Abstract
Skeletal muscle adapts to various forms of exercise depending on the force, speed and duration characteristics of the contraction pattern. The stresses and signals associated with each contraction pattern are likely to specifically activate a network of signal transduction pathways that integrate this information. These pathways include the calcineurin, Calcium/calmodulin-dependent protein kinase (CaMK), mitogen-activated protein kinase (MAPK), protein kinase C (PKC), nuclear factor kappa B (NF-κB), AMP-dependent protein kinase (AMPK), insulin signalling and developmental pathways. Activated signal transduction pathways activate or increase the expression of transcription factors via various mechanisms. Skeletal muscle genes are usually regulated by combinatorial control exerted by several transcription factors and possibly other mechanisms. In addition, adaptations such as an increase in mitochondrial biogenesis or the activation of satellite cell proliferation involve distinct regulatory mechanisms.
Key Words: Adaptation, calcineurin myosin heavy chain, mitochondrial biogenesis, satellite cells
Abstract
İskelet kası egzersizin değişik biçimlerine kasılma modelinin güç, hız ve süre özelliklerine bağlı bir şekilde uyum sağlar. Her kasılma biçimi ile ilişkili sinyal ve stres muhtemelen spesifik olarak sinyal transduction yolunun bir ağını aktive eder. Bu yollar, kalkineurin, Kalsiyum/kalmodulin-bağımlı protein kinaz (CaMK), mitojen-activated protein kinaz (MAPK), protein kinaz C (PKC), nuklear faktör kappa B (NF-κB), AMP-dependent protein kinaz (AMPK), insulin sinyal ve gelişme yollarını içermektedir. Aktive edilmiş transduction yolları değişik mekanizmalar yolu ile kopyalama (transcription) faktörlerinin ifadelerini artırır veya aktive eder. İskelet kası genleri genellikle değişik kopyalama faktörleri tarafından kullanılan birleştirici kontrol ile veya muhtemelen diğer mekanizmalar tarafından düzenlenir. Ayrıca satelayt hücre proliferasyonunun aktivasyonu veya mitokondriyal biogenesizdeki bir artış gibi adaptasyonlar farklı düzenleme mekanizmalarını içermektedir.
Introduction
"How do muscles adapt to exercise?" is one of the key questions in sports science. Here, we aim to give an update on the answer to this question, which is work in rapid progress. There are barriers for sports scientists to this research because it is cellular and molecular biology and we therefore recommend introductory reading on DNA, genes, promoters, enhancers, transcription factors, transcription, translation and gene regulation in order to fully appreciate this review.
Classical research on skeletal muscle adaptation to exercise
Skeletal muscle adapts specifically to chronic exercise or inactivity with well-known changes in protein isoforms and quantities (Saltin and Gollnick, 1983). In their seminal cross-reinnervation experiment, (Buller et al., 1960) demonstrated that the contraction properties of a slow muscle changed from slow to fast after reinnervation by a nerve that normally innervated a fast muscle. Numerous studies since that time have demonstrated the ability of terminally differentiated muscle fibres to adapt to a change in contractile activity. Reduced activity levels as a result of unloading, spaceflight, immobilisation or paraplegia cause atrophy and a shift in gene expression towards a faster, glycolytic skeletal muscle phenotype (Booth and Gollnick, 1983; Talmadge et al., 1995). Increased activity causes specific skeletal muscle adaptation depending on the force, speed and duration characteristics of the contraction pattern. Key adaptations to chronic low-frequency electrical stimulation of fast muscle include an upregulation of mitochondrial biogenesis, oxidative enzymes, fat metabolism enzymes and slow MHC isoforms and a downregulation of glycolytic enzymes and fast MHC isoforms (Chi et al., 1986; Henriksson et al., 1986; Brown et al., 1985; Pette and Vrbova, 1999). Endurance training, a moderate physiological version of chronic stimulation, causes less dramatic changes in a similar direction (Holloszy and Coyle, 1984; Salmons and Henriksson, 1981; Saltin and Gollnick, 1983). In contrast, high force and speed contraction patterns cause muscle hypertrophy predominantly via an increase in the fast twitch fibre area (Tesch, 1988). However, adaptations to strength and power training appear to be bi-directional because type I and the very fast IIX (or IIB) MHCs change towards the intermediate IIA isoform (Andersen et al., 1994; Allemeier et al., 1994). Finally, some skeletal muscle fibres will suffer apoptosis (Podhorska-Okolow et al., 1998) or necrosis (Hikida et al., 1983) as a result of high-impact exercise.
Overview over exercise-induced signal transduction and gene regulation
The vast majority of the classical skeletal muscle adaptation research has been descriptive and did not explain the signalling processes that link contraction patterns to specific gene regulation events and adaptation. The increasing adoption of cellular and molecular techniques by researchers interested in sport and exercise are the main reason for a rapid progress in uncovering the signal transduction events that regulate skeletal muscle adaptation to exercise. A general, schematical overview of exercise-induced signal transduction and gene regulation is shown in figure 1.
Figure 1.
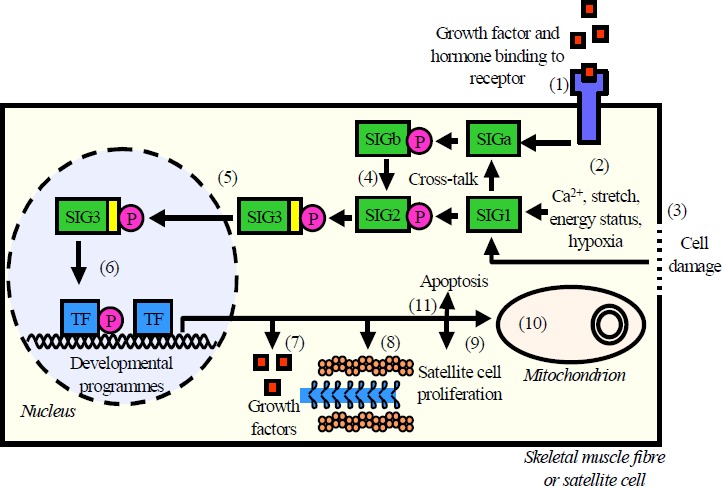
An overview of exercise-induced adaptation signalling and gene regulation in skeletal muscle. Exercise activates signal transduction pathways via (1) growth factors and hormones binding to membrane and nuclear receptors, (2) Ca2+ concentration changes, mechanical stretch, energy status, hypoxia and (3) cell damage/injury. Numerous, sometimes cross-talking signal transduction proteins signal mainly but not exclusively by covalent phosphorylation (4). In a final step, a nuclear localisation signal (yellow) is activated and translocated into the nucleus (Cyert, 2001). Inside the nucleus (6), the signalling protein either acts as a transcription factor itself or it activates transcription factors that will bind regulatory DNA sequences. Skeletal muscle genes are controlled by several regulatory elements and a cross-talk with developmental programmes is likely. Exercise-induced effects include (7) expression of growth factors, (8) contractile proteins, enzymes and other functional proteins, (9) satellite cell proliferation and donation of nuclei to skeletal muscle fibres, (10) mitochondrial biogenesis, and (11) apoptosis.
