Abstract
The Kinetoplastida are flagellated protozoa evolutionary distant and divergent from yeast and humans. Kinetoplastida include trypanosomatids, and a number of important pathogens. Trypanosoma brucei, Trypanosoma cruzi and Leishmania spp. inflict significant morbidity and mortality on humans and livestock as the etiological agents of human African trypanosomiasis, Chagas' disease and leishmaniasis respectively. For all of these organisms, intracellular trafficking is vital for maintenance of the host–pathogen interface, modulation/evasion of host immune system responses and nutrient uptake. Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) are critical components of the intracellular trafficking machinery in eukaryotes, mediating membrane fusion and contributing to organelle specificity. We asked how the SNARE complement evolved across the trypanosomatids. An in silico search of the predicted proteomes of T. b. brucei and T. cruzi was used to identify candidate SNARE sequences. Phylogenetic analysis, including comparisons with yeast and human SNAREs, allowed assignment of trypanosomatid SNAREs to the Q or R subclass, as well as identification of several SNAREs orthologous with those of opisthokonts. Only limited variation in number and identity of SNAREs was found, with Leishmania major having 27 and T. brucei 26, suggesting a stable SNARE complement post-speciation. Expression analysis of T. brucei SNAREs revealed significant differential expression between mammalian and insect infective forms, especially within R and Qb-SNARE subclasses, suggesting possible roles in adaptation to different environments. For trypanosome SNAREs with clear orthologs in opisthokonts, the subcellular localization of TbVAMP7C is endosomal while both TbSyn5 and TbSyn16B are at the Golgi complex, which suggests conservation of localization and possibly also function. Despite highly distinct life styles, the complement of trypanosomatid SNAREs is quite stable between the three pathogenic lineages, suggesting establishment in the last common ancestor of trypanosomes and Leishmania. Developmental changes to SNARE mRNA levels between blood steam and procyclic life stages suggest that trypanosomes modulate SNARE functions via expression. Finally, the locations of some conserved SNAREs have been retained across the eukaryotic lineage.
Keywords: Trypanosoma, SNARE, Molecular evolution, Vesicle trafficking
Graphical abstract
Highlights
-
•
SNARE proteins are essential components of intracellular transport.
-
•
These proteins exhibit considerable conservation across pathogenic trypanosomes.
-
•
Some trypanosome SNARE families are expanded or lost.
-
•
Developmental changes in trypanosome SNARE expression are apparent.
-
•
Orthologous SNAREs demonstrate conserved locations and hence function.
1. Introduction
Kinetoplastids are flagellated protozoa of the Excavata supergoup and evolutionarily distant from model eukaryotes such as fungi, animals and plants [1]; the order contains many pathogenic species. Major kinetoplastid pathogens include the African trypanosomes, represented by Trypanosoma brucei, causing African trypanosomiasis in humans and nagana in livestock and largely restricted to sub-Saharan Africa, the American trypanosome, Trypanosoma cruzi, the etiological agent of Chagas' disease, and also the Leishmania species, that cause various forms of leishmaniasis in Southern Europe, Africa, Asia and America [2]. Globally, approximately 25 million people are affected by trypanosomatid infections, while the number at risk exceeds 250 million [3]. Available kinetoplastid genome sequences indicate significant conservation of gene complement and synteny [4], but different lineages cause highly distinct diseases and survive in discrete biological environments; for example T. brucei is exclusively extracellular while T. cruzi and Leishmania major invade host cells [5].
Intracellular trafficking is responsible for the transport and sorting of lipid and protein cargo between membrane-bound intracellular compartments. Trafficking requires spatially and temporally co-ordinated protein–protein interactions and is fundamental to cell growth and differentiation, nutrient uptake, immune evasion, signaling and many other processes [6]. In trypanosomes, intracellular trafficking is especially important in evading the mammalian host immune system and maintaining the surface proteome. Specifically the copy numbers of proteins and other molecules that participate directly in immune defense or other pathogenesis associated events are significantly varied during life cycle progression. A potent example of this phenomenon is T. brucei, where antigenic variation [7] requires high-level surface expression of the variant surface glycoprotein, but in addition, immune evasion is augmented by recycling of surface antigens and immunoglobulin degradation via the endocytic pathway [8,9].
Among the key proteins mediating intracellular trafficking are the Rab and ARF small GTPases, vesicle coat proteins and soluble N-ethylmaleimide-sensitive factor attachment protein receptors or SNAREs [10]. SNAREs are 10–30 kDa, subcellular compartment-specific, type II membrane proteins, characterized by a highly conserved SNARE motif, a ~ 70 amino acid block comprising hydrophobic heptad repeats [11,12]. The SNARE motif, usually located towards the C-terminus and connected to a trans-membrane domain by a short linker, is critical for forming the SNARE complex during membrane fusion [13]. Many SNARE proteins also contain additional domains at the N-terminus, that serve to regulate SNARE complex assembly, and some SNAREs deviate from this prototypical organization. For example, Homo sapiens SNAP-23, SNAP-25, SNAP-29, Syn11 and Saccharomyces cerevisiae Ykt6 all lack a trans-membrane domain but are membrane anchored via prenylation or palmitoylation [14,15]. Human SNAP-25, which contains two SNARE motifs, attaches to membranes by non-covalent association with trans-membrane domain SNAREs [16,17].
Classification of SNAREs is based on the conservation of an amino acid residue in the central polar layer of the coiled-coil SNARE complex [18]. This residue is either a glutamine (Q) or an arginine (R), and defines Q- and R-SNARE subclasses [19]. Based on the relative positions of these critical residues within the SNARE complex, Q-SNAREs are further sub-classified into Qa- (syntaxins), Qb- and Qc-SNAREs [11]. Q-SNAREs are also differentiated by their N-terminal organization. Syntaxins and a few Qb- and Qc-SNAREs contain an Habc domain three-helix bundle [20] that is thought to act as a binding site for regulatory SM proteins [19]. The Habc domain may also fold back onto the SNARE domain to give a ‘closed’ conformation, preventing interaction of cognate SNARE partners [21]. R-SNAREs are sub-classified into short vesicle-associated membrane proteins (VAMPs; brevins) and long VAMPs (longins) based on the presence of a short and variable domain or a conserved longin domain at the N-terminus respectively [22].
Comparative genomic and phylogenetic analyses have, to some degree, defined a SNARE complement for the last eukaryotic common ancestor (LECA) and thus set expectations for the complement likely present in a given eukaryotic genome. Five Qa-SNARE subfamilies appear to be ancient [54]: Syntaxin 5, 16, 18, as well as the SynPM and SynE clades, which have undergone lineage-specific expansions in animals and yeast [55,56]. The LECA Qb-SNARE complement consists of at least Vti1, Gos1, Bos1 and Sec20, while the Qc complement holds Syntaxin 6, 8, and Bet 1 as a minimum [57]. Finally, the R-SNARE complement consists of three longin subfamilies Sec22, Ykt6 and Vamp7. Vamp7 is expanded in several eukaryote lineages [55,58], and also gave rise to the brevins, Vamp1-6, 8 and Snc1/2, which are believed to be opisthokont-specific [59].
