Abstract
Reverse genetics allows the generation of recombinant viruses entirely from cDNA. One application of this technology is the creation of reporter-expressing viruses, which greatly increase the detail and ease with which these viruses can be studied. However, there are a number of challenges when working with reporter-expressing viruses. Both the reporter protein itself as well as the genetic manipulations within the viral genome required for expression of this reporter can result in altered biological properties of the recombinant virus, and lead to attenuation in vitro and/or in vivo. Further, instability of reporter expression and purging of the genetic information encoding for the reporter from the viral genome can be an issue. Finally, a practical challenge for in vivo studies lies in the attenuation of light signals when traversing tissues. Novel expression strategies and the continued development of brighter, red and farred shifted reporters and the increased use of bioluminescent reporters for in vivo applications promise to overcome some of these limitations in future. However, a “one size fits all” approach to the design of reporter-expressing viruses has thus far not been possible. Rather, a reporter suited to the intended application must be selected and an appropriate expression strategy and location for the reporter in the viral genome chosen. Still, attenuating effects of the reporter on viral fitness are difficult to predict and have to be carefully assessed with respect to the intended application. Despite these limitations the generation of suitable reporter-expressing viruses will become more common as technology and our understanding of the intricacies of viral gene expression and regulation improves, allowing deeper insight into virus biology both in living cells and in animals.
Keywords: reverse genetics, luciferase, reporter, green fluorescent protein, in vivo imaging, mononegaviruses
1. Introduction
Reverse genetics systems are based on the generation and subsequent replication and transcription of full-length virus RNA genomes or truncated genome analogues from cDNA. They encompass both life cycle modelling systems, which allow the modelling and dissection of discrete parts of the virus life cycle, and full-length clone systems, which allow the generation of recombinant viruses from cDNA (Hoenen et al., 2011). Among negative-sense RNA viruses, the first virus to be rescued entirely from cDNA was a recombinant Rabies virus in 1994 (Schnell et al., 1994). Since then, full-length clone systems have become available for almost all negative-sense RNA viruses. An obvious application for these systems is the generation of mutated viruses in order to study the impact of those mutations on virus biology. For example, the first recombinant Rabies viruses generated were used to investigate the role of the Rabies ψ pseudogene region (Schnell et al., 1994). Also a common application of full-length clone systems is to create chimeric viruses in order to identify pathogenic determinants. For example, the role of the ebolavirus glycoprotein GP as a pathogenic determinant has recently been investigated by exchanging the GP gene between different ebolavirus species of varying pathogenicity (Groseth et al., 2012). Another application, which will be discussed in this review, is the generation of viruses that express foreign reporter proteins. These viruses can be used to investigate virus biology; however, they also have tremendous potential in the screening of antivirals, the visualization of infection in animal models in order to increase our understanding of pathogenesis and transmission, and for the rational attenuation of viruses for vaccine purposes, as well as for use as oncolytic agents. This review will discuss the current state of the art for reporter-expressing negative-sense RNA viruses, as well as challenges in their development and perspectives to overcome these challenges, with a special focus on the order Mononegavirales.
2. Genome structures of mononegaviruses
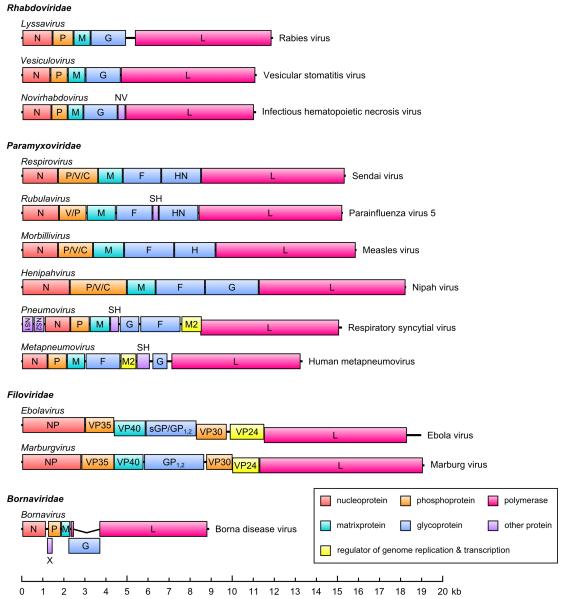
There are four families within the order Mononegavirales: Rhabdoviridae, Paramyxoviridae, Filoviridae and Bornaviridae (in addition, Nyaminiviridae has been proposed as a fifth family within this order). Despite the fact that these families encompass a vast number of viruses, rhabdoviruses, paramyxoviruses and filoviruses share many features in terms of their genome structure and basic virus biology (Lamb and Griffith, 2007; Lyles and Rupprecht, 2007; Sanchez et al., 2007). As the name implies, the genomes of all mononegaviruses (members of the order Mononegavirales) are non-segmented, single-stranded RNA genomes in negative orientation. The general gene order is 1) a nucleoprotein (called N or NP), 2) a phosphoprotein (P or VP35), 3) a matrix protein (M or VP40), 4) one or several glycoproteins (G, sGP, GP1,2, F, H or HN) and 5) a viral polymerase (L) (Fig. 1). Some of the viruses also carry additional genes (VP30, VP24, NV, SH, M2, NS1, NS2), as shown in Figure 1. As a strategy to increase their coding capacity some mononegavirus genes encode for additional proteins that are expressed from alternative open reading frames (ORFs). These additional ORFs are either accessed from alternative start codons, or through editing of the mRNA by insertion of non-templated nucleotides, leading to a frameshift that results in access to the alternative ORF. The first mechanism gives rise to the C proteins for paramyxoviruses and some rhabdoviruses, whereas the second mechanism produces the paramyxovirus V, W, I and D gene products, as well as the ebolavirus GP1,2 and ssGP proteins.
Figure 1. Genome structure of mononegaviruses.
The genes (or in the case of Borna disease virus the coding sequences) of representative members of virus genera are shown as boxes, with the gene names indicated. Non-transcribed regions are shown as a black bar. Gene overlaps are indicated as steps. Genes and genomes are drawn to scale, with the exception of the first part of the Borna disease virus L coding sequence (before the splice site), which is enlarged for the purposes of clarity.
Genes are transcribed from the 3′ end of the genome in a linear fashion, with the polymerase stopping at gene end signals, and then reinitiating at nearby gene start signals. Reinitiation has been shown to be imperfect for rhabdoviruses (Iverson and Rose, 1981), leading to a transcriptional gradient with 3′ proximal genes being transcribed at higher rates than 3′ distal genes. This mechanism is presumed to be a general feature of mononegavirus transcription.
While bornaviruses show similarities to the other mononegaviruses in terms of general gene order, there are significant differences in their expression strategy (Lipkin and Briese, 2007). The bornavirus genome is transcribed into three transcripts, the first of which contains the coding sequence for the nucleoprotein, the second the coding sequences for the P protein and the X protein, whose ORF overlaps with that of the P protein, and the third the coding sequences for the remainder of the viral proteins (M, G and L). The mRNAs encoding for these three proteins are produced through splicing from this third transcript. This approach is unique among the mononegaviruses and is made possible due to the feature that bornavirus replication occurs in the cell nucleus, which is unusual for mononegaviruses.
