Abstract
Background & objectives:
Trisomy 21 is the most common chromosomal aneuploidy in live born infants. Recently, the over expression of chromosome 21-derived microRNAs (miR-99a, let-7c, miR-125b-2, miR-155 and miR-802) in human fetal hippocampus and heart samples from individuals with Down syndrome was observed. Therefore, concentrations and expression profile of extracellular chromosome 21-derived microRNAs were studied to verify their ability to distinguish noninvasively between pregnancies bearing euploid fetuses and those affected with Down syndrome.
Methods:
RNA enriched for small RNAs was isolated from plasma samples of 12 pregnant women with high risk of bearing Down syndrome foetuses (median gestation 18.5 wk), 12 women with normal course of gestation and 10 non-pregnant women. MicroRNA transcribed into cDNA using specific stem-loop primer was detected using real-time PCR assay. Simulation experiments using RNA pools of healthy non-pregnant individuals and aneuploid amniotic fluid samples in descending dilution ratio ranging from 1:1 to 1000:1 were used to test the detection limit of the technique for overexpressed chromosome 21-derived microRNAs specific for Down syndrome. The expression profile of the gene encoding microRNA was studied through the relative gene expression using the comparative Ct (threshold cycle) method. Concentrations of individual microRNAs were subtracted from the calibration curves in the course of analyses and expressed as pg of total RNA per milliliter of plasma.
Results:
Four of the five extracellular chromosome 21-derived microRNAs (miR-99a, let-7c, miR-125b-2 and miR-155) were reliably detected in plasma samples. Simulation experiments revealed the detection limit of aneuploidy at a ratio 100:1 for let-7c, miR-125b-2 and miR-155, and a ratio of 1000:1 for miR-99a. Overexpression of extracellular miR-99a, miR-125b-2 and miR-155 was observed in pregnant women compared to non-pregnant women. Similarly, increased concentrations of extracellular miR-99a and miR-125b-2 were detected in pregnant women than in non-pregnant women. The concentrations and relative gene expression of extracellular chromosome 21-derived microRNAs did not differ between the cohorts of pregnancies bearing euploid foetuses and those affected with Down syndrome.
Interpretation & conclusions:
Analysis of extracellular chromosome 21-derived microRNAs has no benefit for screening programmes and non-invasive diagnosis of Down syndrome.
Keywords: Aneuploidy, chromosome 21, Down syndrome, maternal plasma, microRNAs, real-time PCR, screening
Down syndrome (DS) occurs with an incidence of approximately 1 in 800 births in the general population1, but this risk increases to 1 in 35 term births for women 45 yr of age2. Screening programmes to identify women at increased risk of bearing a foetus with DS are based on maternal age, ultrasound findings3 and/or biochemical markers4. First trimester screening for DS is usually performed in gestational week 10-14 using serum markers such as pregnancy associated plasma protein A (PAPP-A) and the free β-form of human chorionic gonadotrophin (β-hCG), combined with a measurement of the nuchal translucency thickness5. However, this approach still results in a high proportion of false positives (2-4%) and false negatives (5-10%)5. Screening during the second trimester based on the examination of biochemical markers like α-foetoprotein (AFP), unconjugated estriol-3 (uE3) and β-hCG detects 68-75 per cent of pregnancies complicated by foetal DS, with a 5 per cent false-positive rate4. The involvement of additional ultrasound markers (reduction or absence of the nasal bone6; presence of tricuspid regurgitation)7 and biochemical markers8,9 usually leads to improved screening performance in the first and/or second trimester of gestation.
Currently, the definitive diagnosis of DS is based on an examination of the foetal genome after acquisition of foetal cells directly from the uterus using invasive procedures, such as chorionic villous sampling (CVS) and/or amniocentesis. Unfortunately, these procedures are still associated with a 1 per cent risk of miscarriage10. Therefore, much effort is devoted to the development of reliable non-invasive methods for prenatal diagnosis (NIPD) based on the detection of extracellular nucleic acids (DNA11, mRNA12,13 and microRNA14,15 of foetal and/or placental origin in maternal circulation. In general, extracellular nucleic acids are those which are present in various biological fluid samples, including serum, plasma, saliva, urine, amniotic fluid, semen, milk and bronchial lavage16.
The discovery of cell-free foetal DNA11 and placental mRNA12 in maternal plasma was swiftly followed by non-invasive clinical applications for foetal gender12 and blood Rhesus factor status17, exclusion and/or confirmation of paternally inherited alleles for various single gene disorders18 and eventually assessment of chromosomal aneuploidies using massively parallel sequencing19. Extracellular pregnancy associated microRNAs have been identified in maternal circulation throughout gestation15. Our group has20,21 introduced a profile of extracellular placental specific microRNAs capable of reliably differentiating between pregnant and non-pregnant women and the diagnostic potential to predict placental insufficiency related pregnancy complications.
