Abstract
Introduction
Dysregulated NOTCH receptor activity has been implicated in breast cancer but the mechanisms by which NOTCH contributes to transformation are not yet clear, as it has context-dependent effects on the properties of transformed cells.
Methods
We have used various in vitro and in vivo carcinogenic models to analyze the impact of Notch signaling in the onset and progression of breast tumors.
Results
We found that ectopic expression of the Notch1 intracellular domain (N1ICD) in MCF-7 breast adenocarcinoma cell line caused reduction and delocalization of E-CADHERIN levels and increased migratory and invasive abilities. Notch inhibition in the invasive breast cancer cell line MDA-MB-231 resulted in increased E-CADHERIN expression and a parallel reduction in their invasive capacity. The growth of subcutaneous xenografts produced with MCF-7 cells was boosted after N1ICD induction, in a cell autonomous manner. In vivo Notch1 activation in the mammary gland using the MMTV-Cre driver caused the formation of papillary tumors that showed increased Hes1 and Hey1 expression and delocalized E-cadherin staining.
Conclusions
These results confirm NOTCH1 as a signal triggering epithelial-mesenchymal transition in epithelial cancer cells, which may have implications in tumor dissemination, metastasis and proliferation in vivo. The identification of specific factors interacting with NOTCH signaling could thus be relevant to fully understanding the role of NOTCH in breast neoplasia.
Keywords: Mammary tumor, MCF-7, HT-29, MDA-MB-231, NOTCH, E-CADHERIN, EMT, migration, growth
Introduction
Notch is a fundamental signaling pathway that regulates embryonic cell fate specification, proliferation and patterning [1,2]. In addition to its central role in development, Notch signaling is deregulated in a number of cancers [3]. Notch1 mutations lead to oncogene expression in certain T cell acute lymphoblastic leukemias [4] and a subset of breast carcinomas [5]; deregulated Notch activity might also affect cell transformation [6], regulation of the cell cycle [7], progenitor/stem cell maintenance [3] and the outcome of breast cancer [8].
The mammalian Notch proteins (Notch1 to 4) are membrane-bound type I receptors with a large extracellular domain involved in ligand binding, and a cytoplasmic domain responsible for signal transduction. The Notch ligands Delta-like 1, 3 and 4 and Jagged 1 and 2 are also membrane-bound. Ligand-receptor interactions between neighboring cells trigger Notch signaling, which leads to a sequence of proteolytic cleavage events in the receptor. The last of these is mediated by γ-secretase activity, generating the Notch intracellular domain (NICD), which translocates to the nucleus and binds the CSL transcription factor. The NICD/CSL complex induces expression of target genes, including those of the hairy/enhancer of split (Hes) family [1,2], the cell cycle regulator p21 [9] and cyclin D1 [7].
Many studies focus on the role of Notch1 in mammary tumorigenesis. Hyperactivated Notch1 signaling was first implicated in mammary tumorigenesis in studies of the MMTV model, which showed that N1ICD expression in MMTV-Neu mammary tumors is due to an MMTV insertion in the Notch1 locus [10]. Other reports indicated that transgenic activation of N1ICD in mammary glands leads to development of lactation-dependent tumors that regress at weaning [11,12]. These findings link aberrant Notch activation in the murine mammary gland to adenocarcinoma. Experimental evidence shows that altered Notch1 signaling leads to direct transcriptional regulation of c-myc, which is crucial in MMTV-N1ICD-induced murine mammary tumorigenesis [13]. NOTCH1 is also involved in human mammary tumorigenesis as a downstream effector of oncogenic Ras [14].
Here we used various in vitro and in vivo models to analyze the impact of Notch signaling in breast tumor onset and progression. We find that stable or inducible N1ICD expression in the poorly invasive MCF-7 breast adenocarcinoma cell line causes a reduction and delocalization of E-CADHERIN levels, suggesting a disassembly of adherens junctions that correlates with enhanced cell migratory and invasive abilities. These properties may be extended to other epithelial tumor cell lines as we have made similar observations in the colon cancer cell line HT-29 stably expressing N1ICD. To the contrary, Notch inhibition in the highly invasive cell line MDA-MB-231 resulted in increased E-CADHERIN expression and a parallel reduction in their invasive capacity. Notch1 activation in the mouse mammary gland using the MMTV-Cre driver caused the formation of papillary tumors that showed increased Hes1 and Hey1 and delocalized E-cadherin expression. We also found that the growth of subcutaneous xenografts produced with MCF-7 cells was boosted after N1ICD induction, in a cell autonomous manner. These results confirm Notch1 as an epithelial-to-mesenchymal transition (EMT) inducer in breast cancer cells, which may have implications in tumor dissemination and metastasis.
Methods
Cell lines
The human breast cancer cell lines MCF-7 (ATCC® HTB-22™) and MDA-MB-231 (ATCC® HTB-26™), and the human colorectal adenocarcinoma cell line HT-29 (ATCC® HTB-38™) were used. For culture conditions see Additional file 1, Supplementary Materials and methods.
Transfection of MCF-7 and HT-29 cells
A cDNA fragment encoding the active version of mouse Notch1 (N1ICDΔOP) was used [15]. The Tet-Off system was employed to obtain transfectants of MCF-7 with inducible N1ICD expression. In this system, gene expression is turned on when doxycycline (DOXY; a tetracycline derivative) is removed from the culture medium. For details see Additional file 1, Supplementary Materials and methods.
Western blot analysis
For details see Additional file 1, Supplementary Materials and methods.
Semi-quantitative RT-PCR and real-time quantitative PCR
Total RNA was extracted with Trizol reagent (Life Technologies, NY, USA) and cDNA was synthesized with SuperScript III First Strand kit (Life Technologies, NY, USA). N-Cadherin primers were 5´-CACCCAACATGTTTACAATCAACAATGAGAC-3 (forward) and 5´-CTGCAGCAACAGTAAGGACAAACATCCTATT-3 (reverse) [16]. Commercial β-actin primers were used (Stratagene, La Jolla, CA, USA). Quantitative PCR was performed with Power SYBR Green Master Mix (Applied Biosystems, NY, USA, 4367659) and commercial primers for HEY1, HES1, cMYC, NOTCH1, NOTCH4, SNAI1, ECAD, VIMENTIN and HPRT1 (Sigma, St. Louis, MO, USA) were used.
Promoter activity assays
Hes1-Luc promoter activity [17] was measured in MCF-7 cells expressing N1ICD in a constitutive or inducible manner. The activity of the artificial promoter 10XCBF1 [18] was measured after transient transfection of MDA-MB-231 cells. Briefly, cells were co-transfected with the plasmid containing the promoter 10XCBF1-Luc and pcDNA3-CBF1-VP16 or pcDNA3-DN-CBF1/RBPJK. The plasmid pTK-RL (Promega, Madison, WI, USA)
was also included as a control of transfection efficiency. When indicated, cells were treated for the indicated period of time with DOXY 2 μg/ml or with the γ-Secretase Inhibitors DAPT (N-(N-(3,5-Difluorophenacetyl)-L-alanyl)-S-phenylglycine t-butyl ester, 10 to 50 μM; 565770, Calbiochem, Millipore, MA, USA) and RO4929097 ((2,2-dimethyl-N-(S)-6-oxo-6,7-dihydro-5H-dibenzo(b,d)azepin-7-yl)-N'-(2,2,3,3,3-pentafluoro-propyl)-malonamide)), 10 to 20 μM; S1575, Selleckchem, Houston, TX, USA) for 48 h. After transfection cells were lysed with passive lysis buffer (Promega, Madison, WI, USA) and firefly and renilla luciferase were measured with the "Dual-luciferase reporter assay" (Promega, Madison, WI, USA). The activity in MCF-7 clones or in MDA-MB-231 treated cells was referred to the activity in control cells or cells transfected with the empty vector (pcDNA3).
Immunofluorescence and immunohistochemistry
For details see Additional file 1, Supplementary Materials and methods.
In situ hybridization
In situ hybridization was performed as described in [19]. Details of probes will be provided on request.