Given that intracellular membrane transport is so critical for immune evasion and other cellular processes in trypanosomes, a detailed understanding of the process is clearly of importance. The roles of many proteins in trafficking in T. brucei and additional trypanosomatids have been described [23,24], but the contributions made by members of the SNARE repertoire remain to be elucidated. Building on an earlier investigation of L. major SNAREs [25], we identified and classified the putative SNARE complement in predicted proteomes of T. brucei and T. cruzi. These, together with L. major and opisthokont reference sequences, allow a classification for trypanosome SNAREs to be derived. Additionally, we predicted the domain structures and investigate the expression profile of the T. brucei SNAREs. Finally, by determining the subcellular location of a select cohort of the SNAREs that are conserved between trypanosomes, animals and fungi, we provide evidence for retention of a similar location of orthologous SNAREs across the eukaryota.
2. Materials and methods
2.1. Genome searches for candidate SNARE open reading frames
The predicted proteomes of T. brucei and T. cruzi were obtained from EuPathDB (http://eupathdb.org/eupathdb/) and formatted into BLAST searchable databases. Validated Leishmania major SNAREs [25] were used to query the formatted databases using BLASTP [26] with cut-off E-value of 0.0001, given the short length of the proteins. Domain content predictions for the retrieved sequences were generated at the PFAM [27] and PROSITE [28] domain databases. Only sequences predicted to contain the SNARE domain were retained as potential homologues. These sequences were aligned using MUSCLE (62) and manually edited using JALVIEW (63) and subsequently used to create a Hidden Markov Model (HMM) profile that was used to exhaustively reinterrogate the T. brucei and T. cruzi genomes for distant homologues using the HMMER package [29]. Additionally, in cases where one kinetoplastid ortholog of a clade was not initially identified, BLASTp searches using the relevant sequences of the other trypanosomatids were performed. Trans-membrane (TM) domain topology prediction was performed using SMART [60]. Fold recognition was performed using the fold threading software PHYRE (www.sbg.bio.ic.ac.uk/~phyre).
2.2. Sequence alignment and phylogenetic reconstruction
Multiple sequence alignments were generated using MUSCLE [30] and manually edited in MacClade v4.08 to only retain unambiguously aligned regions [31]. Phylogenetic reconstruction was performed using two separate methods. To obtain the best Bayesian trees, topology and posterior probability values, the program MrBayes v3.2.1 [32] was used with the following run parameters; prset aamodelpr = fixed(WAG); mcmc ngen = 10,000,000; samplefreq = 1000; nchains = 4; startingtree = random; sump burnin = 2500; sumt burnin = 2500. Posterior probabilities were used as a measure of node robustness. All calculations were checked for convergence by running the analysis to split frequencies of < 0.1. Maximum-likelihood analysis was performed using the program PhyML v3.0 [33] with the following parameters; nb bootstrapped datasets = 100; substitution model = LG; proportion invariable sites = 0.0; and nb categories = 4. The model of sequence evolution prior to each PhyML analysis was determined using Prot-Test v3.2.1 [34] and included corrections for rate variation used to determine the best substitution model and invariable sites where applicable. Trees were rendered using FigTree v1.2 [35]. To identify SNAREs that are conserved between trypanosomes, humans and yeast, opisthokont landmark sequences were included in the analyses. In some cases selected opisthokont-specific duplications of subfamilies were excluded to alleviate phylogenetic artifact. For R-SNAREs, only longin landmark sequences were used.
2.3. Trypanosome cell culture
Bloodstream form cells of T. brucei Lister 427 (wild-type 427, WT427) and the derived single marker bloodstream (SMB) line [36] were cultured in HMI-9 complete medium (Gibco) [37] supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Biosera), 100 U/mL penicillin, 100 U/mL streptomycin (Gibco) and 2 mM l-glutamine (Gibco), maintained at 37 °C with 5% CO2 in a humid atmosphere in non-adherent culture flasks with vented caps. Cells were maintained at densities between 105 and 5 × 106 cells/mL. Ectopic expression of plasmid constructs was maintained using G418 antibiotic selection at 2.5 μg/mL [38].
2.4. Recombinant DNA constructs
Putative trypanosome SNAREs Tb927.9.3820 (TbQc1B), Tb10.70.7410 (TbVAMP7C), Tb927.10.14200 (TbSyn5) and Tb09.211.3920 (TbSyn16B) were PCR amplified from trypanosome 427 genomic DNA using Vent DNA polymerase (New England BioLabs). For hemagglutinin (HA)-tag fusion constructs of Tb927.9.3820 (TbQc1B), and Tb10.70.7410 (TbVAMP7C), the PCR products were cloned into the BSF expression vector pXS5, containing sequence for a C-terminal HA-epitope, using HindIII and ApaI or HindIII and ClaI using the following primers: Tb927.9.3820-F5′-GCAAGCTTATGTCGGATGTAAAAGGG and Tb927.9.3820-R3′-GCGGGCCCCCTAGACATGTTGTATATCGC; Tb10.70.7410-F5′-GCAAGCTTATGCAGGGAGGAACAAAA and Tb10.70.7410-R3′-GCGGGCCCCTTCTTTTCCTCTTTTTT. For hemagglutinin (HA)-tag fusion constructs of Tb927.10.14200 and Tb09.211.3920, the PCR products were cloned into the BSF expression vector pHD1034, containing sequence for a C-terminal HA-epitope, using HindIII restriction site and the following primers: Tb927.10.14200-F5′-ATCGAAGCTTTTATGGTTGTAGAGCG and Tb927.10.14200-R5′-AACAGGATCCCTAGCGCACAACG; Tb09.211.3920-F5′-ATATAAGCTTTTATGGCGACCCGTGACC and Tb09.211.3920-R5′-GAGCGGATCCTTAAGACAAGCATC. All constructs were verified by standard sequencing methods (Geneservice Ltd) and linearized with NotI, XhoI or BsmI as appropriate, prior to transfection into cells. Clonal transformants were selected by resistance to 2.5 μg/mL G418 (Sigma) and/or 0.2 μg/mL puromycin.
2.5. Transfection of T. brucei
Transfections were performed using the Amaxa human T-Cell Nucleofector® kit (Amaxa, Koeln, Germany) following the manufacturer's guidelines with a few modifications. Briefly, 3 × 107 log phase cells were harvested at 800 ×g for 10 min at 4 °C and re-suspended in 100 μL of ice-cold Amaxa Human T-Cell solution. Linearized DNA plasmid (10 μg) was placed in a cuvette to which the cells were immediately added. The sample was transfected using the Amaxa Human Nucleofector®II running program X-001. Electroporation mixtures were immediately transferred to flasks containing pre-warmed HMI-9 complete medium. After 12 h, selection antibiotic was added to each and the culture was distributed into a 24-well plate and subsequently incubated at 37 °C. Positive transformants were selected on the 5th or 6th day after transfection.