Signals and Stresses Associated With Exercise Activate Numerous Signal Transduction Pathways
Signal transduction is used to describe the transfer of signals and stresses from the outside or inside of the cell usually by kinase or phosphatase cascades or other signalling processes to cytosolic or nuclear targets. These targets either change cellular processes such as energy metabolism or regulate genes, which results in a changed muscle phenotype. In the following paragraphs, we discuss the role of the calcineurin and MAPK pathways in the exercise response in detail and briefly the role of other contraction-responsive signal transduction pathways.
Calcineurin pathway: Signalling via calcium, dephosphorylation and nuclear translocation
Chin et al. (1998) were the first to identify a signal transduction pathway that could link the increased calcium levels during endurance exercise to the fast-to-slow muscle fibre phenotype changes that occur as a result of exercise. They used cyclosporine A to block the calcium-responsive calcineurin signal transduction pathway. This intervention resulted in an increase in fast muscle fibres in mouse skeletal muscle in vivo suggesting that the activated calcineurin pathway could be responsible for the well characterised fast-to-slow change in fibre phenotype that results from chronic exercise. The authors hypothesized that the calcineurin pathway would be "a molecular mechanism by which different patterns of motor activity promote selective changes in gene expression to establish the specialized characteristics of slow and fast myofibres" (Chin et al., 1998). Calcineurin is a protein phosphatase that is activated by increased intracellular free calcium levels. Activated calcineurin dephosphorylates the transcription factor NFAT and this dephosphorylation reveals the nuclear localization sequence of NFAT. Increased levels of free calcium are therefore a pivotal step in bringing about the nuclear translocation of NFAT (Meissner et al., 2001). Nuclear NFAT binds DNA at NFAT enhancer sequences found in the regulatory regions of a number of "slow" skeletal muscle genes (Chin et al., 1998). However, it was subsequently shown that the calcineurin pathway could increase the expression of some fast genes as well (Swoap et al., 2000) which contradicts Chin et al.'s hypothesis. In addition, there is evidence that the calcineurin pathway is involved s keletal muscle hypertrophy induced by the growth factor IGF-1 (Semsarian et al., 1999) which is not in line with the assumption that the calcineurin pathway mediates fast-to-slow transformations. To conclude, the calcineurin pathway appears to play a role in adaptation to exercise but further studies are needed to clarify its true function in vivo.
MAPK pathways: Signalling via kinase cascades and nuclear translocation
MAPK signal transduction pathways are well known from research on different organisms, tissues and pathologies. MAPK pathways are kinase cascades that use protein phosphorylation as their signalling mechanism. The three main MAPK pathways, ERK1/2, p38 (Yu et al., 2001; Boppart et al., 2000) and JNK (Aronson et al., 1998) have been shown to be activated by various forms of contraction (Widegren et al., 2000) suggesting that they might regulate some of the skeletal muscle genes that change their expression rate as a response to exercise. Concrete evidence for a role of the ERK1/2 signal transduction pathway in exercise adaptation can be found in the study by Murgia et al. (2000). The authors demonstrated an increased slow fibre percentage during injury-repair in vivo when the ERK1/2 pathway was activated by transfection (Murgia et al., 2000). In addition, we (Higginson et al., 2002) found that a pharmacological blockade of the ERK1/2 pathway with the MEK1/2 inhibitor U0126 caused an upregulation in the fast MHC IIB and IIX isoforms and down-regulation of the slow MHC I (ß) isoform in primary skeletal muscle cell culture (figure 2). These observations support the hypothesis that exercise-activation of the ERK1/2 signal transduction pathway pathway may promote a fast-to-slow change in skeletal muscle. However, activities of metabolic enzymes did not change as expected in our study, suggesting that the ERK1/2 pathway is not responsible for all adaptations to endurance exercise.
Figure 2.
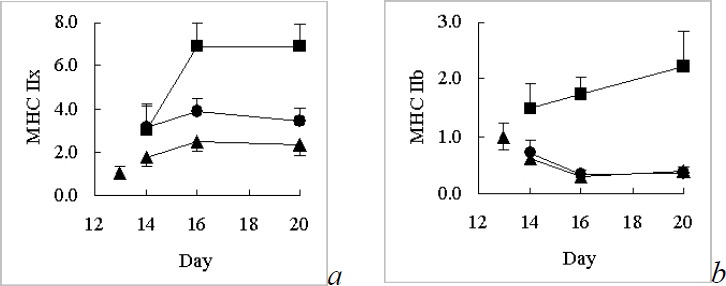
Relative amounts of myosin heavy chain (MHC) isoform IIx (a) and IIb (b) mRNA (mean ± SEM, n=4) in cultured rat myocytes (muscle cells) measured with Northern blotting (Higginson et al., 2002). ▲ Control (no pharmacological treatment). ● Cyclosporin A (calcineurin pathway inhibitor) treated. ■ U0126 (ERK1/2 pathway inhibitor) treated. Blockade of the calcineurin pathway resulted in significantly higher levels of MHC IIx while blockade of the ERK1/2 pathway resulted in significantly higher levels of MHC IIx and MHC IIb. These results suggest that the calcineurin pathway suppresses the MHC IIx isoform when activated by exercise. The ERK1/2 pathway suppresses the MHC IIx and MHC IIb isoforms when activated by exercise.
The research on the calcineurin and MAPK pathways suggests that contraction-responsive signal transduction pathways are either redundant or do only regulate a part of the adaptation response and that adaptation signalling is likely to involve more than one signal transduction pathway.
Brief overview over other contraction-responsive signal transduction pathways
There are now several other examples for signal transduction pathways that are both contraction-responsive and regulate genes as a response to exercise or inactivity. We briefly discuss some of these pathways and their cross-talk in order to develop our argument that adaptation signalling is likely to involve a network of signal transduction pathways capable of integrating numerous signals and stresses associated with exercise. Exercise has been shown to activate the signal transduction kinase PKC in rat skeletal muscle (Richter et al., 1987; Cleland et al., 1989). However, it seems unlikely that calcium is the stimulus because the major PKC isoform in skeletal muscle is PKCθ which lacks a calcium binding motif (Osada et al., 1992; Donnelly et al., 1994). PKC signalling has been shown to regulate skeletal muscle fibre phenotype in avian muscle (DiMario, 2001) but it is unclear whether it has a similar role in mammalian muscle. The transcription factor NF-κB is activated by contraction in rat skeletal muscle (Hollander et al., 2001) and is involved in myogenesis (Lehtinen et al., 1996;Canicio et al., 2001). The growth factor IGF-II has been found to activate NF-κB and effect myogenesis (Kaliman et al., 1999). Other studies demonstrated that NF-κB mediates the protein loss induced by TNFα in skeletal muscle myotubes (Li and Reid, 2000). Moreover, a NF-κB DNA regulatory element has been found in the myostatin gene suggesting that NF-κB is involved in the regulation of skeletal muscle growth (Ma et al., 2001). The calcium-dependent signal transduction kinase CaMK is believed to be activated by exercise in skeletal muscle (Wu et al., 2002) and it is involved in the regulation of the transcription factor MEF2 (Wu et al., 2000). Constitutively activated CaMK in transgenic mice increases mitochondrial biogenesis and increases the percentage of muscle fibres with a slow phenotype in vivo (Wu et al., 2002) suggesting a key role of the CaMK pathway in fast-to-slow fibre phenotype changes. The signal transduction kinase AMPK is activated by AMP and inhibited by phosphocreatine and therefore sensitive to the energy status of the muscle fibre (Winder, 2001). It plays a role in glucose import and there is evidence for a role in activating mitochondrial biogenesis (Bergeron et al., 2001). The PI 3-kinase/AKT/mTOR signal transduction pathway pathway has been shown to be essential for inducing skeletal muscle hypertrophy (Rommel et al., 2001). Integrins are receptor proteins that bind and respond to the extracellular matrix and have been identified as possible inducers of hypertrophy signalling in response to mechanical stresses (Carson and Wei, 2000). This brief overview shows that numerous signal transduction pathways participate in the exercise response.