3. Applications of reporter-expressing viruses
Reporter-expressing viruses are recombinant viruses that express a foreign reporter protein (Fig. 2), resulting in numerous in vitro and in vivo applications. In vitro, the relative ease with which infections with these viruses can be detected, and also quantified, makes them very well suited for high throughput applications where large numbers of samples have to be evaluated for the extent of infection in an automated fashion, e.g. during large scale antiviral drug screening. Indeed, reporter-expressing viruses have been developed and applied for the purpose of antiviral drug screening (Hoenen et al., 2013; Johansen et al., 2013; Panchal et al., 2010; Panchal et al., 2012), as well as for rapid neutralization tests measuring antibody titers (Hu et al., 2012; Zhou et al., 2013). Reporter-expressing viruses have also been used in vitro to address a number of basic research questions. Viruses expressing reporters fused to viral proteins are obviously well suited to studying the spatiotemporal distribution of these proteins in infected cells, and have been successfully used for this purpose (Chambers and Takimoto, 2010; Das et al., 2006; Hoenen et al., 2012; Klingen et al., 2008). However, also those viruses that encode reporters not fused to viral proteins, but rather expressed on their own, have been successfully used for basic research. For example, these viruses were used to demonstrate the preferential translation of viral RNAs over host RNAs during vesicular stomatitis virus (VSV) infection (Whitlow et al., 2006), to investigate the modulation of dendritic cell function by Ebola virus proteins (Lubaki et al., 2013), and to study the susceptibility of different cell lines to measles virus infection (Hashimoto et al., 2002).
Figure 2. Examples of reporter expressing viruses.
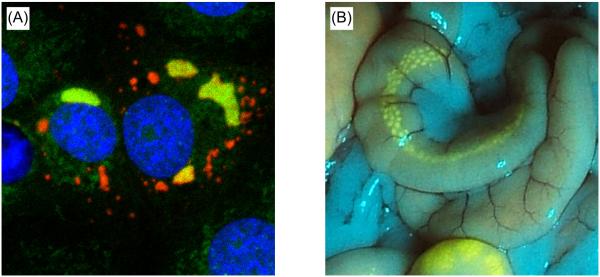
(A) Confocal image of Vero E6 cells infected with an Ebola virus expressing an L-mCherry fusion protein (red). Cells were subjected to nascent RNA staining (green) after treatment with Actinomycin D for inhibition of cellular RNA synthesis, to identify sites of viral genome replication / transcription. Nuclei were stained with DAPI (blue). Image reproduced from (Hoenen et al., 2012) with permission of the copyright holder. Copyright © American Society for Microbiology, J Virol. 86, 2012, 11779–11788, doi: 10.1128/JVI.01525-12. (B) B-cell germinal centers in a Peyer’s patch in the ileum of a ferret infected with an eGFP-expressing canine distemper virus. Image kindly provided by Martin Ludlow and Paul Duprex (Boston University).
In vivo, much of the work with reporter-expressing mononegaviruses has focused on pathogenesis – primarily determining host cell tropism and extending this information to describe how viruses disseminate through the host. Multiple paramyxoviruses expressing various fluorescent reporters have been used to gain insight into viral pathogenesis (Banyard et al., 2010; de Swart et al., 2007; de Vries et al., 2010; Ludlow et al., 2012; Rudd et al., 2006; von Messling et al., 2004), and a human metapneumovirus expressing green fluorescent protein (GFP) was used to evaluate the attenuating effect of altering N-glycosylation sites on various virus proteins both in vitro and in vivo for the development of live attenuated vaccines (Zhang et al., 2011). For Bornaviruses, dissemination throughout the nervous system was studied using a Borna disease virus expressing GFP (Ackermann et al., 2010). A Sendai virus expressing firefly luciferase was used to localize virus replication in both resistant and susceptible mice, and to demonstrate that replication in the upper respiratory tract and trachea supports efficient transmission, even in the absence of disease (Burke et al., 2011).
Some novel in vivo uses of reporter viruses have also been established. Confirmation of exclusive tumor targeting following localized or systemic delivery of oncolytic virotherapies has been demonstrated with a luciferase-expressing recombinant Newcastle disease virus (Wei et al., 2012) and a measles virus expressing the human thyroidal sodium ion symporter (which concentrates radioisotopes inside cells) allowing for PET or SPECT/CT imaging in mice (Msaouel et al., 2009). GFP has been used to demonstrate that a modified Borna disease virus vector may have utility for stable gene expression in the central nervous system (Daito et al., 2011), and GFP-expressing Rabies viruses have been instrumental in studies on neuroarchitecture (Wickersham et al., 2007a; Wickersham et al., 2007b). Based on experiences with other viruses, reporter-expressing mononegaviruses also have the potential for additional uses in vivo such as monitoring the effectiveness of anti-viral therapies and vaccine studies in real time or more detailed pathogenesis studies (both real time and post-mortem) - including determining the effects of viral factors, host genetics, host age, immune status, environmental and inoculation conditions on infection dynamics and transmission (Burke et al., 2011). Finally, reporters can be used as a marker for vaccination to distinguish vaccinated from infected animals (Walsh et al., 2000).
4. Challenges in the development and application of reporter-expressing viruses
There are three major challenges in developing reporter expressing mononegaviruses. First, introduction of a foreign gene or modification of existing viral genes can alter the biological properties of these viruses, which can for example become apparent in reduced virulence of these viruses and can be a significant problem particularly for animal studies. Second, such alterations can create an evolutionary pressure on the virus to purge the genetic information encoding for the reporter protein, leading to loss of reporter gene expression. Third, a technical problem that applies to in vivo work is the attenuation of light signals when traversing tissues, making detection of reporter activity in vivo challenging.
4.1 Altered biological properties of reporter-expressing viruses
Both the reporter protein itself and the changes to the viral genome necessary for expression of the reporter can alter the biological properties of reporter-expressing viruses. One striking example of a direct effect of the reporter protein is a recombinant bunyavirus expressing GFP fused to the N-terminus of Gc, which was highly attenuated (i.e. showed reduced end-point titers and delayed growth kinetics), whereas a similar virus, in which GFP was exchanged for mCherry, showed only slight attenuation (Shi et al., 2010). It has been proposed that in this case GFP was inhibiting the function of Gc by forming inappropriate disulfide bonds with Gc, whereas mCherry does not contain any cysteine residues, which would be required for the formation of such disulfide bonds (Shi et al., 2010). As another example, a respiratory syncytial virus (RSV) expressing Renilla luciferase was attenuated in vivo; however the same virus expressing Katushka2 did not show attenuation, suggesting that the observed attenuation was caused by the reporter protein itself rather than the changes to the virus genome (Hotard et al., 2012). Unfortunately, very few studies have systematically compared different reporters in otherwise identical recombinant viruses, making it difficult to predict in advance which reporters will be tolerated in a given system.
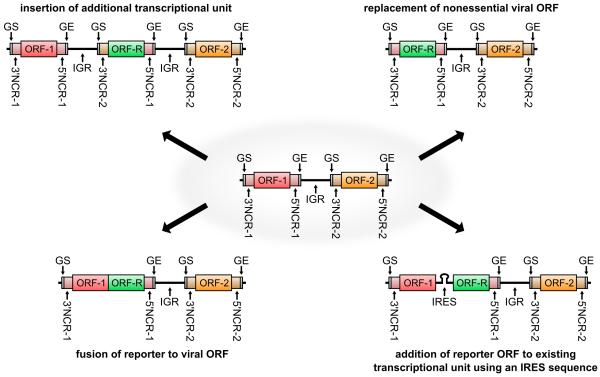
There are several expression strategies (Fig. 3) for altering the virus genome to achieve expression of a foreign reporter protein, each of which can impact biological properties of the reporter expressing virus. The most frequently used approaches rely either on the insertion of an additional transcriptional unit (ATU) into the viral genome, from which the reporter is expressed as a separate protein, or the expression of the reporter as a fusion with a viral protein, with or without expression of the wild-type viral protein. However, a number of other strategies have been recently employed, including direct replacement of non-essential gene products, as well as the use of an internal ribosome entry sequence (IRES) to direct translation of a separate reporter protein from an mRNA also encoding a viral protein.
Figure 3. Expression strategies for reporter genes.
Different expression strategies are depicted. Viral open reading frames (ORF-1, ORF-2) and a reporter open reading frame (ORF-R) and flanking non-coding regions (NCRs) are shown in color. Gene start (GS) and gene end (GE) signals are shown in gray. Viral intergenic regions (IGRs) and an internal ribosomal entry site (IRES) sequence are shown as black lines.
