Abstract
Background
Lung adenocarcinoma is a highly heterogeneous disease with various etiologies, prognoses, and responses to therapy. Although genome-scale characterization of lung adenocarcinoma has been performed, a comprehensive somatic mutation analysis of EGFR/KRAS/ALK-negative lung adenocarcinoma in never-smokers has not been conducted.
Methods
We analyzed whole exome sequencing data from 16 EGFR/KRAS/ALK-negative lung adenocarcinomas and additional 54 tumors in two expansion cohort sets. Candidate loci were validated by target capture and Sanger sequencing. Gene set analysis was performed using Ingenuity Pathway Analysis.
Results
We identified 27 genes potentially implicated in the pathogenesis of lung adenocarcinoma. These included targetable genes involved in PI3K/mTOR signaling (TSC1, PIK3CA, AKT2) and receptor tyrosine kinase signaling (ERBB4) and genes not previously highlighted in lung adenocarcinomas, such as SETD2 and PBRM1 (chromatin remodeling), CHEK2 and CDC27 (cell cycle), CUL3 and SOD2 (oxidative stress), and CSMD3 and TFG (immune response). In the expansion cohort (N = 70), TP53 was the most frequently altered gene (11%), followed by SETD2 (6%), CSMD3 (6%), ERBB2 (6%), and CDH10 (4%). In pathway analysis, the majority of altered genes were involved in cell cycle/DNA repair (P <0.001) and cAMP-dependent protein kinase signaling (P <0.001).
Conclusions
The genomic makeup of EGFR/KRAS/ALK-negative lung adenocarcinomas in never-smokers is remarkably diverse. Genes involved in cell cycle regulation/DNA repair are implicated in tumorigenesis and represent potential therapeutic targets.
Background
Lung cancer is the leading cause of cancer deaths worldwide [1]. In 2008, 1.38 million deaths were attributed to lung cancer, accounting for approximately 20% of cancer-related deaths. Lung cancer is a highly heterogeneous disease with regard to its etiology, prognosis, and response to therapy, complicating both prevention and treatment [1]. Non-small cell lung cancer accounts for approximately 85% of newly diagnosed lung cancers and can be classified into two major histologic subtypes: adenocarcinoma (approximately 50% of cases) and squamous cell carcinoma (approximately 30%). Although the majority of lung cancer cases are attributed to tobacco smoke, approximately 25% of lung cancer cases worldwide occur in never-smokers. Lung cancers in never-smokers have distinct clinicopathologic characteristics and clinical outcomes [2,3].
The discovery of driver mutations, such as epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK), has led to remarkable improvements in personalized therapies for lung adenocarcinoma [4]. For example, erlotinib and gefitinib has been particularly efficacious in patients with EGFR mutations, and crizotinib in patients with ALK fusions [5,6]. Actionable genetic alterations that are treatable with therapeutic agents have been identified in approximately 50% of lung adenocarcinomas and include mutations in EGFR, ERBB2, KRAS, ALK, BRAF, PIK3CA, AKT1, ROS1, NRAS, and MAP2K1[4]. Therefore, identification of novel druggable targets in the remaining 50% of lung adenocarcinomas is a top research priority.
Comprehensive genomic approaches are being undertaken to accelerate the identification of new molecular targets and increase our understanding of the critical cellular and molecular mechanisms underlying lung cancer [7-13]. The first comprehensive mutation profiling of 623 genes in 188 adenocarcinomas identified 26 significantly mutated genes, including known oncogenes (KRAS, EGFR, ERBB2, ERBB4, EPHA3 and other ephrin receptor genes, KDR, and FGFR4) and tumor suppressor genes (TP53, CDKN2A, STK11, NF1, ATM, RB1, and APC) [7]. Recent studies using next-generation sequencing identified new candidate driver mutations, including RET1 rearrangements [8,14] and mutations in CSMD3[9], MXRA5[10], U2AF1, RBM10, and ARID1A[11], and DACH1, CFTR, RELN, ABCB5, and HGF[12]. Previous studies focused primarily on the identification of new driver genes according to mutation frequency and pattern; systematic pathway-based analysis has not been performed. Moreover, sample sizes in previous studies were insufficient to resolve rare driver mutations in lung adenocarcinoma.
In this study, we analyzed exome sequencing data from 16 EGFR/KRAS/ALK-negative tumors and paired normal samples and an applicable expansion cohort of 54 EGFR/KRAS-negative lung adenocarcinomas to identify novel somatic mutations in lung adenocarcinomas of never-smokers.
Methods
Preparation of clinical samples
Tumor and adjacent normal lung fresh tissues were obtained by surgical procedures. Clinical information including age, sex, smoking history, tumor histology, and tumor stage based on the seventh edition of the American Joint Committee on Cancer staging system was collected. Never-smokers were defined as patients who had a lifelong history of smoking fewer than 100 cigarettes. All patients provided informed consent. The study was approved by the Institutional Review Board of Severance Hospital (4-2011-0891) and conducted in accordance with the Helsinki Declaration [15].
EGFR/KRAS mutations were verified by Sanger sequencing and ALK rearrangement was detected by hybridization probes. To screen for EGFR and KRAS mutations by Sanger sequencing, we used the following primer sequences: 5′-CAGATGTTATCGAGGTCCGA-3′ and 5′-CAAGCAGAAGACGGCATACG-3′ to detect deletions in exon 19 of EGFR, 5′-CAAGCAGAAGACGGCATACG-3′ and 5′-GACCACCGAGATCTACACTC-3′ to detect the L858R mutation in exon 21 of EGFR, and 5′-GTGACATGTTCTAATATAGTCAC-3′ and 5′-TCTATTGTTGGATCATATTCGTC-3′ to detect mutations in codons 12 and 13 of KRAS. ALK rearrangements were verified using break-apart fluorescence in situ hybridization probes (Vysis LSI ALK Dual Color, Break Apart Rearrangement Probe; Abbott Molecular, Abbott Park, IL, USA). DNA was extracted from tissues using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA).