Evidence for cross-talk
There is now abundant evidence for connections between signal transduction pathways. A signal transduction between two distinct signal transduction pathways is termed cross-talk and limited evidence exists for cross-talk in skeletal muscle signal transduction. For example, there is cross-talk between the signal transduction kinase MEK1 and the developmental transcription factor MyoD (Perry et al., 2001). There is also evidence for cross-talk between the insulin and p38 MAPK signal transduction pathways (Blair et al., 1999) and in the development of insulin resistance in skeletal muscle cells (Storz et al., 1999). There is only a limited number of examples for cross-talk in skeletal muscle but if skeletal muscle signalling was comparable to the signalling in the functionally related and better researched heart, then numerous examples for cross-talk could be expected (Molkentin and Dorn II, 2001). Based on this research, we suggest that adaptation signalling is likely to be mediated by an interlinked network of several contraction-responsive signal transduction pathways.
Gene Regulation, Mitochondrial Biogenesis and Satellite Cell Activation
Signal transduction pathways are capable of regulating non-genomic changes such as glucose import and of regulating transcription or translation, which results in changed protein concentrations or isoforms. Here, we discuss the transcription factor MEF2 as an example for the complex regulation of transcription factors. We then review the regulation of the myosin heavy chain gene family, mitochondrial biogenesis and the activation of satellite cells to show the diversity of responses to signal transduction processes.
MEF2: Double function as transcription and co-factor that is regulated by several pathways in development and as a response to exercise
MEF2 is a transcription factor which has four different isoforms termed MEF2A-D. Active MEF2 binds to a CTA(A/T)4TAG regulatory DNA sequence and it has been shown to be involved in the regulation of the majority of genes expressed in skeletal muscle including creatine kinase, myoglobin and GLUT4 (Yang et al., 1998; McKinsey et al., 2002). Here, we discuss the example of MEF2 to show that transcription factors can be regulated by a number of diverse mechanisms and pathways. On function of MEF2 is its role as an essential co-regulator of muscle development or myogenesis via interaction with the developmental MRFs (Molkentin et al., 1995; Black and Olson, 1998). Myogenesis is primarily regulated by the four MRFs called MyoD, Myf-5, myogenin and MRF4, which are transcription factors that promote the development towards mature skeletal muscle fibres (Buckingham, 2001; Arnold & Winter, 1998). The distance between the MRF and MEF2 DNA binding sites is constant suggesting that this is a requirement for an interaction between both factors (Fickett, 1996). Because MRF mRNA and protein levels chance as a response to denervation and electrical stimulation (Hu et al., 1997; Walters et al., 2000; Jacobs-El et al., 1995; Voytik et al., 1993) and because MEF2 is activated by various exercise-responsive pathways as will be discussed below, the combination of changed MRFs levels and activated MEF2 is probably important for the adaptation to exercise. However, the role of MRFs in adaptation to exercise is unclear. For example, denervation causes an increase in myogenin expression (Voytik et al., 1993 and Walters et al., 2000) while low-frequency stimulation and static stretch does cause no changes in myogenin expression (Jacobs-El et al., 1995). An overexpression of myogenin, however, results in increased activities of oxidative enzymes and decreased activities of glycolytic enzymes (Hughes et al., 1999) which is opposite to what would be expected.
MEF2 itself is activated by calcium and possibly other signals inside the exercising skeletal muscle (Wu et al., 2001). It is dephosphorylated by calcineurin (Wu et al., 2000; Wu et al., 2001) and phosphorylated by CaMK (McKinsey et al., 2002) and the p38 MAPK (Yang et al., 1998; Zetser et al., 1999; Black and Olson, 1998). There is some evidence that the energy-status sensitive AMPK signal transduction kinase might increase MEF2 DNA binding (Zheng et al., 2001). CaMK, however, also signals via a second, unique mechanism to MEF2 that involves histone deacetylases. Histone deacetylases deacetylate tails on histones which results in tighter packing of the DNA so that the DNA is not accessible (McKinsey et al., 2002). Histone deacetylases also bind to MEF2 and inhibit gene expression (McKinsey et al., 2001). Activated CaMK phosphorylates histone deacetylases resulting in detachment from MEF2 and the histone deacetylases are also transported out of the nucleus (McKinsey et al., 2000; McKinsey et al., 2002). Antagonist histone acetylases do then acetylate the histone tails and the DNA becomes more accessible (McKinsey et al., 2001). The MEF2 example highlights that transcription factors can be activated by several signal transduction pathways.
Myosin heavy chain isoform regulation: Genetic location related to function and numerous regulatory elements
The isoforms of the contractile MHC proteins are key determinants of the contraction characteristics of skeletal muscle fibres. Fibre types are usually named after the MHC isoform that is predominantly expressed inside a fibre. In humans, the "slow" αMHC is found in heart and the ß/I MHC isoform is found in heart and slow skeletal muscle fibres. The genes for both isoforms are located at 21.3 Mb on chromosome 14 (human genome data search in www.ensembl.org). In contrast, the developmental and fast MHC isoforms, namely perinatal, embryonal, IIa, IId/x, IIb and extraocular, are situated in a narrow region around 11.8 and 12.0 Mb on chromosome 17 (human genome data search in www.ensembl.org; Weiss et al., 1999). Such clustering of related genes occurs in some globin, homeobox, T cell receptor genes. In contrast, other contractile proteins with different isoforms such as myosin light chains, actin and troponin I and genes of enzyme isoforms such as creatine kinase, lactate dehydrogenase are not clustered (human genome data search in www.ensembl.org; Weiss et al., 1999). The clustering of the MHC heavy chain isoform genes opens the possibility that these MHC gene clusters are controlled by locus control regions or epigenetic mechanisms (Weiss et al., 1999). If this is the case, the cluster would be highly accessible and capable of recruiting chromatin-modifying, coactivator and transcription complexes as has been discussed for the globin isoform gene cluster (Levings and Bungert, 2002). It is an interesting hypothesis that the calcium-CaMK-dependent export of HDAC which might result in increased histone acetylation might open "slow" DNA regions, so that transcription could take place (McKinsey et al., 2000). In addition, each MHC isoform gene has multiple enhancers that are regulated by the binding of transcription factors. In mice, the promoter regions of the fast MHC IIa, IId and IIb genes include enhancers for NFAT, E-boxes, TATA boxes, AT-rich and CarG regions, often with more than one copy (Allen et al., 2001). Transcription factors will bind to these enhancers and increase the transcription of the gene. It has been shown that the developmental transcription factors MyoD or myf-5 preferentially activate the fast MHC IIb isoform gene while calcineurin preferentially activates the intermediate MHC IIa isoform gene in vitro (Allen et al., 2001). In addition, we found that an ERK1/2 pathway blockade caused significant upregulation of the MHC IIa and IIb isoforms (see Figure 2; Higginson et al., 2002).