2.6. Quantitative real-time polymerase chain reaction
1 × 108 cells were harvested at 800 ×g for 10 min at 4 °C and washed with ice-cold PBS and quick frozen in dry ice for 1 min. RNA was extracted using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions and quantified using a ND-1000 spectrophotometer and Nanodrop software (Nanodrop Technologies). qRT-PCR was performed using iQ-SYBRGreen Supermix on a MiniOpticon Real-Time PCR Detection System (Bio-Rad). Quantification was done using Opticon3 software (Bio-Rad).
2.7. Western blot analysis
Cells were harvested at 800 ×g for 10 min at 4 °C and washed once with ice-cold phosphate-buffered saline (PBS). Samples were then re-suspended in 20 μl 2 × sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer, heated to 95 °C for 10 min, and then subjected to SDS-PAGE. Separated proteins were then electroblotted onto Immobilon-P membrane (Millipore Corp.). Membranes were then blocked with 5% skimmed milk in TBS (137 mM NaCI, 2.7 mM KCI, 25 mM Tris base, pH 7.4, 0.2% Tween 20) for 1 h at room temperature. Probing with the primary antibody (mouse anti-HA epitope immunoglobulin G (IgG) at 1:10,000 dilution; Santa Cruz Biotechnology) was then carried out overnight at 4 °C. Membranes were washed twice with TBS and probed with secondary antibody (rabbit anti-mouse peroxidase-conjugate at 1:10,000 dilution) for 1 h at room temperature. Bound antibodies were detected by enhanced chemiluminescence using Biomax MR-1 films (Kodak). Films were scanned and, where relevant, quantitated using ImageJ software (NIH).
2.8. Immunofluorescence analysis (IFA)
For immunofluorescence analysis, bloodstream parasites in exponential growth were harvested by centrifugation at 800 ×g for 10 min at 4 °C and washed with ice-cold Voorheis's-modified phosphate-buffered saline (vPBS; PBS supplemented with 10 mM glucose and 46 mM sucrose, pH 7.6). Cells were then fixed in 3% parafomaldehyde in vPBS for 10 min at 4 °C. Fixed cells were then applied to poly-lysine microscope slides (VWR International) sectioned with an ImmEdge Pen (Vector Laboratories) for 40 min. For permeabilization, cells were incubated with 0.1% Triton-X100 in PBS for 5 min at room temperature and washed three times for 5 min with PBS. Samples were then blocked with 20% FCS in PBS for 1 h at room temperature. Fixed cells were incubated with primary antibodies for 1 h followed by three washes for 5 min in PBS. Secondary antibodies were then applied for 1 h at room temperature and washed again three times with PBS. Samples were then dried and coverslips were mounted using Vectashield mounting medium supplemented with DAPI (Vector Laboratories, Inc.). Coverslips were sealed with nail varnish (Max Factor Inc.). Both the primary (mouse anti-HA epitope immunoglobin G (IgG); Santa Cruz Biotechnology Inc.) and the secondary (anti-mouse Oregon Green; Molecular Probes or anti-mouse Alexafluor-red as appropriate) were used at a dilution of 1:1000. Cells were examined on a Nikon Eclipse 400 epifluorescence microscope fitted with a Hamamatsu CCD digital camera. Image acquisition was performed with Metamorph software (Molecular Devices, Version 6). Images were processed for presentation with Adobe Photoshop (Adobe Systems Inc.). Quantitation was performed on the raw image data with no prior processing.
3. Results and discussion
3.1. Evolutionary relationships of trypanosomatid SNAREs
Homology searching of the predicted proteomes of T. brucei [39] and T. cruzi yielded a putative SNARE complement of 26 in both cases. Similar searches into L. major yielded a complement of 27 SNAREs, consistent with the previous analysis by Besteiro and co-workers [25]. By contrast, Yoshizawa and co-workers [57], using a different methodology, identified 58 SNAREs in T. cruzi. The discrepancy is likely due to their use of an earlier and lower quality release of the genome database, which is also known to be partially polyploid and with frequent duplications.
Phylogenetic analysis was undertaken to ascertain evolutionary relationships between the predicted SNAREs of T. brucei, L. major and T. cruzi, as well as to classify the proteins into established eukaryotic SNARE subfamilies. A landmark set of SNAREs from H. sapiens and S. cerevisiae was included, as the functions of the majority of the SNAREs in these two organisms have been described. An initial analysis including SNAREs from these five organisms robustly segregated into four subclasses [11,18,40], but with poor intraclade resolution (Qa, Qb, Qc and R, data not shown). To improve resolution, SNAREs from each subclass were subsequently analyzed independently.
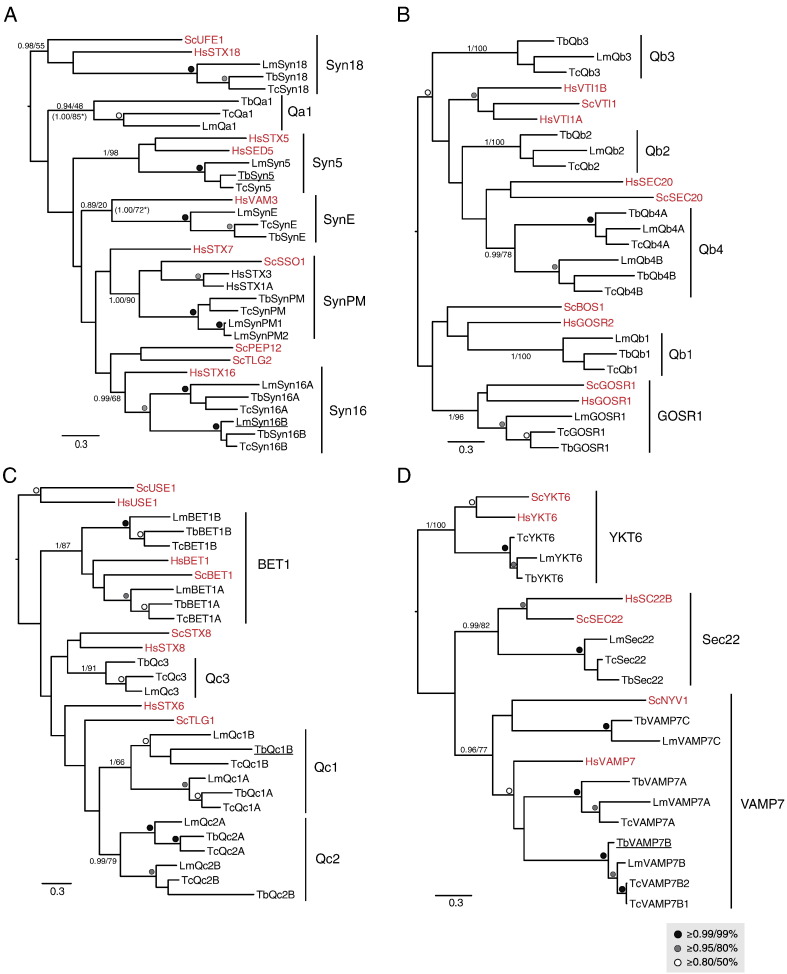
The Qa-SNARE tree (Fig. 1A) identified a set of five opisthokont SNAREs with well supported kinetoplastid orthologs: the endoplasmic reticulum (ER) Syn18 [41], Golgi localized Syn5 and Syn16, endosomally-associated SynE [42] and plasma membrane localized SynPM [41,43]. Other kinetoplastid Qa-SNAREs fell into well-supported clades, but these lack clear opisthokont members. Additionally, we observed an L. major-specific duplication of the SynPM Qa SNARES (LmSynPM1 and LmSynPM2). In the Qb-SNARE tree (Fig. 1B) only the GOSR1 clade resolved with robust support as containing both kinetoplastid and opisthokont sequences. Other tritryp SNAREs in this subclass form well-defined 1:1:1 orthologous relationships, but without identifiable opisthokont affiliation. In the Qc-SNARE tree (Fig. 1C), a clade uniting the opisthokont Bet1 sequences with two robustly supported kinetoplastid subclades was reconstructed, although without internal resolution. Additionally, we observed three Qc clades (Qc-1-3) for which opisthokont orthologs could not be robustly assigned. Qc1 and Qc2 were also reconstructed as encompassing two separate subclades each containing the three trypanosomatids examined. In the R-SNARE tree (Fig. 1D), three opisthokont SNAREs formed clades with trypanosomatid sequences; ER-Golgi Sec22, involved in anterograde transport from the ER, the Golgi-vacuole localized Ykt6, and endosomal Vamp7. Additionally, the clade of R1 contained proteins from all three trypanosomatids, but was not robustly assignable to an opisthokont ortholog (data not shown).