In vitro cell chemotaxis
Cell migration was performed in Transwell (Corning, Tewksbury, MA, USA) with 8 μm pore filters coated with 20 μg/ml collagen type IV (Sigma, St. Louis, MO, USA). Cells were pretreated for the time indicated with DOXY (MCF-7) or DAPT/RO4929097/DMSO (MDA-MB-231), trypsinized and added to the upper chamber in basal medium with 0.5% BSA and the additives. The lower chamber was replenished with basal medium with BSA and the chemoattractant (IGF-1, 50 ng/ml, R&D Systems, Minneapolis, MN, USA or SDF1α, PeProTech (New Jersey, USA). After 18 h incubation, the upper chamber was emptied and cells remaining are removed. Cells in the filter are fixed with PFA and then stained with violet crystal (Sigma-Aldrich). Cell counts were obtained by counting two (MDA-MB-231) or four (MCF-7) grids using a microscope fitted with a grid eyepiece at a total magnification of 100X.
Flow cytometry
For details see Additional file 1, Supplementary Materials and methods.
In vivo experiments with mice
All animal procedures were approved by the Institutional Committee for the Care and Use of Laboratory Animals of the Centro Nacional de Investigaciones Cardiovasculares (CNIC, Madrid, Spain) and Centro Nacional de Biotecnología (CNB-CSIC, Madrid, Spain). Animal procedures conformed to EU Directive 2010/63EU and Recommendation 2007/526/EC, regarding the protection of animals used for experimental and other scientific purposes, enforced in Spanish law under Real Decreto 1201/2005.
Tumorigenic assays
MCF-7/TetOff and B12, M5 and M20 derivatives' clones, growing in culture without DOXY for 25 days, were inoculated s.c in both flanks (1.5 or 2.4 × 106 cells) in BALBc/SCID mice treated with 17α-ethylenestradiol 1 μg/ml (Sigma) provided in the drinking water from one week before cells were injected. Tumor size was monitored weekly and tumor volume estimated with a caliper by measuring the width (a) and the length (b) and applying the formula (a2 × b)/2. Once finished with the period of treatment, mice were sacrificed and tumors were extracted for further analysis. MCF-7/TetOff and B12 were transduced with recombinant retrovirus to express luciferase activity. Plasmid pRV-luc-IRES-CopGreen was used to obtain the retroviral supernatants (Genetrix S.L., Madrid, Spain) and transduced cells were sorted according to the associated green fluorescence. BALBc/SCID mice (Harlan Laboratories, Indianapolis, IN, USA) were injected in the two inguinal mammary glands with 2.5 × 106 cells and mice were treated as above. After injection, half of the mice were treated also with DOXY 2 mg/ml provided in the drinking water. Tumoral growth rate was analyzed by bioluminescence at different weeks after cell inoculation. Briefly, mice were injected with luciferin with the general anesthetic and luciferase activity expressed by cells was detected with a CCD camera placed in a dark box (Hamamatsu Photonics, Shizuoka, Japan). Images were processed with the software provided and luminescence units were represented. Tumor size was estimated as above and once finished with the period of treatment, tumors were excised, weighted and preserved adequately to make further analysis.
Transgenic N1ICD expression in the mammary gland
The transgenic lines MMTV-Cre [20] and Rosa26N1ICD [21] were bred to generate MMTV-Cre/+; Rosa26N1ICD/+ double transgenic mice. For primers and conditions of mouse genotyping see [20,21]. Mice were subjected to several rounds (a median of four) of pregnancy and lactation, and when a breast tumor arose, mice were euthanized and the breast tumor excised and processed for further analysis. Tumor samples were fixed with 10% buffered formalin (Sigma-Aldrich) for 48 h and afterward were paraffin-embedded. Staining of Hes1, ERα, p63, E-cadherin and Ki67 was performed in 5 μm sections of paraffin samples following standard techniques. For details see Additional file 1, Supplementary Materials and methods.
Results
N1ICD expression enhances the invasive capacity of the breast cancer cell line MCF-7
To gain an insight into the role of Notch in breast cancer we used the breast cancer cell line MCF-7 that has several features of differentiated mammary epithelium [22]. These cells show low levels of N1ICD expression by Western blot when compared with the metastatic breast cancer cell line MDA-MB-231 (Figure 1A). This observation fits with the idea that high NOTCH signaling is associated with the expression of basal breast cancer markers [23].
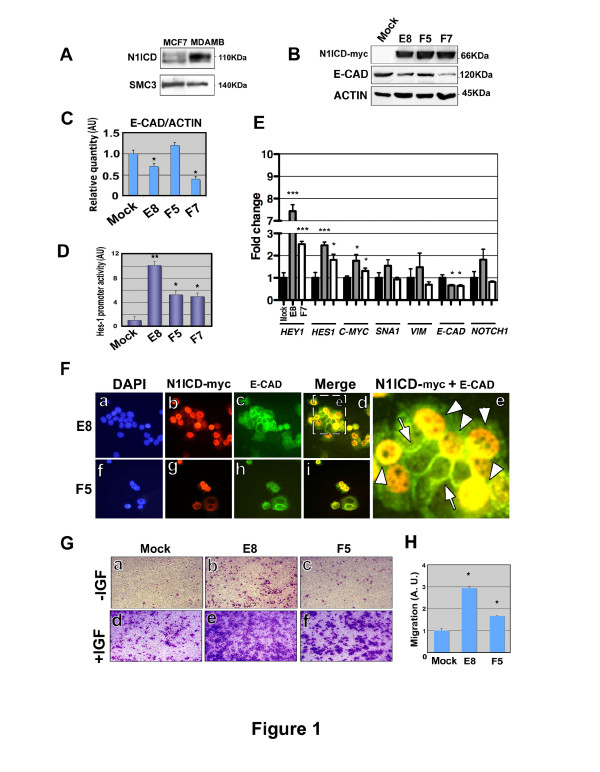
Figure 1.
Stable N1ICD expression induces migration of MCF-7 cells. (A) Western blot showing expression of endogenous N1ICD in MCF-7 and MDABM231 cells. SMC3 (Structural Maintenance of Chromomosomes-3 protein) was used as a loading control. (B) Expression of N1ICD and E-CADHERIN in the stable MCF-7 clones E8, F5 and F7 besides with mock MCF-7 cells; β-ACTIN was used as a loading control. (C) The Western blot in (B) was quantified by densitometry and the ratio of E-CADHERIN/β-ACTIN calculated. (D) NOTCH activity was determined with a Hes-1 promoter fragment. The value in the clones is represented relative to the value in mock cells. Data are mean ± SEM of quadruplicates in three independent experiments (*P <0.05 determined by Student's t test). (E) Expression of HEY1, HES1, C-MYC, SNAI1, VIMENTIN, E-CADHERIN and NOTCH1 in the stable MCF-7 clones E8 and F7, measured by qPCR. Data are mean ± SEM of triplicates in two independents experiments (*P <0.05, ***P <0.001 determined by Student's t-test). (F) Staining of N1ICD (red), E-CADHERIN (green) and nuclei (DAPI, blue) in E8 and F5 MCF-7-N1ICD clones. Arrows indicate the intercellular staining of E-CADHERIN and arrowheads its intracellular accumulation. (G, H) Effect of ectopic N1ICD expression on the chemotaxis of E8 and F5 MCF-7 cell clones. (G) Representative images of transmigrated cells in basal (-IGF-1) and after IGF-1 addition (+IGF). (H) Quantification of the transwell assays. The migration index was calculated for each condition, and then referred to that of mock (considered as 1). Data are mean ± SEM of triplicates in three independent experiments (*P <0.05 determined by Student's t test). DAPI, 4,6-Diamidino-2-phenylindol; MCF-7, Michigan Cancer Foundation-7 breast cancer cell line; N1ICD, Notch one intracellular domain
We generated MCF-7 clones stably expressing a myc-tagged N1ICD version (Figure 1B). MCF-7 cells have a typical cobblestone phenotype (not shown) and express the epithelial cell marker E-CADHERIN (Figure 1B, C). N1ICD expression caused a reduction in total E-CADHERIN levels in MCF-7 clones E8 and F7 but not in clone F5 (Figure 1B, C). The levels of Notch activity in MCF-7/N1ICD cells measured by a luciferase reporter assay using a fragment of the mouse Hes1 promoter [17], revealed an evident activation of the Notch pathway in comparison with control, mock-transfected MCF-7 cells (Figure 1D). qPCR analysis revealed a marked up-regulation of the NOTCH target genes HEY1, HES1 and C-MYC while the epithelial marker E-CADHERIN was down-regulated and SNAIL1 and VIMENTIN were not significantly changed (Figure 1E). We also examined NOTCH1 and NOTCH4 expression because of their role in mouse breast cancer malignancy [24] and their overexpression in triple-negative breast cancer subtypes [25]. NOTCH4 expression was almost undetectable in MCF-7 cells (Additional file 2, Figure S1A) while NOTCH1 expression was unaffected (Additional file 2, Figure S1B and not shown), as previously reported [26]. Semi-quantitative RT-PCR analysis of various MCF-7-N1ICD expressing clones revealed no variation in JAG1 and TWIST1 expression (Additional file 2, Figure S1B). The lack of response of TWIST1 to N1ICD expression is in agreement with previous findings showing that during developmental EMT, Twist1 is induced by Bmp2 [27] but does not respond to Notch [28]. Immunofluorescence analysis confirmed that forced N1ICD expression caused a reduction in membranous E-CADHERIN staining (Figure 1Fa-i). Cells with strong nuclear N1ICD staining showed mostly nuclear E-CADHERIN expression, suggesting a disassembly of adherens junctions [29], which contrasted with its accumulation in the membrane at the cell-cell contacts of MCF-7 cells that did not express N1ICD (Figure 1Fe). Concomitant to the reduction in E-CADHERIN levels, N1ICD expression endowed MCF-7 cells with increased chemotactic ability towards IGF-1 (Figure 1G, H), a chemo-attractant for this cell line [30]; some N1ICD-expressing MCF-7 clones showed an increased migratory capacity, even in basal medium (Figure 1Gb, c). To test if the reduction of E-CADHERIN upon N1ICD expression could be extended to other epithelial tumor cell lines, we transfected N1ICD into the HT-29 colon adenocarcinoma cell line and generated stable clones. We chose HT-29 cells because, similarly to the mammary gland, they derive from a tissue in which NOTCH has an oncogenic role [31,32]. Additional file 3, Figure S2A, B shows that HT-29 cells stably expressing N1ICD down-regulate E-CADHERIN expression.
