleles lost during the transition from T to D. A consequence of this restoration phenomenon was the establishment of remarkable genetic similarity between the T and C polyploids20. Similar results reporting non-Mendelian emergence of bands present in previous generations but not in the immediate ancestors has been reported by Song et al. (1995)21 for Brassica synthetic allopolyploids. Mecchia et al (2007)20 proposed that plants belonging to the “back and forth” series had a genetic structure typical of their ploidy level, with the potential to be specifically restored via an unknown mechanism. Further analysis showed that the epigenetic landscape and the transcriptome of the plants appeared to be also characteristics of the ploidy22. The transition from T to D was characterized by a general modification in the epigenetic landscape, which was restored to a large degree during the transition from D to C22. These results as a whole indicate that there are specific genetic, epigenetic, and expression landscapes associated with a particular ploidy level. Moreover, the results presented here suggest that the epigenetic landscape restoration observed during the transition from D to C22 was still progressing during the period 2003–2007. A new comparison performed among years 2007, 2011 and 2013 showed a clear tendency to increase the similarity of both plants at this level. During these years the plants were still changing but were closer than in 2003. The corresponding similarity coefficients were 0.71 and 0.85 in 2003 and 2013 respectively.
A major point to be considered is the ploidy homogeneity of plant C, which was obtained after colchicine treatment of seeds from self-pollinated line D. The possibility of plant C of being a ploidy chimera was analyzed carefully by flow cytometry and chromosome counting in previous work16. Homogeneity was further demonstrated in order to register C as a new plant variety in the RNC (Registro Nacional de Cultivares, National Cultivars Database, Buenos Aires, Argentina) at INASE (Instituto Nacional de Semillas, National Institute of Seeds, Argentina) (Registration number: RC9193, 2006–2026). It is important to note that E. curvula tetraploids and the diploid plant UNST1122 are easy to differentiate at first sight in the greenhouse. Diploids show wider leaves with a typical pubescence on the base of the leaf adaxial surface and are taller than tetraploids16. None of the morphological characteristics of diploid plants were ever observed in the colchiploid plant C, which showed morphology that was essentially analogous to that of the other tetraploids. If line C was a chimera, we would have expected that at least parts of the plant would have displayed morphology characteristic of the diploids. Moreover, from 2002–2012, numerous vegetative tillers were separated from the original plant C in order to propagate the genotype. In many of the derived plants, ploidy levels were determined by flow cytometry and chromosome counting. In all cases, they were found to be tetraploid. In the period from 2002 to 2007 the meiotic behaviour of plant C (male meiosis) was carefully analyzed in anthers originating from several panicles. A tetraploid configuration with normal meiosis was always observed. In 2007, reduced and unreduced embryo sacs were always detected in the same inflorescences. Based on the above-mentioned considerations, the hypothesis of plant C being a chimera was discarded.
The modification of the apomixis rate in plant C occurred along with genetic and epigenetic modifications, the latter involving a higher rate of methylation when compared to control plant T. The variability detected involved retrotransposons and gene sequences, as revealed by cloning and sequencing of the polymorphic fragments. Moreover, plant C expressed more transcripts related to retrotransposons. Our results are in agreement with those reported by Ochogavía et al.23, who detected high rates of Gypsy retrotransposon expression in tetraploid sexual genotypes of Paspalum notatum when compared with apomictic genotypes of the same ploidy. Further work should analyze the role of these transposable elements in the transition from sexuality to apomixis.
Methods
Plant material
An Eragrostis curvula euploid “back-and-forth” plant series of different ploidy levels and reproductive modes was used in this work16. The series consisted of: 1) an obligate apomictic tetraploid plant T (cv. Tanganyika, 2n = 4x = 40); 2) a diploid plant D (experimental code UNST1122, 2n = 2x = 20, fully sexual), generated from tetraploid T by inflorescence in vitro culture; and 3) a tetraploid plant C (experimental code UNST1131, 2n = 4x = 40, initially classified as highly sexual), obtained by colchicine duplication of R1 seeds of diploid genotype D (Fig. 1). DNA samples were collected from leaves of plants T and C in 2003, 2007, 2011 and 2013. Three additional adult plants belonging to the natural cv. Tanganyika, obtained from seeds, were used as controls in order to analyze the methylation differences among plants. The plants analyzed were grown under the same routine glasshouse conditions.