Mitochondrial biogenensis: Co-ordination of nuclear and mitochondrial gene expression
One key adaptation to endurance exercise is an increase in oxidative capacity. The skeletal muscle contribution to this change is a higher mitochondrial content as a result of increased mitochondrial biogenensis (Holloszy and Coyle, 1984). Mitochondrial biogenesis involves the parallel expression of genes that are encoded in the nuclear and mitochondrial DNA. In addition, the mitochondrial DNA needs to be replicated so that each new mitochondrion has its own DNA copies. Exercise-induced mitochondrial biogenesis can be described as a four-step process: (1) The initial step involves the activation of signal transduction pathways by exercise and the activation or expression of transcription factors that regulate the expression of genes encoding mitochondrial proteins. Both the AMPK and CaMK signal transduction pathways are believed to be contraction-responsive and have been shown to regulate the expression of mitochondrial transcription factors and increase mitochondrial biogenesis. AMPK is activated by AMP and inhibited by phosphocreatine (Winder and Hardie, 1996). Activated AMPK is likely to be responsible for increases in NRF-1, cytochrome c and mitochondrial density, suggesting that AMPK activates mitochondrial biogenesis when cellular energy metabolism is stressed (Bergeron et al., 2001). In addition, transgenic mice with constitutively active CaMK IV showed increased mitochondrial DNA replication and mitochondrial biogenesis (Wu et al., 2002). Constitutively active CaMK IV also increased the expression of the coactivator PGC-1 which is a key mediator of mitochondrial biogenensis (Wu et al., 2002). However, the CaMK IV isoform is not normally expressed in skeletal muscle (Means et al., 1997) and therefore, these results have to be interpreted with care. Two other studies show that PGC-1 is upregulated as a result of exercise and that pharmacological activation of AMPK does increase PGC-1 expression as well (Goto et al., 2000; Terada et al., 2002) suggesting that PGC-1 can be activated by more than one contraction-responsive signal transduction pathway. Finally, overexpression of the transcription factor PGC-1 upregulates not only mitochondrial genes but also extramitochondrial genes such as myoglobin and slow troponin (Lin et al., 2002) indicating a wider role for PGC-1 in the adaptation to endurance exercise. (2) In the second step, genes for mitochondrial proteins encoded in nuclear DNA need to be expressed. Specific transcription factors activate the expression of mitochondrial proteins encoded in the nucleus. Examples for these transcription factors are NRF-1, NRF-2 (Scarpulla, 1997) which are both induced by PGC-1 (Wu et al., 1999) and other transcription factors such as c-jun, c-fos and the ubiquitous Sp1 (Hood et al., 2000). (3) Thereafter, the genes in the mitochondrial DNA need to be expressed and the mitochondrial DNA needs to be replicated. It has been shown that the mitochondrial transcription factor TFAM is capable of activating mitochondrial DNA replication and expression (Parisi and Clayton, 1991). TFAM expression increases as a response to chronic exercise, suggesting that TFAM is indeed involved in activation of mitochondrial biogenensis as a result of endurance exercise (Gordon et al., 2001). TFAM knock-out mouse embryos die because of a complete absence of oxidative phosphorylation and a severe depletion of mitochondrial DNA, highlighting the essential role of TFAM in mitochondrial DNA replication (Larsson et al., 1998). In addition, the mitochondrial transcription factors TFB1M and TFB2M have recently been shown to be necessary for transcription of mammalian mitochondrial DNA and they interact with the mitochondrial RNA polymerase (Falkenberg et al., 2002) (4) Finally, mitochondrial proteins encoded in the nucleus need to be imported into the nascent mitochondria and protein complexes that sometimes consist of mitochondrion-encoded and nucleus-encoded proteins need to be assembled (Poyton and McEwen, 1996). These four steps are necessary for the increase in mitochondrial density that is a well-known result of chronic endurance exercise.
Satellite cell activation: Key role in skeletal muscle injury repair and hypertrophy
Skeletal muscle fibres are very large cells with hundreds of nuclei that are derived through fusion of mononucleated, myogenic cells during muscle development which is also called myogenesis. The nuclei in mature skeletal muscle fibres are unable to replicate and therefore, nuclei and DNA content would be "diluted" in hypertrophying fibres if there was no mechanism to increase the number of nuclei parallel to the growth of the fibre. There is, however, an alternative mechanism that is able to increase the number of nuclei in skeletal muscle fibres during injury repair or hypertrophy. This mechanism involves mononucleated muscle stem cells termed satellite cells by Mauro (1961). Satellite cells can be found under the basement membrane of muscle fibres. Satellite cells are usually dormant but they start to proliferate and donate their nuclei to existing muscle fibres in a response to growth factors such as FGF and IGF-1 (Bischoff, 1994). Satellite cells are likely to be pre-differentiated towards the fibre phenotype of the fibre because satellite cells derived from predominantly fast and slow avian and mouse skeletal muscle do exhibit fast and slow features when cultured in vitro (Feldman and Stockdale, 1991; Rosenblatt et al., 1996). Satellite cell proliferation is essential for skeletal muscle hypertrophy because muscle hypertrophy does not occur if satellite cells are sterilized by irradiation (Rosenblatt et al., 1994). It is likely that the negative skeletal muscle growth factor myostatin plays an important role in the regulation of satellite cell proliferation (Thomas et al., 2000). Myostatin knock-out mice show pronounced skeletal muscle hypertrophy and hyperplasia (McPherron et al., 1997). Myostatin does respond to changes in contractile activity as shown by studies demonstrating a myostatin increase in human muscle atrophy (Reardon et al., 2001) and downregulation in regenerating muscle fibres (Kirk et al., 2000; Mendler et al., 2000). Myostatin prevents myoblast proliferation (Thomas et al., 2000) and it is therefore an attractive hypothesis that hypertrophy training causes a decrease in myostatin expression which in turn relieves the myostatin breaking/inhibitory effect on satellite cell proliferation. The regulatory DNA regions in the vicinity of the myostatin gene include enhancers and silencers responsive to glucocorticoid, androgen and thyroid hormones, myogenic differentiation factor 1, MEF2 2, peroxisome proliferator-activated receptor and NF-κB (Ma et al., 2001). This suggests that several known skeletal muscle growth pathways regulate atrophy/hypertrophy via controlling the expression of myostatin. It has been argued that satellite cells are also involved in adaptation to endurance exercise (Yan, 2000) especially if apoptosis (Podhorska-Okolow et al., 1998) or necrosis occur (Hikida et al., 1983) as a result of the exercise and injury repair is necessary. In addition, bupivacaine hydrochloride induced muscle injury and chronic electrical stimulation cause a greater fast-to-slow fibre type change than chronic electrical stimulation alone (Takahashi et al., 1993). An explanation for this finding might be that the increased involvement of satellite cells may accelerate adaptation because new nuclei donated by fused satellite cells are increasingly expressing a different set of genes. Finally, bone-marrow derived cells are also capable of donating nuclei during muscle fibre repair (Ferrari et al., 1998) indicating that non-muscle adult stem cells can transdifferentiate into skeletal muscle and contribute to its regeneration.
