Abstract
The tumor microenvironment can promote tumor growth and reduce treatment efficacy. Tumors can occur in many sites in the body, but how surrounding normal tissues at different anatomical sites affect tumor microenvironments and their subsequent response to therapy is not known.
We demonstrated that tumors from renal, colon, or prostate cell lines in orthotopic locations responded to immunotherapy consisting of three agonist antibodies, termed Tri-mAb, to a much lesser extent than the same tumor type located subcutaneously. A tissue-specific response to Tri-mAb was confirmed by ex vivo separation of subcutaneous (SC) or orthotopic tumor cells from stromal cells, followed by reinjection of tumor cells into the opposite site. Compared with SC tumors, orthotopic tumors had a microenvironment associated with a type 2 immune response, related to immunosuppression, and an involvement of alternatively activated macrophages in the kidney model. Orthotopic kidney tumors were more highly vascularized than SC tumors. Neutralizing the macrophage- and Th2-associated molecules chemokine (C-C motif) ligand 2 or interleukin-13 led to a significantly improved therapeutic effect. This study highlights the importance of the tissue of implantation in sculpting the tumor microenvironment. These are important fundamental issues in tumor biology and crucial factors to consider in the design of experimental models and treatment strategies.
Introduction
In addition to cancer cells, tumors contain multiple cell types that together comprise the stroma. Stromal cells, in particular leukocytes, can secrete a range of growth factors and cytokines, which contribute to the tumor microenvironment and further can promote tumor growth and inhibit effective antitumor immune responses. The types of leukocytes in the stroma can include regulatory T cells, myeloid-derived suppressor cells, and alternatively activated macrophages (AAMs), which can express immunomodulatory factors such as transforming growth factor β, interleukin (IL)-10, and arginase-1 (1,2,3). These factors can suppress an immune response or divert it from a type 1 immune response, which is able to eliminate aberrant or infected cells, to a type 2 response geared toward neutralizing extracellular microorganisms.
The importance of these regulatory cell types in promoting tumor growth is evident from studies demonstrating that depletion of these cells in mouse cancer models can reduce tumor growth (4,5,6). Furthermore, correlations between a higher degree of tumor infiltration by these cell types have been associated with poorer prognosis in humans with some cancer types (7,8,9,10,11,12). Currently, the tumor microenvironment is known to be crucial in tumor development and its response to treatment (13,14). Furthermore, extrinsic factors and determinants from host tissue microenvironments contribute to create a “metastatic niche” (15,16). Indeed, cancer cells disseminating from primary tumors are dependent on the niche microenvironment encountered at secondary sites for their implantation and growth (17). Tumors can occur in many sites in the body, but how tissues surrounding the site of tumor initiation or implantation at specific anatomical locations affect the tumor microenvironment and the subsequent response to therapy is yet to be elucidated.
Genomic and proteomic profiling has previously identified differing gene expression profiles in tumor cells from different locations, and in this way, genes thought to be important in metastasis have been identified (18). In addition, studies on gene expression in primary tumors have revealed genes associated with poor prognosis (19,20). It is thought that, as tumors are genetically unstable and heterogeneous, genetic variants suited to growth in different tissues arise and colonize distant sites. In other words, the tumor cells themselves can be different in different sites (21,22,23), making it difficult to distinguish the contributions of tumor cells and host tissue in generating the tumor microenvironment. Thus, the role of the normal tissue at the site of tumor implantation in shaping the tumor microenvironment, as distinct from the role of tumor genetic variants, has not been determined before.
In this study, we used a transplantable tumor to inoculate a genetically similar pool of tumor cells in different anatomical sites. The aim was to allow the assessment of its contributions to the microenvironment and therapy response from those of the surrounding normal tissue in isolation of genetic evolution. This was not possible using a spontaneous metastasis model because spontaneous metastases in different sites can vary genetically. We used three mouse tumor models of varying cancer types, including a renal cell carcinoma, a colon carcinoma, and prostate carcinoma, injected either subcutaneously or in orthotopic sites.
In considering which immunotherapy to apply in these tumor models, we decided to use a therapy that was highly effective against a range of subcutaneous (SC) tumors and whose mechanisms of action involved typically important immune components. We had previously demonstrated that a combination of three monoclonal antibodies specific for death receptor 5 (DR5), CD40, and CD137 (4-1BB) (Tri-mAb) was a highly effective immunotherapy against SC tumors. Indeed, we demonstrated that established SC tumors of various types in mice could be eradicated using Tri-mAb, and a type 1 immune response involving CD8+ T cells and interferon-γ was necessary for eradication of SC tumors (24). However, in subsequent studies, we also observed that orthotopic renal tumors responded less than SC tumors to the same therapy (25).
In this study, we therefore decided to extend our previous observations to investigate the nature of the microenvironment of tumors growing in different anatomical sites. Our investigations suggested that the normal tissue surrounding implantation had a major impact on differences in the microenvironment of tumors in different sites, which contributed to the differential responses to therapy.
Results
Visceral tumors respond less than SC tumors to Tri-mAb therapy
We sought to test Tri-mAb therapy in several orthotopic disease models in mice, where cancer cell lines were injected into the tissue of origin of the cell lines compared with SC injection. Three orthotopic models were used, which involved injection of cancer cells into the kidney (intrakidney (IK)) (Renca, renal cell carcinoma), cecum (intracecum (IC)) (CT26, colon carcinoma), or prostate (intraprostrate) (RM-1, prostate carcinoma). Injection into orthotopic sites was chosen instead of spontaneous metastases, because metastases can vary genetically, thereby rendering it difficult to investigate the contribution from the tissue of implantation in isolation of genetic diversity. Tri-mAb treatment began when tumors had grown to ~20 mm2 in size at all sites. In all three models, we found that orthotopic tumors were much less responsive to Tri-mAb than SC tumors (Figure 1a–c). A similar observation was made when Renca was injected in the liver, a site frequently associated with metastasis in renal cancer (Figure 1d). The majority of Tri-mAb–treated SC tumors responded rapidly, whereas tumors grew progressively in control mice (Figure 1e and Supplementary Figure S1a,b). In some experiments, to extend our previously published observations (25), Renca cells expressing luciferase were used, enabling kidney or hepatic tumor growth to be monitored. In these cases, both kidney and hepatic localized tumors responded minimally during and after treatment, and mice died with large tumors (Figure 1f,g and Supplementary Figure S1c).
Figure 1.