4.1.1 Expression of reporters from additional transcriptional units
The most commonly used strategy to introduce reporters employs ATUs. To this end, the coding region of the reporter gene (or a casette consisting of coding regions of two reporter genes joined by an IRES sequence (Marschalek et al., 2009)) is flanked by gene start and stop signals, which are in general highly conserved within a given viral genome. For paramyxoviruses it is essential that this reporter casette does not cause phase-shifts in the viral genome, so that the modified genome of the reporter-expressing virus follows the “rule of six” (Kolakofsky et al., 1998). This casette is then inserted either at one of the viral genome ends, or between existing viral genes (Mebatsion et al., 1996). Due to the transcriptional gradient (i.e. the higher expression of 3′ proximal genes than 3′ distal genes), this potentially has an impact on the viral life cycle through attenuation of the expression of downstream viral gene products. Indeed, for many viruses there seems to be a correlation between the 3′ proximity of the inserted ATU and attenuation of the recombinant virus in vitro and/or in vivo. Tokusumi et al. have shown in a study where they inserted the sequence encoding for secreted alkaline phosphatase into all possible genome positions that this seems to be the case for Sendai virus (Tokusumi et al., 2002). Also, this idea is further supported by other studies in paramyxoviruses comparing different insertion positions (Falchieri et al., 2013; von Messling et al., 2004; Zhao and Peeters, 2003). However, in a study with recombinant VSV, where the authors inserted a 661 nucleotide long non-coding ATU at each of the gene-junctions, the authors only observed a strong attenuation when the ATU was inserted between the N and P gene (first gene junction), and no attenuation in vitro when any of the other gene junctions were used (Wertz et al., 2002). In contrast to this, a recombinant novirhabdovirus expressing GFP from the N-P junction was not attenuated in comparison to viruses expressing GFP from the P-M or the M-G junction (Kim et al., 2013). Maybe even more surprising, for filoviruses the first gene-junction (between the NP and VP35 genes) is actually the most commonly used insertion position, and viruses with ATUs in this site seem to be quite genetically stable, and show very little attenuation in tissue culture, even though they are strongly attenuated in vivo (Ebihara et al., 2007; Hoenen et al., 2013; Martinez et al., 2008; Towner et al., 2005). A possible explanation for this discrepancy between rhabdoviruses and filoviruses is that for filoviruses, in addition to VP35, VP30 is also involved in viral gene expression, and thus might be performing parts of the function of P for other mononegaviruses. Therefore, modulation of the VP35 expression level, due to insertion of an upstream ATU, might have a less pronounced negative effect on viral genome replication and transcription than does modulation of the rhabdovirus P expression level. Unfortunately, apart from the papers already mentioned above there are relatively few studies that directly compare different positions for ATU insertions, and while many studies that feature recombinant viruses with ATUs provide growth kinetics, these are very difficult to compare, since experimental details such as the infectious dose differ between the studies. Maybe even more problematic for the purpose of comparison is the fact that the cell lines used for growth kinetics often differ between studies, and that the attenuation due to insertion of ATUs that is observed in vitro seems to vary between cell lines (Ebihara et al., 2007). Also, in terms of assessing and comparing attenuation of reporter-expressing viruses in vivo, it has to be kept in mind that particularly in disease models that show only mild symptoms it can be quite challenging to make absolute statements regarding the extent of, or the absence of, attenuation. Nevertheless, what is striking is that there seem to be exceptions from the general rule that attenuation correlates with 3′ proximity of the ATU. For example, Measles viruses that express GFP from an ATU upstream of N, which is usually one of the most attenuating positions, show very little attenuation in vitro as well as in vivo (de Swart et al., 2007; Hashimoto et al., 2002). Similarly, recombinant RSVs expressing reporter proteins from an ATU before NS1 (the first gene in the RSV genome) show very little attenuation in tissue culture (Hotard et al., 2012), whereas an RSV expressing chloramphenicol acetyltransferase from an ATU inserted between F and G was attenuated by 2 to 3 log10 in tissue culture (Bukreyev et al., 1996). While the reason for this lack of attenuation is unknown, it is tempting to speculate that this is due to the fact that insertion before the first gene preserves the expression gradient of all viral genes and, therefore, the molar ratios of all viral proteins; however, it remains unclear why for other viruses ATUs in this position are not well tolerated.
Importantly, not only the position of the ATU, but also the flanking non-coding regions can be used to modulate attenuation, as well as greatly influence reporter gene expression. This was systematically studied for Newcastle disease virus, where GFP was flanked by the non-coding regions of different viral genes and inserted either between the P and M genes or the HN and F genes (Kim and Samal, 2010). Similarly, Burke et al. introduced an optimized gene start sequence before the F gene, and thus counteracted the attenuation observed in a Sendai virus expressing firefly luciferase from an ATU between the M and F gene (Burke et al., 2011).
4.1.2 Fusion protein-based approaches
An alternate expression strategy is to fuse the reporter protein to one of the viral proteins. This approach has been used very extensively for a number of paramyxoviruses, where GFP has been inserted into a flexible hinge region in the viral polymerase. In minigenome assays it was shown that such a fusion protein exhibits reduced activity for genome replication and transcription (Duprex et al., 2002; Silin et al., 2007). Further, in most cases recombinant viruses expressing these L fusions were attenuated in vitro as well as in vivo (Brown et al., 2005; Duprex et al., 2002; Silin et al., 2007), even though, at least in tissue culture, similar end titers were usually reached. The same strategy has been used for both rhabdoviruses and filoviruses, but with different outcomes. For rhabdoviruses a recombinant virus expressing L-GFP was highly attenuated in vitro at 37°C, although it showed little attenuation at 33°C, and the fusion proteins exhibited highly impaired transcriptional activity (Ruedas and Perrault, 2009). In contrast, while introducing mCherry into the hinge region of the filovirus polymerase had some impact on genome replication and transcription in minigenome assays, this effect could be completely reversed by increasing the expression level of L-mCherry, and a recombinant virus expressing this protein showed very little attenuation in vitro (Hoenen et al., 2012). Unfortunately, this virus has not yet been assessed in vivo. Also, it is currently unclear whether these differences in attenuation between paramyxo- and rhabdoviruses expressing L-GFP and filoviruses expressing L-mCherry are due to differences in the reporter protein, differences in the precise location of the insertion, differences in the insertion strategy (i.e. the use of amino acid linkers), or due to inherent biological differences in replication and transcription between filoviruses and other mononegaviruses. Another fusion involving the L protein was reported by Chambers et al., who fused GFP to the C-terminus of Sendai virus L, with the resulting virus showing relatively little attenuation in tissue culture (Chambers and Takimoto, 2010).
Other proteins have also been used as fusion partners for reporters, albeit with mixed results. Both recombinant rabies and measles viruses expressing GFP fused to the N-terminus of P could be rescued (Devaux and Cattaneo, 2004; Finke et al., 2004), whereas similar attempts to rescue a recombinant measles virus with GFP fused to the C-terminus failed (Devaux and Cattaneo, 2004). However, these viruses grew to about 100 fold lower titers in tissue culture, and in the case of measles no C protein was expressed by this virus. Interestingly, expressing C from an ATU, in addition to expressing the GFP-P fusion protein, restored end-point titers of this recombinant virus to wild-type levels; however, growth of the virus was still delayed (Devaux and Cattaneo, 2004). In VSV P a flexible hinge region has also been identified, and used as an insertion site for GFP, with the resulting virus expressing this fusion protein being somewhat attenuated, and showing reduced P as well as L incorporation into particles (Das et al., 2006).