Inducible N1ICD expression in MCF-7 cells leads to E-cadherin down-regulation and increased migratory capacity
To study more precisely the effect of Notch expression in MCF-7 cells, we generated N1ICD-inducible clones using the TetOFF system, so that gene expression was induced when the antibiotic doxycycline (DOXY) was removed from the culture medium. Figure 2A shows N1ICD-myc staining of three inducible clones (B12, M5 and M20) cultured in the presence (OFF condition) or absence (ON condition) of DOXY. There was some leaky N1ICD expression in the OFF condition in these three clones (especially for B12; Figure 2Aa-f), but 48 h after DOXY withdrawal there was a clear induction of N1ICD expression (Figure 2Ag-l), although not in 100% of the cells. Examination after seven days of induction also revealed a non-homogenous N1ICD expression in these MCF-7 clones (data not shown).
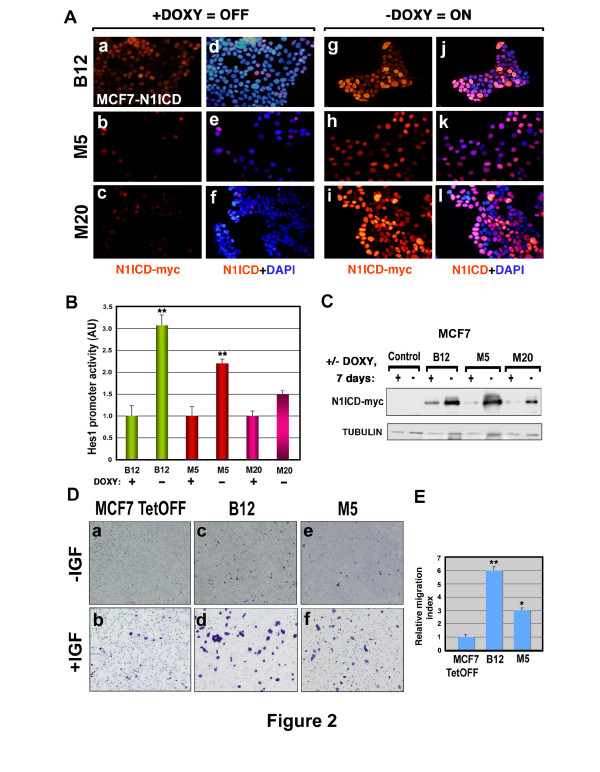
Figure 2.
Inducible N1ICD overexpression in MCF-7 cells promotes a migratory behavior. (Aa-f) General view of the MCF-7 Tet-Off clones (B12, M5 and M20) cultured in the presence of DOXY. a-c, anti-myc staining reveals leaky myc-N1ICD expression; d-f, anti-myc and DAPI staining overlapping. (Ag-l) General view of these clones cultured in the absence of DOXY. g-i, anti-myc staining reveals N1ICD induction in these cells; j-l, anti-myc and DAPI staining. (B) Quantification of Hes1 promoter activity in MCF-7 uninduced and induced clones. Data are mean ± SEM of quadruplicates in three independent experiments (**P <0.005 determined by Student's t-test). (C) Western blot analysis of myc-N1ICD expression in MCF-7 control cells and MCF-7 clones after seven days of culture with or without DOXY. (D) Representative images of transmigrated cells in basal (-IGFI) and IGF-I supplemented (+IGF) cells. (a, b) control, (c, d) N1ICD-induced B12 and (e, f) M5 MCF-7 cells. (E) The relative migration index for each cell type is represented. Data are mean ± SEM of duplicates in three independent experiments (*P <0.05, **P <0.005 determined by Student's t test). DAPI, 4,6-Diamidino-2-phenylindol; DOXY, doxycycline; MCF-7, Michigan Cancer Foundation-7 breast cancer cell line; N1ICD, Notch one intracellular domain
The effect of DOXY retrieval in N1ICD induction was measured by luciferase assay upon transfection of a Hes1 reporter. There was clear reporter activation after N1ICD induction in the different clones studied, especially in clone B12 at 48 h (Figure 2B). This enhanced N1ICD-induced transcriptional activity correlated with the increase of N1ICD expression in the different clones upon doxycycline withdrawal (Figure 2C). Concomitantly to N1ICD induction, there was an increase in the migratory capacity of these cells (Figure 2D, E).
To investigate the possible correlation between the enhancement in the migratory capacity of MCF-7-TetOFF-N1ICD clones and the reduction of E-CADHERIN expression in these cells, we performed an immunofluorescence analysis of clones M5, M20 and B12 after 20 days of culture in the absence of DOXY. Induction of N1ICD expression coincided with a reduction of membranous E-CADHERIN staining in these clones (Figure 3A-D). Western blot analysis after 20 days of induction revealed a marked reduction in E-CADHERIN expression in clone B12 that was not so apparent in clones M5 and M20 (Figure 3E). We then carried out a time-course of N1ICD repression/induction in clone B12 and its effect on E-CADHERIN expression. As Figure 3F shows, there was an inverse correlation between N1ICD and E-CADHERIN expression. MCF-7-TetOFF-N1ICD cells of clone B12 cultured seven days in the absence of DOXY showed strong N1ICD expression (Figure 3F). After one day in culture in the presence of DOXY, N1ICD expression was progressively reduced and in parallel, E-CADHERIN expression was increased throughout seven days of culture (Figure 3F). To the contrary, when cells from clone B12 were grown in the absence of DOXY, N1ICD expression was increased and E-CADHERIN expression was reduced, and this effect was clear after seven days of culture without DOXY (Figure 3F).
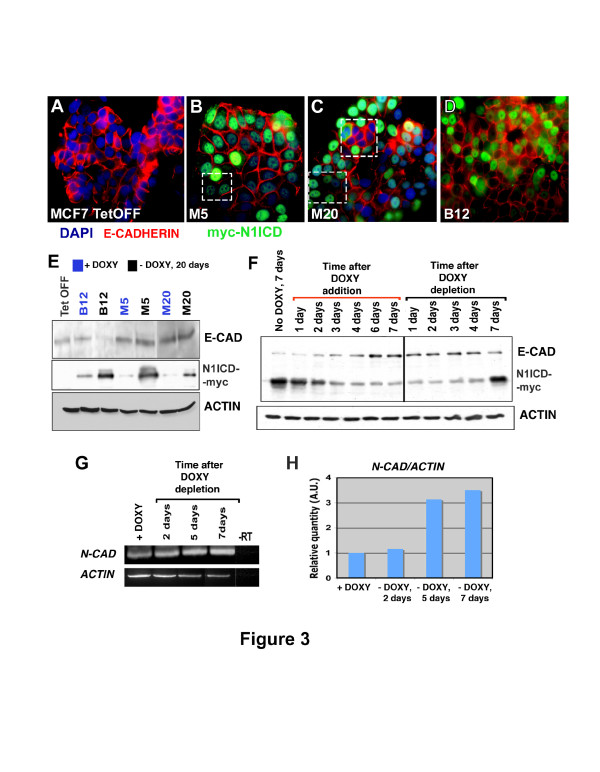
Figure 3.