Tri-mAb inhibits orthotopic tumors less than subcutaneous (SC) tumors. (a) Survival of mice injected with Renca, renal carcinoma cells, subcutaneously or orthotopically into the right kidney cortex (intrakidney (IK)) (n = 7–8 per group, representative experiment of 5). (b) Survival of mice injected with CT26, colon carcinoma cells, subcutaneously or orthotopically into the cecum (intracecum (IC)) (n = 7–9 per group, representative experiment of 2). (c) Tumor weight at day (D) 12 of RM-1, prostate carcinoma cells, injected SC or into the prostate (intraprostrate (IPr)) (n = 7–9 per group, representative experiment of 2). (d) Survival of mice injected with Renca cells in SC sites or into one liver lobe (intrahepatically (IH)) (n = 6–7 per group). (e) Tumor growth, following SC injection with Renca-cherry-luciferase (Renca-Ch-Luc), was monitored using calipers (n = 6, representative experiment of 3). (f,g) Bioluminescence emission imaging was used to monitor Renca IK tumor development (n = 6, representative experiment of 3). Mice were treated with Tri-mAb or isotype controls (Ctls) when tumors were established (~20–30 mm2). *P < 0.05. **P < 0.005. ***P < 0.0005.
To assess whether differences in tumor size could explain the differential response to therapy, we measured tumors from SC and abdominal organs immediately before starting treatment, and observed that SC and visceral tumors were of similar size and weight (Supplementary Figure S2). Thus, the different responses to Tri-mAb could not be explained by a simple difference in tumor size between SC and visceral tumors before treatment.
Renca cells retain their similarity after reisolation from different sites
Because the population of cancer cells injected at different sites was of the same genetic makeup as those injected subcutaneously, it seemed that the normal tissue surrounding the site of implantation was influencing the response of the tumor to therapy. However, despite the use of a transplantable tumor model that placed genetically similar cells in different sites, it remained possible that preexisting genetic variants within the mixed population had a predilection to grow at individual sites and that these genetic variants were solely responsible for differences in microenvironment and differential responses to Tri-mAb. To investigate this possibility we re-isolated Renca-cherry-luciferase (Renca-Ch-Luc) tumor cells from SC and IK sites at day 12, cultured them for 4 weeks, and compared their phenotype. During the culture period, tumor stromal components diminished to negligible levels leaving predominantly Renca cells, identified by their expression of the cherry fluorochrome. Cell cultures from the two sites were indistinguishable with respect to morphology, growth rate, Cherry expression (data not shown), and the expression of several representative cell surface markers, such as MHC I and the molecular targets of Tri-mAb, DR5, CD40, and CD137 (Figure 2a). To further investigate whether implantation into different tissue sites selected for distinct subsets of tumor cells with an inherent ability to grow in distinct sites, crossover experiments were performed in which cultured Renca-Ch-Luc cells, isolated from SC or IK sites, were reinjected into the same and opposite sites from initial isolation (subcutaneously derived cells transplanted SC and IK, and IK-derived cells transplanted SC and IK). There was no significant difference in SC tumor growth between parental, SC, and IK tumor cell lines, and all mice rejected their SC tumors to a similar extent after treatment (Figure 2b). Furthermore, mice with tumors derived from both SC and IK cell lines responded to therapy in a tissue-dependent manner, surviving longer after Tri-mAb injection when tumors were located SC compared with IK (Figure 2c). Although we cannot absolutely exclude the presence of subclones of Renca isolated from individual tumor sites, the data from the crossover experiments strongly suggested that the host normal tissue surrounding tumor implantation played an important role in determining the response of tumors to therapy.
Figure 2.
The host normal tissue contributes to directing the composition of the tumor microenvironment. (a) After 4 weeks in culture, Renca-cherry-luciferase (Renca-Ch-Luc) tumor cells isolated from subcutaneous (SC) and intrakidney (IK) tumors at day 12 (three of each) were stained for the markers listed. Cells were analyzed using flow cytometry relative to the original parental Renca-Ch-Luc cell line maintained in vitro. (b) Tumor growth (±SEM) and (c) survival of mice that received SC and IK Renca cell lines injected at SC sites (SC inj.) or IK (IK inj.). Mice were treated with Tri-mAb or control (Ctl) antibodies. (n = 7–8. ***P < 0.005 for (b) Tri-mAb–treated versus Ctl groups).
The tumor microenvironment varies with anatomical site
In considering how normal tissue at the site of tumor implantation affected the responses of tumors to therapy, we hypothesized that it caused differences in the tumor microenvironment at different sites, which impacted on the relative efficacy of Tri-mAb. To investigate the relative microenvironments of tumors at the two sites, we initially focused on the Renca tumor model and examined the phenotype and frequency of infiltrating leukocytes in established tumors before treatment, 10–12 days after inoculation of tumor cells. The overall composition of leukocytes was similar in both tumor sites, with similar percentages of myeloid and lymphoid subsets observed (Figure 3a–g). However, closer examination of macrophage phenotype revealed a relatively higher percentage of F4/80hiCD11bint cells in kidney tumors and a lower percentage of F4/80intCD11bhi cells (Figure 3h–j). Because the difference in cell infiltrate of SC and kidney tumors was restricted to F4/80-expressing cells and because cells of this phenotype have previously been described as containing a relatively immunosuppressive subset of macrophages termed alternatively activated macrophages (26,27,28), we decided to determine the expression of the AAM-associated molecule, CD206 (mannose receptor) in the F4/80+ subsets (29). We observed that the F4/80hiCD11bint cells expressed CD206, suggesting that these macrophages were of the AAM subset known to be immunosuppressive (29) (Figure 3k–m). AAMs have also been previously demonstrated to express the scavenger receptor, Mgl1, and arginine-depleting enzyme, Arg1 (30), and we therefore wished to determine the expression of these molecules in the F4/80hiCD11bint cell subset. Using reverse transcriptase–polymerase chain reaction (PCR) analysis on both F4/80hi cells and F4/80int cells sorted from IK tumor, we confirmed that F4/80hi cells also overexpressed Mgl1 and Arg1 (Figure 3n). These results raised the possibility that renal tissue had preferentially induced a relatively immunosuppressive microenvironment including AAMs, which rendered the associated tumors refractory to immunotherapy.
Figure 3.
Kidney tumors contain a higher proportion of F4/80hi / CD206+ macrophages. (a–g) Similar leukocytes infiltrate intrakidney (IK) and subcutaneous (SC) tumors. SC and IK Renca tumors were removed on day 10–12 after inoculation. (h,i) Plot of macrophage populations from representative SC and IK tumors after staining for the macrophage markers F4/80 and CD11b. (j) The relative frequency of F4/80hi and F4/80int macrophages in SC and IK tumors. (k,l) Representative flow cytometry plots and (m) quantitative data of CD206 expression on F4/80hi and F4/80int macrophages from IK tumors at day 12 after inoculation. (Average ± SEM, n ≥ 5, representative of at least three experiments for all panels). *P < 0.05. ***P < 0.0005. (n) Agarose gel from reverse transcriptase–polymerase chain reaction on RNA from F4/80hi and F4/80int macrophages, sorted from IK tumors at day 12 on the basis of their level of F4/80 expression using alternatively activated macrophage–associated genes Mgl1(670 bp), Arg1 (881 bp) and the housekeeping gene HPRT (250 bp) as depicted. Tregs, regulatory T cells.