In contrast to P, the matrix protein seems to be a poor choice as a fusion partner for reporter proteins. A recombinant Chandipura virus in which M was replaced by a GFP-M fusion protein grew to about 2 log10 lower titers, and resulted in a small-plaque phenotype (Marriott and Hornsey, 2011). Similarly, a fusion protein of red fluorescent protein and M was not incorporated into virus particles (Das et al., 2009). Glycoproteins have also been used as fusion partners for reporters with some mononegaviruses, again with mixed success. For example, GFP was fused to the C-terminus of VSV G (Dalton and Rose, 2001). While a virus expressing this fusion protein from an ATU did not appear attenuated, even though G-GFP was incorporated into virions in the form of hetero-trimers together with wild-type G, a virus in which G-GFP replaced G was not genetically stable, and quickly abolished GFP expression (Dalton and Rose, 2001). Similarly, for Sendai virus GFP was fused to the C-terminus of F; however, the resulting fusion protein was not expressed from the original F transcription unit, but rather from an ATU at the beginning of the genome, from which the wild-type F was deleted (Miyazaki et al., 2010). Not surprisingly, this virus was attenuated in vitro; however, it remains unclear to what extent the ATU at the 3′ genome end or the fusion of GFP to F each contributed to this attenuation.
4.1.3 Other expression strategies
While for most mononegaviruses all viral genes are essential both in vitro and in vivo, for novirhabdoviruses it has been possible to replace the non-essential NV gene with GFP in both viral hemorrhagic septicemia virus and infectious hematopoietic necrosis virus (Ammayappan et al., 2011; Novoa et al., 2006; Romero et al., 2008). These viruses are highly attenuated in vivo, and considered promising vaccine candidates for these economically important fish diseases. A somewhat similar approach was chosen for GFP-expressing Rabies viruses that were used for synaptic tracing. In these viruses the (essential) G gene was replaced with GFP and the G protein was provided in trans through pseudotyping (Wickersham et al., 2007a). While these viruses were restricted to a single round of infection, this was actually a desired feature in these studies, since it allowed transsynaptic tracing of a single synaptic step (Wickersham et al., 2007b).
A final expression strategy is to express the reporter from an existing transcriptional unit encoding a viral protein, but separated from the coding region of that viral protein by an IRES. This strategy was successfully used for Sendai virus, where a Renilla luciferase ORF was inserted into the 5′ non-coding region of the N gene with several repeats of a 9 nucleotide long IRES-like sequence between the N coding-region and the luciferase coding region (Touzelet et al., 2009). Interestingly, this strategy did not significantly disturb the transcriptional gradient, and the recombinant viruses rescued using this approach did not show any attenuation in tissue culture. Unfortunately, until now recombinant viruses generated using this strategy (i.e. expression of a reporter protein from an existing viral transcriptional unit by joining the reporter coding sequence to the viral coding sequence through an IRES sequence) have, to the best of our knowledge, not been evaluated in animals. As a result it is currently unclear whether these viruses will be attenuated in vivo; nevertheless, this is an extremely promising approach for overcoming the problems that exist with currently used expression strategies with respect to the altered biological properties of reporter-expressing viruses.
4.2 Loss of reporter gene expression
A potential consequence of the attenuation of viruses due to expression of reporters is purging of the genetic information encoding the reporter, although it has to be noted that many reporter-expressing viruses actually show a very high degree of stability of reporter gene expression. Depending on the expression strategy, several purging mechanisms have been observed. In the case of the above-mentioned VSV containing a 661 nucleotide long non-coding ATU between N and P the ATU was rapidly silenced by the acquisition of mutations that disrupted the upstream gene end signal, thus abolishing transcription of the ATU (Wertz et al., 2002). This seems to be a common mechanism used by viruses to purge attenuating ATUs (Quinones-Kochs et al., 2001). Another mechanism that has been observed for a recombinant bunyavirus expressing GFP fused to Gc was deletion of the GFP coding region as well as part of the Gc coding region from the virus genome (Shi et al., 2010). Similarly, a Chandipura virus expressing an M-GFP fusion protein quickly obtained large deletions in the M gene deleting either most of or in some cases the entire GFP coding region (Marriott and Hornsey, 2011).
Interestingly, in the case of fusion protein-based reporter expression there are instances where the genetic information for the reporter was not deleted, but rather point mutations were acquired. For C-terminal fusions of the reporter to the viral protein, mutations have been observed giving rise to stop codons, which allow expression of the viral portion of the fusion protein, but which abolish expression of the reporter (Dalton and Rose, 2001). Alternatively, in some cases mutations have been observed that seem to specifically negate the biological impact of the reporter. For example, a recombinant VSV expressing a P-GFP fusion protein in the place of P quickly attained a single point mutation in the GFP coding region that destroyed fluorescence of the protein (Dinh et al., 2012). Interestingly both wild-type and mutated P-GFP fusion proteins were produced to same levels, but the mutated P-GFP protein was more active in supporting viral genome replication and transcription (Dinh et al., 2012), although it is currently not known how the point mutation in GFP aided the function of P-GFP in these processes.
4.3 Attenuation of reporter signal in tissues
One significant problem for in vivo work with reporter-expressing viruses is the attenuation of reporter signal in tissues. The majority of currently available reporter-expressing viruses feature (e)GFP as the reporter gene, and GFP is also the most widely used reporter for in vivo studies. GFP is relatively small, lacks toxicity, can be continuously synthesized during virus replication and is straightforward to image and quantify without further manipulation (Agungpriyono et al., 1999). Unfortunately, the excitation/emission spectrum of GFP overlaps with hemoglobin, melanin and water (Shcherbakova and Verkhusha, 2013), resulting in poor signal penetration in in vivo situations (Deliolanis et al., 2008). This restricts real time non-invasive imaging to small animals, primarily mice. Therefore, GFP is not optimal for in vivo imaging, unless virus replication occurs at or near the surface of infected animals, such as the skin or mucosae (Ludlow et al., 2012). Despite this, GFP has been very successfully used for ex vivo imaging (Daito et al., 2011; de Swart et al., 2007; von Messling et al., 2004). Recently, red and far red fluorescent reporters are being increasingly used, including mCherry (Hoenen et al., 2012; Shi et al., 2010), (t)dTomato (Ludlow et al., 2012; Wiener et al., 2010), dsRed (Daito et al., 2011; Takeda et al., 2006), red fluorescent protein (RFP) (Dinh et al., 2012; Kim et al., 2013) and Katushka2 (Hotard et al., 2012). The availability of these proteins might significantly improve our ability to perform in vivo imaging studies, as their excitation and emission wavelengths allow greater tissue penetration, and some of them are also brighter than GFP (Table 1) (Deliolanis et al., 2008); however, this remains to be demonstrated. Additional modified reporters that are even more suitable for in vivo imaging also continue to be developed. In particular, the near-infrared region (from 650 to 900 nm) demonstrates less absorption by hemoglobin, melanin and water than green and red fluorescent reporters, thus providing better characteristics for optical imaging. Until recently the availability of reporters in this region that were suitable for in vivo imaging was limited to iRFP (Filonov et al., 2011), but now the development of additional fluorescent proteins based on iRFP could allow multiple color imaging in this region. For example, iRFP670 and iRFP720 will allow two color near-infrared imaging using spectral unmixing (Shcherbakova and Verkhusha, 2013). Despite improved spectral properties, these reporters are still less bright than GFP-based reporters (Table 1). Also, tissue auto-fluorescence is a significant problem for all currently available fluorescent reporters due to the presence of endogenous fluorophores (e.g. keratin, collagen and NAD(P)H), thus decreasing the signal-to-noise ratios that can be achieved. Importantly, GFP can be less problematic in terms of tissue autofluorescence than other reporters. As an example, in a recent study dTomato showed much more intense fluorescence during examination conducted at necropsy, while GFP was more suited to microscopic examination of brain sections due to its lower autofluorescence in this tissue (Ludlow et al., 2012). Thus, there is so far no one optimal fluorescent label for all applications, rather the properties of each fluorescent protein make them more or less suitable for some specific applications than for others.