E-CADHERIN and N-CADHERIN levels are modified in MCF-7 cells by overexpression of N1ICD. (A) Immunofluorescence staining of E-CADHERIN expression (red) in MCF-7 Tet-Off cells. (B-D) E-CADHERIN staining is lost in some cells where N1ICD (green) is induced. Nuclei are counterstained with DAPI (blue). (E) Western blot analysis of E-CADHERIN in MCF-7 mock and MCF-7 inducible clones in the presence (blue) or absence (black) of doxyxycline. (F) Time-course analysis of E-CADHERIN expression by Western blot after DOXY addition and depletion in clone B12. (G, H) N-CADHERIN transcription measured by RT-PCR in clone B12 after DOXY depletion. DAPI, 4,6-Diamidino-2-phenylindol; DOXY, doxyxycline; MCF-7, Michigan Cancer Foundation-7 breast cancer cell line; N1ICD, Notch one intracellular domain
These data indicated that N1ICD expression in MCF-7 cells, either in an inducible or stable manner, leads to a reduction in E-CADHERIN levels, suggesting that these cells began to lose their epithelial phenotype. During EMT there is a progressive cadherin switch, such that E-cadherin expression is reduced and N-cadherin expression is increased [33]. Figure 3G shows a semi-quantitative RT-PCR analysis of N-CADHERIN expression in clone B12. Upon DOXY withdrawal there was a progressive increase in N-CADHERIN expression that was at a maximum after seven days of culture (Figure 3G, H), suggesting that B12 cells acquired a mesenchymal phenotype. Moreover, we found that N1ICD induction enhanced VIMENTIN expression although it did not change Twist1 mRNA levels (Additional file 4, Figure S3).
NOTCH inhibition reduces the migratory ability of MDA-MB-231 cells
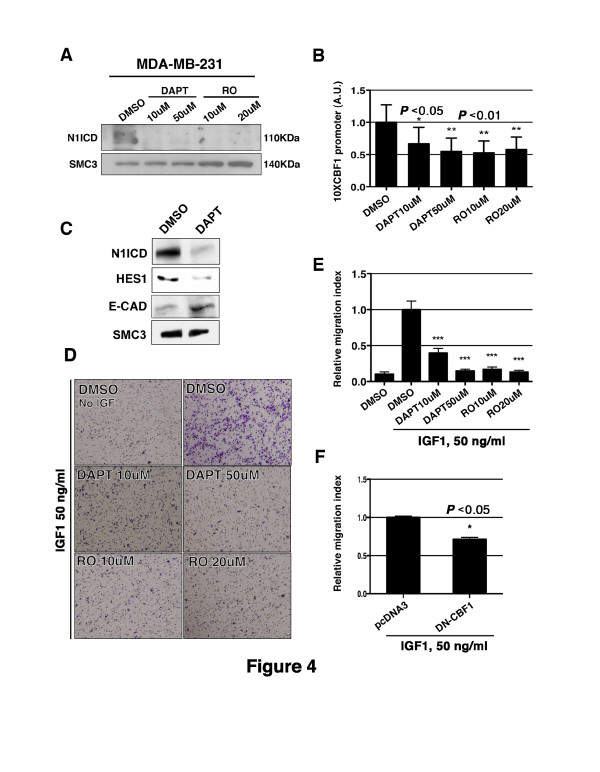
To gain further insights on the role of NOTCH in breast tumor progression and the potential use of NOTCH inhibitors as therapeutic agents against breast cancer, we inhibited NOTCH activity in the adenocarcinoma cell line MDAMB-231 [34], which is more tumorigenic and invasive than the MCF-7 cell line. MDA-MB-231 cells endogenously expressed N1ICD (Figures 1A and 4A) and E-CADHERIN levels in MDA-MB-231 were reduced compared to those of MCF-7 cells (Additional file 5, Figure S4). We used the γ-secretase inhibitors DAPT [35] and RO4929097 [36], to prevent the generation of NICD and thus inhibit Notch activity. After 48 h we could readily detect a drastic reduction in N1ICD levels in DAPT- and RO-treated cells (Figure 4A). This effect could also be measured by the reduction in the activity of a CBF1 reporter (Figure 4B). NOTCH inhibition also resulted in an up-regulation of E-CADHERIN paralleled by a reduction in HES1 expression (Figure 4C). The migratory response of MDA-MB-231 cells towards IGF-1 was reduced around 80% when cells were cultured with DAPT or RO (Figure 4D, E). MDA-MB-231 cells were also transfected with a dominant-negative version of CBF1 (DN-CBF1); we observed a 25% reduction in the migratory capacity of DN-CBF1-expressing MDA-MB-231 cells compared to controls (Figure 4F). The weaker inhibition of migration in comparison with the DAPT or RO treatments was likely due to the fact that MDA-MB-231 cells were transiently transfected and only a subpopulation expressed the DN-CBF1 construct (data not shown).
Figure 4.
NOTCH signaling inhibition decreases the migratory behavior of MDA-MB-231 cells. (A) Western blot showing Notch one intracellular domain (N1ICD) expression in MDA-MB-231 cells after 48 h of treatment with DAPT or RO4929097 (RO). Equal amounts of protein loading were validated with an anti-SMC3 antibody. (B) Notch activity measured with the 10 × CBF1 reporter. The value for DAPT and RO4929097 treated cells is relative to the activity in cells treated with DMSO. Data are mean ± SEM of quadruplicates in three independent experiments (*P <0.05, **P <0.005 determined by Student's t-test). (C) Representative images of migration towards IGF-1 50 ng/ml of MDA-MB-231 cells with DAPT or RO4929097 or the vehicle DMSO. (D) Migration index obtained in both conditions. Data are mean ± SEM of duplicates in three independent experiments (***P <0.001 determined by Student's t-test). (E) Migration index quantification of MDA-MB-231 cells transfected with an empty vector (pCDNA3) or a dominant negative version of CBF1 (DN-CBF1) towards IGF-1 (50 ng/ml). Data are mean ± SEM of duplicates in three independent experiments (*P <0.05 determined by Student's t-test). CBF1, CSL, Suppressor of Hairless; DAPT, N-(N-(3,5-difluorophenacetyl)-l-alanyl)-S-phenylglycine t-butyl ester; DMSO, dimethyl sulfoxide; MCF-7, Michigan Cancer Foundation-7 breast cancer cell line; MDA-MB-231, Breast cancer cell line derived from metastatic site (pleural effusion); N1ICD, Notch one intracellular domain
Inducible Notch1 activation in MCF-7 cells stimulates tumor growth in vivo
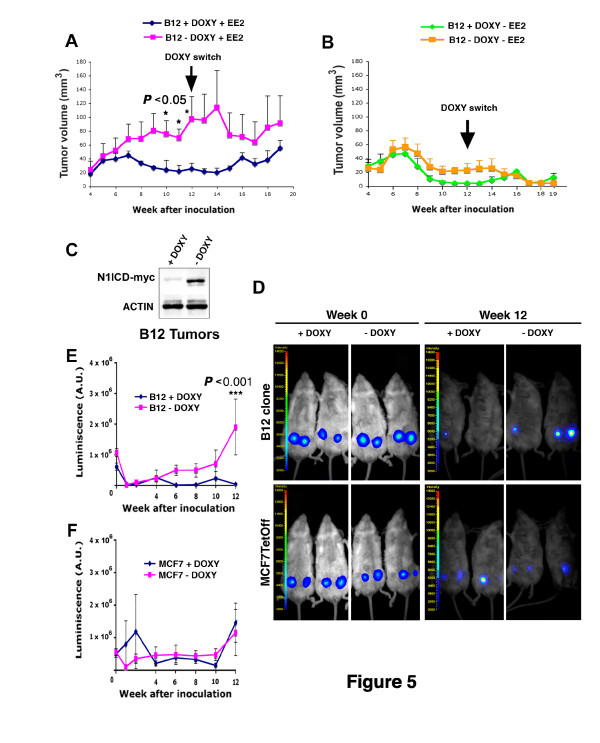
To examine the tumorigenic ability of MCF-7-N1ICD inducible clones we focused on the B12 clone as it showed a clear phenotypic change as a consequence of N1ICD induced overexpression. Growth of MCF-7 xenografts is estrogen-dependent [37], so we injected MCF-7-Tet-Off-N1ICD B12 cells and added 17α-ethynyl estradiol to the drinking water. When small size tumors were evident in all injection points, mice were divided into two groups. In the first group N1ICD expression was "turned on" (no DOXY) and in the second one, N1ICD expression was turned "Off" by addition of DOXY to the drinking water. As Figure 5A shows, the tumors in which N1ICD expression was turned off did not grow significantly, while tumors in which N1ICD expression was maintained for 12 weeks continued growing and were significantly larger than those generated by control MCF-7 (data not shown) or by un-induced cells (Figure 5A). After 12 weeks, treatments were switched between both groups of mice (Figure 5A, arrow), and tumors were monitored for seven weeks more. The result was a reduction in the differences between both groups, leading to similar tumor size (Figure 5A). Thus, tumor growth was clearly dependent on turning "On" or "Off" N1ICD expression.