Whole exome sequencing
Extracted DNA was sheared and a genomic library prepared using the NEBNext kit (New England BioLabs, Inc., Ipswich, MA, USA) according to the manufacturer’s instructions. Exon sequences were captured using the TruSeq Exome Enrichment Kit (Illumina, San Diego, CA, USA). Whole exome sequencing was performed using the Illumina HiSeq2000 platform. Sequencing data are accessible at Sequence Read Archive ([16] accession number [SRA:SRP022932]).
Exome data analyses
The analysis flow chart is illustrated in Additional file 1: Figure S1. The references for software packages used for exome data analyses are summarized in Additional file 1. Briefly, all sequenced reads were aligned to the human reference genome National Center for Biotechnology Information build 37 (hg19) using Novoalign. Local re-aligning around indels and pair-end fixing was performed by GATK (version 1.4-21) and Picard, and PCR duplicates were removed using Picard. Quality scores were recalibrated by GATK.
Three variant calling programs were used to call single nucleotide variants: muTect (1.0.287783), VarScan (version 2.2.11), and GATK Unified Genotyper. Indels were called by the GATK Somatic Indel Detector with default parameters in paired sample mode.
Mutated loci were annotated using ANNOVAR and Polyphen2. Only non-synonymous single nucleotide variants and indels in coding exons and splicing sites were included. Known single nucleotide polymorphisms with minor allelic frequency >5% in the 1000 Genome Project Phase I East Asian (2012 April) and NHLBI Exome Sequencing Project 6500 (2012 Oct) were annotated and removed by ANNOVAR.
Validation by molecular-inversion probe capture
The molecular-inversion probe (MIP) capture method was used to validate 1,401 candidate loci identified in whole exome sequencing. We designed 3,726 probes to capture the candidate loci (Additional file 2: Table S7). The microarray-based MIP preparation and capture experiment followed MIP standard-operating procedures with modifications in the preparation of probes (manuscript in preparation) [17].
For MIP probe hybridization, 1 μg of genomic DNA, 1.5 μl of Ampligase buffer (Epicentre, Madison, WI, USA), 1 μl of probe mixture (genomic DNA to probe ratio, 1:90), and distilled H2O were combined to give a total volume of 15 μl. The reaction was carried out for 5 minutes at 95°C and then the temperature was decreased to 60°C at a rate of 0.1°C per second followed by incubation for 24 hours at 60°C. After addition of 0.2 μl of Phusion polymerase (New England BioLabs Inc.), 1 μl of deoxyribonucleotide triphosphate (New England BioLabs, Inc.), 0.2 μl of Ampligase buffer (Epicentre), 4 units of Ampligase DNA ligase (Epicentre), and 0.3 μl of distilled H2O, the mixture was incubated for an additional 24 hours.
PCR products were purified with a Qiagen gel extraction kit, mixed equally based on concentrations determined using a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA, USA), and sent for Illumina HiSeq2000 sequencing.
Molecular-inversion probe capture data analysis
The raw data were aligned to the human reference genome (hg19) by Novoalign. Aligned data on the position of candidate loci were selected and transformed to pileup format by SAMtools. The candidate loci were defined as validated loci if the variant base was the same as that of whole exome sequencing and the following criteria were satisfied: variant allele frequency in tumor ≥5%, reads supporting variant allele in normal ≤2.
Validation by Sanger sequencing
Candidate driver mutations and randomly selected validated loci were chosen for additional validation and appropriate primer pairs for Sanger sequencing were designed (Additional file 2: Table S8). PCR products were purified and sent for Sanger sequencing (Macrogen, Seoul, Korea). Sequencing data were analyzed with SeqMan (DNASTAR, Madison, WI, USA).
Canonical pathway analysis
The pathway analysis was performed through the use of Ingenuity Pathway Analysis (Ingenuity® Systems [18]). Pathways associated with a set of focus genes were identified from the Ingenuity Pathways Analysis library of canonical pathways. The P-value was measured to decide the likelihood that the association between focus genes and a given pathway was due to random chance. The more focus genes involved, the more likely the association is not due to random chance, and thus the more significant the P-value. A right-tailed Fisher’s exact test was used to calculate a P-value determining the probability of an association between the focus genes and the canonical pathway.
Statistical analyses
Continuous clinical data (for example, age) were compared using independent Student’s t-tests. Categorical data (for example, sex, ethnicity, stage, mutation frequency) were compared using a chi-squared test. The false-discovery rate was corrected for multiple comparisons using the method of Benjamini and Hochberg. All statistical tests were two-tailed, and a P-value ≤0.05 was considered statistically significant. Data analyses were performed using R statistical software version 2.15.3 [19].
Results
Exome sequencing of EGFR/KRAS/ALK-negative tumors in never-smokers
We screened 230 surgically resected lung adenocarcinoma samples to identify EGFR/KRAS/ALK-negative tumors in never-smokers. A total of 16 tumors (7% of all non-small cell lung cancers) were eligible for exome sequencing. Tumor and normal samples had an average sequencing depth of 51.9× and 52.0× respectively with average coverage of 94.6% for each (Additional file 1: Table S1). Somatic variants were validated by target-capture sequencing (154-fold depth; average coverage of 94%) and Sanger sequencing, which had a concordance rate of 94% with exome sequencing (Additional file 1: Tables S2 and S3). We detected a median number of 10 non-synonymous mutations and indels per tumor (range 3 to 27; Additional file 1: Table S4). The median rate of non-synonymous mutation was 0.32 mutations per megabase, which was comparable to that in previous reports for never-smokers [11,12]. The average ratio of transitions to transversions was 1.95; G:C → A:T transitions (37%) were the most frequent followed by A:T → G:C transitions (21%), consistent with a previous lung cancer exome study [11]. Validated loci were further analyzed for functional prediction of amino acid changes using four different prediction algorithms (SIFT, Polyphen2, LRT, and Mutation Taster) (Additional file 1).