Table 1. Selected properties of fluorescent reporters.
| Fluorescent reporter |
MW (kDa) | Excitation (nm) | Emission (nm) | Brightnessa |
|---|---|---|---|---|
| GFP | 26.9 | 475 | 509 | 16 |
| eGFP | 26.9 | 488 | 507 | 34 |
| (t)dTomato | 54.2 | 554 | 581 | 95 |
| dsRed | 26.8 | 554 | 586 | 3.5 to 59 |
| RFP | 27 | 555 | 584 | 48 |
| mCherry | 28.8 | 587 | 610 | 16 |
| Katushka2 | 25.9 | 588 | 635 | 22 |
| iRFP | 29.5 | 690 | 713 | 6.2 |
Brightness was calculated as (extinction coefficient)×(quantum yield)/1000
As an alternative to fluorescence-based imaging, bioluminescent imaging using various luciferases (Table 2) has been applied to in vivo imaging in small animals, and allows the determination of sites of virus replication, monitoring of dissemination of virus throughout the host, and antiviral and/or vaccine efficacy in the context of the entire animal over time (Luker and Luker, 2008). Luciferases have a number of advantages over fluorescent reporters and will likely become the primary reporters for real time in vivo imaging in the future. Fluorescent imaging relies on the excitation of a fluorophore with a light source, in order to generate a longer wavelength emission, which is then detected. However, at both the excitation and emission wavelengths the penetration is reduced greatly by increasing depth of the tissue (which has to be traversed twice, once by the excitation light, and once by the emitted light). In contrast, bioluminescent imaging uses enzymes, most commonly luciferases, to catalyze reactions that produce visible light directly within the animal. The light emitted by these reactions includes wavelengths in the range above 600 nm at body temperature, which significantly decreases light scattering and absorption, resulting in improved detection. Further, since there is no intrinsic autoluminescence, signal-to-noise ratios are improved compared to fluorescence, and facilitate the detection of even very low level emission signals. Currently, firefly luciferase is well suited to in vivo imaging as its substrate (D-luciferin) is readily soluble, well distributed (in particular, it can cross the blood-brain barrier) and has favorable kinetics. Other luciferases (e.g. Gaussia and Renilla) can be used but require the use of a coelenterazine substrate which is less soluble and does not distribute as readily (Barry et al., 2012). Unfortunately, the large size of firefly luciferase is a disadvantage in the context of the relatively small genomes of mononegaviruses. Recent research has focused on the development of “red-shifted” analogues of luciferase that show increased tissue penetration (Caysa et al., 2009), variants of luciferase that allow dual-color imaging (Mezzanotte et al., 2011), and the development of nanoLuc, a novel luciferase, which features a very small size but is extremely bright (Hall et al., 2012).
Table 2. Select properties of bioluminescent reporters.
| Bioluminescent reporter |
MW (kDa) | Substrate | Emission (nm) | Relative light value |
|---|---|---|---|---|
| Firefly | 61 | luciferin | 565 | 6.8 × 104 |
| Renilla | 36 | coelenterazine/ViViRen | 480 | 6.8 × 105 |
| Gaussia | 19.9 | coelenterazine | 485 | 1.7 to 6.2 × 107 |
| nanoLuc | 19.1 | furimazine | 460 | 1.7 × 106 |
5. Design considerations for reporter-expressing viruses
The two principal decisions when designing a reporter-expressing virus are the choice of the reporter, and the choice of the expression strategy. As outlined above, both these choices can significantly influence the biological properties of the reporter-expressing virus. This can lead to attenuation of the recombinant virus and, as a consequence, potentially to loss of reporter gene expression. Also, both choices directly impact the suitability of the recombinant virus for specific scientific applications and questions, and must be made with a specific scientific use in mind.
5.1 In vitro applications
Two primary application fields can be distinguished for in vitro uses of reporter-expressing viruses: They can be used as tools for screening systems, e.g. for antiviral screening or for the detection of neutralizing antibodies, or they can be used for basic research applications. When used as screening tools, the technical properties of the viruses used are of the greatest importance, particularly the strength of the reporter signal (which determines the signal-to-noise ratio of the assays) and the kinetics of reporter expression (which determines the time required for the assays). In contrast, considerations such as attenuation of the reporter-expressing virus are of limited concern for such applications.
In such a situation, expression of the reporter from an ATU is the most common choice, since this is a relatively straight forward approach. Importantly, the position of the ATU has a significant impact not only on attenuation of the recombinant virus (see section 4.1.1), but also on the expression level of the reporter. Due to the transcriptional gradient (i.e. the higher expression of 3′ proximal genes than 3′ distal genes due to imperfect reinitiation of transcription at gene borders), reporter gene expression can be expected to be higher if the gene is placed closer to the 3′ genome end. Experimentally this was confirmed in studies for both paramyxoviruses and rhabdoviruses, where identical ATUs were placed at all possible positions within the viral genome, with a transcriptional gradient clearly observed (Tokusumi et al., 2002; Wertz et al., 2002). Other studies have since provided further support for this correlation (Kim et al., 2013; von Messling et al., 2004). In any case, codon optimization of the reporter gene should be considered to maximize reporter expression, and this has been a very common strategy in currently employed reporter-expressing viruses.
In terms of reporter, the choice should be made depending on the planned applications. Luminescent reporters facilitate very sensitive cell-based reporter assays (Thorne et al., 2010) and, therefore, allow for earlier detection, improving assay times (Hoenen et al., 2013). Also, they eliminate the problem of compound fluorescence (Simeonov et al., 2008). However, they require a substrate, which increases assay costs, and unless secreted luciferases are used, require an additional lysis step, rendering the assay an endpoint assay. In contrast, fluorescent reporters do not require additional assay reagents, and allow continuous monitoring of reporter expression. Also, particularly when used in conjunction with high-content imaging, they allow multiple parameters to be assessed at once (Pegoraro et al., 2012); however, in this case equipment costs are significantly higher than for the relatively simple luminescence-based assays.
For basic research applications, for example to study the subcellular localization of viral proteins, the technical properties, such as reporter expression level and kinetics, remain important. However, alterations to the biological properties of the recombinant viruses, such as overall attenuation or impaired function of viral fusion proteins, also have to be taken into consideration. Due to the multitude of scientific questions and the wide range of requirements no generalized advice for designing reporter expressing-viruses can be given; rather, the approach used and the suitability of a given virus must be carefully evaluated with regard to the specific scientific questions these viruses are intended to study.
5.2 In vivo applications
For in vivo applications, likely the most significant obstacle in the construction of reporter-expressing viruses is the presence of the reporter itself resulting in attenuation. With comparatively small genomes, many reporter-expressing mononegaviruses appear to be somewhat attenuated in vivo. Attenuation has been observed to be variable depending on the location of the reporter in the viral genome, but also varies between different reporters when inserted at identical locations. Importantly, multiple studies report a disconnect between evaluation of attenuation in vitro and in vivo, and demonstrate that a lack of attenuation in vitro is not predictive of a lack of attenuation in vivo. When using ATUs for reporter expression, in general there seems to be a correlation between the 3′ proximity of the inserted ATU and attenuation of the recombinant virus; however, there are also many exceptions to this rule, so that the generation of multiple viruses using different positions for the ATU and their evaluation in the relevant animal model might be prudent. Alternatively, the use of an already existing transcriptional unit, which encodes the reporter in addition to a viral protein, but with both open reading frames separated by an IRES-like sequence, constitutes a very promising approach that might help overcome problems associated with reporter expression from an ATU.