Fig. 1.
Phylogenetic relationships of Trypanosomatid SNAREs. In all panels, the best Bayesian topology is shown, with support values for nodes defining clades of interest given in the order of posterior probabilities (MrBayes) and bootstrap values (PhyML). All values for all other nodes above the threshold of 0.8/50% are iconized as inset. (A) Qa SNARE sub-family analysis. Note the orthology with opisthokont orthologs for Syn18, 5, E, PM, and 16. The Syntaxin16 clade includes two paralogues for each trypanosomatid species. Support values from additional phylogenetic analyses, with long-branching taxa removed, are indicated by asterisks. (B) Qb SNARE sub-family analysis showing orthology with Gos1 and four trypanosomatid Qb clades. (C) Qc SNARE sub-family analysis. Bet1 orthologs plus three additional trypanosomatid clades were reconstructed. (D) R-SNARE analysis. Orthologs for opisthokont sub-families were identified with an expansion in the Vamp7 clades in trypanosomatids. An additional clade of R-SNARE-related trypanosomatid proteins had an unstable position in the phylogeny when the sequences were included (data not shown), therefore, these sequences were removed. Accession numbers for all trypanosomatid sequences are shown in Table S1. Underlined sequences were localized in this study (see Figs. 4–6).
From these reconstructions we observed a few cases of genome-specific expansion and also of failure to identify a particular ortholog. However, overall we largely found a 1:1:1 ortholog among the trypanosomatid SNAREs, indicating general stability of the SNARE complement. This contrasts with the Rab GTPases which are represented by a larger cohort in T. cruzi and L. major than in T. brucei. In just under 50% of the cases, we were unable to identify an opisthokont ortholog for a particular clade of kinetoplastid SNAREs. Whether this is due to true biological novelty or failure of the phylogenetic methodology to resolve relationships between distantly related proteins awaits more in depth analysis, possibly with improved phylogenetic methods when they become available. Nonetheless, we were able to identify ortholog relationships of trypanosomatid SNAREs with opisthokont sequences in 10 of 19 cases; these trypanosome SNAREs are candidates for assuming equivalent cellular functions.
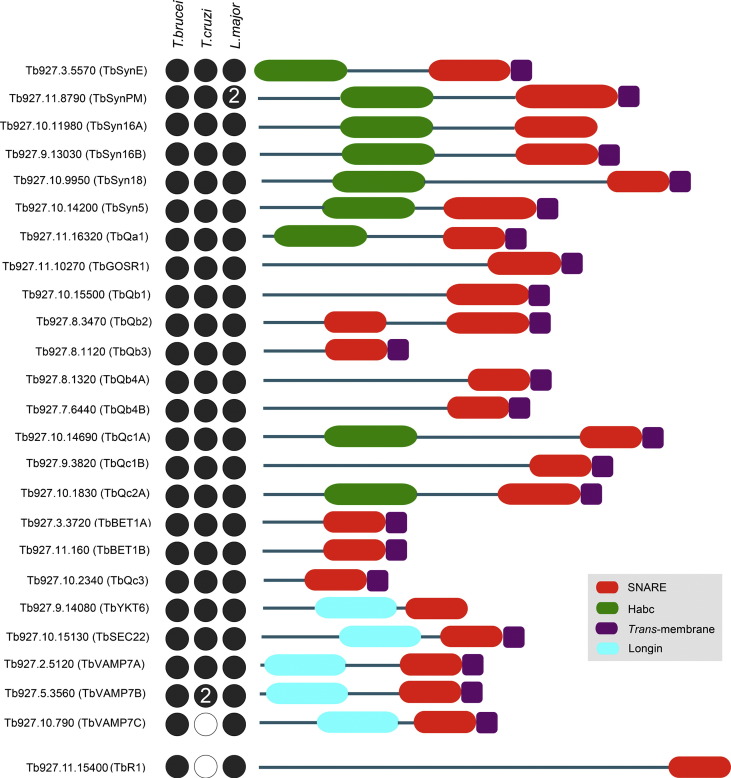
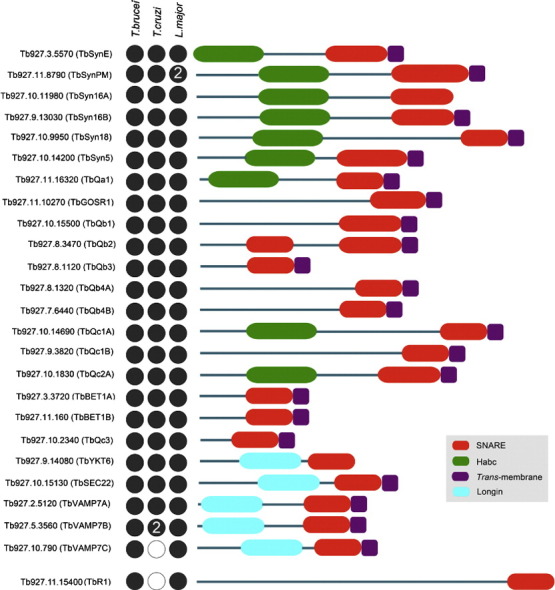
3.2. T. brucei SNARE architecture
The majority of T. brucei, T. cruzi and L. major SNAREs exhibit prototypic SNARE features, i.e. a C-terminal trans-membrane domain linked to a SNARE motif by a short linker, plus, in several, a helical N-terminal domain (Fig. 2). However, several SNAREs in both T. brucei and T. cruzi do not conform to this standard architecture. One of the non-prototypic T. brucei candidates, Tb927.8.3470 (TbQb2), is predicted to contain two putative SNARE domains at the N- and C-termini respectively. This is a unique finding given that such an architecture of N- and C-terminal SNARE domains has been reported for SNAP-23, SNAP-25, SNAP-29, Sec9p and Spo20p, but these are mainly restricted to animals, higher plants, fungi, and ciliates [61]. Further investigation of this T. brucei SNARE is warranted given that the L. major homologue (LmjF.23.1740 (LmQb2)) appears to only contain the N-terminal domain [25].