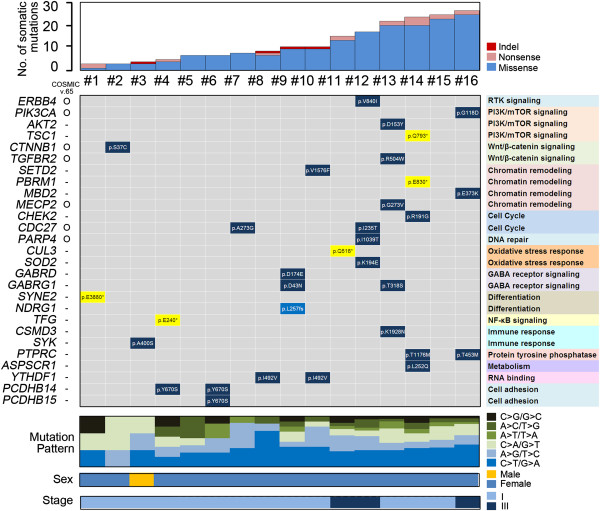
Non-synonymous somatic mutations in 16 EGFR/KRAS/ALK-negative tumors are summarized in Figure 1 (full mutation information is provided in Additional file 1: Table S5). Overall, 14 of 16 patients (87%) had a putative non-synonymous mutation. Although EGFR/KRAS/ALK-negative tumors harbored heterogeneous mutation profiles, somatically altered genes were functionally classified as follows: major mitogenic and targetable pathways such as PI3K/mTOR signaling (TSC1, PIK3CA, AKT2), receptor tyrosine kinase signaling (ERBB4), protein tyrosine phosphatase (PTPRC), cell cycle (CHEK2, CDC27), and DNA repair (PARP4); tumor suppressor pathways including chromatin remodeling (SETD2, PBRM1, MBD2, MECP2), Wnt signaling (CTNNB1, TGFBR2), and NF-κB signaling (TFG); oxidative stress response (CUL3, SOD2) and differentiation (SYNE2, NDRG1) (similar to lung squamous cell carcinoma [20]); pathways not previously highlighted in carcinogenesis such as gamma-aminobutyric acid receptor signaling (GABRD, GABRG1) and immune response (CSMD3, SYK); as well as YTHDF1 (suggested role in RNA binding) and PCDHB14 (role in cell adhesion).
Figure 1.
Mutation summary of 16 EGFR/KRAS/ALK-negative lung adenocarcinomas.
Investigation of functional domains in altered genes
Of 32 loci shown in Figure 1, seven were found in the Catalogue of Somatic Mutations in Cancer (COSMIC v.65): ERBB4 V840I, PIK3CA G118D, CTNNB1 S37C, TGFBR2 R504W, MECP2 G273V, CDC27 A273G, and PARP4 I1039T. In addition, PIK3CA G118D (in squamous cell carcinoma), CTNNB1 S37C (in adenocarcinoma), and MECP2 G273V (in adenocarcinoma) were previously reported in cases of lung cancer.
To explore the functional effects of somatic variants, we investigated functional domains in altered loci. Seventeen loci of 27 genes shown in Figure 1 were located in functional domains, including kinase domains (ERBB4 (tyrosine kinase domain), AKT2 (serine/threonine kinase domain), TGFBR2 (tyrosine kinase domain), SYK (protein kinase domain)) or domains involved in oncogenic kinase activation (TSC1 (TPR/MLP1/MLP2-like protein)); in histone modification domains (SETD2 (SET domain), PBRM1 (bromo domain), MBD2 (methyl CpG binding domain), MECP2 (methyl CpG binding domain)); in oxidative stress response/differentiation domains (CUL3 (cullin domain), SOD2 (superoxide dismutase domain), NDRG1 (Ndr family domain)); in ion-channel domains (GABRD (ion-channel binding domain), GABRG1 (ion-channel transmembrane domain)); or were for cell cycle checkpoint proteins (CHEK2 (fork head associated domain)).
Somatic mutations in an expansion cohort of EGFR/KRAS-negative tumors in never-smokers
To validate and expand our mutation analysis in lung adenocarcinoma in never-smokers, we collected an expansion dataset from five available lung adenocarcinoma studies [9-13] and a The Cancer Genome Atlas lung carcinoma study [21], with no overlap with the study of Imielinski et al. [11]. Clinical information of all patients including sex, age, tumor stage, and ethnicity is given in Table 1. A total of 54 EGFR/KRAS-negative tumors from never-smokers were analyzed. Information on non-synonymous and splicing site mutations were extracted from a pooled dataset. The median rate of non-synonymous mutations in EGFR/KRAS-negative never-smokers was approximately 0.65 mutations per megabase and the median number of non-synonymous mutations per patient was 19.0. The average ratio of transitions to transversions was 1.07 and G:C → A:T transitions (40%) were the most frequent, consistent with our data.
Table 1.