The specific reporter to be used is often dictated by the scientific application, with luminescent reporters providing higher sensitivity and no problems with autofluorescence, but with the necessity to provide a substrate in vivo, which can be cost-prohibitive, particularly when using larger animals. In contrast, when using fluorescent reporters, attenuation in tissues as well as autofluorescence have to be considered, both of which differ between different fluorescent proteins, and both of which might be of varying importance for the scientific questions to be addressed. Also, as pointed out above, different reporters can have different influences on virus attenuation; however, at the moment no clear information are available that would allow reliable predictions to be made, thus further studies in this direction are a necessity.
6. Future directions
While reporter-expressing mononegaviruses have been developed and studied for more than a decade, there are numerous questions that remain regarding their characteristics, and thus the potential for rational design. There are a number of strategies to use reporters to rationally attenuate viruses, for instance by introducing GFP into the hinge region of L, or by introducing reporter ATUs in genome regions where they disrupt the transcriptional gradient in a way that is disadvantageous for the viral life cycle. However, the extent of attenuation is difficult to predict, and challenges remain in ensuring that recombinant viruses do not escape attenuation by disrupting the ATUs or mutating the fusion proteins. Nonetheless, this remains a promising approach for the rational generation of vaccines in future.
One of the greatest challenges at the moment is the ability to generate reporter-expressing viruses that show little or no attenuation in vivo, which is critical for pathogenesis studies. In this regard it will be important to better understand the regulatory elements involved in viral genome replication and gene expression, in order to be better able to fine tune the expression of viral genes in the presence of ATUs and achieve protein expression ratios similar to that of wild type viruses. A better understanding of the effect of different reporters (e.g. GFP vs. mCherry or luciferase) on virus biology might also be very helpful when designing reporter-expressing viruses, particularly if the reporters are fused to viral proteins. This will require a better understanding of the structure-function relationship of viral proteins. Another avenue that is certainly worth pursuing is the expression of reporters using IRES sequences, since this strategy circumvents the need for ATUs altogether. However, recombinant viruses using this strategy will first need to be evaluated in vivo, and this strategy will have to be shown to be transferable to mononegaviruses other than Sendai virus. In addition, other strategies such as the use of a foot and mouth disease virus 2A sequence to achieve expression of several proteins from a single gene might constitute an interesting alternative to the use of IRES sequences (de Felipe et al., 2006).
Finally, practical advancements will be required in order to enhance our ability to visualize infection in animals other than mice. Novel reporters such as nanoLuc (which might also be technically superior to other luciferases due to its small size) or far red-shifted luciferases are very promising in this respect, but still have to be adapted for in vivo application. Despite all these challenges, reporter-expressing viruses are an exciting field of research that bridges basic and applied science, and which holds tremendous promise for the future.
Reporter-expressing viruses have numerous applications both in vitro and in vivo.
The major design considerations are the choice of reporter and expression strategy.
Technical aspects and attenuation determine the suitability of these viruses.
These viruses should be specifically designed for each scientific application.
Acknowledgements
This work was supported by the Intramural Research Program of the NIH, NIAID. The authors are very grateful to Martin Ludlow and Paul Duprex (Boston University) for providing image material.
Abbreviations
- ATU
additional transcriptional unit
- GFP
(enhanced) green fluorescent protein
- IRES
internal ribosomal entry site
- RSV
respiratory syncytial virus
- VSV
vesicular stomatitis virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ackermann A, Guelzow T, Staeheli P, Schneider U, Heimrich B. Visualizing viral dissemination in the mouse nervous system, using a green fluorescent protein-expressing Borna disease virus vector. Journal of virology. 2010;84:5438–5442. doi: 10.1128/JVI.00098-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agungpriyono DR, Yamaguchi R, Tohya Y, Uchida K, Tateyama S. Pathogenicity of Sendai viruses adapted into polarized MDCK cells. The Journal of veterinary medical science / the Japanese Society of Veterinary Science. 1999;61:1299–1307. doi: 10.1292/jvms.61.1299. [DOI] [PubMed] [Google Scholar]
- Ammayappan A, Kurath G, Thompson TM, Vakharia VN. A reverse genetics system for the Great Lakes strain of viral hemorrhagic septicemia virus: the NV gene is required for pathogenicity. Marine biotechnology. 2011;13:672–683. doi: 10.1007/s10126-010-9329-4. [DOI] [PubMed] [Google Scholar]
- Banyard AC, Simpson J, Monaghan P, Barrett T. Rinderpest virus expressing enhanced green fluorescent protein as a separate transcription unit retains pathogenicity for cattle. The Journal of general virology. 2010;91:2918–2927. doi: 10.1099/vir.0.023598-0. [DOI] [PubMed] [Google Scholar]
- Barry MA, May S, Weaver EA. Imaging luciferase-expressing viruses. Methods in molecular biology. 2012;797:79–87. doi: 10.1007/978-1-61779-340-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Rima BK, Allen IV, Baron MD, Banyard AC, Barrett T, Duprex WP. Rational attenuation of a morbillivirus by modulating the activity of the RNA-dependent RNA polymerase. Journal of virology. 2005;79:14330–14338. doi: 10.1128/JVI.79.22.14330-14338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A, Camargo E, Collins PL. Recovery of infectious respiratory syncytial virus expressing an additional, foreign gene. Journal of virology. 1996;70:6634–6641. doi: 10.1128/jvi.70.10.6634-6641.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke CW, Mason JN, Surman SL, Jones BG, Dalloneau E, Hurwitz JL, Russell CJ. Illumination of parainfluenza virus infection and transmission in living animals reveals a tissue-specific dichotomy. PLoS pathogens. 2011;7:e1002134. doi: 10.1371/journal.ppat.1002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caysa H, Jacob R, Muther N, Branchini B, Messerle M, Soling A. A redshifted codon-optimized firefly luciferase is a sensitive reporter for bioluminescence imaging. Photochemical & photobiological sciences: Official journal of the European Photochemistry Association and the European Society for Photobiology. 2009;8:52–56. doi: 10.1039/b814566k. [DOI] [PubMed] [Google Scholar]
- Chambers R, Takimoto T. Trafficking of Sendai virus nucleocapsids is mediated by intracellular vesicles. PloS one. 2010;5:e10994. doi: 10.1371/journal.pone.0010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daito T, Fujino K, Honda T, Matsumoto Y, Watanabe Y, Tomonaga K. A novel borna disease virus vector system that stably expresses foreign proteins from an intercistronic noncoding region. Journal of virology. 2011;85:12170–12178. doi: 10.1128/JVI.05554-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton KP, Rose JK. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology. 2001;279:414–421. doi: 10.1006/viro.2000.0736. [DOI] [PubMed] [Google Scholar]