Figure 5.
N1ICD induction promotes a tumorigenic behavior. (A) Growth of xenografts derived from MCF-7 B12 clone with induced N1ICD (-Doxy) or without N1ICD (+Doxy) expression. Mice also had 17α-ethynyl estradiol (+EE2) in the drinking water. The arrow indicates the 12th week when DOXY treatment was switched between both groups of mice. (B) Growth of N1ICD-induced B12 cells in the absence of estrogen (+EE2) administration. The arrow indicates the week 12th when DOXY treatment was switched. Data are mean ± SEM of n = 8 (four mice and two tumors per mice; *P <0.05 determined by two-way ANOVA with Bonferroni post-test). (C) Western blot showing expression of myc-tagged N1ICD in B12 tumors at 12th week of cell inoculation. (D) MCF-7 Tet-Off or the MCF-7 B12 clone engineered to express luciferase injected in the mammary gland of BALBc/SCID mice were monitored by bioluminescence emission at the time of cell inoculation and after 12 weeks as indicated. Selected images of two independents experiments, four animals per group. (E) Growth dynamics measured by luminescence of MCF-7 B12 clone with (-DOXY) or without (+DOXY) N1ICD expression. (F) Growth of control MCF-7 clones in the same conditions. Data are mean ± SEM of n = 8 (four mice and two tumors per mice; ***P <0.001 determined by two-way ANOVA with Bonferroni post-test), representative of two independents experiments. DOXY, doxyxycline; MCF-7, Michigan Cancer Foundation-7 breast cancer cell line; N1ICD, Notch one intracellular domain
The growth curves shown in Figure 5A, B suggest also that estrogens may be a limiting factor in Notch-mediated tumor formation and growth. Western blot analysis of xenografts generated after 12 weeks of N1ICD induction revealed strong N1ICD expression (Figure 5C).
Next, we analyzed growth of orthotopic tumors formed by the clone B12 transduced with a luciferase-expressing retrovirus to monitor tumor evolution by chemoluminiscence. After cell injection, mice were separated in two groups, receiving (N1ICD off) or not (N1ICD on) DOXY in the drinking water. In agreement with our previous results (Figure 5A), clone B12 with induced N1ICD expression gave rise to tumors significantly larger than those generated when N1ICD was not expressed (Figure 5D, E). DOXY treatment did not affect the growth of tumors formed by control MCF-7 cells (Figure 5D, F). These results suggested that N1ICD induction might be directly responsible for MCF-7 tumor formation.
N1ICD expression in the mammary gland leads to tumor formation and reduction in estrogen receptor and E-cadherin expression
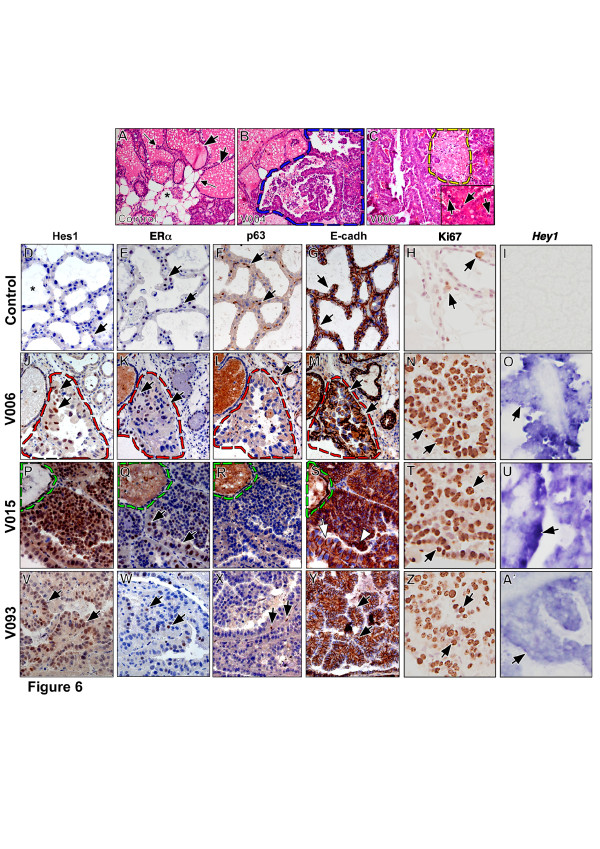
To test in vivo the effect of NOTCH activation in the mammary gland we bred the mouse mammary tumor virus (MMTV)LTR-Cre transgenic line (MMTV-Cre) [20] with the Rosa26N1ICD line that expresses the active form of Notch1 (N1ICD) in a conditional manner [21]. With the MMTV-Cre driver we targeted N1ICD expression to the secretory epithelium of the mammary gland of pregnant and lactating females. MMTV-Cre/+;N1ICD/+ double transgenic mice developed normally and were born at Mendelian ratios (data not shown). Adult MMTV-Cre/+; N1ICD/+ females showed high incidence of papillary breast carcinoma (>90%; Additional file 6, Table S1). Figure 6A shows a wildtype lactating breast, with greatly expanded secretory lobules composed of multiple distended acini. Figure 6B, C shows papillary tumors developed in lactating transgenic females (V004 and V006) after three to four rounds of pregnancy and lactation. Tissue architecture was disorganized and large necrotic areas were observed (Figure 6B). Also, frequent mitotic figures and cytological atypia were common (Figure 6C, inset). We analyzed the expression of the Notch targets Hes1 and Hey1, estrogen receptor, the myoepithelial marker p63, which stains basal/myopithelial preserved cells in normal or non-malignant breast tissue [38], the epithelial marker E-cadherin and the cell proliferation marker Ki67. Hes1 expression was low and restricted to a few cells in normal breast epithelial tissue (Figure 6D), like that of estrogen receptor (Figure 6E), while p63 stained around 30% of cells, as expected for a normal tissue (Figure 6F). E-cadherin was strongly expressed in the membrane of normal breast epithelial cells (Figure 6G). Ki67 was expressed only in a few cells in the normal breast (Figure 6H) and Hey1 was undetectable (Figure 6I). In the tumors generated in double transgenic MMTV-Cre;N1ICD mice (Additional file 6, Table S1) three patterns could be distinguished. The first one, represented by female V006 showed moderate but widespread Hes1 expression (Figure 6J), moderate estrogen receptor staining (Figure 6K), undetectable p63 expression (Figure 6L), normal E-cadherin distribution (Figure 6M), and a marked up-regulation of Ki67 and Hey1 expression (Figure 6N, O) in the tumor area. The second one, exemplified by female V015, showed strong and widely distributed Hes1 expression (Figure 6P), relatively high estrogen receptor expression (Figure 6Q) and undetectable p63 expression (Figure 6R) in the tumor. E-cadherin was found in both membrane and cytoplasm (Figure 6S), while Ki67 and Hey1 expression was strong (Figure 6T, U). The third pattern was represented by female V093, in which Hes1 expression was widespread but not as much as in the V015 female (Figure 6V), moderate estrogen receptor expression (Figure 6W), very low or absent p63 staining (Figure 6X), with relatively normal E-cadherin expression (Figure 6Y) and Ki67 and Hey1 expression was moderate (Figure 6Z, A'). In general, our data showed that Hes1 and Hey1, as markers of Notch1 activation in epithelial tissues, coexisted with higher estrogen receptor expression. These results are compatible with those obtained with the xenografts of inducible MCF-7-N1ICD cells, whose growth was dependent on N1ICD and estrogens (Figure 4A, B). This observation, together with the fact that tumors were only observed after three to four pregnancies, suggested that the involvement of Notch in breast tumor formation was strongly dependent on estrogens and that Notch expression could lead to changes in estrogen receptor expression.