Chromosome 21 harbours more than 500 annotated genes22, of which, 170 are protein coding, and five represent microRNA genes, including miR-99a, let-7c, miR-125b-2, miR-155 and miR-80223. We hypothesized that chromosome 21-derived miRNAs of foetal origin may be detected in maternal circulation throughout gestation, given that foetal cells are continuously trafficking across the placenta24 and the placenta itself is actively releasing microvesicles, exosomes and apoptotic bodies of placental trophoblasts which contain extracellular nucleic acids of foetal origin25. This hypothesis was partially built on the observation of Elton et al26, who demonstrated abundant expression of miR-99a and miR-125b in human tissues inclusive of placental tissue. Furthermore, we hypothesized that the presence of redundant chromosome 21 and an overexpression of chromosome 21-derived miRNAs in the tissues of patients with Down syndrome might enable differentiation between plasma samples derived from pregnancies with euploid and aneuploid foetuses. Initially, we performed Down syndrome simulation experiments by testing artificial mixtures of RNA isolated from plasma samples of healthy individuals and RNA enriched from the foetus affected by Down syndrome in a step-by-step descending representation of aneuploid genetic material in a mixed sample. The purpose of these initial experiments was to assess the detection limit of aneuploidy for individual chromosome-21 derived microRNAs using real-time PCR. Further, we investigated concentrations and expression profile of chromosome 21-derived microRNAs in maternal plasma and the ability of this approach to distinguish between euploid foetus pregnancies and those affected with Down syndrome.
Material & Methods
The study was performed in retrospective manner at the Department of Moelcular Biology and Cell Pathology (Third Faculty of Medicine, Charles University, Prague).
Amniotic fluids were collected at the Department of Biology and Medical Genetics (University Hospital Motol, Prague) from pregnant patients for whom invasive procedures were indicated due to higher risks (maternal age greater than 35 yr and positive results from second trimester biochemical and ultrasound screenings). Five pregnancies with fetuses identified as possessing the karyotype associated with Down syndrome (47, XX, +21 and 47, XY, +21) were involved in the study.
Individual plasma samples were collected at cooperating gynaecological clinics (from September 2005 to June 2006, Clinic of Obstetrics and Gynaecology, First Faculty of Medicine, Charles University, Prague; from February 2008 to November 2010, Institute for the Care of the Mother and Child, Prague) from patients identified to be at a higher risk of bearing Down syndrome fetuses, and these samples were kept frozen at -80°C until further usage. Simultaneously, plasma samples were also collected from normal pregnancies and non-pregnant women (Clinic of Obstetrics and Gynaecology, University Hospital Motol, Prague). Plasma samples with approximately equal storage times were selected for use in the study: normal pregnancies (storage period: range 16-75 months; median storage time 59.0 months); DS-affected pregnancies (storage period: range 17-79 months; median storage time 65.5 months); non-pregnant individuals (storage period: range 16-75 months; median storage time 59.0 months).
Finally, plasma samples from 12 pregnant women aged 26-42 (mean 34.2 yr) with Down syndrome foetuses were collected within 14-22 wk of gestation (mean 18.2 wk; median 18.5 wk) before the performance of any invasive diagnostic procedure. The control group consisted of 12 maternal age-matched and gestational age-matched plasma women with uncomplicated singleton pregnancies, who gave birth to healthy neonates. Plasma samples derived from 10 healthy age-matched non-pregnant women were also included in the study. All women signed informed consent and the study protocol was approved by the University Ethics Committee of the Third Faculty of Medicine, Charles University in Prague.
Processing of samples and RNA extraction and enrichment for microRNAs: Peripheral blood samples (9 ml) were collected into EDTA tubes and centrifuged twice at 1200 g for 10 min at room temperature. Total RNA from non-pregnant women, normal and DS-affected pregnancies was extracted from 1 ml of plasma and 25 mg of normal placental tissue preserved in RNAlater (Ambion, Austin, TX, USA). An enrichment procedure for small RNAs was performed using a mirVana microRNA Isolation kit (Ambion, Austin, TX, USA). Trizol LS reagent (Invitrogen, Carlsbad, CA, USA) was used to isolate total RNA in plasma samples. RNA from cultured amniotic fluid-derived cells was isolated using QIAamp RNA Blood Mini Kit (Qiagen, Hilden, Germany). DNase treatment, with 5 μl of DNase I with a concentration of 1 unit/μl (Fermentas International, Ontario, Canada) for 30 min at 37 °C, was carried out to remove contaminating DNA.