Cytoembryological studies
Inflorescences were collected at the beginning of anthesis, when it is possible to observe all of the embryo sac developmental stages, as described in Meier et al. (2011). Briefly, after fixation in FAA (50% ethanol, 5% acetic acid, 10% formaldehyde and distilled water), individual spikelets were dehydrated in a tertiary butyl alcohol series, embedded in paraplast (Johansen 1940), and then sectioned at 10 mm and stained with safranin-fast green. Observations were carried out with a Nikon Eclipse TE300 light transmission microscope (Tokyo, Japan).
Amplified fragment length polymorphisms (AFLPs)
AFLP studies were performed on samples extracted from leaves, according to Vos et al.17. Genomic DNA (600 ng) samples were double-digested with the enzymes EcoRI and MseI. The resulting fragments were then ligated to EcoRI and MseI adaptors to produce template for further amplifications (primer and adaptor sequences are shown in Supplementary Table S7). PCR amplifications using technical duplicates were carried out with 14 AFLP primer combinations for the comparisons between years 2003 and 2007. PCR products were separated on 6% (w/v) denaturing polyacrylamide gels, silver-stained, and digitized for analysis. Bands with identical migration amplified from different plant/year samples were considered to represent a marker. The standard used to select a particular marker to be included in the analysis was that all representative bands should have produced consistent patterns (presence/absence) in both technical replicates. Variation of less than 5% between technical replicates was required for a primer combination to be included in the analysis. Genetic similarities between the samples were calculated using Jaccard's coefficient, and UPGMA cluster analysis was performed using the NTSyS software package.
Methylation-sensitive amplified polymorphisms (MSAPs)
Detection of cytosine methylation pattern modifications was carried out by performing MSAP studies on DNA samples extracted from leaves, according to Xu et al.18. The methylation-sensitive isoschizomers HpaII and MspI were selected as frequent-cutting enzymes, and EcoRI was chosen as a rare-cutting enzyme (primer and adaptor sequences are shown in Supplementary Table S7). PCR amplifications were carried out with 14 and 6 MSAP primer combinations (for comparisons between years 2003 and 2007 and among years 2007, 2011 and 2013, respectively), using technical duplicates. PCR products were separated on 6% (w/v) denaturing polyacrylamide gels, silver-stained, and digitized for analysis. Bands were counted only when they were present in both replicates. Variation of less than 5% between technical replicates was required for a primer combination to be included in the analysis. MSAP data derived from the four samples were converted into a binary matrix, with 0 representing monomorphic patterns (00 and 11) and 1 representing polymorphic patterns (01 and 10). Matrices were analyzed to determine similarity coefficients between pairs of individuals and for group clustering. Jaccard's coefficient (J) was used as the similarity index, and UPGMA cluster analysis was performed using the NTSyS software package.
Isolation, cloning and sequencing of polymorphic DNA fragments
Bands of interest were moistened with distilled sterile water and excised from polyacrylamide gels using a scalpel, then cut into smaller sections and eluted with buffer solution (0.5 M ammonium acetate, 1 mM EDTA pH 8.0) overnight at 37°C. The DNA was ethanol-precipitated and re-amplified using the same PCR conditions described for AFLP and MSAP assays. The resulting fragments were cloned using the pGEM-T Easy Vector System (Promega) and sequenced by Macrogen (Korea).
Sequence data analysis
Sequence annotation was performed by means of sequence similarity analysis using the BLAST tools available at NCBI (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/) and MIPS at PlantsDB (http://mips.helmholtz-muenchen.de/plant/genomes.jsp).
In silico detection of retrotransposons in E. curvula cDNA libraries
The BLASTN algorithm was used to detect sequences potentially corresponding to retroelements in two cDNA E. curvula libraries constructed from inflorescences of genotypes T and C. The libraries were named as follows: Ec02 (Tanganyika, apomictic tetraploid, T) and Ec04 (UNST1131, sexual colchiploid, C). The libraries were constructed in 200319. Reference sequences used for search queries were obtained from a public non-redundant database (http://wheat.pw.usda.gov/ITMI/Repeats/). Matching sequences with E-value ≤ 1 e-08 were considered to be similar. Results were expressed as percentages for each library.
Author Contributions
D.Z., I.G., S.P. and V.E. wrote the main manuscript text. A.O. performed the AFLP and MSAP, J.R.R. performed in silico detection of retrotransposons and J.M.R. and M.M. performed the cytoembryological studies. All authors reviewed the manuscript.