Figure 6.
MMTV-Cre; N1ICD transgenic mice develop papillary tumors. (A-C) H&E stainings; (D-H, J-N, P-T, V-Z) immunostainings; (I, O, U, A') in situ hybridization. In (D-G, J-M, P-S,V-Y), sections are counterstained with toluidine blue; in (H, N, T, Z) sections are counterstained with Mayer's Hematoxylin. (A) Normal lactating breast. Greatly expanded secretory lobules (arrows) containing milky secretion are surrounded by breast epithelium (small arrows). Adipose tissue containing lipid vesicles (asterisk) is also abundant. (B) Transgenic female V004. Tumor tissue coexisting with normal tissue is demarcated by a blue dashed line. Breast architecture is widely disorganized. (C) Transgenic female V006. Large necrotic areas (yellow dash line) are observed. Inset shows mitosis (arrows) in tumor tissue. (D) Hes1 expression (brown nuclei, arrow) is rare in normal breast epithelium. Asterisk indicates milky secretion. (E, F) estrogen receptor (brown nuclei, arrows in E) and p63 expression (brown nuclei, arrows in F) in normal breast epithelium. (G) E-cadherin expression in the membrane of breast epithelial cells. (H) Few signs of proliferation in normal lactating breast as indicated by Ki67 staining in only a few cells (brown nuclei, arrows). (I) Normal lactating breasts show no Hey1 expression. (J-O) V006 transgenic female. Tumor area is demarcated by a red dashed line. Note the increased Hes1 expression (J, brown nuclei, arrows). (K) estrogen receptor expression (arrows). (L) p63 expression is observed in the non-pathological tissue (arrow) surrounding the tumor. (M) E-cadherin is expressed in membrane of epithelium. (N) Proliferation can be observed throughout the tumor by Ki67 staining (brown nuclei, arrows). (O) Expression of Hey1 in tumor tissue (arrow). (P-U) V015 transgenic female. Normal tissue is demarcated by green dashed line. (P) Hes1 expression is strongly up-regulated in tumor cells. (Q) Estrogen receptor (arrows) expression is more widespread. (R) p63 expression is very reduced in tumor tissue. (S) E-cadherin is expressed in membrane (arrow) but also in cytoplasm (arrowheads) of tumor cells. (T) Strong proliferation shown by Ki67 staining (arrows). (U) Hey1 expression in tumor (arrows). (V-A') V093 transgenic female. (V) Widespread Hes1 expression in tumor (brown nuclei, arrows). Estrogen receptor (W, arrows) and p63 expression (X, arrows) in tumor. (Y) E-cadherin expression in tumor (arrows). (Z) Strong proliferation in tumor shown by Ki67 staining (arrows). (A') Expression of Hey1 in tumor (arrow).
Discussion
There is considerable recent interest in understanding how NOTCH signaling affects the development or progression of breast cancer. Notch is critical in mammary gland development, probably by regulating mammary stem cell function [39]. In addition, NOTCH activity has been associated with a number of pro-tumorogenic activities in breast cancer cell lines, and could cause mammary hyperplasia and carcinogenesis in mice [11,40-42]. This evidence strongly pinpoints NOTCH receptors and/or ligands as targets in breast cancer.
Here we used three different in vitro and in vivo models to analyze the impact of NOTCH signaling in the onset and progression of breast tumors. We found a positive association of NOTCH activity with cancer growth or initiation. In agreement with others [43-46], one of the most consistent observations along our study was the association between NOTCH1 activity and E-CADHERIN down-regulation. First, stable expression of activated Notch (N1ICD) was associated with a reduction and delocalization of E-CADHERIN in most of the MCF-7 cell clones analyzed; second, the data with the inducible MCF-7-N1ICD clone B12 clearly established a causal relationship between active NOTCH1 and reduced E-CADHERIN levels; third, inhibition of endogenous NOTCH activation with DAPT in MDA-MB-231 cells, a highly invasive cell line that expresses high NOTCH1 levels, resulted in an increase of E-CADHERIN expression; and fourth, papillary tumors raised in MMTV-Cre/+;N1ICD/+ transgenic mice expressing high levels of Hes1 also showed, at least, a delocalization of E-cadherin in the epithelium. Repression and/or delocalization of E-CADHERIN is usually associated with adherens junctions disassembly [29] and enhanced cell invasiveness [33]. Concurrently, we observed that N1ICD-induced E-CADHERIN repression correlates with enhanced motility in transwell assays, whereas inhibition of Notch signaling, either by DAPT or RO inhibitors treatment or DN-CBF1 overexpression, reduced the motility of the invasive MDA-MB-231 cells. Collectively, these results indicate that NOTCH1 activation could induce EMT in epithelial tumor cells and, consequently, to favor tumor metastasis [47].
The role of Notch as a critical inducer of EMT has been demonstrated during the formation of the cardiac valve primordium [48]. In this process, Notch activates Snail expression that in turn down-regulates VE-cadherin [48]. We analyzed Snail1 expression in the MCF-7 clones expressing N1ICD, and found no clear correlation among N1ICD expression, E-CADHERIN down-regulation and SNAIL1 expression. We observed a markedly increased HES1 and HEY1 expression in response to NOTCH1 activation that correlated with a reduction in E-CADHERIN expression in both our cellular and animal models. Interestingly, both HES1 and HEY1 have been implicated as part of the hypoxic response associated to breast cancer progression [45].
We also analyzed whether NOTCH1 affects the growth and/or the onset of breast tumors. NOTCH signaling regulates the balance between cell proliferation, differentiation and apoptosis [2] and different reports have demonstrated that NOTCH triggers the proliferation of breast cancer cells [46,49]. In line with these observations, we found that the growth of subcutaneous and orthotopic xenografts produced with MCF-7-B12 cells was boosted after the induction of N1ICD (Figure 5). This effect was cell autonomous, since silencing of N1ICD-expressing tumors by administration of DOXY stopped the growth of tumors whereas induction of N1ICD by DOXY withdrawal boosted tumor growth, with kinetics compatible with N1ICD induction.
Although induction of N1ICD in MCF-7 fosters tumor growth, this effect was only observed when mice were treated with estrogens; indeed, MCF-7 xenografts did not grow in the absence of estrogens, independently of the induction of N1ICD. These results suggest that N1ICD cooperates with the estrogen receptor (ER) on tumor growth, as recently reported [50]. In agreement with this conclusion, MMTV-Cre/+;N1ICD/+ mice only developed mammary tumors after repeated pregnancies. It is noteworthy to mention that these breast tumors appeared in the lactating gland and regressed after gland involution; the regression was independent of N1ICD activity as determined by the Hes1 expression level. Our results thus resemble those obtained by Kiaris et al. [11], and contrast with the formation of non-regressing mammary neoplasm in transgenic mice expressing the active forms of Notch1, -3 and -4, reported by others [12,41,42]. In summary, our results confirm NOTCH1 as an EMT inducer in breast cancer cells, which may have implications in tumor growth, dissemination and metastasis. The identification of specific factors interacting with NOTCH signaling could thus be relevant to fully understand the role of NOTCH in breast neoplasia.
Conclusions
NOTCH1 activation attenuates E-CADHERIN expression and favors the motility and invasive ability of epithelial human breast cancer MCF-7 cells in vitro. In xenografts and in transgenic mice, NOTCH1 activation caused tumors whose increased growth is NOTCH- and estrogen receptor-dependent. To the contrary, NOTCH inhibition leads to increased E-CADHERIN expression and attenuates the migratory properties of invasive MDA-MB-231 breast cancer cells. Our findings in these mammary tumor models point to NOTCH1 as a potential therapeutic target in breast cancer onset and progression.