Reverse transcription reaction using a stem-loop primer: Chromosome 21-derived microRNAs (miR-99a, let-7c, miR-125b-2, miR-155 and miR-802) and selected reference microRNAs (miR-16 and let-7d)21. were reversely transcribed using microRNA-specific stem-loop RT primers (TaqMan MicroRNA Assays, Applied Biosystems, USA) in a total reaction volume of 20 μl using 7500 real-time PCR system (Applied Biosystems, USA) with the following thermal cycling parameters: 30 min at 16°C, 30 min at 42°C, 5 min at 85°C, and then held at 4°C. The recovery of total RNA enriched for small RNAs differed between plasma samples (range 1.1-7.4 ng/μl). Finally, 6.7 μl of the RNA template was used for each RT reaction. The following TaqMan MicroRNA assays were used for the study: miR-99a assay ID: 000435; let-7c assay ID: 000379; miR-125b-2 assay ID: 000449; miR-155 assay ID: 002623; miR-802 assay ID: 002004; miR-16 assay ID: 000391 and let-7d assay ID: 002283.
Absolute and relative quantification of extracellular microRNAs: A volume of 4.4 μl of the RT product was used as a template for each real-time PCR. cDNA (complementary DNA), corresponding to the particular microRNA was detected using TaqMan MicroRNA assay and the TaqMan Universal PCR Master Mix (Applied Biosystems, USA) in a total reaction volume of 35 μl on a 7500 Real-Time PCR System. Analyses were performed in three replicates. A sample was considered positive if the amplification signal occurred before the 40th threshold cycle (Ct <40).
Randomly selected euploid placental tissue derived from gestation with normal course was used as a reference sample for absolute and relative quantification. An RNA fraction highly enriched for small RNAs was isolated from the foetal part of the placenta. The calibration curves plotted the threshold cycle (Ct) of individual microRNA against known concentrations of serially diluted RNA samples. Concentrations of individual microRNAs were expressed as pg of total RNA enriched for small RNAs per milliliter of plasma.
Comparative Ct method compares the Ct values of the samples of interest with a reference sample such as RNA sample from normal tissue. The Ct values of both the reference sample and the samples of interest are normalized to an appropriate endogenous housekeeping gene. The comparative Ct method is also known as the 2-[delta][delta]Ct method, where
[delta][delta]Ct = [delta] Ct, sample - [delta] Ct, reference
Here, [delta] CT, sample is the Ct value for any sample normalized to the endogenous housekeeping gene and [delta] Ct, reference is the Ct value for the reference sample also normalized to the endogenous housekeeping gene.
The relative gene expression of particular microRNA in plasma samples was determined using the comparative Ct method27 (2-(∆∆Ct)) relative to normalization factor (geometric mean of ubiquitous miR-16 and let-7d)21. Comparative Ct method compares the Ct values of the samples of interest with a reference sample such as RNA sample from normal placental tissue that was used throughout the whole study.
Statistical analysis: Absolute and relative quantification of particular microRNAs was compared between individual groups using the Mann-Whitney U-test (Statistica software, StatSoft, Inc., USA). Since the Bonferroni correction was used to address the problem of multiple comparisons, the significance level was established at a P<0.025.
Correlation between variables including absolute quantification of individual extracellular microRNAs and storage life was calculated using the Spearman's rank correlation coefficient (ρ). Significant levels of correlation were assumed at P <0.05.
Results
Failure of TaqMan assay for miR-802: In RNA samples isolated from placental tissues chromosome 21-derived microRNAs (let-7c, miR-99a, miR-125b-2 and miR-155) were amplified as expected, shortly afterwards ubiquitous microRNAs (miR-16 and let-7d), within the range of 16.8-24.0 threshold cycle (let-7c: range Ct 20.4-21.6; miR-99a: range Ct 17.4-18.1; miR-125b-2: range Ct 16.8-17.4 and miR-155: range Ct 23.1-24.0, respectively). However, miR-802 was amplified in placental tissues much later (range Ct 32.1-32.6) than other chromosome 21-derived microRNAs.
Plasma samples derived from normal pregnancies produced amplification curves in let-7c, miR-99a, miR-125b-2 and miR-155 within the Ct 26.5-33.4 (let-7c: range Ct 27.5-32.6; miR-99a: range Ct 26.9-33.4; miR-125b-2: range Ct 26.5-32.5 and miR-155: range Ct 29.2-33.4, respectively). In plasma samples derived from normal pregnancies, miR-802 produced amplification curves beyond Ct 40. Therefore, plasma samples were considered negative for miR-802. All consecutive attempts to optimize the conditions for miR-802 commercial assay (increase of RNA input and reaction volume, etc.) failed; as a result miR-802 was excluded from further testing.