Fig. 2.
Schematic illustration of the structural organization of T. brucei SNAREs and representation among the TriRyps. Red ellipses represent the C-terminal SNARE motif, the trans-membrane domain is represented by dark purple rectangles. The Habc domain is represented by green ellipses while the N-terminus longin domain of R-SNAREs is represented by cyan ellipses. Designations are taken from GeneDB accessions. The N-terminus of the protein is drawn towards the left. Dots represent presence (black) or absence (white) from a detectable ortholog in T. brucei, L. major and T. cruzi. A numeral within a circle represents the presence of more than one ortholog. TbR1 is shown spaced from the main body as this SNARE could not be assigned using phylogenetics, but only on BLAST and domain searches.
Several T. brucei SNAREs, Tb927.9.14080 (TbYKT6), Tb927.11.15400 (TbR1) and Tb927.10.11980 (TbSyn16A), lack a C-terminal trans-membrane domain, necessitating an alternate mechanism for membrane association, for example by acylation [44]. CSS-Palm [45] and PrePS [46] algorithms predict C-terminal palmitoylation sites for TbYKT6 (Cys 201 and 202) and Tb927.11.16320 (TbQa1) (Cys 282). The T. cruzi and L. major orthologs of TbYKT6 are also predicted to be palmitoylated, at Cys201 and Cys202 respectively, while the Tb927.11.16320 (TbQa1) orthologs (TcCLB.506211.230 (TcQa1) and LmjF.19.0120 (LmQa1)) are predicted to be palmitoylated at Cys294 and 272 respectively. TbR1 is also predicted to be palmitoylated at a central residue (Cys996). In addition to acylation, SNAREs lacking a trans-membrane domain may insert into membranes via hydrophobic interactions with proteins possessing a trans-membrane motif as has been reported for SNAP-25 [16].
All T. brucei Qa-SNAREs were predicted to contain the N-terminal Habc domain (Fig. 2). This domain regulates SNARE activity by preventing coiled-coil formation. Although generally restricted to the Qa-SNAREs, the Habc domain was putatively identified in several Qc-SNAREs (TbQc1A, TbQc2A and TbQc3). Finally, the R-SNAREs appeared to possess the canonical domain structure for this subclass. Only in TbR1 did we fail to predict a longin domain.
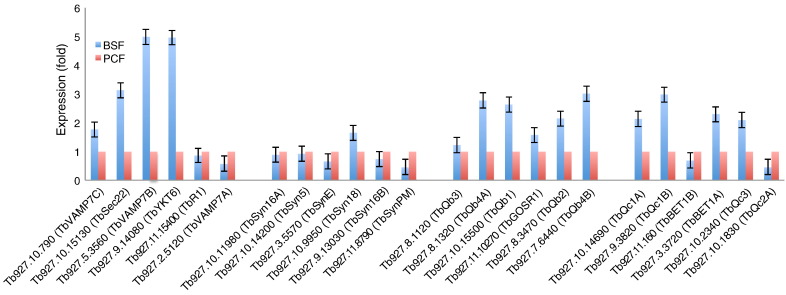
3.3. Differential expression of T. brucei SNAREs
To investigate if the identified T. brucei SNARE genes are transcribed, real-time PCR was performed, using gene-specific primers, against total RNA from both the bloodstream (BSF) and procyclic forms (PCF) of the parasite. Significant levels of transcription were found for the entire cohort. While our transcriptome data suggests that TbSyn5, TbR1 TbSyn16A and TbQb2A are constitutively expressed, a subset of T. brucei SNAREs are differentially expressed at the mRNA level between lifecycle stages. Further, consistent with earlier data [47], we also find that the SNAREs analyzed in this study are differentially expressed, with the majority being up-regulated in the BSF relative to the PCF (Fig. 3). This dynamic expression is also consistent with the earlier study by Bestiero et al. [25], which demonstrated that L. major SNAREs are differentially regulated, suggesting that this may be a general phenomenon of the trypanosomatid SNARE cohort. As membrane trafficking requirements are variable between life stages, these transcriptional changes may reflect significant changes to individual transport steps. In T. brucei, SNAREs must play a critical role in recycling of VSG, a process that requires both high rates of endocytosis as well as recycling/exocytosis. While we did observe strong up-regulation of TbVAMP7B, we saw little evidence for changes in the expression of the remaining cohort of putative endosome-associated SNAREs. By contrast to the endosomal SNAREs, there is prominent up-regulation in the BSF of TbSec22 and TbYKT6 which suggests potential modulation of specific ER exit pathways, and which may be coupled to the presence of two Rab1 orthologs and a Rab 2 ortholog in T. brucei and hence complexity in ER exit [48].
Fig. 3.
Steady state mRNA levels of T. brucei SNAREs. Triplicate RNA samples from wild type BSF and PCF cells were subjected to qRT-PCR. BSF and PCF expression levels are represented by red and blue bars respectively. Data normalization for RNA was relative to β-tubulin and telomerase reverse transcriptase (TERT) proteins. Note error bars are absent from the PCF data set as this is set at 1.0 and variance was less than 5% throughout.
3.4. Subcellular localization of trypanosome SNAREs
The sequences of several differentially expressed T. brucei SNAREs that were also found to have an ortholog in either H. sapiens or S. cerevisiae. TbSyn16B, TbVAMP7C, TbQc2A, TbVAMP7A, TbSyn5 and TbQc1B were chosen for genomic tagging in order to identify the subcellular location of the protein [47]. Multiple attempts to fuse a C-terminal hemagglutinin (HA) epitope tag to TbVAMP7A and TbQc2A were unsuccessful, but the remaining four SNAREs were successfully tagged and expressed. Intracellular localization of the HA-tagged SNARE proteins was assessed by staining with an anti-HA antibody and by co-staining cells using a selection of markers, including early endosomal epsinR, the lysosome marker p67, the plasma membrane and endosomal markers ISG65 and ISG75 and the endosomal/post-Golgi proteins clathrin, Rab5 and Golgi-located GRASP [49–53].
Immunofluorescence revealed juxtaposition between TbVAMP7C and ISG65, clathrin, epsinR and Rab5A, with the majority of the immunoreactivity localized to the region between the nucleus and kinetoplast (Fig. 4). These co-localizations indicate a possible endosomal localization for TbVAMP7C, consistent with the phylogenetic analysis. TbQc1B demonstrated a location very close to the posterior face of the nucleus, but expression levels were rather low and as a consequence localization was equivocal (Fig. 5). TbSyn5 is juxtaposed to GRASP (Fig. 6), suggesting localization to Golgi-associated structures. This was expected given the orthologous relationship with the Golgi located human Syn5 (Fig. 1A). Additionally, LmSyn5 has been experimentally localized at the Golgi [25], while TbSyn16B is also juxtaposed to the Golgi (Fig. 6). This was expected given the orthologous relationship with the Golgi localized human STX16 (Syn16) (Fig. 1A). These data suggest that for three SNAREs where orthologous relationship could be established, the locations of the trypanosome proteins suggest retention of targeting specificity with their mammalian and yeast orthologs.