Conclusions
There is growing evidence that adaptation to exercise is not mediated by one signal transduction pathway as has been previously proposed by (Chin et al., 1998). Instead, the key features that characterise exercise induced signal transduction and gene regulation are:
1) It is likely that the signals and stresses associated with exercise do specifically activate a network of signal transduction pathways that integrates this information.
2) Exercise induced gene regulation in skeletal muscle may involve the activation of transcription factors by more than one signal transduction pathway or mechanism. Skeletal muscle genes are regulated via combinatorial control usually exerted by several transcription factors binding to enhancers and silencers. A control of gene clusters by locus control regions is possible.
3) Adaptation to exercise involves diverse regulatory mechanisms, including the regulation of mitochondrial biogenesis and satellite cell activation.
The complexity of signalling and gene regulation that characterizes the exercise response has been discussed as a general feature of regulation in higher eukaryotes by Ptashne and Gann (2002). Research strategies need to address this complexity. Therefore, methods that allow the assessment of the activation status of numerous signal transduction pathways or microarrays that allow the investigation of the expression of thousands of genes (Campbell et al., 2001) will be key methods in this field.
Glossary of Terms
- Apoptosis:
Programmed cell death (cellular suicide) as opposed to uncontrolled death as a result of injury.
- Cross-talk:
Signal transduction between two distinct signal transduction pathways. E. g. if a kinase in signal transduction pathway A is also able to phosphorylate a kinase in a different signal transduction pathway.
- Enhancer, silencer:
DNA sequence where transcription factors bind to and either increase (enhancer) or decrease (silencer) transcription of a gene (examples are E-boxes, TATA boxes and AT-rich regions).
- Histone:
Proteins that regulate DNA packaging
- Kinase:
An enzyme that adds a phosphate group to another molecule.
- Myostatin:
Negative skeletal muscle growth factor
- mRNA:
messenger RNA encodes a protein (amino acid sequence) and is essentially a copy of a gene (a gene codes a protein).
- Phosphatase:
An enzyme that dephosphorylates another molecule.
- Translation:
Synthesis of a protein (amino acid sequence) on a template of mRNA.
- Transcription:
Synthesis of mRNA on a template of DNA is a first step during protein synthesis.
- Transcription factor:
Proteins that regulate transcription by binding to specific DNA sequences.
Abbreviations
- AMPK;
AMP-dependent protein kinase (signal transduction kinase).
- AKT (PKB);
AKT kinase or Protein kinase B (signal transduction kinase).
- CaMK;
Calcium/calmodulin-dependent protein kinase (signal transduction kinase).
- c-jun, c-fos, Sp1;
Transcription factors.
- ERK;
Extracellular signal regulated kinase (signal transduction kinase, translocates into nucleus when phosphorylated)
- FGF;
Fibroblast growth factor (growth factor).
- GLUT4;
Glucose transporter 4.
- IGF-1;
Insulin-like growth factor 1 (growth factor).
- JNK;
c-Jun-Amino terminal kinase (signal transduction kinase, translocates into nucleus when phosphorylated).
- MAPK;
Mitogen-activated protein kinase (class of signal transduction kinases; ERK, p38 and JNK belong to this class).
- MEF2;
Myocyte enhancer factor-2 (transcription factor).
- MEK1;
MAPK/ERK kinase (signal transduction kinase upstream ERK1/2).
- MHC;
Myosin heavy chain; major subunit of the myosin contractile protein. Several isoforms exist and determine the contraction speed of skeletal muscle fibres.
- MRF;
Myogenic regulatory factors (class of proteins that control muscle development; MyoD, Myf-5, myogenin and MRF4 proteins are MRFs).
- mTOR;
Mammalian target of rampamycin (signal transduction kinase; rampamycin, a pharmacological inhibitor, blocks mTOR).
- NFAT;
Nuclear factor of activated T cells (transcription factor that translocates into the nucleus and activates genes when dephosphorylated).
- NF-κB;
Nuclear factor kappa B (transcription factor that translocates into nucleus when activated)
- NRF-1, NRF-2;
Nuclear respiratory factor (regulatory protein involved in mitochondrial biogenesis).
- PGC-1;
Peroxisome proliferator-activated receptor gamma coactivator 1 (regulatory protein involved in mitochondrial biogenesis).
- PI3K;
Phosphoinositide 3 kinase (signal transduction kinase).
- PKC;
Protein kinase C (signal transduction kinase).
- PPAR;
Peroxisome proliferator-activated receptor (nuclear receptor).
- p38;
p38 mitogen activated protein kinase (signal transduction kinase, translocates into nucleus when phosphorylated).
- TFAM;
Mitochondrial transcription factor A (regulatory protein involved in mitochondrial biogenesis).
- TFB1M, TFB2M;
Mitochondrial transcription factors B1 and B2 (regulatory proteins involved in mitochondrial biogenesis).
- TNFα;
Tumour necrosis factor α .
Biographies
Henning WACKERHAGE
Employment:
Senior Lecturer in Sport Physiology at the University of Central Lancashire in Preston, UK.
Degrees:
PhD.
Research interests:
Exercise energy metabolism and exercise induced signal transduction and gene regulation in skeletal muscle.
E-mail: hwackerhage@uclan.ac.uk
Niall M. WOODS
Employment:
Senior Lecturer in Physiology at the University of Central Lancashire in Preston, UK.
Degrees:
PhD.
Research interests:
Calcium signalling in excitable and non-excitable cells and exercise induced signal transduction and gene regulation in skeletal muscle.