Patient characteristics (N = 70)
| Characteristic | This study | Expansion cohort a | TCGA-LUAD a |
|---|---|---|---|
| Number of patients |
16 |
40 |
14 |
| Age (years) |
|
|
|
| Median |
58.5 |
63 |
69 |
| Range |
47 to 79 |
36 to 87 |
52 to 79 |
| Sex (%) |
|
|
|
| Male |
1 (6) |
7 (17) |
2 (14) |
| Female |
15 (94) |
29 (73) |
12 (86) |
| n/a |
0 |
4 (10) |
0 |
| Clinical stage (%) |
|
|
|
| I |
13 (81) |
24 (60) |
9 (65) |
| II |
0 |
6 (15) |
2 (14) |
| III |
3 (19) |
6 (15) |
3 (21) |
| IV |
0 |
1 (2) |
0 |
| n/a |
0 |
3 (8) |
0 |
| Ethnic group (%) |
|
|
|
| Caucasian |
0 |
16 (40) |
14 (100) |
| Asian | 16 (100) | 24 (60) | 0 |
aNever-smokers with EGFR/KRAS-negative adenocarcinoma. Mb, megabase; n/a, not available; SD, standard deviation; TCGA-LUAD, The Cancer Genome Atlas - Lung adenocarcinoma.
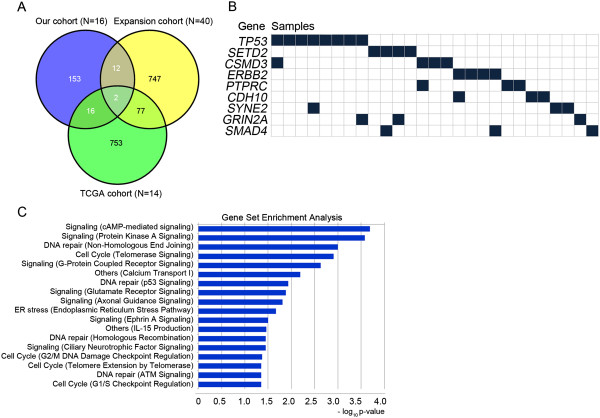
Comparison of altered genes among the three cohorts is shown in Figure 2. SETD2 and CSMD3 were altered in all three cohorts (Figure 2A). Commonly altered genes with information on affected loci, amino acid changes, and functional predictions are summarized in Table 2 (full information is provided in Additional file 1: Table S6). The most frequently mutated gene was TP53, which was altered in 11% of tumors, followed by SETD2 (6%, 4 of 70 cases), CSMD3 (6%, 4 of 70 cases), and ERBB2 (6%, 4 of 70 cases). PTPRC, SYNE2, GRIN2A, CDH10, and SMAD4 were each altered in 3 of 70 cases (4%). SETD2 interacts with p53 and regulates genes downstream of p53 in addition to increasing p53 stability [22]. Mutations in SETD2 were nonsense mutations in three cases and missense mutation in one case. The missense mutation V1576F is located in the SET domain; one nonsense mutation, R839*, is a truncating mutation upstream of the SET domain, and two nonsense mutations, Q1981* and K2067*, are truncating mutation upstream of the WW domain. In addition to known cancer driver genes such as ERBB2 (6% of cases), NRAS (3%), MET (3%), PIK3CA (1%), AKT2 (1%), TSC1 (1%), and ERBB4 (1%), several putative cancer genes were identified, such as PTPRC[23], SYNE2[24], GRIN2A[25], and CDH10[26]. The mutation pattern is summarized in Figure 2B.
Figure 2.
Distribution and pathway analysis of somatic mutations. (A) Venn diagram plot comparing somatically altered genes among our cohort (N = 16), the expansion cohort (N = 40), and TCGA-LUAD cohort (N = 14). (B) Gene profiles across EGFR/KRAS-negative tumors from never-smokers. (C) Pathway analysis of altered genes in EGFR/KRAS-negative lung adenocarcinoma from never smokers (N = 70). The most significant functions are shown. TCGA-LUAD, The Cancer Genome Atlas - Lung adenocarcinoma.
Table 2.
Mutated genes and loci information for EGFR / KRAS -negative lung adenocarcinomas from our cohort, the expansion cohort, and The Cancer Genome Atlas cohort
| Gene | Mutation frequency in our set (N = 16) | Mutation frequency in expansion set (N = 40) | Mutation frequency in TCGA set (N = 14) | Total (N) | AA substitution | COSMIC v.65 | Number of damage algorithms |
|---|---|---|---|---|---|---|---|
|
TP53 |
- |
6 of 40 |
2 of 14 |
8 of 70 |
K93R |
O |
4 of 4 |
| 122_122del |
O |
n/a |
|||||
| Y124C |
O |
4 of 4 |
|||||
| C203F |
O |
4 of 4 |
|||||
| G206S |
O |
4 of 4 |
|||||
| R209W |
O |
4 of 4 |
|||||
| E219* |
O |
2 of 4 |
|||||
| G295A |
O |
4 of 4 |
|||||
|
SETD2 |
1 of 16 |
2 of 40 |
1 of 14 |
4 of 70 |
R839* |
- |
1 of 2 |
| V1576F |
- |
3 of 3 |
|||||
| Q1981* |
- |
2 of 2 |
|||||
| K2067* |
- |