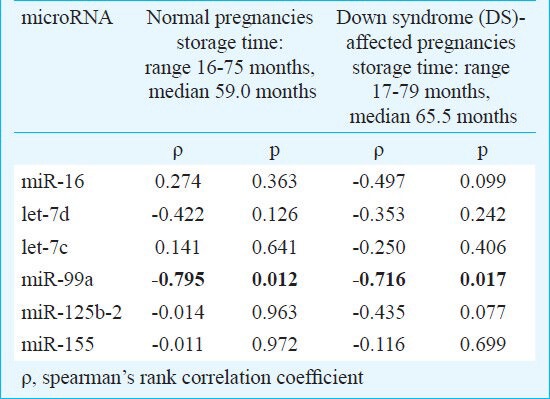
Stability of extracellular microRNAs: Initially, the stability of extracellular microRNAs was investigated. The patients were subdivided into individual groups: aneuploid (Down syndrom-affected) pregnancies, euploid pregnancies and non pregnant women. No effect of the long-term storage on the levels of extracellular chromosome 21-derived and ubiquitous microRNAs was indicated in any studied group except for miR-99a, where a strong negative correlation between plasma concentrations in the cohort of euploid and aneuploid pregnancies and advancing storage time was observed (Table). While the plasma levels of the five studied microRNAs remained stable, a significant decline of extracellular miR-99a associated with the long-term storage of plasma samples was observed in the cohort of euploid and aneuploid pregnancies. In the cohort of non-pregnant individuals, similar results were also obtained (data not shown).
Table.
Stability of extracellular microRNAs
Simulation experiments of Down syndrome - identification of redundant foetal derived chromosome 21 specific microRNAs in euploid maternal plasma throughout gestation: The expression profile of chromosome 21-derived microRNAs in genetic material of foetuses affected with Down syndrome was studied. Chromosome 21-derived microRNAs were overexpressed in cultured amniotic fluid-derived cells from foetuses affected with DS (miR-99a: range 1.34-3.77 fold; let-7c: range 2.65-3.86 fold; miR-125b-2: range: 1.22-2.5 fold; miR-155: range 76.9–274.3 fold) when compared with the reference (euploid placental tissue) used throughout the entire study to calculate 2-(∆∆Ct) relative gene expression. The expression of chromosome 21-derived microRNAs in the amniotic fluid of euploid foetuses was equal compared to the reference euploid placental tissue in all examined cases: the values of 2-(∆∆Ct) were around 1.0.
Minimal expression of chromosome 21-derived microRNAs was identified in plasma samples derived from healthy non-pregnant individuals compared with reference euploid sample (miR-99a: range 0.0001-0.0007 fold; let-7c: range 0.02-0.07 fold; miR-125b-2: range: 0.0005-0.0009 fold; miR-155: range 0.01-0.03 fold).
Artificial mixtures were prepared from RNA samples isolated from plasma of healthy non-pregnant women and cultured amniotic fluid-derived cells from foetuses affected with DS. The RNA samples derived from healthy non-pregnant women were always used in invariant concentrations, while RNA samples of DS foetuses were diluted stage-by-stage using the decimal scale. The main aim of these experiments was to assess the detection limit of the technique. The maximal dilution of DS derived RNA samples that would always produce reliable detection of trisomy 21 was established.
Relative quantification approach reliably differentiated between plasma samples derived from healthy non-pregnant women and mixed samples containing cultured amniotic fluid-derived cells from DS foetuses up to a ratio of 100:1 for let-7c, miR-125b-2 and miR-155, and up to 1000:1 for miR-99a. In case of miR-155 significant overexpression was observed in cultured amniotic fluid-derived cells from foetus affected with DS (76.9 fold) when compared with reference sample. On the other hand, no miR-155 expression was found in plasma samples derived from non-pregnant women (0.01 fold) when compared with reference sample. Artificial mixed samples containing always the equal amount of euploid RNA sample and the variable amount of aneuploid RNA sample demonstrated overexpression of miR-155. Thus, 31.56- fold expression of miR-155 was detected in a mixture containing the equal amount of euploid and aneuploid sample (ratio 1:1). Ten-fold dilution of aneuploid RNA sample showed 3.28 fold of miR-155 expression (ratio 10:1). In case of 100-fold dilution of aneuploid RNA sample (ratio 100:1) miR-155 expression value was 0.27. On the other hand, the ratio 1000:1 was not able to detect redundant chromosome 21 specific microRNA in artificial mixture. MiR-155 expression reached the comparable value (0.018) as plasma sample derived from non-pregnant individual (0.015).