Fig. 4.
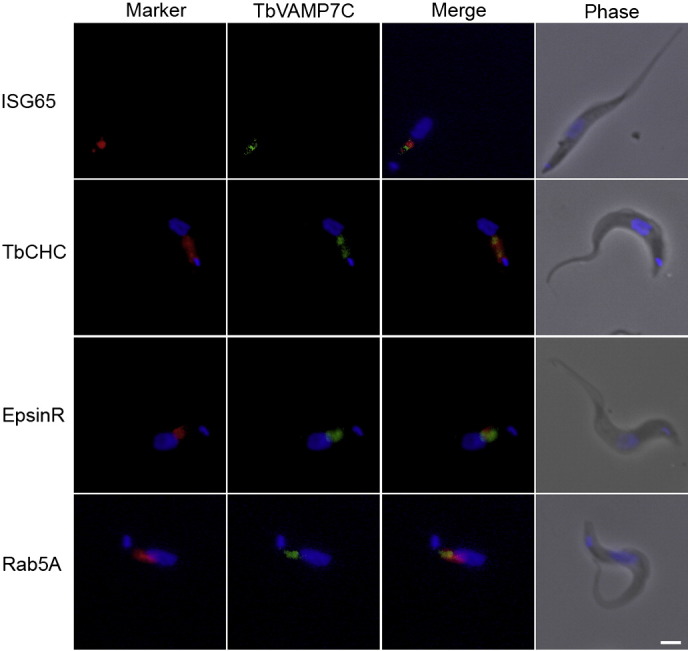
Subcellular localization of HA-tagged Tb927.10.790 (TbVAMP7C) protein in the bloodstream form of T. brucei. Shown is the localization of Tb927.10.790 (TbVAMP7C) relative to organelle markers ISG65, clathrin, epsinR and Rab5A. The tagged protein was visualized with a mouse monoclonal anti-HA antibody (green). Organelles were stained with rabbit polyclonal antibodies against specific trypanosome marker proteins (red). The nucleus and kinetoplast were stained blue with DAPI. Scale bar: 2 μm.
Fig. 5.
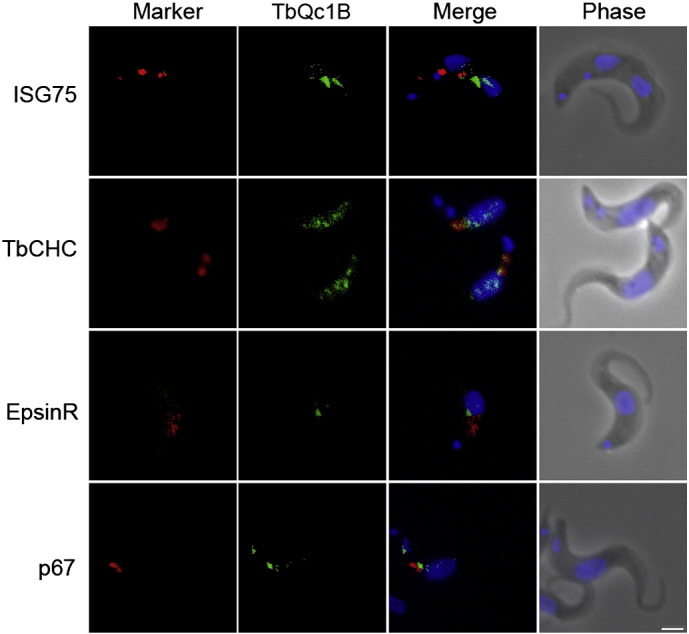
Localization of HA-tagged Tb927.9.3820 (TbQc1B) protein in the bloodstream form T. brucei. Shown is the localization of Tb927.9.3820 (TbQc1B) relative to known organelle markers ISG75, clathrin, epsinR and p67. The tagged protein was visualized with a mouse monoclonal anti-HA antibody (green). Organelles were stained with rabbit polyclonal antibodies against specific trypanosome marker proteins (red). The nucleus and kinetoplast (blue) were stained with DAPI. Scale bar: 2 μm.
Fig. 6.
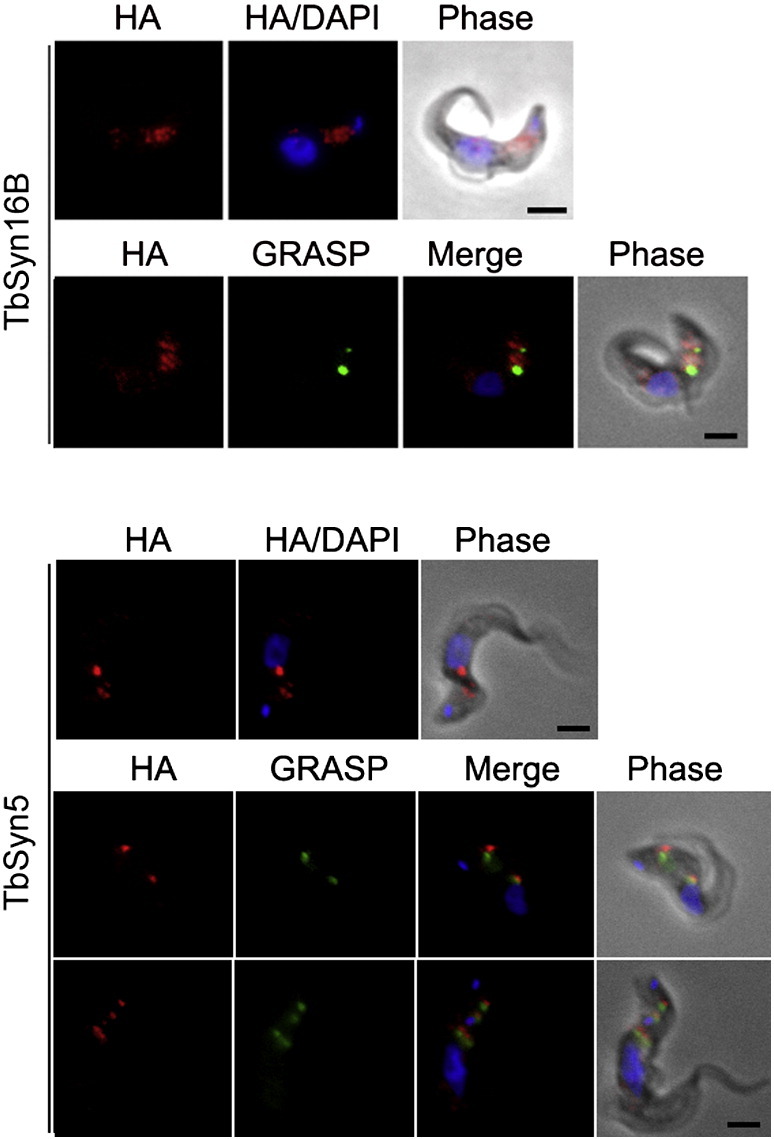
Localizations of Tb927.10.1420 (TbSyn5) and Tb927.9.13030 (Syn16B) proteins in the bloodstream form of T. brucei. Shown are the localizations of Tb927.10.1420 (TbSyn5) and Tb927.9.13030 (TbSyn16B) relative to DAPI or DAPI and GRASP. The tagged protein was visualized with a mouse monoclonal anti-HA antibody (red). Organelles were stained with rabbit polyclonal antibodies against specific trypanosome marker proteins (green). The nucleus and kinetoplast (blue) were stained with DAPI. Scale bar: 2 μm.