AAM-associated molecules are expressed more highly in kidney tumors
To gain a wider view of potential differences in the microenvironment, we used DNA microarray to compare gene expression on tumor tissue dissected from the two sites, SC and IK. We designated genes as differentially expressed, if their fold change was ±1.5 with a P value of <0.05 because these values have been frequently used previously to determine functionally relevant differences in gene expression (31,32). A total of 106 genes were significantly upregulated by 1.5-fold or higher in kidney tumors (Supplementary Figure S3). As our therapeutic approach involved immunotherapy, we initially focused on the differential expression of immune-related genes. Expression of 30 immune-related genes were increased in renal tumors compared with that of SC tumors (P < 0.05), 18 of which are associated with macrophages (Supplementary Table S1). Included in these genes were those encoding for Arg1 and Mgl1 previously mentioned (Figure 3n), and CD163 that are markers for AAMs (30). Other genes found to be upregulated in kidney tumors included IL-6, chemokine (C-C motif) ligand 2 (CCL2), and leukemia inhibitory factor, which have been demonstrated to play a role in skewing the differentiation of macrophages toward an AAM phenotype (33,34).
To confirm gene upregulation found in the microarray and to extend this to protein levels, we used reverse transcriptase–PCR array and protein array on the SC and IK tumors. Increased expression of a variety of macrophage-associated molecules in kidney tumors was detected (Figure 4a,b and Supplementary Figure S4a,b). In addition, the Th2-associated cytokine IL-10 and CCL2 were secreted by cultured F4/80hi macrophages isolated from tumors (Figure 4c and Supplementary Figure S4c). IL-6 protein was also demonstrated to be expressed at higher levels in renal tumors (Supplementary Figure S4d). Strikingly, greater levels of the AAM-associated chemokine CCL2 were found in the serum of mice bearing kidney tumors (Figure 4d). This also supports the strong involvement of this chemokine in the differential immune response generated against kidney tumors as compared with SC tumors.
Figure 4.
Increased gene expression and protein in intrakidney (IK) and subcutaneous (SC) tumors and associated macrophages. (a) Gene expression significantly upregulated in IK tumors (left panel, white bars) or SC tumors (right panel, black bars), following analysis of tumor RNA using an reverse transcriptase–polymerase chain reaction array. (b) Protein significantly upregulated in IK tumors (left panels) or SC tumors (right panel) following analysis of tumor lysates using a protein array. a,b) represent three Renca tumors, isolated at day 10–12 after inoculation (before treatment), from three different mice. (c) Cytometric bead array analysis of macrophage, isolated from IK tumors at day 12 on the basis of their level of F4/80-CD11b expression, supernatants from overnight culture. Cytokines produced by F4/80hi or F4/80int macrophages (results pooled from two experiments, each with duplicate wells). (d) Serum from mice bearing IK or SC tumors at day 12 was analyzed for levels of cytokines and chemokines using cytometric bead array, only significant difference between both sera (CCL2) is depicted (average ± SEM, n = 5). *P < 0.05, ***P < 0.0005. CCL, CC chemokine ligand.
Taken together, these data suggested that an association between macrophages and Th2 cytokines existed preferentially in kidney tumor microenvironments in a tissue-specific manner, which may have led to a relatively ineffective antitumor immune response following the administration of Tri-mAb.
To extend the observation that different microenvironments are generated in tumors in different locations, we compared gene expression in another model, using CT26 colon carcinoma cells. RNA-seq was used to compare gene expression of SC and cecum CT26 tumors. Significant upregulation of a wide range of immune-related genes was detected in SC tumors with a predominance of Th1-associated genes (e.g., IL-12, interferon-γ, and tumor necrosis factor) (Supplementary Table S2). In cecum tumors, there was a predominance of Th2 or immunosuppressive genes including arginase-1, transforming growth factor β, IL-6, and IL-23 (Supplementary Table S2).
Neutralizing CCL2 or IL-13 enhances Tri-mAb therapy of orthotopic kidney tumors
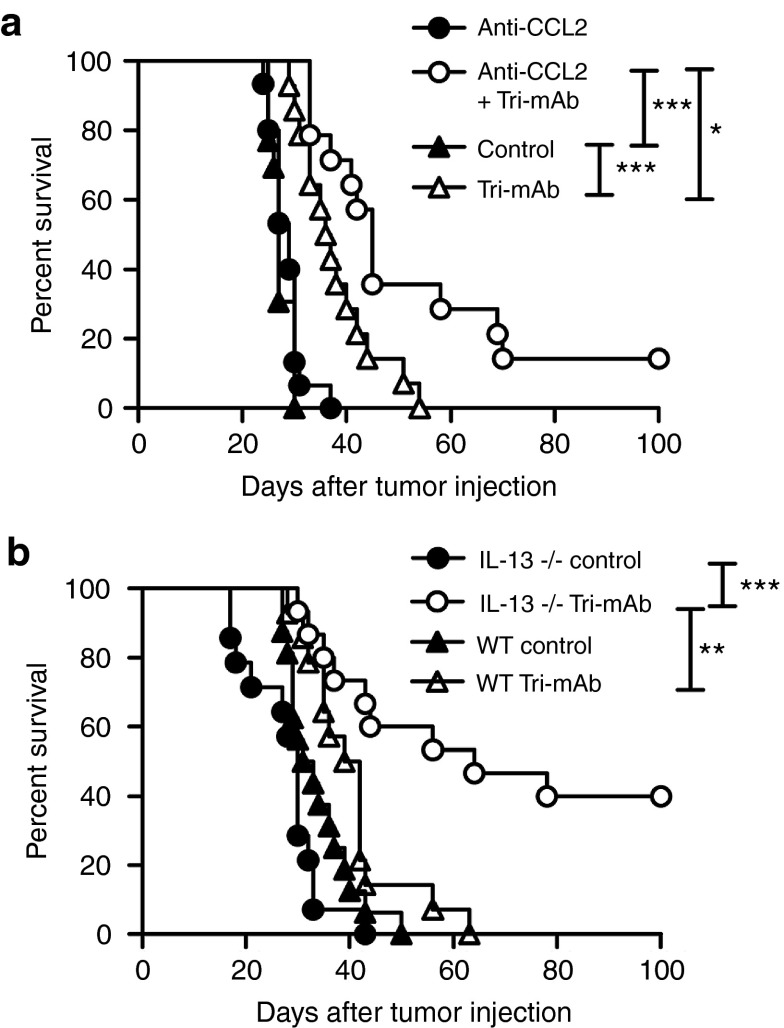
Molecules important in the recruitment and differentiation of AAMs include the chemokine CCL2 and the cytokines IL-4 and IL-13 (35). We, therefore, investigated the role of these molecules in the differential tumor responses using a neutralizing antibody for CCL2 and mice deficient in the genes for IL-4 or IL-13. Although kidney tumors responded poorly to Tri-mAb when CCL2 was present, responses were significantly enhanced when a CCL2 blocking antibody was administered before and during Tri-mAb treatment (Figure 5a). Similarly, kidney tumors in IL-13–deficient mice responded significantly better than tumors in wild-type mice (Figure 5b), whereas no effect was observed in IL-4–deficient mice (data not shown). These data, and the association between CCL2, IL-13, and AAM, provided supporting evidence for a role for this macrophage subset in the differential responses of tumors in the two anatomical locations.