0 of 1 |
|||||
|
CSMD3 |
1 of 16 |
1 of 40 |
2 of 14 |
4 of 70 |
P667S |
O |
3 of 4 |
| M1440I |
- |
3 of 4 |
|||||
| K1928N |
- |
3 of 4 |
|||||
| Y2028C |
- |
3 of 4 |
|||||
|
PTPRC |
2 of 16 |
1 of 40 |
- |
3 of 70 |
Y444N |
O |
3 of 4 |
| T453M |
- |
3 of 4 |
|||||
| T1176M |
- |
4 of 4 |
|||||
|
SYNE2 |
1 of 16 |
2 of 40 |
- |
3 of 70 |
2579_2580del |
- |
n/a |
| E3880* |
- |
n/a |
|||||
| E3903K |
O |
3 of 4 |
|||||
|
GRIN2A |
- |
3 of 40 |
- |
3 of 70 |
L307Q |
- |
3 of 4 |
| N886S |
O |
0 of 4 |
|||||
| T1069M |
O |
3 of 4 |
|||||
|
CDH10 |
- |
2 of 40 |
1 of 14 |
3 of 70 |
E171K |
O |
4 of 4 |
| D315H |
O |
1 of 4 |
|||||
| R472C |
O |
4 of 4 |
|||||
|
ERBB2 |
- |
- |
3 of 14 |
3 of 70 |
E740delinsEAYVM (2) |
- |
n/a |
| G746delinsVC |
O |
n/a |
|||||
|
SMAD4 |
- |
- |
3 of 14 |
3 of 70 |
S242* |
- |
1 of 2 |
| R361S |
O |
4 of 4 |
|||||
| D493N |
O |
3 of 4 |
|||||
|
CDC27 |
2 of 16 |
- |
- |
2 of 70 |
I235T |
- |
4 of 4 |
| A273G |
O |
3 of 4 |
|||||
|
GABRG1 |
2 of 16 |
- |
- |
2 of 70 |
D43N |
- |
2 of 4 |
| T318S |
- |
3 of 4 |
|||||
|
YTHDF1 |
2 of 16 |
- |
- |
2 of 70 |
I492V (2) |
- |
4 of 4 |
|
PCDHB14 |
2 of 16 |
- |
- |
2 of 70 |
Y670S (2) |
- |
2 of 3 |
|
MECP2 |
1 of 16 |
- |
1 of 14 |
2 of 70 |
R168L |
- |
2 of 3 |
| G273V |
O |
3 of 3 |
|||||
|
NRAS |
- |
2 of 40 |
- |
2 of 70 |
Q61H |
O |
2 of 4 |
| Q61K |
O |
3 of 4 |
|||||
|
MET |
- |
- |
2 of 14 |
2 of 70 |
Y1021* |
- |
2 of 2 |
| 1027_1028del |
- |
NA |
|||||
|
EPHA2 |
- |
2 of 40 |
- |
2 of 70 |
P147L |
O |
4 of 4 |
| P278S |
O |
2 of 4 |
|||||
|
CHD2 |
- |
2 of 40 |
- |
2 of 70 |
S391fs |
- |
n/a |
| splicing(c.5153) |
O |
n/a |
|||||
|
MST1 |
- |
- |
2 of 14 |
2 of 70 |
406_407del |
- |
n/a |
| R535G |
- |
3 of 4 |
|||||
|
ERBB4 |
1 of 16 |
- |
- |
1 of 70 |
V840I |
O |
4 of 4 |
|
PIK3CA |
1 of 16 |
- |
- |
1 of 70 |
G118D |
O |
2 of 3 |
|
AKT2 |
1 of 16 |
- |
- |
1 of 70 |
D153Y |
- |
4 of 4 |
|
TSC1 |
1 of 16 |
- |
- |
1 of 70 |
Q793* |
- |
2 of 2 |
|
CTNNB1 |
1 of 16 |
- |
- |
1 of 70 |
S37C |
O |
4 of 4 |
|
TGFBR2 |
1 of 16 |
- |
- |
1 of 70 |
R504W |
O |
4 of 4 |
|
PBRM1 |
1 of 16 |
- |
- |
1 of 70 |
E830* |
- |
2 of 2 |
|
MBD2 |
1 of 16 |
- |
- |
1 of 70 |
E373K |
- |
4 of 4 |
|
CHEK2 |
1 of 16 |
- |
- |
1 of 70 |
R191G |
- |
3 of 4 |
|
PARP4 |
1 of 16 |
- |
- |
1 of 70 |
I1039T |
O |
4 of 4 |
|
CUL3 |
1 of 16 |
- |
- |
1 of 70 |
Q516* |
- |
2 of 2 |
|
SOD2 |
1 of 16 |
- |
- |
1 of 70 |
K194E |
- |
3 of 4 |
|
GABRD |
1 of 16 |
- |
- |
1 of 70 |
D174E |
- |
4 of 4 |
|
NDRG1 |
1 of 16 |
- |
- |
1 of 70 |
L257fs |
- |
n/a |
|
TFG |
1 of 16 |
- |
- |
1 of 70 |
E240* |
- |
1 of 1 |
|
SYK |
1 of 16 |
- |
- |
1 of 70 |
A400S |
- |
4 of 4 |
|
ASPSCR1 |
1 of 16 |
- |
- |
1 of 70 |
L252Q |
- |
3 of 3 |
| PCDHB15 | 1 of 16 | - | - | 1 of 70 | Y670S | - | 3 of 3 |
COSMIC, Catalogue of Somatic Mutations in Cancer; n/a, not available; TCGA, The Cancer Genome Atlas.
Pathway analysis of 1,760 genes that were altered in 70 EGFR/KRAS-negative tumors of never-smokers revealed alterations in genes related to DNA repair and the cell cycle, including components of p53/ATM signaling, G1/S or G2/M checkpoint regulation, and non-homologous end joining (Figure 2C). The most significantly enriched pathway was cAMP-dependent protein kinase A signaling, which can activate the mitogen-activated protein kinase cascade in lung adenocarcinoma [27]. Other enriched functions of altered genes were calcium transport (P = 0.006), axonal guidance (P = 0.015), and Ephrin A signaling (P = 0.031).