4. Conclusions
The SNARE repertoire appears well conserved between L. major, T. brucei and T. cruzi, with a restricted number of losses or expansions between these organisms. It is therefore unlikely that the SNARE complement plays a major role in defining the highly divergent life styles and specific pathogenesis and immune evasion mechanisms of these parasites. This contrasts with a more restricted Rab protein repertoire in African trypanosomes compared with T. cruzi and Leishmania, and further underscores the importance of Rab proteins in mediating evolution of new trafficking pathways. Any contribution from SNAREs to adaptation of the trypanosomatid trafficking system is likely in expression levels, specific amino acid changes and/or precise mechanistic aspects. Endocytosis is significantly developmentally regulated in African trypanosomes, but significantly we observed little up-regulation of SNAREs assigned as endocytosis orthologs. Experimental investigation of the three SNAREs conserved between trypanosomatids and opisthokonts suggests that the subcellular locations of the orthologs are conserved. This mirrors the conservation observed among the vast majority of Rab GTPases, and while location and function need not been fully concordant, this evidence does suggest a likely functional equivalence has been retained, in at least some aspects; direct experimental evidence is needed to verify this hypothesis. Further our phylogenetic evidence indicates that a substantial proportion of trypanosome SNAREs may be orthologous with those in other eukaryotes and consequently possibly perform similar functions. SNAREs could therefore serve as excellent cellular markers in many organisms for the definition of intracellular compartments.
The following is the supplementary data related to this article.
Accessions and assignments for TriTryp SNAREs. Assignments, details to the relevant phylogenetic reconstruction and orthology between the TriTryp SNAREs are detailed. New accession numbers for T. brucei are in bold. Note that SNAREs that have been localized are in red.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.parint.2013.11.002.
Acknowledgements
We thank Amanda O'Reilly for extensive informatics work and producing a validated set of candidate trypanosomatid SNARE ORFs. This work was supported by the South African Research Chairs Initiative, Department of Science and Technology and National Research Foundation of South Africa (to AC), the Wellcome Trust (program grant 082813 to MCF), and Canada Research Chairs program and Alberta Innovates Technology Futures (to JBD). LDB was supported by an award from the National Science and Engineering Research Council of Canada.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution–NonCommercial–No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Joel B. Dacks, Email: dacks@ualberta.ca.
Mark C. Field, Email: m.c.field@dundee.ac.uk.
Alan Christoffels, Email: alan@sanbi.ac.za.
References
- 1.Adl S.M., Simpson A.G., Lane C.E., Lukeš J., Bass D., Bowser S.S. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett M.P., Croft S.L. Management of trypanosomiasis and leishmaniasis. Br Med Bull. 2012;104:175–196. doi: 10.1093/bmb/lds031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.http://www.who.int/trypanosomiasis_african/en/
- 4.Ghedin E., Bringaud F., Peterson J., Myler P., Berriman M., Ivens A. Gene synteny and evolution of genome architecture in trypanosomatids. Mol Biochem Parasitol. 2004;134:183–191. doi: 10.1016/j.molbiopara.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 5.El-Sayed N.M., Myler P.J., Bartholomeu D.C., Nilsson D., Aggarwal G., Tran A.N. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 6.Alsford S., Field M.C., Horn D. Receptor-mediated endocytosis for drug delivery in African trypanosomes: fulfilling Paul Ehrlich's vision of chemotherapy. Trends Parasitol. 2013;29:207–212. doi: 10.1016/j.pt.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Taylor J.E., Rudenko G. Switching trypanosome coats: what's in the wardrobe? Trends Genet. 2006;22:614–620. doi: 10.1016/j.tig.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Morgan G.W., Allen C.L., Jeffries T.R., Hollinshead M., Field M.C. Devolopmental and morphological regulation of clathrin-mediated endocytosis in Trypanosoma brucei. J Cell Sci. 2001;114:2605–2615. doi: 10.1242/jcs.114.14.2605. [DOI] [PubMed] [Google Scholar]
- 9.Engstler M., Pfohl T., Herminghaus S., Boshart M., Wiegertjes G., Heddergott N. Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell. 2007;131:505–515. doi: 10.1016/j.cell.2007.08.046. [DOI] [PubMed] [Google Scholar]
- 10.Cai H., Reinisch K., Ferro-Novick S. Coats, Tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 11.Bock J.B., Matern H.T., Peden A.A., Scheller R.H. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- 12.Zwilling D., Cypionka A., Pohl W.H., Fasshauer D., Walla P.J., Wahl M.C. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J. 2007;26:9–18. doi: 10.1038/sj.emboj.7601467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jahn R., Sudhof T.C. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- 14.Fukasawa M., Varlamov O., Eng W.S., Sollner T.H., Rothman J.E. Localization and activity of the SNARE Ykt6 determined by its regulatory domain and palmitoylation. Proc Natl Acad Sci U S A. 2004;101:4815–4820. doi: 10.1073/pnas.0401183101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prekeris R., Klumperman J., Scheller R.H. Syntaxin 11 is an atypical SNARE abundant in the immune system. Eur J Cell Biol. 2000;79:771–780. doi: 10.1078/0171-9335-00109. [DOI] [PubMed] [Google Scholar]
- 16.Vogel K., Cabaniols J.P., Roche P.A. Targeting of SNAP-25 to membranes is mediated by its association with the target SNARE syntaxin. J Biol Chem. 2000;275:2959–2965. doi: 10.1074/jbc.275.4.2959. [DOI] [PubMed] [Google Scholar]
- 17.Bonifacino J.S., Glick B.S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 18.Fasshauer D., Sutton R.B., Brunger A.T., Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci U S A. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malsam J., Kreye S., Sollner T.H. Membrane fusion: SNAREs and regulation. Cell Mol Life Sci. 2008;65:2814–2832. doi: 10.1007/s00018-008-8352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez I., Ubach J., Dulubova I., Zhang X., Sudhof T.C., Rizo J. Three dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 21.Dulubova I., Sugita S., Hill S., Hosaka M., Fernandez I., Sudhof T.C. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi V., Banfield D.K., Vacca M., Dietrich L.E.P., Ungermann C., D'Espotio M. Longins and their longin domains: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem Sci. 2004;29:682–688. doi: 10.1016/j.tibs.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Field M.C., Carrington M. The trypanosome flagellar pocket. Nat Rev Microbiol. 2009;7:775–786. doi: 10.1038/nrmicro2221. [DOI] [PubMed] [Google Scholar]