Figure 5.
CCL2 and IL-13 are involved in the differential responses of tumors in the two anatomical locations. (a) Survival of mice with 10- to 12-day established intrakidney (IK) tumors, treated with Tri-mAb or control antibodies (day 10, 14, and 18) in the presence or absence of a blocking antibody specific for CCL2 (day 6, 10, 14, and 18). Data pooled from two experiments. n = 14 per group. (b) Survival of IL-13–deficient or wild-type (WT) BALB/c mice with 10- to 12-day established IK tumors, treated with Tri-mAb or control antibodies (day 10, 14, and 18). Data pooled from two experiments. n = 13–15 per group. *P < 0.05. **P < 0.005. *** P < 0.0005. CCL, CC chemokine ligand; IL, interleukin.
Kidney tumors were more highly vascularized than SC tumors
While the tumor microenvironment can impact on immunity, it can also affect morphologic and other stromal qualities of the tumors. Of particular interest was the level of vascularization of tumors, because macrophages can stimulate angiogenesis (36), and because we had demonstrated a relatively greater expression of the angiogenic markers CXCR2 (IL-8R), CXCL1 (KC), and endothelial cell–specific molecule-1 in kidney tumors using DNA microarray (Supplementary Table S1).We found that renal tumors were more highly vascularized as evidenced by a higher frequency of CD31-positive endothelial cells (Figure 6a,b). This information raised the possibility that AAMs contributed to angiogenesis, and kidney tumor growth may have been more robust with a greater blood supply. Another possibility was that the tumors varied in their permeability leading to differences in penetration by components of the Tri-mAb regimen. However, using the analysis of tumor permeability following intravenous injection of Evan's blue, tumors in the skin and kidney were found to be similarly infused with the dye (Figure 6c), suggesting no obvious differences in vessel permeability before the administration of therapy.
Figure 6.
Intrakidney (IK) tumors have a greater density of CD31+ cells. (a) Sections of Renca tumors, taken from mice at day 12 after injection and stained with anti-CD31 or isotype control antibodies, one representative view per tumor (IK and SC), scale bar = 100 µm (b) CD31 expression depicted as pixel density for five IK and five SC tumors (three sections per tumor, ten fields of each). (c) Diffusion of Evans blue dye into IK and SC tumors following intravenous injection in mice 30 minutes before. Results pooled from two experiments ±SEM, n = 19 per group. (d–f) Tri-mAb localization in (d) SC and IK tumors, (e) blood, and (f) skin following I-125 radiolabeled Tri-mAb antibodies injected intraperitoneally. Tissues were analyzed in cohorts of mice at 4, 24, 48, and 72 hours after Tri-mAb injection. Results represented as ±SEM, n = 4–5 tumors per group. *P < 0.05. **P < 0.005.
To further investigate this aspect of the microenvironment, we radiolabeled Tri-mAb and determined its localization to SC and kidney tumors periodically after the start of treatment. There was no significant difference between the localization of Tri-mAb to SC and kidney tumors immediately after (4 hours) administration of the antibodies (Figure 6d), confirming the Evans blue finding. However, longer-term analysis of Tri-mAb localization to tumor in the days following the start of therapy revealed that less Tri-mAb was present in kidney tumors compared with that in SC tumors (Figure 6d). The reduction in Tri-mAb tissue localization in kidney tumor-bearing mice was only observed for tumor, and similar amounts of Tri-mAb were found in the blood and skin of mice irrespective of tumor location (Figure 6e,f).
Discussion
The above observations indicate that tumors in different anatomical sites vary in their response to immunotherapy. They can also differ in their microenvironment, despite the injection of genetically matched cancer cells, suggesting that the host normal tissue surrounding the site of tumor implantation can have a decisive role in determining the composition of the tumor microenvironment. Tumors in a visceral orthotopic site had a microenvironment associated with a type 2, relatively immunosuppressive, immune response.
It should be noted that in this study, we used transplantable tumor models that involved the injection of tumor cells in a small amount of saline. This differs from the process of natural tumor initiation and metastatic implantation and may amplify the contribution of surrounding normal tissues to the subsequent tumor microenvironment. We used the transplantable models because spontaneous tumors and their metastases differ genetically, making it difficult to discern the contribution of genetic composition and surrounding tissue to differing tumor microenvironments. Nevertheless, our results have direct relevance to the use of various tumor models in investigating immunotherapies for cancer.
Although in this study we refer to the tumor microenvironment in visceral tumors as immunosuppressive, it may be more accurate to specify it as a different class to that observed in the SC tumors. Recent arguments for tissue-appropriate classes of immune response have merit (37), and this may well be the case in the systems described in this article.
At present, the sequence of events leading to the different tumor microenvironments is not clear. There could be an initial contribution from a tissue-specific component that may include cytokines such as leukemia inhibitory factor (33) and IL-33 (38), which was upregulated in kidney tumors. We demonstrated that a higher frequency of macrophages with an immunosuppressive AAM phenotype infiltrated IK tumors. Although the identification of the factors responsible for the increased frequency of AAMs in kidney tumors await future investigations, our observations of increased expression of the AAM-associated molecules CCL2 (MCP1), CSF1 (GM-CSF), leukemia inhibitory factor, and IL-6 in renal tumors, and their previously described role in skewing the differentiation of macrophages to an AAM phenotype (33,34), suggest that these molecules may play a role in sculpting the tumor microenvironment to favor AAMs. In addition, previous observations that AAMs aid in the repair of damaged or diseased renal tissue (39,40) suggest that tumor growth in our system tumors may use this repair program in the kidney to its advantage in order to escape the effector immune response.
IL-13, upregulated in orthotopic kidney tumors and produced predominantly by Th2 cells, can induce differentiation of macrophages to an AAM phenotype. This suggests that Th2 cells play a relatively early role in the development of an immunosuppressive microenvironment. The exact nature of the relative T-cell responses of tumors in different sites is not clear at present, and the absence of defined tumor antigens in our models makes this difficult to determine. Our previous work demonstrating a strong CD8+ effector T-cell response in the SC tumor model (24) suggests that a Th1 response is necessary for optimal tumor rejection, but further insight into this question awaits further investigations in the orthotopic models.