Discussion
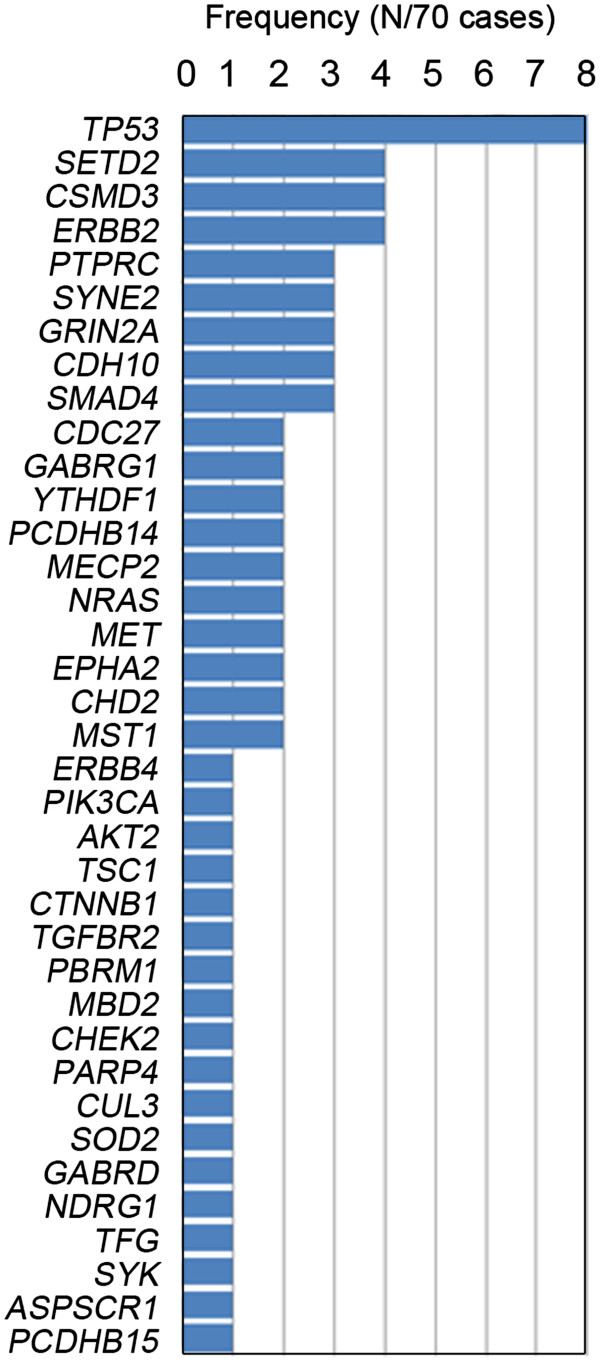
The somatic mutation profile in lung adenocarcinomas lacking targetable EGFR or KRAS mutations or ALK rearrangements in never-smokers is highly complex. Our exome analysis of 70 tumors identified several common mutations involving the known cancer genes TP53, NRAS, ERBB2, PIK3CA, and CTNNB1, but also mutations in SETD2, CSMD3, PTPRC, and SYNE2 (Figure 3).
Figure 3.
Significantly mutated genes in EGFR / KRAS -negative lung adenocarcinomas from never-smokers (N = 70).
SETD2 (mutated in 6% of cases) is a histone methyltransferase that is involved in transcriptional elongation and chromatin remodeling. Interaction with p53 is facilitated by the SET and WW domains and might increase p53 stability [22]. Interestingly, SETD2 and TP53 mutations were mutually exclusive in lung adenocarcinoma of never-smokers (Figure 2B). CSMD3 (mutated in 6% of cases) is a transmembrane protein with CUB and sushi multiple domains that is thought to function in protein-protein interactions and the immune response. Recent studies showed that loss of CSMD3 increases proliferation of airway epithelial cells [9] and may be involved in tumorigenesis in lung cancer. In our study, missense mutations (P667S, M1440I, K1928N, and Y2028C) in CSMD3 were predicted to be deleterious to protein function. PTPRC (mutated in 4% of cases) is a member of the protein tyrosine phosphatase family and regulates a variety of cellular processes including cell growth, differentiation, and tumorigenesis. PTPRC regulates the JAK/STAT signaling pathway and functional defects can activate JAK/STAT signaling [23]. We observed three missense mutations (Y444N, T453M, T1176M) in PTPRC, all of which were predicted to be deleterious. SYNE2 plays a role in cadherin-mediated cell-cell adhesion and regulates the Wnt signaling pathway [24].
We identified several targetable pathways in EGFR/KRAS/ALK-negative lung adenocarcinoma including PI3K/mTOR signaling (TSC1, PIK3CA, AKT2), receptor tyrosine kinase signaling (ERBB4), cell cycle regulation (CHEK2, CDC27), and DNA repair (PARP4). PI3K pathway inhibitors and cell cycle inhibitors are actively under investigation for lung adenocarcinoma in preclinical and early phase clinical trials [28,29]. A current mutation screening program for tailored targeted therapies is also on-going in 1,000 patients with advanced lung adenocarcinoma based on 10 single driver mutations: KRAS (25%), EGFR (23%), ALK rearrangements (6%), BRAF (3%), PIK3CA (3%), MET amplifications (2%), ERBB2 (1%), MEK1 (0.4%), NRAS (0.2%), and AKT1 (<0.1%) [30]. Additional genomic alterations will be incorporated in a comprehensive manner based on next-generation sequencing data.
More than 200 putative cancer-causing genes have been identified in recent genomic landscape studies using next-generation sequencing technology, and several cellular processes not previously implicated in cancer have been revealed, such as chromatin remodeling, splicing, and ubiquitination [31,32]. We identified alterations in genes involved in chromatin remodeling (PBRM1, SETD2), oxidative stress (CUL3, SOD2), immune response (CSMD3, SYK), and gamma-aminobutyric acid receptor signaling (GABRD, GABRG1) in lung adenocarcinoma. Interestingly, although somatic mutation is rare in EGFR/KRAS/ALK-negative lung adenocarcinoma of never-smokers, the PCDHB14 (cell adhesion) Y670S mutation and YTHDF1 (RNA binding) I492V mutations were each found in two cases (12.5%). Future studies to elucidate the role of these newly implicated functions in tumorigenesis are warranted.