E-mail: nmwoods@uclan.ac.uk
References
- Allemeier C. A., Fry A. C., Johnson P., Hikida R. S., Hagerman F. C., Staron R. S. (1994) Effects of sprint cycle training on human skeletal muscle. Journal of Applied Physiology 77,2385-2390 [DOI] [PubMed] [Google Scholar]
- Allen D. L., Sartorius C. A., Sycuro L. K., Leinwand L. A. (2001) Different pathways regulate expression of the skeletal myosin heavy chain genes. Journal of Biological Chemistry 276, 43524-43533 [DOI] [PubMed] [Google Scholar]
- Andersen J. L., Klitgaard H., Saltin B. (1994) Myosin heavy chain isoforms in single fibres from m. vastus lateralis of sprinters: influence of training. Acta Physiologica Scandinavica 151, 135-142 [DOI] [PubMed] [Google Scholar]
- Arnold H. H., Winter B. (1998) Muscle differentiation: more complexity to the network of myogenic regulators. Current Opinion in Genetics & Development. 8, 539-544 [DOI] [PubMed] [Google Scholar]
- Aronson D., Boppart M. D., Dufresne S. D., Fielding R. A., Goodyear L. J. (1998) Exercise stimulates c-Jun NH2 kinase activity and c-Jun transcriptional activity in human skeletal muscle. Biochemical and Biophysical Research Communications 251, 106-110 [DOI] [PubMed] [Google Scholar]
- Bergeron R., Ren J. M., Cadman K. S., Moore I. K., Perret P., Pypaert M., Young L. H., Semenkovich C. F., Shulman G. I. (2001) Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. American Journal of Physiology 281, E1340-E1346 [DOI] [PubMed] [Google Scholar]
- Bischoff R. (1994) The satellite cell and muscle regeneration. Myology, Engel A. G., Franzini-Armstrong C. 97-118 McGraw-Hill [Google Scholar]
- Black B. L., Olson E. N. (1998). Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annual Review of Cell and Developmental Biology 14, 167-196 [DOI] [PubMed] [Google Scholar]
- Blair A. S., Hajduch E., Litherland G. J., Hundal H. S. (1999) Regulation of glucose transport and glycogen synthesis in L6 muscle cells during oxidative stress. Evidence for cross-talk between the insulin and SAPK2/p38 mitogen-activated protein kinase signalling pathways. Journal of Biological Chemistry 274, 36293-36299 [DOI] [PubMed] [Google Scholar]
- Booth F. W., Gollnick P. D. (1983) Effects of disuse on the structure and function of skeletal muscle. Med.Sci.Sports Exerc. 15, 415-420 [PubMed] [Google Scholar]
- Boppart M. D., Asp S., Wojtaszewski J. F., Fielding R. A., Mohr T., Goodyear L. J. (2000) Marathon running transiently increases c-Jun NH2-terminal kinase and p38 activities in human skeletal muscle. Journal of Physiology (London) 526 Pt 3, 663-669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown W. E., Salmons S., Whalen R. G. (1985) Mechanisms underlying the asynchronous replacement of myosin light chain isoforms during stimulation-induced fibre-type transformation of skeletal muscle. FEBS Letters 192, 235-238 [DOI] [PubMed] [Google Scholar]
- Buckingham M. (2001) Skeletal muscle formation in vertebrates. Current Opinion in Genetics & Development. 11, 440-448 [DOI] [PubMed] [Google Scholar]
- Buller A. J., Eccles J. C., Eccles R. (1960) Interactions between motoneurons and muscles in respect of the characteristic speeds of their responses. Journal of Physiology (London) 150, 417-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell W. G., Gordon S. E., Carlson C. J., Pattison J. S., Hamilton M. T., Booth F. W. (2001) Differential global gene expression in red and white skeletal muscle. American Journal of Physiology 280, C763-C768 [DOI] [PubMed] [Google Scholar]
- Canicio J., Ruiz-Lozano P., Carrasco M., Palacin M., Chien K., Zorzano A., Kaliman P. (2001) Nuclear factor kappa B-inducing kinase and Ikappa B kinase-alpha signal skeletal muscle cell differentiation. Journal of Biological Chemistry 276, 20228-20233 [DOI] [PubMed] [Google Scholar]
- Carson J. A., Wei L. (2000) Integrin signalling's potential for mediating gene expression in hypertrophying skeletal muscle. Journal of Applied Physiology 88, 337-343 [DOI] [PubMed] [Google Scholar]
- Chi M. M., Hintz C. S., Henriksson J., Salmons S., Hellendahl R. P., Park J. L., Nemeth P. M., Lowry O. H. (1986) Chronic stimulation of mammalian muscle: enzyme changes in individual fibres. American Journal of Physiology 251, C633-C642 [DOI] [PubMed] [Google Scholar]
- Chin E. R., Olson E. N., Richardson J. A., Yang Q., Humphries C., Shelton J. M., Wu H., Zhu W., Bassel-Duby R., Williams R. S. (1998) A calcineurin-dependent transcriptional pathway controls skeletal muscle fibre type. Genes & Development 12, 2499-2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland P. J., Appleby G. J., Rattigan S., Clark M. G. (1989) Exercise-induced translocation of protein kinase C and production of diacylglycerol and phosphatidic acid in rat skeletal muscle in vivo. Relationship to changes in glucose transport. Journal of Biological Chemistry 264, 17704-17711 [PubMed] [Google Scholar]
- Cyert M. S. (2001) Regulation of nuclear localization during signalling. Journal of Biological Chemistry 276, 20805-20808 [DOI] [PubMed] [Google Scholar]
- DiMario J. X. (2001) Protein kinase C signalling controls skeletal muscle fibre types. Experimental Cell Research. 263, 23-32 [DOI] [PubMed] [Google Scholar]
- Donnelly R., Reed M. J., Azhar S., Reaven G. M. (1994) Expression of the major isoenzyme of protein kinase-C in skeletal muscle, nPKC theta, varies with muscle type and in response to fructose- induced insulin resistance. Endocrinology 135, 2369-2374 [DOI] [PubMed] [Google Scholar]
- Ensembl human genome browser (release 6.28.1) Avaliable from URL: www.ensembl.org.
- Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N. G., Gustafsson C. M. (2002) Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nature Genetics, in press [DOI] [PubMed] [Google Scholar]
- Feldman J. L., Stockdale F. E. (1991) Skeletal muscle satellite cell diversity: satellite cells form fibres of different types in cell culture. Developmental Biology. 143, 320-334 [DOI] [PubMed] [Google Scholar]
- Ferrari G., Cusella-De Angelis G., Coletta M., Paolucci E., Stornaiuolo A., Cossu G., Mavilio F. (1998) Muscle regeneration by bone marrow-derived myogenic progenitors. Science 279, 1528-1530 [DOI] [PubMed] [Google Scholar]
- Fickett J. W. (1996) Coordinate positioning of MEF2 and myogenin binding sites. Gene 172, GC19-GC32 [DOI] [PubMed] [Google Scholar]
- Goto M., Terada S., Kato M., Katoh M., Yokozeki T., Tabata I., Shimokawa T. (2000). cDNA Cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochemical and Biophysical Research Communications 274, 350-354 [DOI] [PubMed] [Google Scholar]
- Gordon J. W., Rungi A. A., Inagaki H., Hood D. A. (2001) Effects of contractile activity on mitochondrial transcription factor A expression in skeletal muscle. Journal of Applied Physiology 90, 389-396 [DOI] [PubMed] [Google Scholar]
- Henriksson J., Chi M. M., Hintz C. S., Young D. A., Kaiser K. K., Salmons S., Lowry O. H. (1986) Chronic stimulation of mammalian muscle: changes in enzymes of six metabolic pathways. American Journal of Physiology 251, C614-C632 [DOI] [PubMed] [Google Scholar]