Supplementary Material
Supplementary dataset number 1
Supplementary Info File #1
Acknowledgments
This project was funded by grants from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, PIP 112-200801-01517), Universidad Nacional del Sur (UNS, PGI 24/A133) and ANPCyT PICT 2011 1269, Argentina.
References
- Nogler G. [Gametophytic apomixis.]. Embryology of angiosperms [Johri, B.] (ed.). [475–518] (Springer Verlag, Berlin, 1984). [Google Scholar]
- Asker S. & Jerling L. Apomixis in Plants (CRC Press, Boca Raton, USA, 1992). [Google Scholar]
- Koltunow A. Apomixis: embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5, 1425–1437 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Aguilar M., Michaud C., Leblanc O. & Grimanelli D. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22, 3249–3267 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M. et al. Sexual and apomictic reproduction in Hieracium subgenus Pilosella are closely interrelated developmental pathways. Plant Cell 15, 1524–1537 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow A. & Grossniklaus U. Apomixis: a developmental perspective. Ann. Rev. Plant Biol. 54, 547–574 (2003). [DOI] [PubMed] [Google Scholar]
- Grimanelli D. Epigenetic regulation of reproductive development and the emergence of apomixis in angiosperms. Curr. Opin. Plant Biol. 15, 57–62 (2012). [DOI] [PubMed] [Google Scholar]
- Koltunow A., Johnson S. & Bicknell R. Apomixis is not conserved in related, genetically characterised Hieracium plants of varying ploidy. Sex. Plant Reprod. 12, 253–266 (2000). [Google Scholar]
- Quarin C. Seasonal changes in the incidence of apomixis of diploid, triploid and tetraploide plants of Paspalum cromyorrhizon. Euphytica 35, 515–522 (1986). [Google Scholar]
- Gounaris E., Sherwood R., Gounaris I., Hamilton R. & Gustine D. Inorganic salts modify embryo sac development in sexual and aposporous Cenchrus ciliaris. Sex. Plant Reprod. 4, 188–192 (1991). [Google Scholar]
- Davies L. & Cohen D. Phenotypic variation in somaclones of Paspalum dilatatum and their seedling offspring. Can. J. Plant Sci. 72, 773–784 (1992). [Google Scholar]
- Streetman L. Reproduction of the lovegrass, the genus Eragrostis-I. E. chloromelas Steud, E. curvula (Schrad.) Nees, E. lehmanniana Nees and E. superba Peyr. Wrightia 3, 41–51 (1963). [Google Scholar]
- Meier M., Zappacosta D., Selva J. P., Pessino S. & Echenique V. Evaluation of different methods for assessing the reproductive mode of weeping lovegrass plants, Eragrostis curvula (Schrad.) Nees. Austral. J. Bot. 59, 253–261 (2011). [Google Scholar]
- Voigt P. &Bashaw E. Facultative apomixis in Eragrostis curvula. Crop Sci. 16, 803–805 (1976). [Google Scholar]
- Voigt P. Discovery of sexuality in Eragrostis curvula (Schrad.) Nees. Crop Sci. 11, 424–425 (1971). [Google Scholar]
- Cardone S. et al. Novel genotypes of the subtropical grass Eragrostis curvula for the study of apomixis. Euphytica 151, 263–272 (2006). [Google Scholar]
- Vos P. et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Li X. & Korban S. AFLP-Based detection of DNA methylation. Plant Mol. Biol. Rep. 18, 361–368 (2000). [Google Scholar]
- Cervigni G. et al. Expressed sequence tag analysis and development of gene associated markers in a near-isogenic plant system of Eragrostis curvula. Plant Mol. Biol. 67, 1–10 (2008). [DOI] [PubMed] [Google Scholar]
- Mecchia M. et al. Genome polymorphisms and gene differential expression in a ‘back-and-forth' ploidy-altered series of weeping lovegrass (Eragrostis curvula). J. Plant Physiol. 164, 1051–1061 (2007). [DOI] [PubMed] [Google Scholar]
- Song K., Lu P., Tang K. & Osborn T. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92, 7719–7723 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochogavía A., Cervigni G., Selva J. P., Echenique V. & Pessino S. Variation in cytosine methylation patterns during ploidy level conversions in Eragrostis curvula. Plant Mol. Biol. 70, 17–29 (2009). [DOI] [PubMed] [Google Scholar]
- Ochogavía A. et al. Characterization of retrotransposon sequences expressed in inflorescences of apomictic and sexual Paspalum notatum plants. Sex. Plant Reprod. 24, 231–246 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary dataset number 1
Supplementary Info File #1