Although in this study we found a more highly immunosuppressive microenvironment in visceral tumors, we do not exclude that immunosuppression can play a role in the immune response of SC tumors. Indeed, many studies have demonstrated that immunosuppressive cells can infiltrate SC tumors (41,42) and that immunosuppressive factors can regulate effector immune responses in SC tumors (43). Furthermore, in our SC tumor model, we observed an infiltration of regulatory T cells and macrophages with an AAM phenotype that probably participate in immunosuppression, but nevertheless, we suggest that the effector immune response dominates and induces tumor regression after Tri-mAb treatment, unlike in the orthotopic kidney site.
Our investigations into the relative ability of Tri-mAb to localize to SC and renal tumors produced some interesting and novel observations. Although similar amounts of Tri-mAb initially enter tumors in both anatomical locations, significantly less Tri-mAb localized to kidney tumors from 24 to 72 hours after the start of therapy. This suggested that the initial responses in tumors from different sites were fundamentally different, potentially influenced by the differing nature of the tumors, and this impacted on the subsequent relative ability of Tri-mAb to localize to tumors. Although the sites of action of Tri-mAb have not been previously determined, these sites may include tumor and the lymphoid compartment, and the reduced localization of Tri-mAb to renal tumors may have contributed to the reduced efficacy of treatment. It will be of interest to gain more insight into this phenomenon in future studies.
Mixed responses to immunotherapy of tumors in different sites have been frequently observed in humans, where some metastases regress while others progress (44,45,46). However, whether the observed site-specific differences in the microenvironment and responses of mouse tumors extend to human tumors remains to be determined. Nevertheless, some support for similar events in patients can be found in studies where tumor-associated macrophages were found in human kidney tumors and could be associated with CCL2 and IL-10 expression (47,48). Similarly, the presence of tumor-associated macrophages in melanoma and colon cancer has correlated with poorer prognosis (9,49). Not surprisingly, metastatic burden correlates with survival of patients, although little has been done previously to analyze patient responses to immunotherapy according to the location of metastases. In mice, two earlier studies have reported differential responses to interferon of tumors in different sites, but no insight into the mechanism was provided (50,51).
The above observations indicate that the host normal tissue has a major impact on the tumor microenvironment, which varies with anatomical site, and may be important to consider in the application of immunotherapy. Further investigations into a range of tumor types in different tissues may help to identify tissue-specific signatures important in regulating the tumor microenvironment. Armed with this knowledge, clinicians could predict responses of individual tumors to various treatment regimens and design appropriate therapies to maximize responses.
Materials and Methods
Cell lines, mice, and tumors. Renca is a renal cell carcinoma cell line (52), CT26 is a N-nitroso-N-methyl-urethane-induced undifferentiated adenocarcinoma of the colon (ATCC, Manassas, VA), both derived from BALB/c mice, and RM-1 is a prostate carcinoma cell line, from C57BL/6 mice (53). The Renca-Ch-Luc cell line was generated by transduction of Renca with a retroviral vector (murine stem cell virus) containing the cDNA for cherry and luciferase. All tumor cell lines were maintained at 37°C and 5% CO2 in RPMI 1640 or Dulbecco's Modified Eagle Medium media (Invitrogen, Melbourne, Australia), supplemented with 10% heat-inactivated foetal calf serum (Moregate Biotech, Brisbane, Australia), 2 mM glutamine (JRH Bioscience, Melbourne, Australia), 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Sigma-Aldrich, Sydney, Australia). BALB/c and C57BL/6 mice were purchased from the Walter and Eliza Hall Institute for Medical Research, Melbourne, Australia and from the Animal Resource Centre, Perth, Australia. BALB/c-IL-13–deficient mice were bred and maintained at the Peter MacCallum Cancer Centre, East Melbourne, Australia. Mice that were 6 to 20 weeks of age were used in accordance with the Peter MacCallum Cancer Centre Animal Ethics Committee guidelines.
To produce SC tumors, mice were injected with 2 × 105 Renca, Renca-Ch-Luc, CT26 cells, and RM-1 cells in 100 µl of phosphate-buffered saline (PBS). Orthotopic tumors were established by injecting 2 × 105 Renca or Renca-Ch-Luc cells IK into the outer cortex of the kidney in 20 µl of PBS, 2 × 105 CT26 cells subserously on the cecum (IC) in 20 µl of PBS, or 2 × 104 RM-1 cells in 10 µl PBS in the anterior lobe of the prostate (intraprostrate). Liver tumors were established by injecting 2 × 105 Renca-Ch-Luc cells intrahepatically. For SC tumors, progression was determined using callipers, and survival was defined as tumors' size exceeding the ethically defined limit of 200 mm2. For kidney, cecum, prostate, and hepatic tumors, survival was defined as overt signs of stress. Progression of luciferase-expressing tumors was monitored using bioluminescent imaging performed with highly sensitive cooled charge-coupled device camera (IVIS Lumina II; Xenogen, Toronto, Canada) and intraperitoneal injection of the substrate D-luciferin (Thermo Fisher Scientific, Melbourne, Australia) in 150 µg/ml of PBS.
Tri-mAb was injected intraperitoneally every 3–4 days for 10–12 days, resulting in a total of three doses. Tri-mAb consisted of a mixture of 50 µg of anti-DR5 (clone MD5.1) and 25 µg of anti-CD40 (clone FGK-45), and 25 µg of anti-CD137 (clone 3H3) per mouse. Some groups (control) received PBS or control rat IgG2a (clone Mac4) and hamster IgG (clone UC8-1B9) instead of Tri-mAb. RM-1 tumors were treated with two doses of Tri-mAb at 25 µg of each component starting on day 5–7 after tumor inoculation. Details of additional methods are presented as supplementary material. Blocking of CCL2 was accomplished using a specific antibody (C-1142, Janssen R&D, Radnor, PA) at 0.2 mg per mouse intraperitoneally every 4–5 days for four doses beginning 2 days before Tri-mAb administration. Therapeutic and control antibodies were produced at Peter MacCallum Cancer Centre or The Walter and Eliza Hall Institute for Medical Research.
Tumor processing, antibodies, and flow cytometry. Tumors were excised from mice, minced finely, and dissociated for 25 minutes in RPMI 1640 containing 1 mg/ml collagenase type 4 (Worthington Biochemical, Lakewood, NJ), 30 units/ml DNase type II, and 100 µg/ml hyaluronidase type V (both from Sigma-Aldrich) at 37°C with agitation. Following dissociation, tumor suspensions were passed through a 70-µm cell strainer and washed twice in PBS. For cell line generation, tumor cells extracted from SC or IK solid tumors were established in culture for at least 4 weeks. For flow cytometric analysis, cells were resuspended in fluorescence-activated cell sorting buffer (PBS 2% fetal calf serum) in the presence of 2.4G2 (anti-CD16/32, to block Fc receptors) and then used for analysis.