Absolute and relative quantification of extracellular microRNAs in non-pregnant individuals, normal pregnancies and those bearing DS foetuses: The concentrations and expression profiles of chromosome 21-derived microRNAs (miR-99a, let-7c, miR-125b-2, miR-155) were determined in plasma samples derived from non-pregnant women, normal pregnancies and those bearing foetuses with DS.
The levels of extracellular miR-99a and miR-125b-2 were significantly higher in pregnant women than in non-pregnant individuals (Fig. 1). Comparable levels of ubiquitous extracellular microRNAs (miR-16 and let-7d) were found in all plasma samples, regardless of the occurrence and the course of gestation. Chromosome 21-derived microRNAs (miR-99a and miR-125b-2) differentiated between pregnant and non-pregnant women. The overexpression of extracellular miR-99a, miR-125b-2 and miR-155 was identified in the group of pregnant women compared with non-pregnant women (Fig. 2). MiR-99a, miR-125b-2 and miR-155 differentiated between pregnant and non-pregnant women. The overexpression of extracellular miR-99a, miR-125b-2 and miR-155 was identified in pregnant women. No difference in chromosome 21-derived microRNA expression profile was observed between the women with DS-bearing pregnancies and gestational-age-matched normal pregnancies with euploid foetus. Unfortunately, both plasma concentrations and expression profiles of chromosome 21-derived microRNAs were unable to differentiate between DS-bearing pregnancies and gestational-age-matched normal pregnancies with euploid foetuses.
Fig. 1.
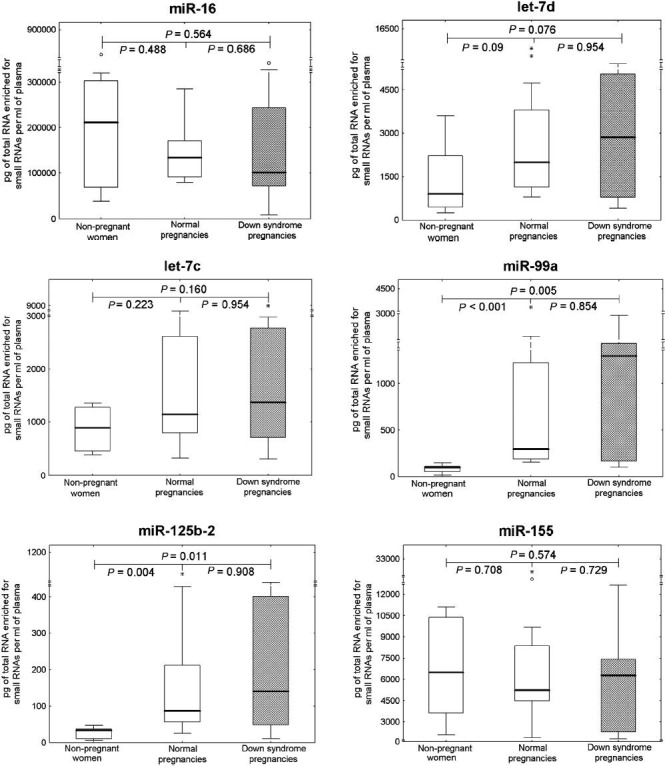
Absolute quantification of ubiquitous and chromosome 21-derived extracellular microRNAs in non-pregnant women, normal pregnancies and those bearing DS foetuses. Concentrations of extracellular microRNAs in plasma samples were subtracted from the standard curve and expressed in pg of total RNA enriched for small RNAs per millilitre of plasma. Absolute quantification of particular microRNAs was compared between the individual groups using the Mann-Whitney U-test (P<0.25). *, extremes; ○, outliers.
Fig. 2.
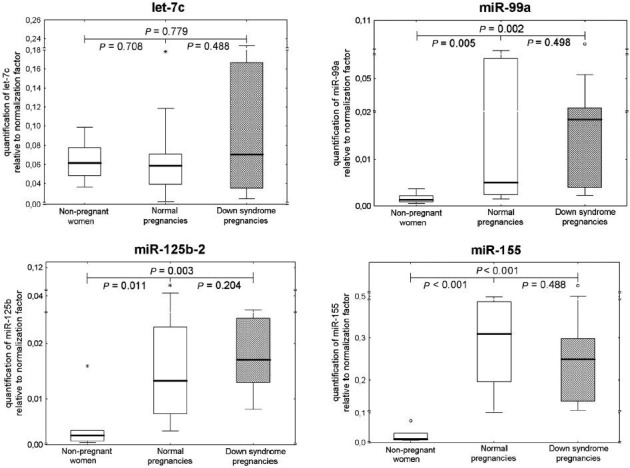
Relative quantification of chromosome 21-derived extracellular microRNAs in non-pregnant women, normal pregnancies and those bearing DS foetuses. Expression profile of chromosome 21-derived microRNAs in plasma samples was determined using comparative Ct method relative to normalization factor (geometric mean of ubiquitous invariable miR-16 and let-7d). The reference sample (euploid placental tissue) was used throughout the whole study to calculate 2-(ΔΔCt) relative gene expression. *, extremes; ○, outliers.