- Higginson J., Wackerhage H., Woods N. M., Schjerling P., Ratkevicius A., Grunnet N., Quistorff B. (2002) Blockades of mitogen activated protein kinase and calcineurin both change fibre type markers in skeletal muscle culture. Pfluegers Archiv. In Press [DOI] [PubMed] [Google Scholar]
- Hikida R. S., Staron R. S., Hagerman F. C., Sherman W. M., Costill D. L. (1983) Muscle fibre necrosis associated with human marathon runners. Journal of the Neurological Sciences 59, 185-203 [DOI] [PubMed] [Google Scholar]
- Hollander J., Fiebig R., Gore M., Ookawara T., Ohno H., Ji L. L. (2001) Superoxide dismutase gene expression is activated by a single bout of exercise in rat skeletal muscle. Pfluegers Archiv. 442, 426-434 [DOI] [PubMed] [Google Scholar]
- Holloszy J. O., Coyle E. F. (1984) Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. Journal of Applied Physiology 56, 831-838 [DOI] [PubMed] [Google Scholar]
- Hood D. A., Takahashi M., Connor M. K., Freyssenet D. (2000) Assembly of the cellular powerhouse: current issues in muscle mitochondrial biogenesis. Exercise and Sport Sciences Reviews 28, 68-73 [PubMed] [Google Scholar]
- Hu P., Zhang K. M., Wright L. D., Spratt J. A., Briggs F. N. (1997). Correlations between MyoD, myogenin, SERCA1, SERCA2 and phospholamban transcripts during transformation of type-II to type-I skeletal muscle fibers. Pflugers Archiv 434, 209-211 [DOI] [PubMed] [Google Scholar]
- Hughes S. M., Chi M. M., Lowry O. H., Gundersen K. (1999) Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. Journal of Cell Biology 145, 633-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs-El J., Zhou M. Y., Russell B. (1995) MRF4, Myf-5, and myogenin mRNAs in the adaptive responses of mature rat muscle. American Journal of Physiology 268, C1045-C1052 [DOI] [PubMed] [Google Scholar]
- Kaliman P., Canicio J., Testar X., Palacin M., Zorzano A. (1999) Insulin-like growth factor-II, phosphatidylinositol 3-kinase, nuclear factor-kappaB and inducible nitric-oxide synthase define a common myogenic signalling pathway. Journal of Biological Chemistry 274, 17437-17444 [DOI] [PubMed] [Google Scholar]
- Kirk S., Oldham J., Kambadur R., Sharma M., Dobbie P., Bass J. (2000) Myostatin regulation during skeletal muscle regeneration. Journal of Cellular Physiology 184, 356-363 [DOI] [PubMed] [Google Scholar]
- Larsson N. G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G. S., Clayton D. A. (1998) Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genetics 18, 231-236 [DOI] [PubMed] [Google Scholar]
- Lehtinen S. K., Rahkila P., Helenius M., Korhonen P., Salminen A. (1996) Down-regulation of transcription factors AP-1, Sp-1, and NF-kappa B precedes myocyte differentiation. Biochemical and Biophysical Research Communications 229, 36-43 [DOI] [PubMed] [Google Scholar]
- Levings P. P., Bungert J. (2002) The human beta-globin locus control region. European Journal of Biochemistry 269, 1589-1599 [DOI] [PubMed] [Google Scholar]
- Li Y. P., Reid M. B. (2000) NF-kappaB mediates the protein loss induced by TNF-alpha in differentiated skeletal muscle myotubes. American Journal of Physiology 279, R1165-R1170 [DOI] [PubMed] [Google Scholar]
- Lin J., Wu H., Tarr P. T., Zhang C. Y., Wu Z., Boss O., Michael L. F., Puigserver P., Isotani E., Olson E. N., Lowell B. B., Bassel-Duby R., Spiegelman B. M. (2002). Transcriptional co-activator PGC-1alpha drives the formation of slow-twitch muscle fibres. Nature 418, 797-801 [DOI] [PubMed] [Google Scholar]
- Ma K., Mallidis C., Artaza J., Taylor W., Gonzalez-Cadavid N., Bhasin S. (2001) Characterization of 5'-regulatory region of human myostatin gene: regulation by dexamethasone in vitro. American Journal of Physiology 281, E1128-E1136 [DOI] [PubMed] [Google Scholar]
- Mauro A. (1961) Satellite cell of skeletal muscle fibres. Journal of Biophysical and Biochemical Cytology 9, 493-495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey T. A., Zhang C. L., Lu J., Olson E. N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey T. A., Zhang C. L., Olson E. N. (2001) Control of muscle development by dueling HATs and HDACs. Current Opinion in Genetics & Development. 11, 497-504 [DOI] [PubMed] [Google Scholar]
- McKinsey T. A., Zhang C. L., Olson E. N. (2002) MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends in Biochemical Sciences. 27, 40-47 [DOI] [PubMed] [Google Scholar]
- McPherron A. C., Lawler A. M., Lee S. J. (1997) Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387, 83-90 [DOI] [PubMed] [Google Scholar]
- Means A. R., Ribar T. J., Kane C. D., Hook S. S., Anderson K. A. (1997) Regulation and properties of the rat Ca2+/calmodulin-dependent protein kinase IV gene and its protein products. Recent Progress in Hormone Research 52, 389-406 [PubMed] [Google Scholar]
- Meissner J. D., Gros G., Scheibe R. J., Scholz M., Kubis H. P. (2001) Calcineurin regulates slow myosin, but not fast myosin or metabolic enzymes, during fast-to-slow transformation in rabbit skeletal muscle cell culture. Journal of Physiology (London) 533, 215-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendler L., Zador E., Ver H. M., Dux L., Wuytack F. (2000) Myostatin levels in regenerating rat muscles and in myogenic cell cultures. Journal of Muscle Research and Cell Motility. 21, 551-563 [DOI] [PubMed] [Google Scholar]
- Molkentin J. D., Black B. L., Martin J. F., Olson E. N. (1995) Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83, 1125-1136 [DOI] [PubMed] [Google Scholar]
- Molkentin J. D., Dorn II G. W. (2001) Cytoplasmic signalling pathways that regulate cardiac hypertrophy. Annual Review of Physiology 63, 391-426 [DOI] [PubMed] [Google Scholar]
- Murgia M., Serrano A. L., Calabria E., Pallafacchina G., Lomo T., Schiaffino S. (2000) Ras is involved in nerve-activity-dependent regulation of muscle genes. Nature Cell Biology 2, 142-147 [DOI] [PubMed] [Google Scholar]
- Osada S., Mizuno K., Saido T. C., Suzuki K., Kuroki T., Ohno S. (1992) A new member of the protein kinase C family, nPKC theta, predominantly expressed in skeletal muscle. Journal of Cell Biology 12, 3930-3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M. A., Clayton D. A. (1991) Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science 252, 965-969 [DOI] [PubMed] [Google Scholar]
- Perry R. L., Parker M. H., Rudnicki M. A. (2001) Activated MEK1 binds the nuclear MyoD transcriptional complex to repress transactivation. Moecular Cell 8, 291-301 [DOI] [PubMed] [Google Scholar]
- Pette D., Vrbova G. (1999) What does chronic electrical stimulation teach us about muscle plasticity? Muscle Nerve 22, 666-677 [DOI] [PubMed] [Google Scholar]
- Podhorska-Okolow M., Sandri M., Zampieri S., Brun B., Rossini K., Carraro U. (1998) Apoptosis of myofibres and satellite cells: exercise-induced damage in skeletal muscle of the mouse. Neuropathology and Applied Neurobiology 24, 518-531 [DOI] [PubMed] [Google Scholar]
- Poyton R. O., McEwen J. E. (1996) Crosstalk between nuclear and mitochondrial genomes. Annual Review of Biochemistry 65, 563-607 [DOI] [PubMed] [Google Scholar]