Abbreviations
CBF1: CSL: Suppressor of Hairless; CSL: CBF1: Suppressor of Hairless: Lag-1; DAB: 3,3'-Diaminobenzidine; DAPI: 4,6-Diamidino-2-phenylindol; DAPT: N-(N-(3,5-difluorophenacetyl)-l-alanyl)-S-phenylglycine t-butyl ester; DMSO: dimethyl sulfoxide; DOXY: doxycycline; ECAD: E-CADHERIN; EMT: epithelial-mesenchymal transition; EE2: 17α-ethynyl estradiol; ERα: estrogen receptor alpha; H&E: Hematoxylin and eosin; Hes1: hairy and enhancer of split 1; Hey1: hairy/enhancer-of-split related with YRPW motif one; HT-29: Human colon adenocarcinoma grade II cell line; IGF-1: Insulin growth factor one; MCF-7: Michigan Cancer Foundation-7 breast cancer cell line; MDA-MB-231: Breast cancer cell line derived from metastatic site (pleural effusion); MMTV-Cre: Mouse mammary Tumor Virus-driven Cre recombinase; N1ICD: Notch one intracellular domain; RO4929097: (2,2-dimethyl-N-(S)-6-oxo-6,7-dihydro-5H-dibenzo(b,d)azepin-7-yl)-N'-(2,2,3,3,3-pentafluoro-propyl)-malonamide); SEM: standard error of the mean; SMC3: Structural Maintenance of Chromomosomes-3 protein; SNA1: SNAIL1; SDF1α: Stromal cell Derived factor 1α; Tet-Off: tetracycline-Off system
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
VB, EM, BMP, GL and MC carried out all the experiments. VB, EM, BMP, MC, CMA, SM and JLdlP designed the experiments and analyzed the data. SM and JLdlP wrote the manuscript. All authors read and approved the manuscript.
Supplementary Material
Supplementary Materials and Methods.
Figure S1. NOTCH4 expression in the stable MCF-7 clones E8 and F7 compared to the mock MCF-7 cells, measured by qPCR. cDNA of HUVEC cells was used as positive control of NOTCH4 expressing cells. Data are mean ± SEM of triplicates in two independents experiments (***P<0.001 determined by student's t test).
Figure S2. (A) Western Blot showing the expression of N1ICD and E-cadherin in the stable HT-29 clones E11, E12, G12 and G9 compared with mock HT-29 cells; b-actin was used as loading control. (B) Western Blot was quantified by densitometry and the ratio of E-CADHERIN/β-ACTIN was calculated and referred to the mock cells.
Figure S3. The inducible clone MCF-7-B12 shows increased VIMENTIN expression while TWIST1 is unaffected upon N1ICD induction. (A) Western blot showing inducible N1ICD expression in MCF-7-B12 cells cultured in the presence (+Doxy) or absence (-Doxy) of doxycycline. VIMENTIN expression is increased. (B) Semiquantitative RT-PCR showing that TWIST1 transcription does not change after 7 days of N1ICD induction.
Figure S4. E-CADHERIN analysis of MCF-7 and MDA-MB-231 cells by flow cytometry. (A) E-CADHERIN staining of MCF-7 cells (black line) and MDA-MB-231 cells (dotted line) is shown. Ten-fold reduction of the mean fluorescence intensity (MnIX) is shown. Gray graph correspond to the staining of both cell lines with the corresponding IgG1 isotype control. (B) Raw data of the MCF-7 analysis (right) compared to the negative control (left). (C) Raw data of the MDA-MB-231 analysis (right) compared to the negative control. Note that there is a ten-fold reduction both in the staining intensity but also in the number of MDA-MB-231 cells staining for E-CADHERIN (79.6%) in comparison with MCF-7 (99.4%).
Table S1. Breast tumor formation in MMTV-Cre/+; N1ICD/+ females.
Contributor Information
Victoria Bolós, Email: vbolos@cnic.es.
Emilia Mira, Email: emira@cnb.csic.es.
Beatriz Martínez-Poveda, Email: bmartinez@cnic.es.
Guillermo Luxán, Email: gluxan@cnic.es.
Marta Cañamero, Email: mcanamero@cnio.es.
Carlos Martínez-A, Email: cmartineza@cnb.csic.es.
Santos Mañes, Email: smanes@cnb.csic.es.
José Luis de la Pompa, Email: jlpompa@cnic.es.
Acknowledgements
We thank Dr. Alberto Muñoz (IIB, CSIC, Madrid, Spain) for the HT-29 cell line and S. Bartlett (CNIC) for English editing. This work was supported by grants from the Spanish Ministry of Economy and Competitiveness (MINECO) or Comunidad de Madrid (CM). The grants were: SAF2010-21205 (MINECO) and MITICP2010/BMD-2502 (CM) to CMA, SAF2011-24453 (MINECO) and INMUNOTHERCAN S2010/BMD-2326 (CM) to SM and SAF2007-62445 and SAF2010-17555 (MINECO) to JLdlP. The CNIC is supported by the MINECO and the Pro-CNIC Foundation.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;15:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocr Rev. 2007;15:339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- Harrison H, Farnie G, Brennan KR, Clarke RB. Breast cancer stem cells: something out of notching? Cancer Res. 2010;15:8973–8976. doi: 10.1158/0008-5472.CAN-10-1559. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;15:649–661. doi: 10.1016/0092-8674(91)90111-B. [DOI] [PubMed] [Google Scholar]
- Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso CS, Lonigro RJ, Quist M, Siddiqui J, Mehra R, Jing X, Giordano TJ, Sabel MS, Kleer CG, Palanisamy N, Natrajan R, Lambros MB, Reis-Filho JS, Kumar-Sinha C, Chinnaiyan AM. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;15:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;15:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- Ronchini C, Capobianco AJ. Notch(ic)-ER chimeras display hormone-dependent transformation, nuclear accumulation, phosphorylation and CBF1 activation. Oncogene. 2000;15:3914–3924. doi: 10.1038/sj.onc.1203719. [DOI] [PubMed] [Google Scholar]
- Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;15:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 2005;15:1485–1495. doi: 10.1101/gad.341405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dievart A, Beaulieu N, Jolicoeur P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 1999;15:5973–5981. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- Kiaris H, Politi K, Grimm LM, Szabolcs M, Fisher P, Efstratiadis A, Artavanis-Tsakonas S. Modulation of notch signaling elicits signature tumors and inhibits hras1-induced oncogenesis in the mouse mammary epithelium. Am J Pathol. 2004;15:695–705. doi: 10.1016/S0002-9440(10)63333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Dievart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;15:973–990. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinakis A, Szabolcs M, Politi K, Kiaris H, Artavanis-Tsakonas S, Efstratiadis A. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc Natl Acad Sci USA. 2006;15:9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, Rudolf M, Siziopikou K, Kast WM, Miele L. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;15:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- Milner LA, Bigas A, Kopan R, Brashem-Stein C, Bernstein ID, Martin DI. Inhibition of granulocytic differentiation by mNotch1. Proc Natl Acad Sci USA. 1996;15:13014–13019. doi: 10.1073/pnas.93.23.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen TJ, Letourneau PC, Costa Lda F, Schubert D. Modifier of cell adhesion regulates N-cadherin-mediated cell-cell adhesion and neurite outgrowth. J Neurosci. 2005;15:281–290. doi: 10.1523/JNEUROSCI.3692-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch [see comments] Nature. 1995;15:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- McKenzie GJ, Stevenson P, Ward G, Papadia S, Bading H, Chawla S, Privalsky M, Hardingham GE. Nuclear Ca2+ and CaM kinase IV specify hormonal- and Notch-responsiveness. J Neurochem. 2005;15:171–185. doi: 10.1111/j.1471-4159.2005.03010.x. [DOI] [PubMed] [Google Scholar]
- Kanzler B, Kuschert SJ, Liu YH, Mallo M. Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development. 1998;15:2587–2597. doi: 10.1242/dev.125.14.2587. [DOI] [PubMed] [Google Scholar]
- Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, Hennighausen L. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;15:545–553. doi: 10.1023/A:1013063514007. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;15:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soule HD, Vazguez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst. 1973;15:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- Reedijk M, Pinnaduwage D, Dickson BC, Mulligan AM, Zhang H, Bull SB, O'Malley FP, Egan SE, Andrulis IL. JAG1 expression is associated with a basal phenotype and recurrence in lymph node-negative breast cancer. Breast Cancer Res Treat. 2008;15:439–448. doi: 10.1007/s10549-007-9805-3. [DOI] [PubMed] [Google Scholar]