Conclusions
We identified novel somatic mutations in EGFR/KRAS/ALK-negative lung adenocarcinoma in never-smokers and investigated the mutation frequency of altered genes. EGFR/KRAS/ALK-negative lung adenocarcinoma in never-smokers is highly heterogeneous at the somatic mutation level. However, most of the altered genes were involved in the cell cycle, and might represent novel therapeutic targets in lung adenocarcinoma. Future research on the functional role of chromatin remodeling, oxidative stress/differentiation, and the immune response will enhance our understanding of the mechanisms of tumorigenesis.
Abbreviations
ALK: anaplastic lymphoma kinase; COSMIC: Catalogue of Somatic Mutations in Cancer; EGFR: epidermal growth factor receptor; MIP: molecular-inversion probe; PCR: polymerase chain reaction; TCGA: The Cancer Genome Atlas.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JWA and HSK analyzed the data and wrote the manuscript. JKY, HJ, SE, and SMH performed the experiments and designed the molecular inversion probes for validation. HSS performed pathologic review. HJK, DJK, JGL, CYL, MKB, and KYC prepared the samples for exome sequencing. JYJ, EYK, SKK, JC, and MGL helped to revise the manuscript. HRK and JHK provided clinical information. BCC, JHL, and DB designed and managed the study. All authors read and approved the final manuscript.
Supplementary Material
Analysis flow chart for exome-sequencing data. Table S1. Summary of depth and coverage of whole exome sequencing. Table S2. Summary of depth and coverage in target capture sequencing for validation. Table S3. Validation results using target capture sequencing and Sanger sequencing. Table S4. Summary of validated somatic exonic mutations in EGFR/KRAS/ALK-negative lung adenocarcinomas. Table S5. Somatic mutations in EGFR/KRAS/ALK-negative lung adenocarcinoma exomes. Table S6. Mutated genes and loci information in EGFR/KRAS-negative lung adenocarcinoma.
Sequences of molecular inversion probes. Table S8. Sequences of primers used for Sanger sequencing.
Contributor Information
Jin Woo Ahn, Email: jinwoo.pinokios89@gmail.com.
Han Sang Kim, Email: modeerfhs@gmail.com.
Jung-Ki Yoon, Email: neododari@gmail.com.
Hoon Jang, Email: nachtstraum2@gmail.com.
Soo Min Han, Email: betsoom@gmail.com.
Sungho Eun, Email: dkrtndhkd@gmail.com.
Hyo Sup Shim, Email: SHIMHS@yuhs.ac.
Hyun-Jung Kim, Email: hjkim7@yuhs.ac.
Dae Joon Kim, Email: KDJCOOL@yuhs.ac.
Jin Gu Lee, Email: CSJGLEE@yuhs.ac.
Chang Young Lee, Email: CYLEECS@yuhs.ac.
Mi Kyung Bae, Email: MKBAE@yuhs.ac.
Kyung Young Chung, Email: KYCHU@yuhs.ac.
Ji Ye Jung, Email: STOPYES@yuhs.ac.
Eun Young Kim, Email: NARAE97@yuhs.ac.
Se Kyu Kim, Email: SEKYUKIM@yuhs.ac.
Joon Chang, Email: CHANG@yuhs.ac.
Hye Ryun Kim, Email: NOBELG@yuhs.ac.
Joo Hang Kim, Email: KJHANG@yuhs.ac.
Min Goo Lee, Email: MLEE@yuhs.ac.
Byoung Chul Cho, Email: cbc1971@yuhs.ac.
Ji Hyun Lee, Email: JIHYNI@yuhs.ac.
Duhee Bang, Email: duheebang@yonsei.ac.kr.
Acknowledgments
This study was supported in part by a grant from the Korea Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI12C1440, BCC) and by a grant from the National Project for Personalized Genomic Medicine, Korea Health 21 R&D Project (A111218-11-PG03). The results published here are in part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at “http://cancergenome.nih.gov”.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Wakelee HA, Chang ET, Gomez SL, Keegan TH, Feskanich D, Clarke CA, Holmberg L, Yong LC, Kolonel LN, Gould MK, West DW. Lung cancer incidence in never smokers. J Clin Oncol. 2007;25:472–478. doi: 10.1200/JCO.2006.07.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Kim JH, Kim SK, Ha SJ, Mok TS, Mitsudomi T, Cho BC. Lung cancer in never smokers: change of a mindset in the molecular era. Lung Cancer. 2011;72:9–15. doi: 10.1016/j.lungcan.2010.12.013. [DOI] [PubMed] [Google Scholar]
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, Varella-Garcia M, Kim WH, Lynch TJ, Fidias P, Stubbs H, Engelman JA, Sequist LV, Tan W, Gandhi L, Mino-Kenudson M, Wei GC, Shreeve SM, Ratain MJ, Settleman J, Christensen JG, Haber DA, Wilner K, Salgia R, Shapiro GI, Clark JW. et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, Kim YT, Kim JI, Kang JH, Seo JS. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–445. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Morrison C, Wang L, Xiong D, Vedell P, Cui P, Hua X, Ding F, Lu Y, James M, Ebben JD, Xu H, Adjei AA, Head K, Andrae JW, Tschannen MR, Jacob H, Pan J, Zhang Q, Van den Bergh F, Xiao H, Lo KC, Patel J, Richmond T, Watt MA, Albert T, Selzer R, Anderson M, Wang J, Wang Y. et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis. 2012;33:1270–1276. doi: 10.1093/carcin/bgs148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong D, Li G, Li K, Xu Q, Pan Z, Ding F, Vedell P, Liu P, Cui P, Hua X, Jiang H, Yin Y, Zhu Z, Li X, Zhang B, Ma D, Wang Y, You M. Exome sequencing identifies MXRA5 as a novel cancer gene frequently mutated in non-small cell lung carcinoma from Chinese patients. Carcinogenesis. 