For flow cytometry, cells were stained with antimouse CD11c-PE-Cy7 (clone N418), TCRβ-PerCP-Cy5.5 (clone H57-597), CD25-APC-eF780 (clone PC61.5), F4/80-PE-Cy7 (clone BM8), FR4-APC (clone 2A5), CD8α-PE-Cy7 (clone 53–6.7), CD4-APC-eF780 (clone RM4-5), CD11b-APC (clone M1/70), CD19-AF647 (clone eBio1D3), CD137-Biotin (clone 17B5), DR5-Biotin (clone MD5.1), Streptavidin-Pe-Cy7 (all from eBioscience, Kensington, Australia), Ly-6G-APC-Cy7 (clone 1A8), H-2K[d]-Biotin (clone SF11-1.1), CD40-Biotin (clone 3/23) (all from BD Biosciences, Sydney, Australia) and CD206 (MMR)-AF647 (clone MR5D3) (Biosearch Technologies, Perth, Australia). Cells were analyzed and sorted on BD FACS CantoII and DIVA SORTER (BD Biosciences). Analysis was performed using the software programs FCS express (De Novo Software, Los Angeles, CA) and Gatelogic308 (eBioscience).
Immunohistochemistry. Immunostaining of the blood vessels was performed following antigen retrieval using trypsin digestion (Trypsin 250, Difco, BD Biosciences). A rat antimouse CD31 antibody (HyCult HM1084), an IgG2a isotype control (BD Pharmingen), and a biotinylated rabbit antirat secondary antibody (DAKO E0468) were used following the Tyramide Signal Amplification kit (PerkinElmer Life and Analytical Sciences, Melbourne, Australia) according to the manufacturer's instructions. Five SC and five IK tumors were taken, with three sections per tumor, and ten fields of each were analyzed. Morphometric analysis of digital images was performed using the MetaMorph Premier (Version 7.6.2) software program (Molecular Devices, Middle Cove, Australia).
Tri-mAb localization. For Evan's blue studies, tumor-bearing mice were injected intravenously in the tail with 200 μl of Evans blue dye (Sigma-Aldrich) (0.2%; 30 mg/kg in PBS) or PBS (control mice). Thirty minutes later, mice were culled, tumors excised, rinsed in PBS, gently blotted, and weighed. Evans blue dye was extracted from the tumor by overnight incubation in dimethylformamide (Merck, Melbourne, Australia). Optical density of the extract was then measured at 620 nm with a microplate reader (VersaMax, Molecular Devices). A standard curve was used to quantify Evans blue dye per mg of tumor.
For radiolabeled antibody studies, 90 µg of Tri-mAb (0.02 mM) was incubated with 500 µCi NaI-125 (0.04 mM) (Perkin Elmer, Melbourne, Australia), 0.4 mM hydrogen peroxide, and 4 ng lactoperoxidase (Sigma) in a 100 µl volume in 0.1 mol/l sodium acetate buffer (pH 5.6) for 15 minutes at 22°C. I-125-labeled antibody was separated from free radioisotope into PBS using a PD-10 desalting column according to the manufacturer's instructions (Sigma-Aldrich). Mice received 2 µg of I-125-labeled Tri-mAb (~13.35 × 106 counts per minute) together with 100 µg of unlabeled Tri-mAb (as above) in 200 µl of PBS by intraperitoneal injection. Tumors and other tissues were taken periodically after antibody administration, and radioactivity was determined using a γ counter (Wallac Wizard 1470).
Gene expression studies. For reverse transcriptase–PCR, total RNA was isolated from single-sorted cells from kidney tumor with the RNeasy minikit (QIAGEN, Melbourne, Australia) following the manufacturer instructions. RNA (0.5–1 μg) was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (Promega, Melbourne, Australia) and Oligo dT primers. The resulting cDNA was used for amplification. The reaction volume was 50 μl and contained 1 μl of cDNA, 5× TAQ PCR mastermix (Promega, Melbourne, Australia) and the following primers: Arg1 primers (Arg1 forward: 5′GCTCCAAGCCAAAGTCCTTAGAGA-TTAT-3′ Arg1 reverse: 5′-GGCTTATGGTTACCCTCCCGTTGAGTTC-3′), Mgl1 primers (Mgl1 forward: 5′GACCCACCTCCTCCTGTTCTCCCTG-3′ Mgl1 reverse:5′AGTCCTCACCTCCTCCCAGTCCGTGTC-3′), and HPRT primers (HPRT forward: 5′-GCTGGTGAAAAGGACCTCT-3′ HPRT reverse: 5′-CACAGGACTAGAACACCTGC-3′). Products were electrophoresed in 1% agarose gel with 0.5 μg/ml of ethidium bromide and photographed under UV light. For microarray, Affymetrix Mouse Gene 1.0 ST chips (Affymetrix, Santa Clara, CA) were used to determine gene expression of cDNA prepared following dissection of Renca tumors from surrounding normal tissue. Quantitative PCR was performed using the mouse cytokine and chemokine PCR array (SABiosciences, Valencia, CA) according to the manufacturer's instructions. Analysis was based on the ΔΔCt method with normalization of the raw data to housekeeping genes. The full data set is available in the Gene Expression Omnibus, Accession Number GSE40679.
Protein array and cytometric bead array. Renca-Ch-luc SC and IK tumors were processed with the protease inhibitors, and protein levels were quantified using BCA Protein Assay kit (Pierce, Rockford, IL). Detection of chemokines and cytokines was performed using the Mouse Cytokine Array Panel A according to the manufacturer's instructions (BioScientific). For cytokine release, sorted macrophages (according to F480 and CD11b expression) were cultured at 1 × 105 cells per well in a 96-well flat bottom plate in complete RPMI media ± lipopolysaccharide (4 µg/ml) (Sigma-Aldrich) for 16 hours. For IL-6 detection in tumors, SC and IK tumors at day 12 were dissociated in PBS and supernatant was analyzed. A mouse inflammation cytometric bead array kit (552364) and the cytometric bead array Flex Systems Kit (both from BD Biosciences) were used to measure the concentration of cytokines according to the manufacturer's instructions.
Statistical analysis. Results are expressed as the mean ± SEM. The difference in tumor growth and variation in survival between different groups was analyzed by two-way ANOVA. Other experiments were analyzed using a Mann–Whitney test. A P value <0.05 was considered significant.