- 24.Bangs J.D. Surface coats and secretory trafficking in African trypanosomes. Curr Opin Microbiol. 1998;1:448–454. doi: 10.1016/s1369-5274(98)80064-7. [DOI] [PubMed] [Google Scholar]
- 25.Besteiro S., Coombs G.H., Mottram J.C. The SNARE protein family of Leishmania major. BMC Genomics. 2006;7:250. doi: 10.1186/1471-2164-7-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Finn R.D., Mistry J., Tate J., Coggill P., Heger A., Pollington J.E. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sigrist C.J.A., Cerutti L., de Castro E., Langendijk-Genevaux P.S., Bulliard V., Bairoch A. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:161–166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 30.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview version 2 — a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2005;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 33.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 34.Abascal F., Zardoya R., Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2004;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 35.http://tree.bio.ed.ac.uk/software/figtree/
- 36.Wirtz E., Leal S., Ochatt C., Cross G.A. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 37.Hirumi H., Hirumi K. Axenic culture of African trypanosome bloodstream forms. Parasitol Today. 1994;10:80–84. doi: 10.1016/0169-4758(94)90402-2. [DOI] [PubMed] [Google Scholar]
- 38.Leung K.-F., Dacks J.D., Field M.C. Evolution of the multi-vesicular body ESCRT machinery; subunit retention and functional equivalence across eukaryotic lineage. Traffic. 2008;9:1698–1716. doi: 10.1111/j.1600-0854.2008.00797.x. [DOI] [PubMed] [Google Scholar]
- 39.Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D.C. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 40.Kloepper T.H., Kienle C.N., Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18:3463–3471. doi: 10.1091/mbc.E07-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hong W. SNAREs and traffic. Biochim Biophys Acta. 2005;1744:493–517. [PubMed] [Google Scholar]
- 42.Burri L., Lithgow T. A complete set of SNAREs in yeast. Traffic. 2004;5:45–52. doi: 10.1046/j.1600-0854.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- 43.Tai G., Lu L., Wang T.L., Tang B.L., Goud B., Johannes L. Participation of the syntaxin 5/Ykt6/GS28/GS 15 SNARE complex in transport from the early/recycling endosome to the trans-Golgi network. Mol Biol Cell. 2004;15:4011–4022. doi: 10.1091/mbc.E03-12-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borgese N., Brambillasca S., Colombo S. How tails guide tail-anchored proteins to their destinations. Curr Opin Cell Biol. 2007;19:368–375. doi: 10.1016/j.ceb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 45.Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel. 2008;21:639–644. doi: 10.1093/protein/gzn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maurer-Stroh S., Koranda M., Benetka W., Schneider G., Sirota F.L., Eisenhaber F. Towards complete sets of farnesylated and geranylgeranylated proteins. PLoS Comput Biol. 2007;3:e66. doi: 10.1371/journal.pcbi.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koumandou V.L., Natesan S.K.A., Sergeenko T., Field M.C. The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics. 2008;9:298. doi: 10.1186/1471-2164-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dhir V., Goulding D., Field M.C. TbRAB1 and TbRAB2 modulate trafficking through the early secretory pathway of Trypanosoma brucei. Mol Biochem Parasitol. 2004;137:253–265. doi: 10.1016/j.molbiopara.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Alexander D.L., Schwartz K.J., Balber A.E., Bangs J.D. Developmentally regulated trafficking of the lysosomal membrane protein p67 in Trypanosoma brucei. J Cell Sci. 2002;115:3253–3263. doi: 10.1242/jcs.115.16.3253. [DOI] [PubMed] [Google Scholar]
- 50.Hall B.S., Pal A., Goulding D., Field M.C. Rab4 is an essential regulator of lysosomal trafficking in trypanosomes. J Biol Chem. 2004;279:45047–45056. doi: 10.1074/jbc.M407271200. [DOI] [PubMed] [Google Scholar]
- 51.Pal A., Hall B.S., Jeffries T.R., Field M.C. Rab5 and Rab11 mediate transferrin and anti-variant surface glycoprotein antibody recycling in Trypanosoma brucei. Biochem J. 2003;374:443–451. doi: 10.1042/BJ20030469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabernet-Castello C., Dacks J.B., Field M.C. The single ENTH-domain protein of trypanosomes; endocytic functions and evolutionary relationship with Epsin. Traffic. 2009;10:894–911. doi: 10.1111/j.1600-0854.2009.00910.x. [DOI] [PubMed] [Google Scholar]
- 53.Morgan G.W., Hall B.S., Denny P.W., Carrington M., Field M.C. The kinetoplastida endocytic apparatus. Part I: a dynamic system for nutrition and evasion of host defences. Trends Parasitol. 2002;18:491–496. doi: 10.1016/s1471-4922(02)02391-7. [DOI] [PubMed] [Google Scholar]
- 54.Dacks J.B., Doolittle W.F. Novel syntaxin genes from Giardia, Trypanosoma and algae: Implications for the ancient evolution of the eukaryotic endomembrane system. J Cell Sci. 2002;115:1635–1642. doi: 10.1242/jcs.115.8.1635. [DOI] [PubMed] [Google Scholar]
- 55.Sanderfoot A. Increases in the number of SNARE genes parallels the rise of multicellularity among green plants. Plant Physiol. 2007;144:6–17. doi: 10.1104/pp.106.092973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dacks J.B., Poon P.P., Field M.C. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc Natl Acad Sci U S A. 2008;105:588–593. doi: 10.1073/pnas.0707318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshizawa A.C., Kawashima S., Okuda S., Fujita M., Itoh M., Moriya Y. Extracting sequence motifs and the phylogenetic features of SNARE-dependent membrane traffic. Traffic. 2006;7:1104–1118. doi: 10.1111/j.1600-0854.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 58.Vedovato M., Rossi V., Dacks J.B., Filippini F. Comparative analysis of plant genomes allows the definition of the “Phytolongins”: a novel non-SNARE longin domain protein family. BMC Genomics. 2009;10:510. doi: 10.1186/1471-2164-10-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rossi V., Banfield D.K., Vacca M., Dietrich L.E.P., Ungermann C., D'Esposito M. Longins and their longin domain: regulated SNAREs and multifunctional SNARE regulators. Trends Biochem Sci. 2004;29:682–688. doi: 10.1016/j.tibs.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 60.Letunic I., Doerks T., Bork P. SMART-7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schilde C., Lutter K., Kissmehl R., Plattner H. Molecular identification of a SNAP-25-like SNARE protein in Paramecium. Eukaryot Cell. 2008;7:1387–1402. doi: 10.1128/EC.00012-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accessions and assignments for TriTryp SNAREs. Assignments, details to the relevant phylogenetic reconstruction and orthology between the TriTryp SNAREs are detailed. New accession numbers for T. brucei are in bold. Note that SNAREs that have been localized are in red.