2012;33:1797–1805. doi: 10.1093/carcin/bgs210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansen S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, Maher CA, Fulton R, Fulton L, Wallis J, Chen K, Walker J, McDonald S, Bose R, Ornitz D, Xiong D, You M, Dooling DJ, Watson M, Mardis ER, Wilson RK. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, Lee J, Jung YJ, Kim JO, Shin JY, Yu SB, Kim J, Lee ER, Kang CH, Park IK, Rhee H, Lee SH, Kim JI, Kang JH, Kim YT. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, Sakamoto H, Tsuta K, Furuta K, Shimada Y, Iwakawa R, Ogiwara H, Oike T, Enari M, Schetter AJ, Okayama H, Haugen A, Skaug V, Chiku S, Yamanaka I, Arai Y, Watanabe S, Sekine I, Ogawa S, Harris CC, Tsuda H, Yoshida T, Yokota J, Shibata T. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Medical Association. Declaration of Helsinki - Ethical Principles for Medical Research Involving Human Subjects. [ http://www.wma.net/en/30publications/10policies/b3/]
- National Center for Biotechnology Information. The Sequence Read Archive (SRA) [ http://trace.ncbi.nlm.nih.gov/Traces/sra/]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- Ingenuity Pathway Analysis. [ http://www.ingenuity.com]
- R statistical software version 2.15.3. [ http://www.r-project.org]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas. Lung adenocarcinoma (LUAD. [ https://tcga-data.nci.nih.gov/tcga/]
- Xie P, Tian C, An L, Nie J, Lu K, Xing G, Zhang L, He F. Histone methyltransferase protein SETD2 interacts with p53 and selectively regulates its downstream genes. Cell Signal. 2008;20:1671–1678. doi: 10.1016/j.cellsig.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Porcu M, Kleppe M, Gianfelici V, Geerdens E, De Keersmaecker K, Tartaglia M, Foa R, Soulier J, Cauwelier B, Uyttebroeck A, Macintyre E, Vandenberghe P, Asnafi V, Cools J. Mutation of the receptor tyrosine phosphatase PTPRC (CD45) in T-cell acute lymphoblastic leukemia. Blood. 2012;119:4476–4479. doi: 10.1182/blood-2011-09-379958. [DOI] [PubMed] [Google Scholar]
- Neumann S, Schneider M, Daugherty RL, Gottardi CJ, Eming SA, Beijer A, Noegel AA, Karakesisoglou I. Nesprin-2 interacts with {alpha}-catenin and regulates Wnt signaling at the nuclear envelope. J Biol Chem. 2010;285:34932–34938. doi: 10.1074/jbc.M110.119651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, Davis S, Program NCS, Stemke-Hale K, Davies MA, Gershenwald JE, Robinson W, Robinson S, Rosenberg SA, Samuels Y. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MM, Ellis SM, Auza MJ, Patel A, Clark P. The intercellular adhesion molecule, cadherin-10, is a marker for human prostate luminal epithelial cells that is not expressed in prostate cancer. Mod Pathol. 2008;21:85–95. doi: 10.1038/modpathol.3800988. [DOI] [PubMed] [Google Scholar]
- Schuller HM. Mechanisms of smoking-related lung and pancreatic adenocarcinoma development. Nat Rev Cancer. 2002;2:455–463. doi: 10.1038/nrc824. [DOI] [PubMed] [Google Scholar]
- Spoerke JM, O'Brien C, Huw L, Koeppen H, Fridlyand J, Brachmann RK, Haverty PM, Pandita A, Mohan S, Sampath D, Friedman LS, Ross L, Hampton GM, Amler LC, Shames DS, Lackner MR. Phosphoinositide 3-kinase (PI3K) pathway alterations are associated with histologic subtypes and are predictive of sensitivity to PI3K inhibitors in lung cancer preclinical models. Clin Cancer Res. 2012;18:6771–6783. doi: 10.1158/1078-0432.CCR-12-2347. [DOI] [PubMed] [Google Scholar]
- Dickson MA, Schwartz GK. Development of cell-cycle inhibitors for cancer therapy. Curr Oncol. 2009;16:36–43. doi: 10.3747/co.v16i2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kris BEJ MG, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Aronson SL, Engelman JA, Shyr Y, Khuri FR, Rudin CM, Garon EB, Pao W, Schiller JH, Haura EB, Shirai K, Giaccone G, Berry LD, Kugler K, Minna JD, Bunn PA. Identification of driver mutations in tumor specimens from 1,000 patients with lung adenocarcinoma: The NCI’s Lung Cancer Mutation Consortium (LCMC) J Clin Oncol. 2011;29:abstr CRA7506. [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis flow chart for exome-sequencing data. Table S1. Summary of depth and coverage of whole exome sequencing. Table S2. Summary of depth and coverage in target capture sequencing for validation. Table S3. Validation results using target capture sequencing and Sanger sequencing. Table S4. Summary of validated somatic exonic mutations in EGFR/KRAS/ALK-negative lung adenocarcinomas. Table S5. Somatic mutations in EGFR/KRAS/ALK-negative lung adenocarcinoma exomes. Table S6. Mutated genes and loci information in EGFR/KRAS-negative lung adenocarcinoma.
Sequences of molecular inversion probes. Table S8. Sequences of primers used for Sanger sequencing.