- Das SC, Nayak D, Zhou Y, Pattnaik AK. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. Journal of virology. 2006;80:6368–6377. doi: 10.1128/JVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SC, Panda D, Nayak D, Pattnaik AK. Biarsenical labeling of vesicular stomatitis virus encoding tetracysteine-tagged m protein allows dynamic imaging of m protein and virus uncoating in infected cells. Journal of virology. 2009;83:2611–2622. doi: 10.1128/JVI.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, Ryan MD. E unum pluribus: multiple proteins from a self-processing polyprotein. Trends in biotechnology. 2006;24:68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- de Swart RL, Ludlow M, de Witte L, Yanagi Y, van Amerongen G, McQuaid S, Yuksel S, Geijtenbeek TB, Duprex WP, Osterhaus AD. Predominant infection of CD150+ lymphocytes and dendritic cells during measles virus infection of macaques. PLoS pathogens. 2007;3:e178. doi: 10.1371/journal.ppat.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RD, Lemon K, Ludlow M, McQuaid S, Yuksel S, van Amerongen G, Rennick LJ, Rima BK, Osterhaus AD, de Swart RL, Duprex WP. In vivo tropism of attenuated and pathogenic measles virus expressing green fluorescent protein in macaques. Journal of virology. 2010;84:4714–4724. doi: 10.1128/JVI.02633-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deliolanis NC, Kasmieh R, Wurdinger T, Tannous BA, Shah K, Ntziachristos V. Performance of the red-shifted fluorescent proteins in deep-tissue molecular imaging applications. Journal of biomedical optics. 2008;13:044008. doi: 10.1117/1.2967184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux P, Cattaneo R. Measles virus phosphoprotein gene products: conformational flexibility of the P/V protein amino-terminal domain and C protein infectivity factor function. Journal of virology. 2004;78:11632–11640. doi: 10.1128/JVI.78.21.11632-11640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh PX, Panda D, Das PB, Das SC, Das A, Pattnaik AK. A single amino acid change resulting in loss of fluorescence of eGFP in a viral fusion protein confers fitness and growth advantage to the recombinant vesicular stomatitis virus. Virology. 2012;432:460–469. doi: 10.1016/j.virol.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duprex WP, Collins FM, Rima BK. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J Virol. 2002;76:7322–7328. doi: 10.1128/JVI.76.14.7322-7328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara H, Theriault S, Neumann G, Alimonti JB, Geisbert JB, Hensley LE, Groseth A, Jones SM, Geisbert TW, Kawaoka Y, Feldmann H. In vitro and in vivo characterization of recombinant Ebola viruses expressing enhanced green fluorescent protein. J Infect Dis. 2007;196(Suppl 2):S313–322. doi: 10.1086/520590. [DOI] [PubMed] [Google Scholar]
- Falchieri M, Lupini C, Cecchinato M, Catelli E, Kontolaimou M, Naylor CJ. Avian metapneumoviruses expressing Infectious Bronchitis virus genes are stable and induce protection. Vaccine. 2013;31:2565–2571. doi: 10.1016/j.vaccine.2013.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov GS, Piatkevich KD, Ting LM, Zhang J, Kim K, Verkhusha VV. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nature biotechnology. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke S, Brzozka K, Conzelmann KK. Tracking fluorescence-labeled rabies virus: enhanced green fluorescent protein-tagged phosphoprotein P supports virus gene expression and formation of infectious particles. Journal of virology. 2004;78:12333–12343. doi: 10.1128/JVI.78.22.12333-12343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A, Marzi A, Hoenen T, Herwig A, Gardner D, Becker S, Ebihara H, Feldmann H. The Ebola virus glycoprotein contributes to but is not sufficient for virulence in vivo. PLoS Pathog. 2012;8:e1002847. doi: 10.1371/journal.ppat.1002847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, Robers MB, Benink HA, Eggers CT, Slater MR, Meisenheimer PL, Klaubert DH, Fan F, Encell LP, Wood KV. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS chemical biology. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Ono N, Tatsuo H, Minagawa H, Takeda M, Takeuchi K, Yanagi Y. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. Journal of virology. 2002;76:6743–6749. doi: 10.1128/JVI.76.13.6743-6749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Groseth A, Callison J, Takada A, Feldmann H. A novel Ebola virus expressing luciferase allows for rapid and quantitative testing of antivirals. Antiviral research. 2013 doi: 10.1016/j.antiviral.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Groseth A, de Kok-Mercado F, Kuhn JH, Wahl-Jensen V. Minigenomes, transcription and replication competent virus-like particles and beyond: reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses. Antiviral Res. 2011;91:195–208. doi: 10.1016/j.antiviral.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Shabman RS, Groseth A, Herwig A, Weber M, Schudt G, Dolnik O, Basler CF, Becker S, Feldmann H. Inclusion bodies are a site of ebolavirus replication. J Virol. 2012;86:11779–11788. doi: 10.1128/JVI.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotard AL, Shaikh FY, Lee S, Yan D, Teng MN, Plemper RK, Crowe JE, Jr., Moore ML. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology. 2012;434:129–136. doi: 10.1016/j.virol.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Chen W, Huang K, Baron MD, Bu Z. Rescue of recombinant peste des petits ruminants virus: creation of a GFP-expressing virus and application in rapid virus neutralization test. Veterinary research. 2012;43:48. doi: 10.1186/1297-9716-43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson LE, Rose JK. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell. 1981;23:477–484. doi: 10.1016/0092-8674(81)90143-4. [DOI] [PubMed] [Google Scholar]
- Johansen LM, Brannan JM, Delos SE, Shoemaker CJ, Stossel A, Lear C, Hoffstrom BG, Dewald LE, Schornberg KL, Scully C, Lehar J, Hensley LE, White JM, Olinger GG. FDA-Approved Selective Estrogen Receptor Modulators Inhibit Ebola Virus Infection. Science translational medicine. 2013;5:190ra179. doi: 10.1126/scitranslmed.3005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Park JS, Kim KH. Optimal place of a foreign gene in the genome of viral haemorrhagic septicaemia virus (VHSV) for development of VHSV-based viral-vectored vaccines. Journal of applied microbiology. 2013;114:1866–1873. doi: 10.1111/jam.12177. [DOI] [PubMed] [Google Scholar]
- Kim SH, Samal SK. Role of untranslated regions in regulation of gene expression, replication, and pathogenicity of Newcastle disease virus expressing green fluorescent protein. Journal of virology. 2010;84:2629–2634. doi: 10.1128/JVI.02049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingen Y, Conzelmann KK, Finke S. Double-labeled rabies virus: live tracking of enveloped virus transport. Journal of virology. 2008;82:237–245. doi: 10.1128/JVI.01342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA, Griffith DP. Paramyxoviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; 2007. pp. 1450–1496. [Google Scholar]
- Lipkin WI, Briese T. Bornaviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; 2007. pp. 1830–1852. [Google Scholar]
- Lubaki NM, Ilinykh P, Pietzsch C, Tigabu B, Freiberg AN, Koup RA, Bukreyev A. The lack of maturation of ebola virus-infected dendritic cells results from the cooperative effect of at least two viral domains. Journal of virology. 2013;87:7471–7485. doi: 10.1128/JVI.03316-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow M, Nguyen DT, Silin D, Lyubomska O, de Vries RD, von Messling V, McQuaid S, De Swart RL, Duprex WP. Recombinant canine distemper virus strain Snyder Hill expressing green or red fluorescent proteins causes meningoencephalitis in the ferret. Journal of virology. 2012;86:7508–7519. doi: 10.1128/JVI.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker KE, Luker GD. Applications of bioluminescence imaging to antiviral research and therapy: multiple luciferase enzymes and quantitation. Antiviral research. 2008;78:179–187. doi: 10.1016/j.antiviral.2008.01.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles DS, Rupprecht CE. Rhabdoviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; 2007. pp. 1364–1409. [Google Scholar]