- Callahan R, Raafat A. Notch signaling in mammary gland tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;15:23–36. doi: 10.1023/A:1009512414430. [DOI] [PubMed] [Google Scholar]
- Speiser J, Foreman K, Drinka E, Godellas C, Perez C, Salhadar A, Ersahin C, Rajan P. Notch-1 and Notch-4 biomarker expression in triple-negative breast cancer. Int J Surg Pathol. 2012;15:139–145. doi: 10.1177/1066896911427035. [DOI] [PubMed] [Google Scholar]
- Clementz AG, Rogowski A, Pandya K, Miele L, Osipo C. NOTCH-1 and NOTCH-4 are novel gene targets of PEA3 in breast cancer: novel therapeutic implications. Breast Cancer Res. 2011;15:R63. doi: 10.1186/bcr2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;15:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Luna-Zurita L, Prados B, Grego-Bessa J, Luxan G, del Monte G, Benguria A, Adams RH, Perez-Pomares JM, de la Pompa JL. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest. 2010;15:3493–3507. doi: 10.1172/JCI42666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud P, Shioi J, Serban G, Georgakopoulos A, Sarner S, Nagy V, Baki L, Wen P, Efthimiopoulos S, Shao Z, Wisniewski T, Robakis NK. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 2002;15:1948–1956. doi: 10.1093/emboj/21.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira E, Lacalle RA, Gonzalez MA, Gomez-Mouton C, Abad JL, Bernad A, Martinez AC, Manes S. A role for chemokine receptor transactivation in growth factor signaling. EMBO Rep. 2001;15:151–156. doi: 10.1093/embo-reports/kve027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D, Wang W, Xie H, Li Y, Dong G, Xu C, Chen D, Zheng J, Li M, Lu Z, Ji G. Notch1 expression in colorectal carcinoma determines tumor differentiation status. J Gastrointest Surg. 2009;15:253–260. doi: 10.1007/s11605-008-0689-2. [DOI] [PubMed] [Google Scholar]
- Noah TK, Shroyer NF. Notch in the intestine: regulation of homeostasis and pathogenesis. Annu Rev Physiol. 2013;15:263–288. doi: 10.1146/annurev-physiol-030212-183741. [DOI] [PubMed] [Google Scholar]
- Peinado H, Portillo F, Cano A. Transcriptional regulation of cadherins during development and carcinogenesis. Int J Dev Biol. 2004;15:365–375. doi: 10.1387/ijdb.041794hp. [DOI] [PubMed] [Google Scholar]
- Cailleau R, Young R, Olive M, Reeves WJ Jr. Breast tumor cell lines from pleural effusions. J Natl Cancer Inst. 1974;15:661–674. doi: 10.1093/jnci/53.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Latimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, Mutter LC, Nietz J, Quinn KP, Sacchi KL, Seubert PA, Shopp GM, Thorsett ED, Tung JS, Wu J, Yang S. et al. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem. 2001;15:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- Huynh C, Poliseno L, Segura MF, Medicherla R, Haimovic A, Menendez S, Shang S, Pavlick A, Shao Y, Darvishian F, Boylan JF, Osman I, Hernando E. The novel gamma secretase inhibitor RO4929097 reduces the tumor initiating potential of melanoma. PLoS One. 2011;15:e25264. doi: 10.1371/journal.pone.0025264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manes S, Mira E, Colomer R, Montero S, Real LM, Gomez-Mouton C, Jimenez-Baranda S, Garzon A, Lacalle RA, Harshman K, Ruíz A, Martínez-A C. CCR5 expression influences the progression of human breast cancer in a p53-dependent manner. J Exp Med. 2003;15:1381–1389. doi: 10.1084/jem.20030580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Silva A, Ramalho LN, Garcia SB, Zucoloto S. Does the correlation between EBNA-1 and p63 expression in breast carcinomas provide a clue to tumorigenesis in Epstein-Barr virus-related breast malignancies? Braz J Med Biol Res. 2004;15:89–95. doi: 10.1590/s0100-879x2004000100013. [DOI] [PubMed] [Google Scholar]
- Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;15:429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Callahan R, Egan SE. Notch signaling in mammary development and oncogenesis. J Mammary Gland Biol Neoplasia. 2004;15:145–163. doi: 10.1023/B:JOMG.0000037159.63644.81. [DOI] [PubMed] [Google Scholar]
- Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, Callahan R. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;15:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- Gallahan D, Jhappan C, Robinson G, Hennighausen L, Sharp R, Kordon E, Callahan R, Merlino G, Smith GH. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 1996;15:1775–1785. [PubMed] [Google Scholar]
- Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, Karsan A. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;15:2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad S, Stanners SR, Yong R, Tang O, Pollock CA. Notch mediated epithelial to mesenchymal transformation is associated with increased expression of the Snail transcription factor. Int J Biochem Cell Biol. 2010;15:1115–1122. doi: 10.1016/j.biocel.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Chen J, Imanaka N, Griffin JD. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br J Cancer. 2010;15:351–360. doi: 10.1038/sj.bjc.6605486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo P, Miao H, D'Souza G, Osipo C, Song LL, Yun J, Zhao H, Mascarenhas J, Wyatt D, Antico G, Hao L, Yao K, Rajan P, Hicks C, Siziopikou K, Selvaggi S, Bashir A, Bhandari D, Marchese A, Lendahl U, Qin JZ, Tonetti DA, Albain K, Nickoloff BJ, Miele L. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;15:5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. Erratum in Cancer Res 2008, 68:7246. Song, Lynda L [added] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon AE, LaBonne C. Tumor metastasis: a new twist on epithelial-mesenchymal transitions. Curr Biol. 2004;15:R719–721. doi: 10.1016/j.cub.2004.08.048. [DOI] [PubMed] [Google Scholar]
- Timmerman LA, Grego-Bessa J, Raya A, Bertrán E, Pérez-Pomares JM, Díez J, Aranda S, Palomo S, McCormick F, Izpisúa-Belmonte JC, de la Pompa JL. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;15:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, Shimizu K, Honma R, Yanagisawa Y, Nishikawa A, Kawamura M, Imai J, Ohwada S, Tatsuta K, Inoue J, Semba K, Watanabe S. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res. 2008;15:1881–1888. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- Hao L, Rizzo P, Osipo C, Pannuti A, Wyatt D, Cheung LW, Sonenshein G, Osborne BA, Miele L. Notch-1 activates estrogen receptor-alpha-dependent transcription via IKKalpha in breast cancer cells. Oncogene. 2010;15:201–213. doi: 10.1038/onc.2009.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods.
Figure S1. NOTCH4 expression in the stable MCF-7 clones E8 and F7 compared to the mock MCF-7 cells, measured by qPCR. cDNA of HUVEC cells was used as positive control of NOTCH4 expressing cells. Data are mean ± SEM of triplicates in two independents experiments (***P<0.001 determined by student's t test).
Figure S2. (A) Western Blot showing the expression of N1ICD and E-cadherin in the stable HT-29 clones E11, E12, G12 and G9 compared with mock HT-29 cells; b-actin was used as loading control. (B) Western Blot was quantified by densitometry and the ratio of E-CADHERIN/β-ACTIN was calculated and referred to the mock cells.
Figure S3. The inducible clone MCF-7-B12 shows increased VIMENTIN expression while TWIST1 is unaffected upon N1ICD induction. (A) Western blot showing inducible N1ICD expression in MCF-7-B12 cells cultured in the presence (+Doxy) or absence (-Doxy) of doxycycline. VIMENTIN expression is increased. (B) Semiquantitative RT-PCR showing that TWIST1 transcription does not change after 7 days of N1ICD induction.
Figure S4. E-CADHERIN analysis of MCF-7 and MDA-MB-231 cells by flow cytometry. (A) E-CADHERIN staining of MCF-7 cells (black line) and MDA-MB-231 cells (dotted line) is shown. Ten-fold reduction of the mean fluorescence intensity (MnIX) is shown. Gray graph correspond to the staining of both cell lines with the corresponding IgG1 isotype control. (B) Raw data of the MCF-7 analysis (right) compared to the negative control (left). (C) Raw data of the MDA-MB-231 analysis (right) compared to the negative control. Note that there is a ten-fold reduction both in the staining intensity but also in the number of MDA-MB-231 cells staining for E-CADHERIN (79.6%) in comparison with MCF-7 (99.4%).
Table S1. Breast tumor formation in MMTV-Cre/+; N1ICD/+ females.