SUPPLEMENTARY MATERIAL Figure S1. Monitoring tumor growth in different locations. Figure S2. Visceral and SC tumors are similar in size and weight prior to treatment. Figure S3. Genes differentially expressed between IK and SC tumors. Figure S4. Genes and protein significantly differentially expressed between IK and SC tumors. Table S1. Relative expression of immune-related genes in kidney tumors compared to subcutaneous tumors, as determined using cDNA microarray. Table S2. Relative expression of immune-related genes in cecum tumors compared to subcutaneous tumors, as determined using RNAseq.
Acknowledgments
This work was supported by a grant from the Cancer Council of Victoria, Australia, No. 1006209. M.H.K. and P.K.D. were supported by Senior Research Fellowships from the National Health and Medical Research Council of Australia. The authors thank Viki Milovac and Sophie Curcio of the Peter MacCallum Cancer Center Flow Cytometry Facility for their excellent technical assistance in this study; members of the Peter MacCallum Cancer Center Animal Facility for their care and maintenance of mice used in this study; and Dr Pavel Lobachevsky for his help with the radiolabeled antibody studies. L.A.S. is employed by Janssen Research and Development who supplied the monoclonal anti-CCL2 antibody for this study.
Supplementary Material
References
- Shevach EM. Biological functions of regulatory T cells. Adv Immunol. 2011;112:137–176. doi: 10.1016/B978-0-12-387827-4.00004-8. [DOI] [PubMed] [Google Scholar]
- Marigo I, Dolcetti L, Serafini P, Zanovello P, Bronte V. Tumor-induced tolerance and immune suppression by myeloid derived suppressor cells. Immunol Rev. 2008;222:162–179. doi: 10.1111/j.1600-065X.2008.00602.x. [DOI] [PubMed] [Google Scholar]
- Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33:119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng MW, Ngiow SF, von Scheidt B, McLaughlin N, Sparwasser T, Smyth MJ. Conditional regulatory T-cell depletion releases adaptive immunity preventing carcinogenesis and suppressing established tumor growth. Cancer Res. 2010;70:7800–7809. doi: 10.1158/0008-5472.CAN-10-1681. [DOI] [PubMed] [Google Scholar]
- Gazzaniga S, Bravo AI, Guglielmotti A, van Rooijen N, Maschi F, Vecchi A, et al. Targeting tumor-associated macrophages and inhibition of MCP-1 reduce angiogenesis and tumor growth in a human melanoma xenograft. J Invest Dermatol. 2007;127:2031–2041. doi: 10.1038/sj.jid.5700827. [DOI] [PubMed] [Google Scholar]
- Robinson-Smith TM, Isaacsohn I, Mercer CA, Zhou M, Van Rooijen N, Husseinzadeh N, et al. Macrophages mediate inflammation-enhanced metastasis of ovarian tumors in mice. Cancer Res. 2007;67:5708–5716. doi: 10.1158/0008-5472.CAN-06-4375. [DOI] [PubMed] [Google Scholar]
- Bates GJ, Fox SB, Han C, Leek RD, Garcia JF, Harris AL, et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol. 2006;24:5373–5380. doi: 10.1200/JCO.2006.05.9584. [DOI] [PubMed] [Google Scholar]
- Gobert M, Treilleux I, Bendriss-Vermare N, Bachelot T, Goddard-Leon S, Arfi V, et al. Regulatory T cells recruited through CCL22/CCR4 are selectively activated in lymphoid infiltrates surrounding primary breast tumors and lead to an adverse clinical outcome. Cancer Res. 2009;69:2000–2009. doi: 10.1158/0008-5472.CAN-08-2360. [DOI] [PubMed] [Google Scholar]
- Jensen TO, Schmidt H, Møller HJ, Høyer M, Maniecki MB, Sjoegren P, et al. Macrophage markers in serum and tumor have prognostic impact in American Joint Committee on Cancer stage I/II melanoma. J Clin Oncol. 2009;27:3330–3337. doi: 10.1200/JCO.2008.19.9919. [DOI] [PubMed] [Google Scholar]
- Medrek C, Pontén F, Jirström K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. 2012;12:306. doi: 10.1186/1471-2407-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen HK, Donskov F, Marcussen N, Nordsmark M, Lundbeck F, von der Maase H. Presence of intratumoral neutrophils is an independent prognostic factor in localized renal cell carcinoma. J Clin Oncol. 2009;27:4709–4717. doi: 10.1200/JCO.2008.18.9498. [DOI] [PubMed] [Google Scholar]
- Li YW, Qiu SJ, Fan J, Zhou J, Gao Q, Xiao YS, et al. Intratumoral neutrophils: a poor prognostic factor for hepatocellular carcinoma following resection. J Hepatol. 2011;54:497–505. doi: 10.1016/j.jhep.2010.07.044. [DOI] [PubMed] [Google Scholar]
- Ko SY, Barengo N, Ladanyi A, Lee JS, Marini F, Lengyel E, et al. HOXA9 promotes ovarian cancer growth by stimulating cancer-associated fibroblasts. J Clin Invest. 2012;122:3603–3617. doi: 10.1172/JCI62229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakasone ES, Askautrud HA, Kees T, Park JH, Plaks V, Ewald AJ, et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 2012;21:488–503. doi: 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer. 2009;9:285–293. doi: 10.1038/nrc2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano D, Zollo M. Tumor microenvironment: a main actor in the metastasis process. Clin Exp Metastasis. 2012;29:381–395. doi: 10.1007/s10585-012-9457-5. [DOI] [PubMed] [Google Scholar]
- Sleeman JP. The metastatic niche and stromal progression. Cancer Metastasis Rev. 2012;31:429–440. doi: 10.1007/s10555-012-9373-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KH, Rhee H, Kang HJ, Yang E, You KT, Lee H, et al. Differential gene expression profiles of metastases in paired primary and metastatic colorectal carcinomas. Oncology. 2008;75:92–101. doi: 10.1159/000155211. [DOI] [PubMed] [Google Scholar]
- Koh SS, Wei JP, Li X, Huang RR, Doan NB, Scolyer RA, et al. Differential gene expression profiling of primary cutaneous melanoma and sentinel lymph node metastases. Mod Pathol. 2012;25:828–837. doi: 10.1038/modpathol.2012.32. [DOI] [PubMed] [Google Scholar]
- Uno T, Takeda K, Kojima Y, Yoshizawa H, Akiba H, Mittler RS, et al. Eradication of established tumors in mice by a combination antibody-based therapy. Nat Med. 2006;12:693–698. doi: 10.1038/nm1405. [DOI] [PubMed] [Google Scholar]