- Marriott AC, Hornsey CA. Reverse genetics system for Chandipura virus: tagging the viral matrix protein with green fluorescent protein. Virus research. 2011;160:166–172. doi: 10.1016/j.virusres.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Marschalek A, Finke S, Schwemmle M, Mayer D, Heimrich B, Stitz L, Conzelmann KK. Attenuation of rabies virus replication and virulence by picornavirus internal ribosome entry site elements. J Virol. 2009;83:1911–1919. doi: 10.1128/JVI.02055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MJ, Biedenkopf N, Volchkova V, Hartlieb B, Alazard-Dany N, Reynard O, Becker S, Volchkov V. Role of Ebola virus VP30 in transcription reinitiation. Journal of virology. 2008;82:12569–12573. doi: 10.1128/JVI.01395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebatsion T, Schnell MJ, Cox JH, Finke S, Conzelmann KK. Highly stable expression of a foreign gene from rabies virus vectors. Proc Natl Acad Sci U S A. 1996;93:7310–7314. doi: 10.1073/pnas.93.14.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzanotte L, Que I, Kaijzel E, Branchini B, Roda A, Lowik C. Sensitive dual color in vivo bioluminescence imaging using a new red codon optimized firefly luciferase and a green click beetle luciferase. PloS one. 2011;6:e19277. doi: 10.1371/journal.pone.0019277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Segawa H, Yamashita T, Zhu Y, Takizawa K, Hasegawa M, Taira H. Construction and characterization of a fluorescent sendai virus carrying the gene for envelope fusion protein fused with enhanced green fluorescent protein. Bioscience, biotechnology, and biochemistry. 2010;74:2293–2298. doi: 10.1271/bbb.100511. [DOI] [PubMed] [Google Scholar]
- Msaouel P, Iankov ID, Allen C, Morris JC, von Messling V, Cattaneo R, Koutsilieris M, Russell SJ, Galanis E. Engineered measles virus as a novel oncolytic therapy against prostate cancer. The Prostate. 2009;69:82–91. doi: 10.1002/pros.20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa B, Romero A, Mulero V, Rodriguez I, Fernandez I, Figueras A. Zebrafish (Danio rerio) as a model for the study of vaccination against viral haemorrhagic septicemia virus (VHSV) Vaccine. 2006;24:5806–5816. doi: 10.1016/j.vaccine.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Panchal RG, Kota KP, Spurgers KB, Ruthel G, Tran JP, Boltz RC, Bavari S. Development of high-content imaging assays for lethal viral pathogens. Journal of biomolecular screening. 2010;15:755–765. doi: 10.1177/1087057110374357. [DOI] [PubMed] [Google Scholar]
- Panchal RG, Reid SP, Tran JP, Bergeron AA, Wells J, Kota KP, Aman J, Bavari S. Identification of an antioxidant small-molecule with broad-spectrum antiviral activity. Antiviral Res. 2012;93:23–29. doi: 10.1016/j.antiviral.2011.10.011. [DOI] [PubMed] [Google Scholar]
- Pegoraro G, Bavari S, Panchal RG. Shedding light on filovirus infection with high-content imaging. Viruses. 2012;4:1354–1371. doi: 10.3390/v4081354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero A, Figueras A, Thoulouze MI, Bremont M, Novoa B. Recombinant infectious hematopoietic necrosis viruses induce protection for rainbow trout Oncorhynchus mykiss. Diseases of aquatic organisms. 2008;80:123–135. doi: 10.3354/dao01932. [DOI] [PubMed] [Google Scholar]
- Rudd PA, Cattaneo R, von Messling V. Canine distemper virus uses both the anterograde and the hematogenous pathway for neuroinvasion. Journal of virology. 2006;80:9361–9370. doi: 10.1128/JVI.01034-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruedas JB, Perrault J. Insertion of enhanced green fluorescent protein in a hinge region of vesicular stomatitis virus L polymerase protein creates a temperature-sensitive virus that displays no virion-associated polymerase activity in vitro. Journal of virology. 2009;83:12241–12252. doi: 10.1128/JVI.01273-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Geisbert TW, Feldmann H. Filoviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 5th ed. Lippincott Williams & Wilkins; 2007. pp. 1410–1449. [Google Scholar]
- Schnell MJ, Mebatsion T, Conzelmann KK. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nature methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, van Mierlo JT, French A, Elliott RM. Visualizing the replication cycle of bunyamwera orthobunyavirus expressing fluorescent protein-tagged Gc glycoprotein. Journal of virology. 2010;84:8460–8469. doi: 10.1128/JVI.00902-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silin D, Lyubomska O, Ludlow M, Duprex WP, Rima BK. Development of a challenge-protective vaccine concept by modification of the viral RNA-dependent RNA polymerase of canine distemper virus. Journal of virology. 2007;81:13649–13658. doi: 10.1128/JVI.01385-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonov A, Jadhav A, Thomas CJ, Wang Y, Huang R, Southall NT, Shinn P, Smith J, Austin CP, Auld DS, Inglese J. Fluorescence spectroscopic profiling of compound libraries. Journal of medicinal chemistry. 2008;51:2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- Takeda M, Nakatsu Y, Ohno S, Seki F, Tahara M, Hashiguchi T, Yanagi Y. Generation of measles virus with a segmented RNA genome. Journal of virology. 2006;80:4242–4248. doi: 10.1128/JVI.80.9.4242-4248.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne N, Inglese J, Auld DS. Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chemistry & biology. 2010;17:646–657. doi: 10.1016/j.chembiol.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokusumi T, Iida A, Hirata T, Kato A, Nagai Y, Hasegawa M. Recombinant Sendai viruses expressing different levels of a foreign reporter gene. Virus research. 2002;86:33–38. doi: 10.1016/s0168-1702(02)00047-3. [DOI] [PubMed] [Google Scholar]
- Touzelet O, Loukili N, Pelet T, Fairley D, Curran J, Power UF. De novo generation of a non-segmented negative strand RNA virus with a bicistronic gene. Virus research. 2009;140:40–48. doi: 10.1016/j.virusres.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Towner JS, Paragas J, Dover JE, Gupta M, Goldsmith CS, Huggins JW, Nichol ST. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology. 2005;332:20–27. doi: 10.1016/j.virol.2004.10.048. [DOI] [PubMed] [Google Scholar]
- von Messling V, Milosevic D, Cattaneo R. Tropism illuminated: lymphocyte-based pathways blazed by lethal morbillivirus through the host immune system. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:14216–14221. doi: 10.1073/pnas.0403597101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh EP, Baron MD, Rennie LF, Monaghan P, Anderson J, Barrett T. Recombinant rinderpest vaccines expressing membrane-anchored proteins as genetic markers: evidence of exclusion of marker protein from the virus envelope. Journal of virology. 2000;74:10165–10175. doi: 10.1128/jvi.74.21.10165-10175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Sun N, Nan G, Wang Y, Liu HQ, Peeters B, Chen ZN, Bian H. Construction of recombinant Newcastle disease virus Italien strain for oncolytic virotherapy of tumors. Human gene therapy. 2012;23:700–710. doi: 10.1089/hum.2011.207. [DOI] [PubMed] [Google Scholar]
- Wertz GW, Moudy R, Ball LA. Adding genes to the RNA genome of vesicular stomatitis virus: positional effects on stability of expression. Journal of virology. 2002;76:7642–7650. doi: 10.1128/JVI.76.15.7642-7650.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlow ZW, Connor JH, Lyles DS. Preferential translation of vesicular stomatitis virus mRNAs is conferred by transcription from the viral genome. Journal of virology. 2006;80:11733–11742. doi: 10.1128/JVI.00971-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nature methods. 2007a;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, Callaway EM. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007b;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener D, Vandevelde M, Zurbriggen A, Plattet P. Investigation of a unique short open reading frame within the 3′ untranslated region of the canine distemper virus matrix messenger RNA. Virus research. 2010;153:234–243. doi: 10.1016/j.virusres.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Zhang J, Dou Y, Wu J, She W, Luo L, Zhao Y, Liu P, Zhao X. Effects of N-linked glycosylation of the fusion protein on replication of human metapneumovirus in vitro and in mouse lungs. The Journal of general virology. 2011;92:1666–1675. doi: 10.1099/vir.0.030049-0. [DOI] [PubMed] [Google Scholar]
- Zhao H, Peeters BP. Recombinant Newcastle disease virus as a viral vector: effect of genomic location of foreign gene on gene expression and virus replication. The Journal of general virology. 2003;84:781–788. doi: 10.1099/vir.0.18884-0. [DOI] [PubMed] [Google Scholar]
- Zhou M, Kitagawa Y, Yamaguchi M, Uchiyama C, Itoh M, Gotoh B. Expeditious neutralization assay for human metapneumovirus based on a recombinant virus expressing Renilla luciferase. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2013;56:31–36. doi: 10.1016/j.jcv.2012.09.014. [DOI] [PubMed] [Google Scholar]