- Ptashne M., Gann A. (2002) Genes and Signals. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Reardon K. A., Davis J., Kapsa R. M., Choong P., Byrne E. (2001) Myostatin, insulin-like growth factor-1, and leukemia inhibitory factor mRNAs are upregulated in chronic human disuse muscle atrophy. Muscle Nerve 24, 893-899 [DOI] [PubMed] [Google Scholar]
- Richter E. A., Cleland P. J., Rattigan S., Clark M. G. (1987) Contraction-associated translocation of protein kinase C in rat skeletal muscle. FEBS Letters 217, 232-236 [DOI] [PubMed] [Google Scholar]
- Rommel C., Bodine S. C., Clarke B. A., Rossman R., Nunez L., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001) Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/ mTOR and PI(3)K/Akt/GSK3 pathways. Journal of Cell Biology 3, 1009-1013 [DOI] [PubMed] [Google Scholar]
- Rosenblatt J. D., Parry D. J., Partridge T. A. (1996) Phenotype of adult mouse muscle myoblasts reflects their fibre type of origin. Differentiation 60, 39-45 [DOI] [PubMed] [Google Scholar]
- Rosenblatt J. D., Yong D., Parry D. J. (1994) Satellite cell activity is required for hypertrophy of overloaded adult rat muscle. Muscle & Nerve 17, 608-613 [DOI] [PubMed] [Google Scholar]
- Salmons S., Henriksson J. (1981) The adaptive response of skeletal muscle to increased use. Muscle & Nerve 4, 94-105 [DOI] [PubMed] [Google Scholar]
- Saltin B., Gollnick P. D. (1983) Skeletal muscle adaptatbility: Significance for metabolism and performance. In Skeletal Muscle, Peachey L. D., Adrian R. H., Geiger S. R.American Physiological Society, Bethesda, MD: 555-631 [Google Scholar]
- Scarpulla R. C. (1997) Nuclear control of respiratory chain expression in mammalian cells. Journal of Bioenergectics and Biomembranes. 29, 109-119 [DOI] [PubMed] [Google Scholar]
- Semsarian C., Wu M. J., Ju Y. K., Marciniec T., Yeoh T., Allen D. G., Harvey R. P., Graham R. M. (1999) Skeletal muscle hypertrophy is mediated by a Ca2+-dependent calcineurin signalling pathway. Nature 400, 576-581 [DOI] [PubMed] [Google Scholar]
- Storz P., Doppler H., Wernig A., Pfizenmaier K., Muller G. (1999) Cross-talk mechanisms in the development of insulin resistance of skeletal muscle cells palmitate rather than tumour necrosis factor inhibits insulin-dependent protein kinase B (PKB)/Akt stimulation and glucose uptake. European Journal of Biochemistry 266, 17-25 [DOI] [PubMed] [Google Scholar]
- Swoap S. J., Hunter R. B., Stevenson E. J., Felton H. M., Kansagra N. V., Lang J. M., Esser K. A., Kandarian S. C. (2000) The calcineurin-NFAT pathway and muscle fibre-type gene expression. American Journal of Physiology 279, C915-C924 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Miyamura H., Eguchi S., Homma S. (1993) The effect of bupivacaine hydrochloride on skeletal muscle fibre type transformation by low frequency electrical stimulation. Neuroscience Letters 155, 191-194 [DOI] [PubMed] [Google Scholar]
- Talmadge R. J., Roy R. R., Bodine-Fowler S. C., Pierotti D. J., Edgerton V. R. (1995) Adaptations in myosin heavy chain profile in chronically unloaded muscles. Basic Applied Myology 5, 17-137 [PubMed] [Google Scholar]
- Terada S., Goto M., Kato M., Kawanaka K., Shimokawa T., Tabata I. (2002). Effects of low-intensity prolonged exercise on PGC-1 mRNA expression in rat epitrochlearis muscle. Biochemical and Biophysical Research Communications 296, 350-354 [DOI] [PubMed] [Google Scholar]
- Tesch P. A. (1988) Skeletal muscle adaptations consequent to long-term heavy resistance exercise. Medicine and Science in Sports and Exercise 20, S132-S134 [DOI] [PubMed] [Google Scholar]
- Thomas M., Langley B., Berry C., Sharma M., Kirk S., Bass J., Kambadur R. (2000) Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. Journal of Biological Chemistry 275, 40235-40243 [DOI] [PubMed] [Google Scholar]
- Voytik S. L., Przyborski M., Badylak S. F., Konieczny S. F. (1993). Differential expression of muscle regulatory factor genes in normal and denervated adult rat hindlimb muscles. Developmental Dynamics 198, 214-224 [DOI] [PubMed] [Google Scholar]
- Walters E. H., Stickland N. C., Loughna P. T. (2000) The expression of the myogenic regulatory factors in denervated and normal muscles of different phenotypes. J.Muscle Res.Cell Motil. 21, 647-653 [DOI] [PubMed] [Google Scholar]
- Weiss A., McDonough D., Wertman B., Acakpo-Satchivi L., Montgomery K., Kucherlapati R., Leinwand L., Krauter K. (1999) Organization of human and mouse skeletal myosin heavy chain gene clusters is highly conserved. Proceedings of the National Academy of Sciences of the U.S.A 96, 2958-2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widegren U., Wretman C., Lionikas A., Hedin G., Henriksson J. (2000) Influence of exercise intensity on ERK/MAP kinase signalling in human skeletal muscle. Pfluegers Archiv 441, 317-322 [DOI] [PubMed] [Google Scholar]
- Winder W. W. (2001) Energy-sensing and signalling by AMP-activated protein kinase in skeletal muscle. Journal of Applied Physiology 91, 1017-1028 [DOI] [PubMed] [Google Scholar]
- Wu H., Kanatous S. B., Thurmond F. A., Gallardo T., Isotani E., Bassel-Duby R., Williams R. S. (2002). Regulation of Mitochondrial Biogenesis in Skeletal Muscle by CaMK. Science 296, 349-352 [DOI] [PubMed] [Google Scholar]
- Wu H., Naya F. J., McKinsey T. A., Mercer B., Shelton J. M., Chin E. R., Simard A. R., Michel R. N., Bassel-Duby R., Olson E. N., Williams R. S. (2000) MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fibre type. EMBO Journal 19, 1963-1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Rothermel B., Kanatous S., Rosenberg P., Naya F. J., Shelton J. M., Hutcheson K. A., DiMaio J. M., Olson E. N., Bassel-Duby R., Williams R. S. (2001). Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO Journal 20, 6414-6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder W. W., Hardie D. G. (1996). Inactivation of acetyl-CoA carboxylase and activation of AMP-activated protein kinase in muscle during exercise. American Journal of Physiology 270, E299-E304 [DOI] [PubMed] [Google Scholar]
- Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., Spiegelman B. M. (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115-124 [DOI] [PubMed] [Google Scholar]
- Yan Z. (2000) Skeletal muscle adaptation and cell cycle regulation. Exercise and Sport Sciences Reviews. 28, 24-26 [PubMed] [Google Scholar]
- Yang C. C., Ornatsky O. I., McDermott J. C., Cruz T. F., Prody C. A. (1998) Interaction of myocyte enhancer factor 2 (MEF2) with a mitogen- activated protein kinase, ERK5/BMK1. Nucleic Acids Research 26, 4771-4777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Blomstrand E., Chibalin A. V., Krook A., Zierath J. R. (2001) Marathon running increases ERK1/2 and p38 MAP kinase signalling to downstream targets in human skeletal muscle. Journal of Physiology 536, 273-282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetser A., Gredinger E., Bengal E. (1999) p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. Journal of Biological Chemistry 274, 5193-5200 [DOI] [PubMed] [Google Scholar]
- Zheng D., MacLean P. S., Pohnert S. C., Knight J. B., Olson A. L., Winder W. W., Dohm G. L. (2001) Regulation of muscle GLUT-4 transcription by AMP-activated protein kinase. Journal of Applied Physiology 91, 1073-1083 [DOI] [PubMed] [Google Scholar]