- Westwood JA, Darcy PK, Guru PM, Sharkey J, Pegram HJ, Amos SM, et al. Three agonist antibodies in combination with high-dose IL-2 eradicate orthotopic kidney cancer in mice. J Transl Med. 2010;8:42. doi: 10.1186/1479-5876-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Lichtnekert J, Thompson LJ, Karna P, Bouabe H, Hohl TM, et al. Resident renal mononuclear phagocytes comprise five discrete populations with distinct phenotypes and functions. J Immunol. 2013;191:3358–3372. doi: 10.4049/jimmunol.1300342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Faunce DE, Stacey M, Terajewicz A, Nakamura T, Zhang-Hoover J, et al. The macrophage F4/80 receptor is required for the induction of antigen-specific efferent regulatory T cells in peripheral tolerance. J Exp Med. 2005;201:1615–1625. doi: 10.1084/jem.20042307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med. 2012;209:139–155. doi: 10.1084/jem.20101387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Peart MJ, Smyth GK, van Laar RK, Bowtell DD, Richon VM, Marks PA, et al. Identification and functional significance of genes regulated by structurally different histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2005;102:3697–3702. doi: 10.1073/pnas.0500369102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, et al. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell. 2008;3:109–118. doi: 10.1016/j.stem.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Duluc D, Delneste Y, Tan F, Moles MP, Grimaud L, Lenoir J, et al. Tumor-associated leukemia inhibitory factor and IL-6 skew monocyte differentiation into tumor-associated macrophage-like cells. Blood. 2007;110:4319–4330. doi: 10.1182/blood-2007-02-072587. [DOI] [PubMed] [Google Scholar]
- Roca H, Varsos ZS, Sud S, Craig MJ, Ying C, Pienta KJ. CCL2 and interleukin-6 promote survival of human CD11b+ peripheral blood mononuclear cells and induce M2-type macrophage polarization. J Biol Chem. 2009;284:34342–34354. doi: 10.1074/jbc.M109.042671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, et al. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008;18:349–355. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat Rev Cancer. 2008;8:618–631. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- Matzinger P, Kamala T. Tissue-based class control: the other side of tolerance. Nat Rev Immunol. 2011;11:221–230. doi: 10.1038/nri2940. [DOI] [PubMed] [Google Scholar]
- Kurowska-Stolarska M, Stolarski B, Kewin P, Murphy G, Corrigan CJ, Ying S, et al. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol. 2009;183:6469–6477. doi: 10.4049/jimmunol.0901575. [DOI] [PubMed] [Google Scholar]
- Lee S, Huen S, Nishio H, Nishio S, Lee HK, Choi BS, et al. Distinct macrophage phenotypes contribute to kidney injury and repair. J Am Soc Nephrol. 2011;22:317–326. doi: 10.1681/ASN.2009060615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Wang C, Zheng D, Wang Y, Lee VW, Wang YM, et al. IL-25 induces M2 macrophages and reduces renal injury in proteinuric kidney disease. J Am Soc Nephrol. 2011;22:1229–1239. doi: 10.1681/ASN.2010070693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marabelle A, Kohrt H, Sagiv-Barfi I, Ajami B, Axtell RC, Zhou G, et al. Depleting tumor-specific Tregs at a single site eradicates disseminated tumors. J Clin Invest. 2013;123:2447–2463. doi: 10.1172/JCI64859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesokhin AM, Hohl TM, Kitano S, Cortez C, Hirschhorn-Cymerman D, Avogadri F, et al. Monocytic CCR2(+) myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer Res. 2012;72:876–886. doi: 10.1158/0008-5472.CAN-11-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalotto-Moreno T, Croci DO, Cerliani JP, Martinez-Allo VC, Dergan-Dylon S, Méndez-Huergo SP, et al. Targeting galectin-1 overcomes breast cancer-associated immunosuppression and prevents metastatic disease. Cancer Res. 2013;73:1107–1117. doi: 10.1158/0008-5472.CAN-12-2418. [DOI] [PubMed] [Google Scholar]
- Dudley ME, Wunderlich JR, Yang JC, Sherry RM, Topalian SL, Restifo NP, et al. Adoptive cell transfer therapy following non-myeloablative but lymphodepleting chemotherapy for the treatment of patients with refractory metastatic melanoma. J Clin Oncol. 2005;23:2346–2357. doi: 10.1200/JCO.2005.00.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolchok JD, Hoos A, O'Day S, Weber JS, Hamid O, Lebbé C, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. 2009;15:7412–7420. doi: 10.1158/1078-0432.CCR-09-1624. [DOI] [PubMed] [Google Scholar]
- Chang E, Rosenberg SA. Patients with melanoma metastases at cutaneous and subcutaneous sites are highly susceptible to interleukin-2-based therapy. J Immunother. 2001;24:88–90. doi: 10.1097/00002371-200101000-00010. [DOI] [PubMed] [Google Scholar]
- Daurkin I, Eruslanov E, Stoffs T, Perrin GQ, Algood C, Gilbert SM, et al. Tumor-associated macrophages mediate immunosuppression in the renal cancer microenvironment by activating the 15-lipoxygenase-2 pathway. Cancer Res. 2011;71:6400–6409. doi: 10.1158/0008-5472.CAN-11-1261. [DOI] [PubMed] [Google Scholar]
- Komohara Y, Hasita H, Ohnishi K, Fujiwara Y, Suzu S, Eto M, et al. Macrophage infiltration and its prognostic relevance in clear cell renal cell carcinoma. Cancer Sci. 2011;102:1424–1431. doi: 10.1111/j.1349-7006.2011.01945.x. [DOI] [PubMed] [Google Scholar]
- Herrera M, Herrera A, Domínguez G, Silva J, García V, García JM, et al. Cancer-associated fibroblast and M2 macrophage markers together predict outcome in colorectal cancer patients. Cancer Sci. 2013;104:437–444. doi: 10.1111/cas.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunda MJ, Sulich V, Bellantoni D. The anti-tumor effect of recombinant interferon alpha or gamma is influenced by tumor location. Int J Cancer. 1987;40:807–810. doi: 10.1002/ijc.2910400617. [DOI] [PubMed] [Google Scholar]
- Marquet RL, Westbroek DL, Jeekel J. Interferon treatment of a transplantable rat colon adenocarcinoma: importance of tumor site. Int J Cancer. 1984;33:689–692. doi: 10.1002/ijc.2910330521. [DOI] [PubMed] [Google Scholar]
- Murphy GP, Hrushesky WJ. A murine renal cell carcinoma. J Natl Cancer Inst. 1973;50:1013–1025. doi: 10.1093/jnci/50.4.1013. [DOI] [PubMed] [Google Scholar]
- Lu X, Park SH, Thompson TC, Lane DP. Ras-induced hyperplasia occurs with mutation of p53, but activated ras and myc together can induce carcinoma without p53 mutation. Cell. 1992;70:153–161. doi: 10.1016/0092-8674(92)90541-j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.