Discussion
The number of pregnant women who are candidates to undergo invasive testing for foetal chromosomal abnormalities, including Down syndrome, has increased with the advance of modern, more complex screening algorithms. There remains the traditional risk group of women who are ≥35 yr at term, and this group has grown significantly during the last 20 years2,3. These women are faced with the decision to undergo an invasive sampling of foetal genetic material, which still carries a risk of miscarriage10. Therefore, to reduce the risk of miscarriage and allow early testing for DS, reliable and practical methods of non-invasive prenatal diagnosis (NIPD) have been a continuous challenge.
Kuhn et al23 demonstrated that mature miR-99a, let-7c, miR-125b-2, miR-155 and miR-802, which are encoded by genes harboured on chromosome 21, were overexpressed in human foetal hippocampus and heart samples in individuals with DS. Because the final result of miRNA-mediated gene regulation is a reduction in the total amount of target protein that is produced, they speculated that trisomy 21-induced overexpression of chromosome 21-derived miRNAs might result in decreased expression of specific target proteins and contribute, in part, to features of the DS phenotype23. This hypothesis was later verified, by the same group, in experiments that identified important chromosome 21-derived microRNA/mRNA target pairs and demonstrated that miR-155 and miR-802 could regulate the expression of methyl-CpG-binding protein (MeCP2) that was underexpressed in DS brain specimens. This repression may contribute to the abnormality in the neurochemistry observed in the brains of DS individuals28.
In this study, we investigated maternal plasma concentrations and expression levels of chromosome 21-derived microRNAs (miR-99a, let-7c, miR-125b-2, miR-155 and miR-802) in DS pregnancies to verify their application for non-invasive DS diagnosis. Initially, we confirmed the presence of all chromosome 21-derived microRNAs in placental tissues. Later, we also demonstrated the occurrence of extracellular chromosome 21-derived microRNAs like let-7c, miR-99a, miR-125b-2 and miR-155 in the circulation of pregnant and non-pregnant women. MiR-802, whose amplification signals occurred in most plasma samples beyond Ct 40.0, was considered negative. Elton et al26 reported abundant expression of let-7c, miR-99a and miR-125b-2 in number of tissues including DS relevant tissues, the brain and the heart. This group also reported abundant expression of miR-99a and miR-125b-2 in placental tissues. In contrast, miR-155 and miR-802 were observed to be expressed at much lower levels with specific expression in the thymus, colon and small intestine, respectively.
Kuhn et al23 observed overexpression of chromosome 21-derived microRNAs in human foetal hippocampus and heart samples from individuals with DS; we confirmed this when we found chromosome 21-derived microRNAs overexpression in DS derived cultured amniotic fluid cells.
To establish the detection limit of real-time PCR technique, simulation experiments were performed in which RNA isolated from plasma samples of healthy non-pregnant women was mixed with cultured amniotic fluid-derived cells from DS foetuses in various dilution ratios ranging from 1:1 to 1000:1. As expected, this approach confirmed decreasing amounts of the tested chromosome 21-derived microRNAs in the artificial mixture with descending representation of overexpressed aneuploid RNA. It was possible to distinguish between plasma samples derived from healthy non-pregnant women and the same samples mixed with RNA from cultured amniotic fluid-derived cells from DS foetuses at a ratio of 100:1 for let-7c, miR-125b-2 and miR-155, and a ratio of 1000:1 for miR-99a.
Our study showed that the plasma concentrations of miR-99a and miR-125b-2 and expression levels of miR-99a, miR-125b-2 and miR-155 were significantly increased in pregnant women compared with non-pregnant individuals. On the other hand, the concentration and expression level of extracellular let-7c remained unchanged signifying that the state of let-7c is probably pregnancy independent. These observations were supported by several independent findings. Gilad et al14 reported that serum microRNAs might be used as promising new biomarkers for differentiation between pregnant and non-pregnant women. These biomarkers primarily involved placental specific microRNAs levels which usually rise as pregnancy progresses due to placental growth14,20,21. The levels of some extracellular ubiquitous microRNAs that are certainly not unique to pregnancy, were also reported to have increased in maternal circulation throughout gestation. For this reason, these may differentiate between pregnant and non-pregnant women as well15,29. Since Elton et al26 reported abundant expression of miR-99a and miR-125b-2 in a number of human tissues (including placental tissues) we expected and finally confirmed that their levels were higher in maternal circulation compared to non-pregnant women. In concordance with Gilad et al14, we observed similar abundance of let-7d (encoded by a gene located on chromosome 9) across the tested pregnant and non-pregnant women and for that reason let-7d was one of the microRNAs chosen for normalization. However, consecutive analysis of extracellular chromosome 21-derived microRNAs did not distinguish between pregnancies with euploid and aneuploid foetuses. It was found that the concentrations and expression levels of chromosome 21-derived microRNAs (miR-99a, let-7c, miR-125b-2, and miR-155) were comparable between maternal plasma samples regardless of the course of gestation and foetal karyotype. We believe that pregnancy itself is associated with upregulation in extracellular chromosome 21-derived microRNAs (miR-99a, miR-125b-2 and miR-155). This phenomenon might cause the inability of extracellular chromosome 21-derived microRNAs to differentiate between pregnancies bearing euploid and aneuploid foetuses solely on the basis of redundant chromosome 21 being present in DS foetuses.
There have been no reports to demonstrate the application of extracellular chromosome 21-derived microRNAs for the purpose of non-invasive prenatal diagnosis of Down syndrome in the foetus. Currently, there are intense efforts to improve the current screening programmes; however, further work needs to be done to detect and diagnose DS non-invasively, especially in the early stages of gestation. The ultimate goal remains the introduction of NIPD tests that are rapid, inexpensive, automated and accurate. Even if extracellular chromosome 21-derived microRNAs have no diagnostic value for the non-invasive prenatal diagnosis of Down syndrome, these do represent promising biomarkers for occult hepatitis B virus infection screening30 and non-small cell lung cancer31.
Acknowledgment
This work was supported by GAUK (Grant Agency of Charles University, Czech Republic) no. 434011 and PRVOUK (Program for Progress of Scientific Fields on Charles University, Czech Republic) P32.
References
- 1.Centers for Disease Control and Prevention. Improved national prevalence estimates for 18 selected major birth defects-United States, 1999-2001. Morb Mortal Wkly Rep. 2006;54:1301–5. [PubMed] [Google Scholar]
- 2.Savva GM, Morris JK, Mutton DE, Alberman E. Maternal age-specific fetal loos rates in Down syndrome pregnancies. Prenatal Diagn. 2006;26:499–504. doi: 10.1002/pd.1443. [DOI] [PubMed] [Google Scholar]
- 3.Nicolaides KH. Screening for chromosomal defects. Ultrasound Obstet Gynecol. 2003;21:313–21. doi: 10.1002/uog.128. [DOI] [PubMed] [Google Scholar]
- 4.Malone FD, Canick JA, Ball RH, Nyberg DA, Comstock CH, Bukowski R, et al. First-trimester or second-trimester screening, or both, for Down's syndrome. N Engl J Med. 2005;353:2001–11. doi: 10.1056/NEJMoa043693. [DOI] [PubMed] [Google Scholar]
- 5.Nicolaides KH, Spencer K, Avgidou K, Faiola S, Falcon O. Multicenter study of first-trimester screening for trisomy 21 in 75 821 pregnancies: results and estimation of the potential impact of individual risk-orientated two-stage first-trimester screening. Ultrasound Obstet Gynecol. 2005;25:221–6. doi: 10.1002/uog.1860. [DOI] [PubMed] [Google Scholar]
- 6.Kagen KO, Cicero S, Staboulidou I, Wright D, Nicolaides KH. Fetal nasal bone in screening for trisomies 21, 18 and 13 and Turner syndrome at 11-13 weeks of gestation. Ultrasound Obstet Gynecol. 2009;33:259–64. doi: 10.1002/uog.6318. [DOI] [PubMed] [Google Scholar]
- 7.Falcon O, Auer M, Gerovassili A, Spencer K, Nicolaides KH. Screening for trisomy 21 by fetal tricuspid regurgitation, nuchal translucency and maternal serum free beta-hCG and PAPP-A at 11+0 to 13+6 weeks. Ultrasound Obstet Gynecol. 2006;27:151–5. doi: 10.1002/uog.2699. [DOI] [PubMed] [Google Scholar]
- 8.Christiansen M, Norgaard-Pedersen B. Inhibin A is a maternal serum marker for Down's syndrome early in the first trimester. Clin Genet. 2005;68:35–9. doi: 10.1111/j.1399-0004.2005.00441.x. [DOI] [PubMed] [Google Scholar]
- 9.Christiansen M, Sorensen TL, Norgaard-Pedersen B. Human placental lactogen is a first-trimester maternal serum marker for Down syndrome. Prenat Diagn. 2007;27:1–5. doi: 10.1002/pd.1600. [DOI] [PubMed] [Google Scholar]
- 10.Smidth-Jensen S, Permin M, Philip J, Lundsteen C, Zachary JM, Fowler SE, et al. Randomised comparison of amniocentesis and transabdominal and transcervical chorionic villus sampling. Lancet. 1992;340:1237–44. doi: 10.1016/0140-6736(92)92946-d. [DOI] [PubMed] [Google Scholar]
- 11.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–7. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 12.Poon LL, Leung TN, Lau TK, Lo YM. Presence of fetal RNA in maternal plasma. Clin Chem. 2000;46:1832–4. [PubMed] [Google Scholar]
- 13.Ng EK, Leung TN, Tsui NB, Lau TK, Panesar NS, Chiu RW, et al. The concentration of circulating corticotropin-releasing hormone mRNA in maternal plasma is increased in preeclampsia. Clin Chem. 2003;49:727–31. doi: 10.1373/49.5.727. [DOI] [PubMed] [Google Scholar]
- 14.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–90. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 16.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ. The microRNA spectrum in 12 body fluids. Clin Chem. 2012;11:1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo YM, Hjelm NM, Fidler C, Sargent IL, Murphy MF, Chamberlain PF, et al. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N Engl J Med. 1998;339:1734–8. doi: 10.1056/NEJM199812103392402. [DOI] [PubMed] [Google Scholar]
- 18.Chiu RW, Lau TK, Cheung PT, Gong ZQ, Leung TN, Lo YM. Noninvasive prenatal exclusion of congenital adrenal hyperplasia by maternal plasma analysis: a feasibility study. Clin Chem. 2002;48:778–80. [PubMed] [Google Scholar]
- 19.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105:16266–71. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotlabova K, Doucha J, Hromadnikova I. Placental-specific microRNA in maternal circulation - identification of appropriate pregnancy-associated microRNAs with diagnostic potential. J Reprod Immunol. 2011;89:185–91. doi: 10.1016/j.jri.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Hromadnikova I, Kotlabova K, Doucha J, Dlouha K, Krofta L. Absolute and relative quantification of placenta-specific microRNAs in maternal circulation with placental insufficiency-related complications. J Mol Diagn. 2012;14:160–7. doi: 10.1016/j.jmoldx.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Gardiner K, Costa AC. The proteins of human chromosome 21. Am J Med Genet C Semin Med Genet. 2006;142C:196–205. doi: 10.1002/ajmg.c.30098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn DE, Nuovo GJ, Martin MM, Malana GE, Pleister AP, Jiang J, et al. Human chromosome 21-derived miRNAs are overexpressed in down syndrome brains and hearts. Biochem Biophys Res Commun. 2008;370:473–7. doi: 10.1016/j.bbrc.2008.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Hahn S, Holzgrave W, editors. Basel: Karger; 2001. Fetal cells and fetal DNA in maternal blood: new developments for a new millennium. 11th Fetal Cell Workshop; 2000 April 15; Basel, Switzerland. [Google Scholar]
- 25.Huppertz B, Kingdom JC. Apoptosis in the trophoblast - role of apoptosis in placental morphogenesis. J Soc Gynecol Investig. 2004;11:353–62. doi: 10.1016/j.jsgi.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Elton TS, Sansom SE, Martin MM. Trisomy-21 gene dosage over-expression of miRNAs results in the haploinsufficiency of specific target proteins. RNA Biol. 2010;5:540–7. doi: 10.4161/rna.7.5.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn DE, Nuovo GJ, Terry AV, Jr, Martin MM, Malana GE, Sansom SE, et al. Chromosome 21-derived microRNAs provide an etiological basis for aberrant protein expression in human Down syndrome brains. J Biol Chem. 2010;285:1529–43. doi: 10.1074/jbc.M109.033407. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Miura K, Miura S, Yamasaki K, Higashijima A, Kinoshita A, Yoshiura K, et al. Identification of pregnancy-associated microRNAs in maternal plasma. Clin Chem. 2010;56:1767–71. doi: 10.1373/clinchem.2010.147660. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Li L, Zhou Z, Wang N, Zhang CY, Zen K. A pilot study of serum microRNA signatures as a novel biomarker for occult hepatitis B virus infection. Med Microbiol Immunol. 2012;3:389–95. doi: 10.1007/s00430-011-0223-0. [DOI] [PubMed] [Google Scholar]
- 31.Yuxia M, Zhennan T, Wei Z. Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. J Cancer Res Clin Oncol. 2012;12:2045–50. doi: 10.1007/s00432-012-1285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]


